Abstract
Aim
Biomarker-based tracking of human stem cells xenotransplanted into animal models is crucial for studying their fate in the field of cell therapy or tumor xenografting.
Materials & methods
Using immunohistochemistry and in situ hybridization, we analyzed the expression of three human-specific biomarkers: Ku80, human mitochondria (hMito) and Alu.
Results
We showed that Ku80, hMito and Alu biomarkers are broadly expressed in human tissues with no or low cross-reactivity toward rat, mouse or pig tissues. In vitro, we demonstrated that their expression is stable over time and does not change along the differentiation of human-derived induced pluripotent stem cells or human glial-restricted precursors. We tracked in vivo these cell populations after transplantation in rodent spinal cords using aforementioned biomarkers and human-specific antibodies detecting apoptotic, proliferating or neural-committed cells.
Conclusion
This study assesses the human-species specificity of Ku80, hMito and Alu, and proposes useful biomarkers for characterizing human stem cells in xenotransplantation paradigms.
Keywords: cell therapy, human-specific biomarker, image quantification, stem cells, xenotransplantation
In humans, organs such as brain, heart, eyes and endocrine pancreas have limited regenerative abilities following injury, infarct or degenerating conditions [1]. Today, the concept of regenerative medicine has pushed the development of stem cell therapies forward. Alternative sources of stem cells, other than embryonic stem cells, have accelerated their preclinical evaluation in diseases lacking efficient treatments, in particular in central nervous system disorders [2,3]. The translation of human stem cells to patients is still critically dependent on preclinical studies, mostly validated through animal models [4]. Before approval by competent authorities, post-mortem analysis addressing safety, biodistribution and fate of transplanted cells is requested and cannot be circumvented. They represent complementary methods to new noninvasive tracking systems [5,6].
Prelabeling human stem cells using membrane permeable-fluorescent dyes (DiI), quantum dots, 5-bromo-2-deoxyuridine loading or insertion of reporter genes (e.g., LacZ, luciferase, red/yellow/green fluorescent protein) allow for easy identification in situ in animal tissues [7,8], but is potentially associated with turning off of the reporter gene, cell toxicity [9], unforeseen consequences on differentiation [10] and/or an immune response [11]. For instance, discrepancies in the expression level of green fluorescent protein between in vitro (high expression) and in vivo (low expression) have already been described in transplantation studies [12,13]. While exogenous labeling of transplanted cells or modifying the cells with reporter transgenes is convenient for research applications, this may produce genetic perturbations of unknown significance jeopardizing the preclinical study or clinical translation approval by regulatory authorities.
Terminal procedures including immunohistochemical methods are routinely carried out as analysis of cell fate upon transplantation. Species-specific antibodies (Ab), gender-specific or human-specific biomarkers are essential tools to track engrafted cells of human origin by immunohistochemistry. Along these lines, the marker `human nuclear antigen' recognizes an epitope of human histone H1 family member 0 and is ubiquitously expressed in all human cell nuclei. Ab generated against human nuclear antigen have been widely used to track human cells xenotransplanted in animal tissues. Unfortunately, most of these studies only focused on frozen sections [14–17], which is a shortcoming for applications on paraffin-embedded or long-stored/shipped specimens. In addition, human-specific Ab recognizing blood antigens such as TRA-1-85 [18,19] or minor/major histocompatibility antigens [20] have also been tested but have not yielded satisfactory results in terms of broad applicability, ubiquitous expression or lasting expression following differentiation.
In the present study, we aimed at characterizing three ubiquitous biomarkers – Ku80, human mitochondria (hMito) and Alu sequences – as tools for tracking human stem cells xenotransplanted into animal models and suitable for paraffin-embedded samples. Using computer-assisted image analysis, we quantified the engraftment of human neural- or glial-precursor cells following transplantation into rat and mouse spinal cord, respectively. Completing such panel, we characterized human-specific Ab detecting apoptotic, proliferative or neural-lineage differentiating cells. Based on immunohistochemistry and in situ hybridization, this methodological paper assesses the human-species specificity and ubiquitous expression of several biomarkers and proposes useful tools to analyze the fate of human stem cells in preclinical studies.
Materials & methods
Ethic statement
Human skin fibroblasts were obtained from the Centre de Ressources Biologiques in Lyon, France, with the approval of competent authorities. A statement of biological samples was made according to French laws formulated by the Ministère de la Recherche and to the Comité de Protection des Personnes, Ile de France (DC 2009–1067). Human glial-restricted precursors (GRP) were obtained from brains of fetal cadavers of gestational age from 17 to 24 weeks. Tissue was procured by Procurement Specialists employed by Advanced Bioscience Resources (Alameda, CA, USA; FEIN 3005208435) following informed consent standard operating procedure and donor medical record review procedures.
Cell culture
Induced pluripotent stem cells (iPS) were prepared as described elsewhere [21,22]. Briefly, iPS were generated following forced expression of OCT4, SOX2, KLF4 and c-MYC transcription factors with retroviral vectors. They were grown on irradiated mouse embryonic fibroblast feeder layers in the following medium (iPS medium): DMEM/F12 containing 20% Knock-Out Serum Replacement (Life Technologies, CA, USA), 10 ng/ml FGF2 (Miltenyi Biotec, Paris, France), 100 μM nonessential amino acids (Life Technologies), 100 μM mercaptoethanol (Life Technologies), 50 U/ml penicillin and 50 mg/ml streptomycin. Cultures were passaged every 5–10 days either manually or enzymatically with collagenase type IV (1 mg/ml; Life Technologies). Human iPS-derived neural precursor cells (NPC) were obtained as previously detailed [22,23]. For neural differentiation, iPS were collected as small clusters and resuspended in iPS medium without FGF2. After 2 weeks, floating clusters were dissociated into single cell suspension with Accumax (PAA Laboratories, Linz, Austria). Cells were further differentiated into neurons for 14 days in DMEM/F12 containing 2% B27 (Life Technologies).
GRP were derived as described by Campanelli's protocol [24]. Briefly, forebrain from human fetus was mechanically and enzymatically dissociated, followed by magnetic-activated cell sorting-based positive selection with the glial progenitor specific cell surface antigen A2B5. Following selection, GRP were cultured on polyornithine-coated flasks in DMEM/F12, with l-glutamine, 15 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (Life Technologies), 1X N1 (Sigma-Aldrich, MO, USA), 0.01% human serum albumin, 20 ng/ml FGF2 and 10 ng/ml PDGF-AA (both from Peprotech, NJ, USA) for 20 days. To prepare pMXs-mRFP1 retrovirus, 293FT cells (Invitrogen, CA, USA) were transfected with plasmids pMXs-mRFP1, pCL-Eco and pCMV-VSV-G (all were purchased from Addgene, MA, USA). After 2 days of culture, pMXs-mRFP1 retrovirus was concentrated from supernatant with PEG-it™ Virus Precipitation Solution (System Biosciences, CA, USA) following manufacturer's protocol. On the day of infection, pMXs-mRFP1 retrovirus solution was added to GRP culture medium at the concentration of 2 × 106 IU/ml, and then the medium was changed in 24 h. After 4–5 days of culture, the red fluorescent signal of red fluorescent protein (RFP) in the infected cells became visible under the florescence microscope. The cells were trypsinized and resuspended with artificial cerebrospinal fluid (Tocris Bioscience, Bristol, UK) at the concentration of 1.5 × 105 cells/μl, in preparation for transplantation. To induce GRP differentiation, the cells were cultured in DMEM/F12 (Invitrogen), N2/B27 (Invitrogen) supplement and 10 ng/ml hBMP4 (Peprotech) for 4 days.
Animals
Male C57BL/6 mice aged from 8 to 12 weeks and female Sprague–Dawley rats weighing 250–300 g were purchased from Taconic Farm (MD, USA). All animals were housed in a humidity-, temperature-and light-controlled animal facility with ad libitum access to food and water. Experimental procedures were reviewed by the institutional animal care and use committee (Laboratory agreement LA1230342, protocol 292N), conducted in accordance with the European Communities Council Directive (2010/63/EU, 86/609/EEC and 87-848/EEC) and the NIH Guide for the Care and Use of Laboratory Animals.
Experimental design & surgery
Mice were anesthetized with ketamine (120 mg/kg) and xylazine (5 mg/kg). The dorsal skin and underlying muscle layers were incised along the midline between the spinous processes of C2 and T1 [25–27]. Following right unilateral laminectomy, mice were administered 100,000 GRP in 2 μl in the ventral horns of both C4 and C5 levels, at a depth of 0.75 mm. Mice were immunosuppressed by intraperitoneal administration of FK-506 and rapamycin (1 mg/kg each per day; LC Laboratories, MA, USA) daily beginning 3 days before grafting and continuously until sacrifice [16].
Anesthesia in rats was induced by ketamine (60 mg/kg) and xylazine (7.5 mg/kg). The dorsal skin and underlying muscle layers were incised along the midline between the spinous processes of L1 and L6. Following laminectomy on L4 and L5 levels, rats received 200,000 iPS-derived NPC in 2 μl targeting the respective ventral horn segments at a depth of 1.5 mm [23]. Rats were immunosuppressed with cyclosporine A (Neoral; Novartis International, Basel, Switzerland) at 10 mg/kg per day, added in the drinking water. The immunosuppression was started 5 days before transplantation and continued until the sacrifice. Sham animals underwent the same surgery, including laminectomy, but received injections of artificial cerebrospinal fluid. At the end of the procedure, overlying muscles were closed in layers with sterile 4–0 silk sutures, and the skin incision was closed using sterile wound clips. Animals were allowed to recover on a circulating water-heating pad until awake and then returned to their home cages. They were daily monitored for 5 days, and measures were taken to avoid dehydration and to minimize any potential pain or discomfort. Animals were sacrificed by exsanguination under anesthesia at different time post-transplantation.
Tissue preparation
Animals were transcardially perfused with saline and then 4% buffered paraformaldehyde. Spinal cord was collected and pot-fixed overnight in paraformaldehyde. Spinal cord was longitudinally split in two hemicords. One segment of hemicord was rinsed in tap water and then was dehydrated in 70% alcohol before paraffin-embedding. The other half was washed in 0.1 M phosphate buffer and cryoprotected in 30% sucrose for 3 days at 4°C followed by embedding in freezing medium.
Species-specific tissue microarray
Tissue microarrays (TMA) were constructed using a Minicore tissue arrayer (Alphelys), as previously detailed [28]. TMA blocks bring together tissue samples from various mice, rats and human organs and included skeletal muscle, trachea, stomach, small intestine, colon, spleen, tonsil, lymph nodes, hematopoietic bone marrow, liver, pancreas, cerebellum, brain (hippocampus), spinal cord, lung, heart, kidney, thymus, prostate, testis, thyroid, mammary gland, ovary, fallopian tube, endometrium, cervix, myometrium and placenta. Additionally, pancreas, spleen, liver, lung, heart, hematopoietic bone marrow and lymph nodes (kindly gifted by Professor Desmecht, ULg, Belgium) from swine pigs were included in the study and processed for paraffin-embedding.
Immunohistochemistry
Immunostainings were performed on Ventana discovery XT™ (Ventana, Roche Diagnostics, Vilvoorde, Belgium) using the DABMap detection system according to manufacturer's recommendations. Briefly, 5 μm-thick tissue sections were deparaffinized and endogenous peroxydase activity was inhibited with 3% hydrogen peroxide. Each primary Ab was added according to Ab concentration specified in Table 1. Next, slides were washed and incubated with biotinylated secondary Ab against mouse or rabbit (1:200, Vector Laboratories, Peterborough, UK) followed by the addition of complex avidin–horseradish peroxidase. Immunostaining was detected by incubation with diaminobenzidine and hydrogen peroxide. Sections were counterstained with hematoxylin and mounted with Entellan®. When hMito immunostaining was performed on mouse tissue, a MoMap Kit (760–137, Ventana) was used according to manufacturer's protocol.
Table 1.
Cross-reactivity of human biomarkers.
| Biomarker | Supplier | Reference | Concentration | Species reactivity |
|---|---|---|---|---|
| Ku80 | Cell Signaling | CST-2180 | 1:100 | Hu† |
| hMito | Millipore | MAB1273 | 1:1000 | Hu, Sw |
| Histone H1 FO | Sigma-Aldrich | HPA000843 | 1:500 | Hu, Ms, Rt, Sw |
| Alu | Roche Ventana | 780-2221 | 100 ng/ml | Hu† |
| Ki67 | Thermo Fisher | MA5-14520 | 1:200 | Hu, Ms, Rt, Sw |
| Ki67 | Dako | M7240 | 1:1000 | Hu, Ms, Sw |
| CyclinB1 | Epitomics | 1495-1 | 1:200 | Hu, Sw |
| c-PARP | Cell Signaling | CST-5625 | 1:200 | Hu† |
| Nestin | Millipore | MAB5326 | 1:200 | Hu |
| Neuron-specific enolase | Dako | N1557 | Ready-to-use | Hu, Ms |
Not mouse, not rat and not swine pig.
hMito: Human mitochondria; Hu: Human; Ms: Mouse; Rt: Rat; Sw: Swine pig.
In situ hybridization
In situ hybridization was carried out on Ventana discovery XT™ using the DABMap Detection System according to manufacturer's recommendations. Five micrometer thick sections were deparaffinized. Antigen retrieval was performed with RiboCC™ (pH 6.0) for 60 min at 95°C followed by digestion with Protease 3 (760–2020, Ventana) for 20 min at 37°C. Alu Positive Control Probe II (780–2221, Ventana) was dropped, slides were warmed at 85°C to denature DNA, hybridization was performed at 47°C for 1 h. Slides were washed twice with sodium saline citrate. Nonspecific binding was impeded by incubation of inhibitor D and endogenous biotins were blocked with avidin–biotin kit (760–050, Ventana). Bound probe was detected by rabbit anti-digoxigenin (780–4335, Ventana) and then tissues were incubated with biotinylated anti-rabbit Ab (Vector Laboratories). Slices were exposed at complex avidin–horseradish peroxidase at room temperature and finally were incubated with diaminobenzidine and hydrogen peroxide.
Immunofluorescence
Cells in culture were fixed with 4% buffered paraformaldehyde, washed in phosphate buffered saline (PBS) and permeabilized 15 min with 0.1% Triton X-100. After washing, primary Ab (diluted at 1:100) were incubated overnight at 4°C in blocking buffer (PBS supplemented with 4% fetal bovine serum). Primary Ab were against Ku80, hMito, nestin, SSEA4 (Chemicon, CA, USA), SMI32 (Covance, Belgium), GFAP (Dako, Heverlee, Belgium). The following day, the cells were washed and secondary Ab incubated for 1.5 h at room temperature. Secondary Ab FITC goat anti-mouse IgG, FITC goat anti-rabbit IgG, TRITC goat anti-rabbit IgG were all purchased from Molecular Probes (Life Technologies) and diluted at 1:200 to recognize the matched primary Ab. Thirty micrometer thick frozen spinal cord sections were air-dried and successively washed and permeabilized with 0.4% Triton X-100 in PBS for 5 min at room temperature, then incubated in blocking solution (PBS containing 10% normal goat serum and 0.4% Triton X-100) for 1 h at room temperature. Sections were labeled overnight at 4°C with Ku80 Ab (1:100) in blocking solution, and after washed three-times with PBS (5 min each time), sections were incubated with the secondary Ab in blocking solution for 1 h in room temperature. After washing twice with PBS (10 min each time), sections were mounted with coverslips.
Computer-assisted image analysis
Immunostained spinal cord slides were scanned using a NanoZoomer slide scanner (Hamamatsu, Japan). For each virtual slide, immunostaining quality was controlled. Using the Visiomorph software package (Visiopharm, HØrsholm, Denmark), hemicord areas were selected manually, discarding tissue folds and other technical artifacts. Quantitative analysis of the injected hemicord was further performed. For each selected area and each staining, we computed the labeling index (LI) [28], which corresponds to the percentage of the immunostained tissue area. For nuclear immunostaining (i.e., Ku80 and Alu probes), the LI corresponds to the percentage of the immunostained nucleus area.
Statistics
Independent groups of quantitative data were compared using nonparametric one-way analysis of variance (Kruskal–Wallis test), followed by post-hoc test (Dunn method) for multiple comparisons. p < 0.05 was considered as significant.
Results
Broad & human-specific expression of biomarkers
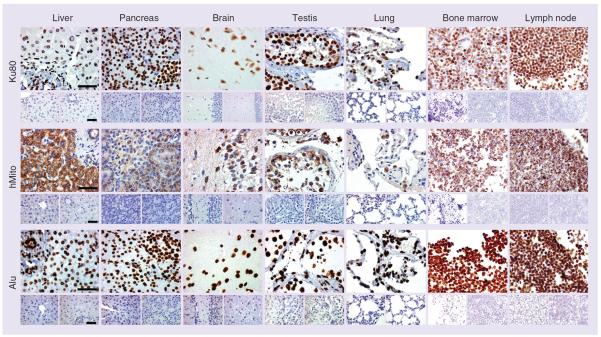
Through TMA screening, we first sought to determine the human specificity of three putatively ubiquitous biomarkers: Ku80, hMito and Alu sequences. Using immunohistochemistry and in situ hybridization, we showed that Ku80, hMito and Alu were broadly expressed throughout the human body. Human liver, pancreas, brain, testis, lung, hematopoietic bone marrow and lymph node were strongly immunoreactive for the three biomarkers (Figure 1), while no signal could be seen in their rat or mouse counterparts (lower panels in Figure 1). As expected, the localization of the immunostaining was confined to the nucleus for Ku80 and Alu, and to the cytoplasmic compartment for hMito. An in-depth analysis aimed at identifying the cellular expression was performed using TMA slides encompassing numerous normal human, mouse, rat and pig organs. As shown in Table 2, Alu was the most intense, the most expressed and the broadest human biomarker in the tissues analyzed, even found in the latest stage of spermatogenesis. Although Ku80 had the same expression pattern and immunoreactivity (Supplementary Figure 1; see online at www.future-medicine.com/doi/suppl/10.2217/rme.14.26) as Alu, it could not be found in the spermatozoa nucleus. Among the three biomarkers, hMito demonstrated the weakest immunoreactivity and, for instance, was not observed in type I pneumocytes or spermatozoa (Table 2). Ku80 and Alu were never detected in mouse, rat or pig tissues, confirming their specificity toward human species (Supplementary Figure 1). Surprisingly, the hMito Ab recognized a homologous protein in pig tissues making it unsuitable for xenotransplantation follow-up in this animal model. hMito did not cross-react with mouse or rat tissues (Figure 1). Although claimed to work on paraffin sections, a commercially available histone H1 Ab did not yield satisfactory results in terms of ubiquitous or human-specific expression (data not shown).
Figure 1. Immunohistochemistry of Ku80, human mitochondria and Alu biomarkers in organs from human, mouse and rat origins.
All three biomarkers are expressed in human liver, pancreas, brain, testis, lung, hematopoietic bone marrow and lymph node (upper panels) but not in mouse (lower left panels) or rat (lower right panels) tissue counterpart. Ku80 and Alu strongly label the nucleus while hMito is confined to the cytoplasmic compartment. In most of the human organs, more than 99% of the cells, regardless of their tissular origin (epithelial, muscular, connective), are immunoreactive for Alu and Ku80 biomarkers. hMito is less broadly expressed and for instance does not label all types of human pneumocytes. A detailed analysis of biomarker expression is shown in Table 2. Scale bar represents 50 μm.
hMito: Human mitochondria.
Table 2.
Expression and distribution of human-specific biomarkers in normal tissue.
| Cell type | Ku80 | hMito | Alu |
|---|---|---|---|
| Nervous system | |||
|
| |||
| Neurons | +++ | ++ | +++ |
| Glial cells | +++ | + | +++ |
|
| |||
| Endocrine system | |||
|
| |||
| Thyrocytes | +++ | ++ | +++ |
|
| |||
| Respiratory system | |||
|
| |||
| Pneumocytes type I | +++ | − | +++ |
| Pneumocytes type II | +++ | +++ | +++ |
|
| |||
| Digestive tract | |||
|
| |||
| Esophageal epithelium | +++ | +++ | +++ |
| Fundic glands | +++ | +++ | +++ |
| Enterocytes | +++ | +++ | +++ |
| Goblet cells | +++ | +++ | +++ |
| Lieberkuhn's glands | +++ | +++ | +++ |
|
| |||
| Digestive organs | |||
|
| |||
| Liver: | |||
| Hepatocytes | +++ | +++ | +++ |
| Bile duct | +++ | +++ | +++ |
| Pancreas: | |||
| Islets of Langerhans | +++ | + | +++ |
| Acinar cells | +++ | ++ | +++ |
|
| |||
| Immune system | |||
|
| |||
| Spleen, lymph node, tonsil: | |||
| Lymphocytes | +++ | ++ | +++ |
| Hematopoietic cells | ++ | +++ | +++ |
|
| |||
| Urinary tract | |||
|
| |||
| Kidney: | |||
| Proximal tubule | +++ | +++ | +++ |
| Loop of Henle | +++ | +++ | +++ |
| Collecting duct | +++ | ++ | +++ |
|
| |||
| Connective tissue | |||
|
| |||
| Fibroblasts | ++ | + | +++ |
| Chondrocytes | +++ | + | +++ |
| Osteoblasts | +++ | + | +++ |
| Osteocytes | +++ | + | +++ |
|
| |||
| Muscle and cardiovascular system | |||
|
| |||
| Smooth muscle | ++ | ++ | +++ |
| Skeletal muscle | +++ | ++ | +++ |
| Cardiomyocytes | +++ | ++ | +++ |
| Endothelial cells | +++ | + | +++ |
|
| |||
| Reproductive system | |||
|
| |||
| Testis: | |||
| Spermatogonia | +++ | ++ | +++ |
| Spermatid | +++ | ++ | +++ |
| Spermatozoa | − | − | +++ |
| Leydig cells | +++ | +++ | +++ |
| Prostate: | |||
| Epithelial cells | +++ | ++ | +++ |
| Uterus: | |||
| Myometrium | +++ | ++ | +++ |
| Endometrial glands | +++ | +++ | +++ |
| Mammary glands | +++ | +++ | +++ |
| Placenta: | |||
| Cytotrophoblast | +++ | ++ | +++ |
| Syncitiotrophoblast | +++ | +++ | +++ |
Labeling for Ku80 and Alu was mostly confined to the nucleus unlike cytoplasm staining for hMito. Immunoreactivity was assessed by semi-quantitative scoring.
−: 0%; +: <30%; ++: 30–60%; +++: >60% of immunoreactive cells throughout the whole organ; hMito: Human mitochondria.
Stable expression of Ku80 & hMito biomarkers along differentiation of human iPS cells
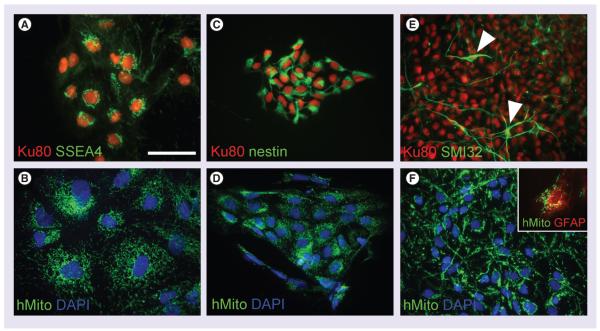
To be considered as a reliable biomarker for long-term follow-up, the biomarker should be expressed in the tracked cells at all stages of cell differentiation. We therefore cultured human iPS and differentiated them toward the neuronal lineage. iPS co-expressed the undifferentiated marker SSEA4 with the nuclear biomarker Ku80 (Figure 2A). The same cells were strongly immunoreactive for hMito (Figure 2B). iPS were further differentiated into NPC which expressed the neural lineage marker nestin concomitantly with Ku80 (Figure 2C). Pure NPC cultures also expressed hMito biomarker (Figure 2D). Finally, NPC cultures were incubated in medium without growth factors, achieving the ultimate differentiation stages. After two weeks, we were able to detect SMI32+ cells expressing Ku80 biomarker (Figure 2E). These SMI32+ cells displayed the typical morphology of mature neurons (arrow in Figure 2E). Cells with a neuronal morphology derived from the same culture condition all expressed hMito biomarker (Figure 2F). Although our protocol of differentiation was being optimized to push NPC toward a neuronal phenotype [22], we sparsely observed GFAP+ expressing likewise hMito biomarker (inset in Figure 2F).
Figure 2. Immunohistochemistry of Ku80 and human mitochondria along the differentiation process of human induced pluripotent stem cells toward neuronal lineage.
(A & B) Ku80 and hMito immunolabeled all clones of induced pluripotent stem cell, whose cells are characterized by the expression of the SSEA4. (C & D) Upon differentiation into neural precursor cells, they start expressing nestin while keeping Ku80 and hMito expression. (E & F) At further differentiation stages, cells with a typical neuronal morphology were observed (white arrowheads in E). SMI32+ neurons were still expressing Ku80 and hMito. Sparse GFAP+ cells were also found immunoreactive for hMito (inset in F). Scale bar represents 50 μm.
DAPI: 4′,6-diamidino-2-phenylindole; hMito: Human mitochondria.
NPC expressed human biomarkers up to 60 days after transplantation in rat spinal cord
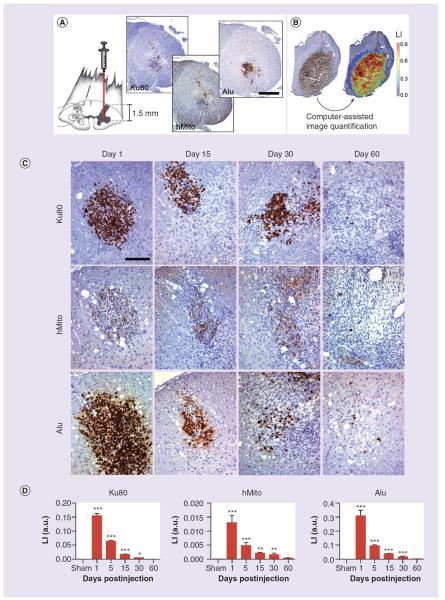
Human iPS-derived NPC were injected in the ventral horn of rat lumbar spinal cord and tracked up to 60 days post-transplantation (Figure 3A). Ku80, hMito and Alu immunoreactive cells were observed in a rat sacrificed 1-h postinjection, suggesting that these biomarkers could be used in vivo to track human stem cells and eventually to follow the cell fate for longer time periods (Figure 3A). Using computer-assisted quantification analysis, we were able to compute an LI for each slide (Figure 3B). Human NPC were visualized in the spinal cord tissue by Ku80, hMito and Alu stainings up to 60 days postinjection (Figure 3C). At gross microscopic analysis, the number of immunoreactive cells in the cord tended to decrease after grafting, which was confirmed by further image quantification. The LI of each immunostaining was significantly higher at 1, 15 and 30 days post-transplantation compared to sham animals but progressively decreased over time likely due to cell death (Figure 3D). We did not detect immunostaining for any of the biomarkers in the sham group receiving injection of artificial cerebrospinal fluid (data not shown).
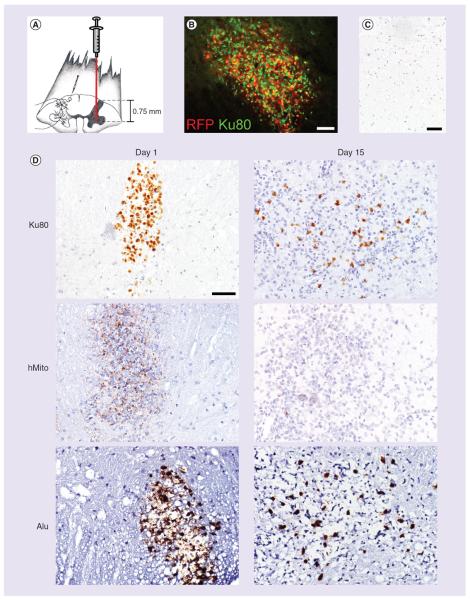
Figure 3. Human neural precursor cells are injected into the rat spinal cord, targeting the ventral horn (facing page).
(A) At 1-day postinjection, Ku80, hMito and Alu immunostainings help visualize the injection site. (B) Computer-assisted image analysis allows to make the difference between the blue staining from hematoxylin counterstaining and the di-amino-benzidine brown precipitates from specific anti-human immunohistochemistry. Hence, LI corresponding to the percentage of the immunostained tissue surface was calculated. (C) Using anti-human Ku80, hMito and Alu immunohistochemistry, neural precursor cell-derived human cells are visualized in the spinal tissue from 1 day to 60 days post-transplantation. (D) Around the injection site, we detect inflammatory reaction and gliosis, most likely causing a loss of engrafted human cells over time. This is confirmed by image quantification showing a progressive decrease of the LI of Ku80, Alu and hMito immunostainings. Scale bars represent 500 μm in (A) and 100 μm in (C). Results are expressed as mean ± standard error of the mean. *p < 0.05, **p < 0.01, ***p < 0.001 for comparisons 1, 5, 15, 30 and 60 days versus sham group.
The number of animals was, respectively, 8, 3, 5, 7, 14 and 5 rats for the experimental conditions: sham, 1, 5, 15, 30 and 60 days.
hMito: Human mitochondria; LI: Labeling index.
Stable expression of Ku80 & Alu biomarkers along differentiation process of human GRP
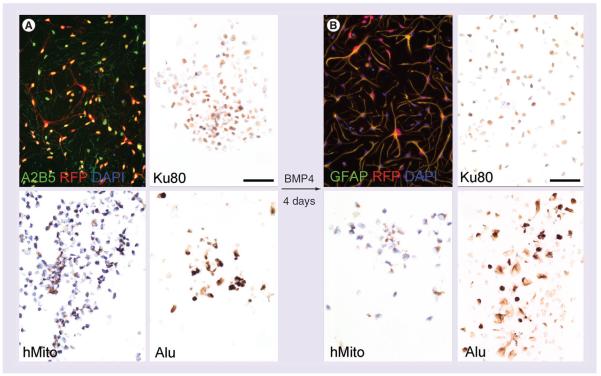
In order to extend these findings, we used a second human progenitor cell type, A2B5+ glial-lineage restricted precursors, which are able to give rise to fully mature astrocytes. These A2B5+ GRP were in vitro transduced with an RFP-carrying viral vector and subcultured in undifferentiated and differentiated conditions. Most undifferentiated GRP expressed both A2B5 and RFP (Figure 4A). They also expressed Ku80 and Alu biomarkers and to a lesser extent hMito (Figure 4A). Differentiation was initiated by addition of BMP4 in the medium, which induced morphological changes toward star-shaped cells. GFAP immunostaining confirmed the differentiation process to an astrocyte phenotype (Figure 4B). Upon differentiation, cells retained their RFP expression, and at least expression of two of the biomarkers: Ku80 and Alu. hMito immunolabeling resulted in a very weak signal in differentiated GRP, suggesting decreasing expression of this biomarker upon differentiation (Figure 4B).
Figure 4. Immunohistochemistry of Ku80, human mitochondria and Alu along the differentiation process of human glial-restricted precursors toward astrocytic lineage.
(A) Undifferentiated glial-restricted precursors (GRP) are isolated from human fetuses based on their A2B5-specific expression. Cells are further transduced with a retroviral vector driving the RFP reporter gene. Undifferentiated GRP preferentially display a rounded shape and express all three biomarkers: Ku80, hMito and Alu. (B) Upon differentiation with BMP4, star-shaped cells are observed close to an astrocyte morphology and showing a strong immunoreactivity for GFAP. Differentiated GRP still express Ku80 and Alu, but the immunostaining for hMito is more faint. Scale bar represents 50 μm.
hMito: Human mitochondria; RFP: Red fluorescent protein.
GRP expressed human biomarkers up to 15 days after transplantation in mouse spinal cord
According to a similar protocol of transplantation, undifferentiated GRP were injected in the mouse cervical spinal cord (Figure 5A) in order to demonstrate the usefulness of Ku80, Alu and hMito biomarkers as immunohistochemical tools for tracking human xenotransplanted progenitor cells. As cells were previously transduced in vitro with the RFP exogenous reporter gene, it was straightforward to identify the transplanted cells under fluorescence on cryostat-processed spinal sections. We demonstrated that RFP+ GRP transplants expressed Ku80 biomarker in vivo at 1 and 15 days post-transplantation (Figure 5B). Using classical immunohistochemistry on paraffin-embedded samples, we tested Ku80, hMito and Alu immunolabeling on sham- and GRP-injected spinal cord. We did not detect immunostaining for any of the biomarkers in the sham group receiving injection of artificial cerebrospinal fluid (Figure 5C). In transplanted mice, we showed that Ku80 and Alu biomarkers were continuously expressed at a high intensity over time and allowed for rapid identification of human cells (Figure 5D). While hMito was significantly expressed in GRP at 1-day post-transplantation, labeling with this biomarker progressively was lost at the later time point. Rare and discrete hMito+ cells were observed containing faint cytoplasmic granulations at 15 days post-transplantation (Figure 5D).
Figure 5. Detection of human glial-restricted precursors injected into the mouse spinal cord.
(A) RFP prelabeled human glial-restricted precursors (GRP) are injected into the mouse spinal cord, targeting the ventral horn. (B) At 1-day postinjection, RFP+ human GRPs are visualized in the injection site of the spinal cord. These cells co-express the human-specific biomarker Ku80. (C) Using anti-human Ku80, hMito and Alu immunohistochemistry, human GRP cells can be tracked in the spinal tissue from 1 day to 15 days post-transplantation. (D) Vehicle-injected animals do not show any biomarker immunoreactivity in the spinal tissue. While Ku80 and Alu immunoreactive cells are still found at 15 days post-transplantation, the signal for hMito is very weak. Scale bar represents 50 μm.
hMito: Human mitochondria; RFP: Red fluorescent protein.
Human-specific biomarkers of apoptotic, proliferative or neural-lineage differentiating cells
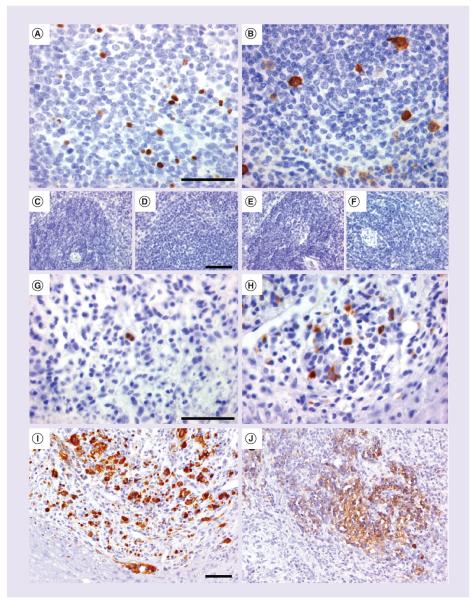
We also found that c-PARP and cyclinB1 Ab are human markers of apoptotic and proliferating cells, respectively (Figure 6A–H), which could be used to further characterize the fate of transplanted stem cells. Immunolabeling could be seen on human spleen samples where apoptotic c-PARP+ cells (Figure 6A) or proliferating cyclinB1+ cells (Figure 6B) were present in the white pulp. Spleens from mouse (Figure 6C & E) and rat (Figure 6D & F) were not immunostained with the human-specific biomarkers. CyclinB1 Ab, but not c-PARP, cross-reacted with spleen sample from pig origin (Table 1). Fifteen days after intraspinal transplantation, some human NPC underwent apoptosis since they were immunoreactive for c-PARP (Figure 6G) and some continued to proliferate as demonstrated by immunoreactivity for cyclinB1 (Figure 6H). Two different Ki67 Ab (data not shown), one of the most commonly used markers of proliferation, were also tested, but neither was specific to the human species (Table 1). We next characterized supplementary biomarkers, nestin and neuron-specific enolase (NSE), which allow for the identification of neural-committed cells and neurons. Their cross-reactivity was tested on human, rat and mouse nervous tissue (Table 1). In the rat model transplanted with human NPC, we observed the presence of nestin+ cells as early as 1-day post-transplantation (Figure 6I) whereas NSE started to be expressed from 30 days post-transplantation, most likely in differentiated NPC (Figure 6J). Both biomarkers did not label brain or spinal cord tissue from rat origin allowing for specific detection of the human cell population.
Figure 6. Human biomarkers of apoptosis, proliferation and neural commitment.
(A) c-PARP and (B) CyclinB1 antibodies are biomarkers detecting cell death and proliferation in human spleen, which do not cross-react with (C–E) mouse or (D–F) rat spleens. (G) Fifteen days post-transplantation of human neural precursor cells, these detected cells undergoing apoptosis or (H) clusters of proliferating human cells into the rat spinal cord tissue. (I) Neural precursor cells express nestin at 1-day postinjection and (J) later on become progressively immunoreactive for a human-specific neuron-specific enolase antibody. Scale bar represents 50 μm.
Discussion & conclusion
In pioneering transplantation clinical trials, dopamine neurons derived from human fetal forebrain were injected into Parkinson patients and were functional for months, showing sustained uptake of dopamine precursor and producing long-lasting clinical improvements. Proof of concept that the graft survived up to 16 years after initial transplantation was shown by post-mortem immunohistochemistry, demonstrating tyrosine hydroxylase immunoreactive dopaminergic cells in the parkinsonian recipient brain [29,30]. Although there is no definite proof that these dopaminergic cells arose from the graft, there is an overall growing consensus among the scientific community that a better understanding of the fate of transplanted cells – biodistribution within or outside of the target tissue, viability over time, integration into host tissue, differentiation into functional cells, tumorigenic potential – will be critical for the long-term approval and ultimate success of these stem cell-based strategies. Before translation to patients, preclinical assessment of a human cell therapy product must address strict regulations. Many crucial parameters such as delivery dose accuracy or engraftment efficiency must be quantified. These important questions can be addressed by immunohistochemical methodology using aforementioned human-specific biomarkers: Ku80, Alu and hMito. In the past, hMito Ab was used for tracking the engraftment of human mesenchymal stem cells in rodent traumatic CNS injury models [31,32] and the fate of neural precursors in a rat model of amyotrophic lateral sclerosis [23]. Mostly on cryostat-processed tissues, Ku80 and Alu probes also allowed for tracking human cells in several xenotransplantation paradigms [33–37].
Our methodology of detection is based on native protein biomarkers expressed by most human cells that could be used to follow the fate of any human xenotransplanted stem cells, from their most undifferentiated state to their final maturation state. Through human tissue microarray screening, we first identified Ku80, hMito antigen and Alu sequences as good immunohistochemical biomarkers, broadly expressed throughout the human body. As they are expressed by most differentiated human cells, they can be applied in a variety of preclinical study settings, assessing human stem cells giving rise to many different cell types: muscles, epithelial cells, fibroblasts, osteoblasts, endocrine cells or nervous cells. Moreover, preliminary results from our laboratory suggested that the aforementioned biomarkers are similarly expressed in human stem cells arisen from various tissues: neural progenitor cells, cardiac progenitor cells and hepatic progenitor cells. Among the limitations, hMito Ab had the lowest sensitivity for some cell types (i.e., pneumocytes, lymphocytes and fibroblasts) and the final stage of male gamete was only labeled with Alu probe. In this work, we demonstrated that Ku80 was stably expressed along the differentiation process of our stem cell populations, from the very undifferentiated iPS cell to mature neurons and from the glial precursor to astrocytes. Surprisingly, hMito staining, unlike Ku80 or Alu, was weak in GRP cells and progressively vanished over the differentiation process of GRP in vitro and in vivo; this highlights the importance of appropriate biomarker selection for specific cell populations and its validation with respect to sensitivity and expression stability along differentiation [18]. Ku80 and Alu biomarkers represent reliable biomarkers to track the long-term engraftment of human stem cells in the mostly used xenotransplantation animal models (rat and mouse). As Ku80 Ab and Alu did not cross-react with any pig tissue, their use can be extended to xenotransplantation paradigms in this species, a well-known model for the study of cardiac disease [38]. The advantage of using these three biomarkers is their applicability on paraffin-embedded/stored tissues compared to previously used common human biomarkers (e.g., human nuclei, human nuclear antigen) supporting only cryostat applications [16,39]. In our quest for universal human biomarkers, we confirmed Ku80, hMito and Alu as being specifically expressed in many human cells and not in normal tissues from mouse, rat or pig origins. However, we cannot rule out whether these biomarkers are present during pathological processes (i.e., inflammation, apoptosis or necrosis) in the mentioned animal species. For instance, Ki67, a widely used marker of proliferation in human diagnostics, has been shown to unexpectedly label apoptotic cells in particular conditions [40,41] and is downregulated during hypoxia [42,43]. We therefore proposed two alternative human-specific markers for labeling proliferative and apoptotic cells, namely CyclinB1 and c-PARP. Surprisingly, we did not observe huge amount of c-PARP+ apoptotic cells in our xenotransplantation paradigm while significant cell loss could be detected over time. A possible explanation for this intriguing result is that the cell loss we observed is not related to apoptosis but mediated by alternative pathways such as necrosis or destruction by host immune cells. For instance, when Schwann cells were transplanted into injured rat spinal cord, it has been shown that six-times more transplanted cells died during the first week after transplantation by necrosis than apoptosis, with the majority of cell death occurring within the first hours [44].
Finally, computer-assisted image analysis allowed us to quantify the cell engraftment and to tridimensionnally reconstruct the site of injection (data not shown). Such quantitative data could serve as valuable parameters when comparing different types of cell delivery (intraparenchymal/intravenous/intraperitoneal/intracavitary injections), vehicle solution or cell populations. Although alternative methods of quantification do exist, most of them are based on the detection of human-specific DNA sequence by real-time PCR [45–47] in xenotransplanted tissues. Our approach has the advantage of allowing both morphological analysis and quantification. Nevertheless, real-time PCR may be seen as a complementary, and possibly more sensitive, method to detect low amounts of engrafted cells, particularly in large organs that would be processed as a whole.
Noninvasive molecular imaging, including MRI and PET, as well as ongoing improvements of these modalities that enhance their spatial and temporal resolution, is likely to increase the prospects of successful tracking in preclinical studies. So far, immunohistochemistry-based post-mortem analysis together with image quantification still provides information on cell engraftment or distribution after transplantation as accurate and quantitative as modern noninvasive techniques, helping at corroborating in vivo findings in the fields of stem cell therapy [32,35] or tumor xenograft [20].
Future perspective
The translation of human stem cells to patients is still critically dependent on preclinical studies, mostly validated through animal models. Before approval by competent authorities, post-mortem immunohistochemistry, using specific biomarkers detecting human engrafted cells, cannot be circumvented and allows for addressing safety, biodistribution and fate of transplanted cells. They represent complementary methods to in vivo noninvasive tracking systems. There is an overall growing consensus among the scientific community that a better understanding of the fate of transplanted cells – biodistribution within or outside of the target tissue, viability over time, integration into host tissue, differentiation into functional cells, tumorigenic potential – will be critical for the long-term approval and ultimate success of these stem cell-based strategies.
Supplementary Material
Executive summary.
Broad & human-specific expression of biomarkers (Ku80, human mitochondria & Alu)
Ku80, human mitochondria (hMito) and Alu can serve as reliable biomarkers to track the long-term engraftment of human stem cells in the mostly used xenotransplantation animal models (rat and mouse). As Ku80 antibody and Alu did not cross-react with any pig tissue, their use can be extended to xenotransplantation paradigms in this species.
Ku80, hMito and Alu are human biomarkers broadly expressed throughout human tissues.
Expression of biomarkers of interest along differentiation of human induced pluripotent stem & glial-restricted precursor cells
In two human stem cell populations (induced pluripotent stem cells and glial-restricted precursors), the expression of Ku80 and Alu biomarkers seemed to be stable over time and does not change upon cell differentiation into more committed cells. This was not the case for hMito.
By allying both immunohistochemistry and computer-assisted image quantification, we were able to sensitively detect the engraftment of human stem cells, their morphology and that of the surrounding environment while generating quantitative data.
Human-specific biomarkers of apoptotic or proliferative cells
Human-specific c-PARP and CyclinB1 can serve as alternative markers of apoptotic and proliferating cells, respectively.
Acknowledgements
The authors are grateful to Audrey Verellen, Sébastien Sauvage and Sabine Paternot for technical assistance, and Song Liu for neurosurgery. The authors thank James T Campanelli (Q Therapeutics, Inc., UT, USA) for providing human glial-restricted progenitors.
This work was supported by grants from Thierry Latran Foundation, INSERM and Institut Pasteur (to D Bohl). This work was also funded by the NIH (1R01NS079702 to AC Lepore) and the Craig H. Neilsen Foundation (#190140 to AC Lepore). X Moles Lopez is supported by the Télévie program of the `Fonds National de la Recherche Scientifique' (FNRS, Brussels, Belgium) and Fonds Yvonne Boël. The CMMI is supported by the European Regional Development Fund and the Walloon Region. C Decaestecker is a Senior Research Associate with the FNRS. Q Therapeutics, Inc., has patents on human GRP and is currently developing these for potential clinical study.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest
- 1.Baddour JA, Sousounis K, Tsonis PA. Organ repair and regeneration: an overview. Birth Defects Res. C Embryo Today. 2012;96(1):1–29. doi: 10.1002/bdrc.21006. [DOI] [PubMed] [Google Scholar]
- 2.Lindvall O, Kokaia Z. Stem cells in human neurodegenerative disorders – time for clinical translation? J. Clin. Invest. 2010;120(1):29–40. doi: 10.1172/JCI40543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitrecic D, Nicaise C, Klimaschewski L, Gajovic S, Bohl D, Pochet R. Genetically modified stem cells for the treatment of neurological diseases. Front. Biosci. (Elite Ed.) 2012;4:1170–1181. doi: 10.2741/449. [DOI] [PubMed] [Google Scholar]
- 4.Glover JC, Boulland JL, Halasi G, Kasumacic N. Chimeric animal models in human stem cell biology. ILAR J. 2009;51(1):62–73. doi: 10.1093/ilar.51.1.62. [DOI] [PubMed] [Google Scholar]
- 5.Bengel FM. Noninvasive stem cell tracking. J. Nucl. Cardiol. 2011;18(5):966–973. doi: 10.1007/s12350-011-9436-2. [DOI] [PubMed] [Google Scholar]
- 6.Kircher MF, Gambhir SS, Grimm J. Noninvasive cell-tracking methods. Nat. Rev. Clin. Oncol. 2011;8(11):677–688. doi: 10.1038/nrclinonc.2011.141. [DOI] [PubMed] [Google Scholar]
- 7.Chu K, Kim M, Chae SH, et al. Distribution and in situ proliferation patterns of intravenously injected immortalized human neural stem-like cells in rats with focal cerebral ischemia. Neurosci. Res. 2004;50(4):459–465. doi: 10.1016/j.neures.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 8.Mitrecic D, Nicaise C, Gajovic S, Pochet R. Distribution, differentiation, and survival of intravenously administered neural stem cells in a rat model of amyotrophic lateral sclerosis. Cell Transplant. 2010;19(5):537–548. doi: 10.3727/096368910X498269. [DOI] [PubMed] [Google Scholar]
- 9.Liu HS, Jan MS, Chou CK, Chen PH, Ke NJ. Is green fluorescent protein toxic to the living cells? Biochem. Biophys. Res. Commun. 1999;260(3):712–717. doi: 10.1006/bbrc.1999.0954. [DOI] [PubMed] [Google Scholar]
- 10.Chen N, Kamath S, Newcomb J, et al. Trophic factor induction of human umbilical cord blood cells in vitro and in vivo. J. Neural. Eng. 2007;4(2):130–145. doi: 10.1088/1741-2560/4/2/013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krestel HE, Mihaljevic AL, Hoffman DA, Schneider A. Neuronal co-expression of EGFP and beta-galactosidase in mice causes neuropathology and premature death. Neurobiol. Dis. 2004;17(2):310–318. doi: 10.1016/j.nbd.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Kurre P, Morris J, Andrews RG, Kohn DB, Kiem HP. Kinetics of fluorescence expression in nonhuman primates transplanted with GFP retrovirus-modified CD34 cells. Mol. Ther. 2002;6(1):83–90. doi: 10.1006/mthe.2002.0623. [DOI] [PubMed] [Google Scholar]
- 13.Rosenzweig M, Connole M, Glickman R, et al. Induction of cytotoxic T lymphocyte and antibody responses to enhanced green fluorescent protein following transplantation of transduced CD34+ hematopoietic cells. Blood. 2001;97(7):1951–1959. doi: 10.1182/blood.v97.7.1951. [DOI] [PubMed] [Google Scholar]
- 14.Rossi SL, Nistor G, Wyatt T, et al. Histological and functional benefit following transplantation of motor neuron progenitors to the injured rat spinal cord. PLoS ONE. 2010;5(7):e11852. doi: 10.1371/journal.pone.0011852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rizvanov AA, Guseva DS, Salafutdinov II, et al. Genetically modified human umbilical cord blood cells expressing vascular endothelial growth factor and fibroblast growth factor 2 differentiate into glial cells after transplantation into amyotrophic lateral sclerosis transgenic mice. Exp. Biol. Med. (Maywood) 2011;236(1):91–98. doi: 10.1258/ebm.2010.010172. [DOI] [PubMed] [Google Scholar]
- 16.Lepore AC, O'Donnell J, Kim AS, et al. Human glial-restricted progenitor transplantation into cervical spinal cord of the SOD1 mouse model of ALS. PLoS ONE. 2011;6(10):e25968. doi: 10.1371/journal.pone.0025968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hodgetts SI, Simmons PJ, Plant GW. Human mesenchymal precursor cells (Stro-1+) from spinal cord injury patients improve functional recovery and tissue sparing in an acute spinal cord injury rat model. Cell Transplant. 2013;22(3):393–412. doi: 10.3727/096368912X656081. [DOI] [PubMed] [Google Scholar]
- 18.Draper JS, Pigott C, Thomson JA, Andrews PW. Surface antigens of human embryonic stem cells: changes upon differentiation in culture. J. Anat. 2002;200(Pt 3):249–258. doi: 10.1046/j.1469-7580.2002.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; • HLA antigens are known to be ubiquitously and broadly expressed throughout the human body. In this study, unexpectedly, Draper et al. showed that the expression of HLA was downregulated upon differentiation of embryonic stem cells. This suggests that HLA antigens are not stable biomarkers of embryonic stem cells during their differentiation and may not be used as reliable biomarkers to follow their fate in xenotransplantation studies.
- 19.Lathia JD, Gallagher J, Myers JT, et al. Direct in vivo evidence for tumor propagation by glioblastoma cancer stem cells. PLoS ONE. 2011;6(9):e24807. doi: 10.1371/journal.pone.0024807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Burkhardt JK, Hofstetter CP, Santillan A, et al. Orthotopic glioblastoma stem-like cell xenograft model in mice to evaluate intra-arterial delivery of bevacizumab: from bedside to bench. J. Clin. Neurosci. 2012;19(11):1568–1572. doi: 10.1016/j.jocn.2012.03.012. [DOI] [PubMed] [Google Scholar]
- 21.Ebert AD, Yu J, Rose FF, Jr, et al. Induced pluripotent stem cells from a spinal muscular atrophy patient. Nature. 2009;457(7227):277–280. doi: 10.1038/nature07677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lemonnier T, Blanchard S, Toli D, et al. Modeling neuronal defects associated with a lysosomal disorder using patient-derived induced pluripotent stem cells. Hum. Mol. Genet. 2011;20(18):3653–3666. doi: 10.1093/hmg/ddr285. [DOI] [PubMed] [Google Scholar]
- 23.Popescu IR, Nicaise C, Liu S, et al. Neural progenitors derived from human induced pluripotent stem cells survive and differentiate upon transplantation into a rat model of amyotrophic lateral sclerosis. Stem Cells Transl. Med. 2013;2(3):167–174. doi: 10.5966/sctm.2012-0042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campanelli JT, Sandrock RW, Wheatley W, et al. Expression profiling of human glial precursors. BMC Dev. Biol. 2008;8:102. doi: 10.1186/1471-213X-8-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lepore AC. Intraspinal cell transplantation for targeting cervical ventral horn in amyotrophic lateral sclerosis and traumatic spinal cord injury. J. Vis. Exp. 2011;(55):3069. doi: 10.3791/3069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicaise C, Putatunda R, Hala TJ, et al. Degeneration of phrenic motor neurons induces long-term diaphragm deficits following mid-cervical spinal contusion in mice. J. Neurotrauma. 2012;29(18):2748–2760. doi: 10.1089/neu.2012.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nicaise C, Hala TJ, Frank DM, et al. Phrenic motor neuron degeneration compromises phrenic axonal circuitry and diaphragm activity in a unilateral cervical contusion model of spinal cord injury. Exp. Neurol. 2012;235(2):539–552. doi: 10.1016/j.expneurol.2012.03.007. [DOI] [PubMed] [Google Scholar]
- 28.Decaestecker C, Lopez XM, D'Haene N, et al. Requirements for the valid quantification of immunostains on tissue microarray materials using image analysis. Proteomics. 2009;9(19):4478–4494. doi: 10.1002/pmic.200800936. [DOI] [PubMed] [Google Scholar]
- 29.Kordower JH, Freeman TB, Snow BJ, et al. Neuropathological evidence of graft survival and striatal reinnervation after the transplantation of fetal mesencephalic tissue in a patient with Parkinson's disease. N. Engl. J. Med. 1995;332(17):1118–1124. doi: 10.1056/NEJM199504273321702. [DOI] [PubMed] [Google Scholar]
- 30.Li JY, Englund E, Holton JL, et al. Lewy bodies in grafted neurons in subjects with Parkinson's disease suggest host-to-graft disease propagation. Nat. Med. 2008;14(5):501–503. doi: 10.1038/nm1746. [DOI] [PubMed] [Google Scholar]; • Patients with Parkinson's disease who had long-term survival of transplanted fetal dopaminergic neurons (11–16 years) developed α-synuclein-positive Lewy bodies in grafted neurons. This is a pioneering transplantation clinical trial demonstrating the long-term survival of human-grafted cells in the CNS. Unfortunately, authors provided evidence that the disease (where α-synuclein protein seeds from one neuron to another one) could propagate from host to healthy grafted cells.
- 31.Sheth RN, Manzano G, Li X, Levi AD. Transplantation of human bone marrow-derived stromal cells into the contused spinal cord of nude rats. J. Neurosurg. Spine. 2008;8(2):153–162. doi: 10.3171/SPI/2008/8/2/153. [DOI] [PubMed] [Google Scholar]
- 32.Li L, Jiang Q, Qu CS, et al. Transplantation of marrow stromal cells restores cerebral blood flow and reduces cerebral atrophy in rats with traumatic brain injury: in vivo MRI study. J. Neurotrauma. 2011;28(4):535–545. doi: 10.1089/neu.2010.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Just L, Timmer M, Tinius J, et al. Identification of human cells in brain xenografts and in neural co-cultures of rat by in situ hybridisation with Alu probe. J. Neurosci. Methods. 2003;126(1):69–77. doi: 10.1016/s0165-0270(03)00065-7. [DOI] [PubMed] [Google Scholar]
- 34.Cummings BJ, Uchida N, Tamaki SJ, et al. Human neural stem cells differentiate and promote locomotor recovery in spinal cord-injured mice. Proc. Natl Acad. Sci. USA. 2005;102(39):14069–14074. doi: 10.1073/pnas.0507063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Guzman R, Uchida N, Bliss TM, et al. Long-term monitoring of transplanted human neural stem cells in developmental and pathological contexts with MRI. Proc. Natl Acad. Sci. USA. 2007;104(24):10211–10216. doi: 10.1073/pnas.0608519104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tamaki SJ, Jacobs Y, Dohse M, et al. Neuroprotection of host cells by human central nervous system stem cells in a mouse model of infantile neuronal ceroid lipofuscinosis. Cell Stem Cell. 2009;5(3):310–319. doi: 10.1016/j.stem.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 37.Salazar DL, Uchida N, Hamers FP, Cummings BJ, Anderson AJ. Human neural stem cells differentiate and promote locomotor recovery in an early chronic spinal cord injury NOD-scid mouse model. PLoS ONE. 2010;5(8):e12272. doi: 10.1371/journal.pone.0012272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dixon JA, Spinale FG. Large animal models of heart failure: a critical link in the translation of basic science to clinical practice. Circ. Heart Fail. 2009;2(3):262–271. doi: 10.1161/CIRCHEARTFAILURE.108.814459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ravindran G, Rao HS. Enriched NCAM-positive cells form functional dopaminergic neurons in the rat model of Parkinson's disease. Stem Cells Dev. 2006;15(4):575–582. doi: 10.1089/scd.2006.15.575. [DOI] [PubMed] [Google Scholar]
- 40.Coates PJ, Hales SA, Hall PA. The association between cell proliferation and apoptosis: studies using the cell cycle-associated proteins Ki67 and DNA polymerase alpha. J. Pathol. 1996;178(1):71–77. doi: 10.1002/(SICI)1096-9896(199601)178:1<71::AID-PATH456>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 41.Baisch H. Elevated Ki-67 expression is correlated with TNFalpha- and IFNgamma-induced apoptosis in tumour cells. Cell Prolif. 2002;35(6):333–342. doi: 10.1046/j.1365-2184.2002.00243.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verheijen R, Kuijpers HJ, Van Driel R, et al. Ki-67 detects a nuclear matrix-associated proliferation-related antigen. II. Localization in mitotic cells and association with chromosomes. J. Cell Sci. 1989;92(Pt 4):531–540. doi: 10.1242/jcs.92.4.531. [DOI] [PubMed] [Google Scholar]
- 43.Wharton SB, Chan KK, Anderson JR, Stoeber K, Williams GH. Replicative Mcm2 protein as a novel proliferation marker in oligodendrogliomas and its relationship to Ki67 labelling index, histological grade and prognosis. Neuropathol. Appl. Neurobiol. 2001;27(4):305–313. doi: 10.1046/j.0305-1846.2001.00333.x. [DOI] [PubMed] [Google Scholar]
- 44.Hill CE, Hurtado A, Blits B, et al. Early necrosis and apoptosis of Schwann cells transplanted into the injured rat spinal cord. Eur. J. Neurosci. 2007;26(6):1433–1445. doi: 10.1111/j.1460-9568.2007.05771.x. [DOI] [PubMed] [Google Scholar]
- 45.Becker M, Nitsche A, Neumann C, Aumann J, Junghahn I, Fichtner I. Sensitive PCR method for the detection and real-time quantification of human cells in xenotransplantation systems. Br. J. Cancer. 2002;87(11):1328–1335. doi: 10.1038/sj.bjc.6600573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee ST, Chu K, Kim EH, et al. Quantification of human neural stem cell engraftments in rat brains using ERV-3 real-time PCR. J. Neurosci. Methods. 2006;157(2):225–229. doi: 10.1016/j.jneumeth.2006.04.019. [DOI] [PubMed] [Google Scholar]; • One of the methods for quantification of human cells engrafted in animal tissues is based on PCR. Lee et al. used real-time PCR targeting the ERV-3 sequence, which is an endogenous retrovirus present with a known copy number in all human cells, but not present in rodent cells, in order to detect human neural stem cells transplanted in rat brains.
- 47.Cheng K, Gupta S. Quantitative tools for assessing the fate of xenotransplanted human stem/progenitor cells in chimeric mice. Xenotransplantation. 2009;16(3):145–151. doi: 10.1111/j.1399-3089.2009.00526.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Using real-time PCR and in situ hybridization, Cheng and Gupta found that specific probes against Charcot-Marie-Tooth disease element, short tandem repeats for HLA regions or human sex-determining region were efficient at quantifying human cells in human–mouse chimeric tissues.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.