Abstract
Clostridium botulinum is a dangerous pathogen that forms the highly potent botulinum toxin, which when ingested causes a deadly neuroparalytic disease. The closely related Clostridium sporogenes is occasionally pathogenic, frequently associated with food spoilage and regarded as the non-toxigenic equivalent of Group I C. botulinum. Both species form highly resistant spores that are ubiquitous in the environment and which, under favourable growth conditions germinate to produce vegetative cells. To improve the control of botulinum neurotoxin-forming clostridia, it is imperative to comprehend the mechanisms by which spores germinate. Germination is initiated following the recognition of small molecules (germinants) by a specific germinant receptor (GR) located in the spore inner membrane. The present study precisely defines clostridial GRs, germinants and co-germinants. Group I C. botulinum ATCC3502 contains two tricistronic and one pentacistronic GR operons, while C. sporogenes ATCC15579 has three tricistronic and one tetracistronic GR operons. Insertional knockout mutants, allied with characterisation of recombinant GRs shows for the first time that amino acid stimulated germination in C. botulinum requires two tri-cistronic encoded GRs which act in synergy and cannot function individually. Spore germination in C. sporogenes requires one tri-cistronic GR. Two other GRs form part of a complex involved in controlling the rate of amino-acid stimulated germination. The suitability of using C. sporogenes as a substitute for C. botulinum in germination studies and food challenge tests is discussed.
Author Summary
Clostridium botulinum is a dangerous pathogen that forms the deadly botulinum neurotoxin. Strains of C. botulinum are present in the environment as spores. Under suitable conditions, the dormancy of the bacterial spore is broken, and germination occurs. Germination is initiated following the recognition of small molecules by a specific germinant receptor (GR) located within spores. Currently, the identification and characterisation of these GRs remains unknown, but is critical if strategies are to be developed to either prevent spore germination altogether, or to germinate all the spores and then inactivate the emergent sensitive vegetative cells. The present study has characterised two functionally active GRs in C. botulinum which act in synergy and cannot function individually, and a related functionally active GR in C. sporogenes. These GRs respond to amino acids. Other GRs appear to form part of a complex involved in controlling the speed of germination, or are not functionally active. This study provides new insights into the mechanisms involved in germination and will allow us to develop new strategies to control this deadly pathogen.
Introduction
Clostridium botulinum and Clostridium sporogenes are closely related anaerobic spore-forming bacteria. C. botulinum is a dangerous pathogen that forms the deadly botulinum neurotoxin. This is the most potent toxin known, as little as 30–100 ng can be fatal [1]. Eight distinct types of botulinum neurotoxin (types A to H), and more than thirty different neurotoxin sub-types (e.g. sub-types A1 to A5) are recognised [2]–[5]. The botulinum neurotoxins are 150 kD proteins with zinc-endopeptidase activity that block acetylcholine transmission in cholinergic nerves, leading to a floppy paralysis known as botulism, that may prove fatal to both humans and animals [6], [7]. The most frequently reported types of human botulism are foodborne botulism, infant botulism and wound botulism [1], [8]. Foodborne botulism is an intoxication caused by consumption of neurotoxin formed by C. botulinum following spore germination and growth of vegetative cells in food. Infant and wound botulism are infections involving spore germination, growth of vegetative cells and neurotoxin formation in the gut of young infants and in deep wounds (often associated with drug abuse), respectively. Botulinum neurotoxins are also important pharmaceuticals used to treat a range of localised conditions e.g. blepharospasm, hemifacial spasm, and for cosmetic purposes [9].
C. botulinum is a heterogeneous species that comprises a complex of four distinct groups of bacteria that share the common property of forming the botulinum neurotoxin [3], [10], [11]. Group I (proteolytic) C. botulinum is associated with foodborne botulism, infant botulism and wound botulism, and forms one or more neurotoxins of types A, B, F or H [3], [5], [10], [11]. Strains of Group I C. botulinum that form type A1 neurotoxin have received the most attention to date because they are often associated with botulism in humans, the extreme potency of the type A1 neurotoxin, and due to the use of type A1 neurotoxin as a pharmaceutical [1], [12]–[15]. Indeed, the first C. botulinum genome to be sequenced was that of Group I C. botulinum type A1 strain ATCC3502 [16].
C. sporogenes is occasionally pathogenic [17], a significant cause of food spoilage [18], and because of its strong physiological similarity to Group I C. botulinum is very widely used as a surrogate for this organism in demonstrating the effectiveness of food preservation processes [19], [20]. Genome sequencing, whole genome analysis using DNA microarrays, and other typing methods have confirmed the close genetic relationship of C. sporogenes and Group I C. botulinum [2], [21]–[23]. The formation of botulinum neurotoxin is used to distinguish strains of Group I C. botulinum from those of C. sporogenes [19].
Strains of Group I C. botulinum and C. sporogenes are both present in the environment as spores. This highly resistant dormant state enables survival in adverse conditions (e.g. absence of nutrients, UV light, heat treatment, radiation, desiccation, high pressure, toxic chemicals) that vegetative cells would not survive, and their formation by Group I C. botulinum and C. sporogenes is a primary reason why these bacteria present a significant food safety and food spoilage problem. Significantly, strains of Group I C. botulinum and C. sporogenes form spores of very high heat resistance, and the “botulinum cook” has been adopted by the canning industry as the standard minimum heat treatment (121°C for 3 min) for low acid canned foods [1]. Under suitable conditions, the dormancy of bacterial spores is broken, and germination occurs. This is often initiated by a germinant receptor (GR) located in the spore inner membrane responding to nutrient germinants, and is followed by the release of dipicolinic acid and partial core hydration. Later, cortex-lytic enzymes degrade peptidoglycan in the spore cortex, enabling further core hydration and core expansion, and this results in the loss of spore resistance [24]. Germination involves pre-formed enzymes located in the dormant spore, and is followed by the initiation of metabolism and macromolecular synthesis, eventually leading to the emergence of a cell that is able to multiply. One approach that has interested microbiologists for many decades is to develop strategies to either prevent spore germination altogether and thereby prevent subsequent growth, or to germinate all the spores and then inactivate the emergent sensitive vegetative cells. Unfortunately the highly heterogeneous nature of spore germination (as observed for example with Group II C. botulinum) [25]–[27] has prevented the development of suitable processes. However, a greater understanding of spore germination in Group I C. botulinum and C. sporogenes may enable the development of novel intervention strategies to prevent or reduce disease and other adverse events such as food spoilage.
Spore germination in Bacillus species generally involves a GR located in the spore inner membrane. The GR is composed of three proteins (A, B and C) that are encoded in a tricistronic operon. The A and B proteins are integral membrane proteins, while the C proteins are lipoproteins [24], [28]. Spore germination in Clostridium species is not as extensively studied as that in Bacillus species, although recently significant advancements have been made with several clostridia including C. perfringens, C. difficile and C. sordellii [24], [29]–[43]. Spore germination in clostridia often proceeds more slowly than that in Bacillus species [1], and recent evidence suggests that although there are many similarities there are also a number of important differences in spore germination between clostridia and Bacillus species [24], [28], [29]. For example, spores of Bacillus species require a complete GR system to germinate effectively, and while this is also the case for some clostridia, spores of C. difficile are able to germinate effectively in the absence of what is classically understood as a GR. Such differences between clostridia probably reflect their wide genetic diversity [1], [24]. Spores of Group I C. botulinum and C. sporogenes germinate when specific germinant nutrients such as a combination of L-alanine and L-lactate (with less efficient germination in response to other amino acids and L-lactate or single amino acids [3], [44]) interact with a GR located in the clostridial spore inner membrane. GR operons are well conserved amongst strains of Group I C. botulinum. Group I C. botulinum type A strains ATCC3502, Hall, ATCC19397, Kyoto and NCTC2012 possess a pentacistronic GR operon (gerXB-XA2-XB2-XC2-XB), two tricistronic GR operons (gerXA1-XB1-XC1 and gerXA3-XB3-XC3), and an orphan gerXB homologue [3], [45]. Gene clusters resembling the pentacistronic GR operon and gerXA3-XB3-XC3 have been characterised in C. sporogenes strain NCIMB701792 and Group I C. botulinum type B strain NCTC7273, respectively [16], [46]. Strains of Group I C. botulinum type B (Okra) and F (Langeland) possess an additional tricistronic GR operon (gerXC4-XA4-XB4) [3], [45]. It is still not known, however, to which nutrient germinant(s) the various specific GRs are responding, or the relative importance of each of the GRs.
The purpose of the present study was to dissect, at the molecular level, spore germination in Group I C. botulinum strain ATCC3502 and in C. sporogenes strain ATCC15579. In particular, the key aims were to establish for each strain which of the multiple nutrient GRs was active in spore germination, and for the active nutrient GRs which nutrient germinant(s) they responded to. This was achieved using a combination of genomic, genetic and physiological approaches. The spore GRs in the two clostridia showed a number of interesting similarities, and this study provided further evidence of differences between spore germination in Clostridium and Bacillus species. Interestingly, subtle differences were also noted in spore germination between that in Group I C. botulinum strain ATCC3502 and that in C. sporogenes strain ATCC15579. This study has provided novel insights into spore germination in these two important clostridia.
Results
Involvement of L-lactate in amino acid induced germination in C. botulinum and C. sporogenes
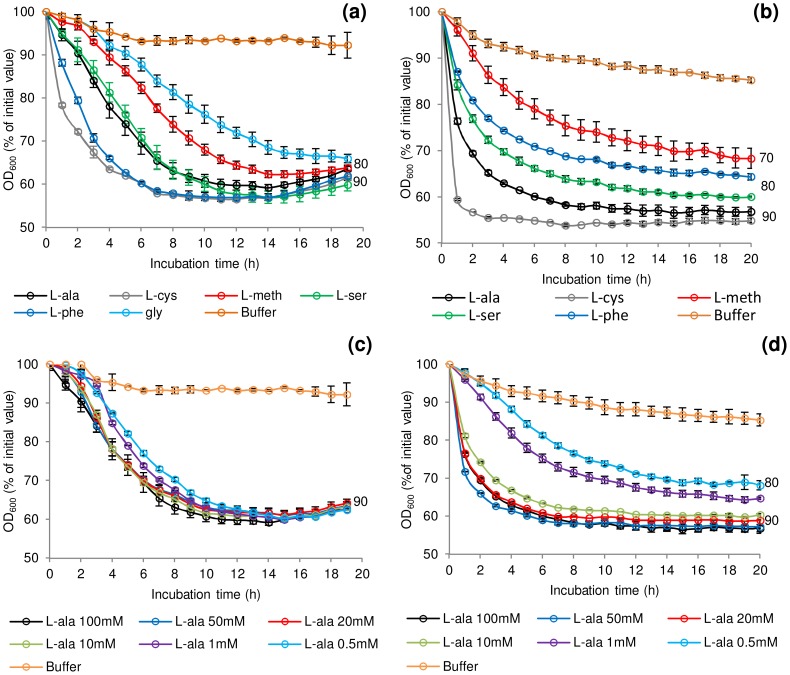
A selection of amino acids at various concentrations (Table S1) were assessed for their individual effect on germination of spores of C. botulinum and C. sporogenes. The majority of the amino-acids tested have previously been reported to contribute as germinants or co-germinants for spores of Clostridium or Bacillus [1]. Initial tests showed that spore germination was similar under aerobic and anaerobic conditions (data not shown). This confirmed previous reports for C. botulinum and C. sporogenes [47], [48]. In the presence of Tris buffer (pH 7.4), L-lactate (50 mM) and NaHCO3 (50 mM) at 30°C the addition at 100 mM of either L-alanine or L-cysteine initiated spore germination in C. botulinum and C. sporogenes, although at differing rates (Figure 1a & 1b). L-lactate was not essential for L-alanine or L-cysteine stimulated germination and had no effect on rate or the overall extent of germination (data not shown). Three amino acids (L-methionine, L-serine, L-phenylalanine) each required L-lactate for inducement of germination in both species. The addition of L-lactate on its own failed to stimulate germination (<10% fall in OD600, equating to <1% germination). Spores of C. botulinum, but not those of C. sporogenes, were also germinated by glycine in combination with L-lactate (Figure 1a). L-cysteine combined with L-lactate produced the most rapid germination of C. botulinum spores (40% of initial OD600, approximately 90% germination after 6 h) and C. sporogenes spores (40% of initial OD600, approximately 90% germination after 4 h). C. sporogenes germination proceeded far more rapidly to completion with all the tested amino acids compared to C. botulinum. A number of other amino acids were tested, but failed to induce spore germination in either C. botulinum or C. sporogenes, both in the presence and absence of L-lactate (Table S1). For both species, the saturation concentration of the amino acids was 50–100 mM with l-lactate at 50 mM (Figure 1c & 1d). For simplicity, L-lactate was included in all subsequent germination studies. The optimum pH range was pH 6–8 for germination with most amino acids + l-lactate. However, spore germination was also observed at pH 10 in the presence of L-serine, and L-alanine + l-lactate. The rich microbiological growth medium, TY medium, was not optimum for spore germination, with less germination observed than in the defined system, although the addition of L-lactate did increase the rate and overall extent of germination in TY medium. Spore germination measured using the Bioscreen system correlated well with direct counts of phase-dark spores by phase-contrast microscopy (data not shown) with an OD600 fall of 40% correlating to >90% germination of spores.
Figure 1. Rate of C. botulinum and C. sporogenes spore germination in the presence of selected amino acids.
Effect of L-alanine, L-cysteine, L-methionine, L-serine, L-phenylalanine and glycine (C. botulinum only) at 100 mM on germination of (a) C. botulinum ATCC3502 and (b) C. sporogenes ATCC15579. The effect of increasing L-alanine concentrations (0.5 mM–100 mM) on germination was also tested on (c) C. botulinum ATCC3502 and (d) C. sporogenes ATCC15579. Tests were conducted in 20 mM Tris buffer, pH 7.4, L-lactate (50 mM) and NaHCO3 (50 mM) at 30°C. Buffer only controls contained 20 mM Tris, pH 7.4, L-lactate (50 mM) and NaHCO3 (50 mM). Data labels (right) refer to percentage germination observed by phase contrast microscopy at the end of the experiment. Error bars represent the standard deviation of 3 independent experiments.
Spore production environment affects rate of germination
The effect of sporulation medium and sporulation temperature on the subsequent germination properties of C. sporogenes spores was determined (Figure 2). Assessment of C. botulinum was precluded by frequent poor sporulation of this strain, which was less than 5% and 1% in Robertson's cooked meat broth (CMB) and TY respectively, compared to the optimum yield of 30% spores on RCM plus skimmed milk (RCM+SM) plates at 37°C. C. sporogenes spores were produced using either RCM+SM, CMB or TY broth at 37°C. Microscopic observations showed sporulation was notably lower (ca. 40%) using CMB and TY compared to RCM+SM. C. sporogenes spores were also produced at 15, 20, 28, 30, 37 and 42°C in CMB to evaluate the effect of temperature. Microscopic observations showed that sporulation was notably lower (ca.50%) at 15°C compared to 37°C in CMB, with sporulation not observed at 42°C. Germination of the spores was then evaluated in the presence of L-alanine + L-lactate (Figure 2). Interestingly, spores produced in CMB at 37°C germinated at a faster rate compared to spores produced in the other media at 37°C. Sporulation temperature also affected germination rates, with spores produced at 37°C germinating more rapidly than spores produced at other temperatures (Figure 2). Subsequently, all spore crops used in germination studies were produced at 37°C in CMB for C. sporogenes and on RCM+SM for C. botulinum.
Figure 2. The effect of spore production environment on subsequent germination of C. sporogenes spores.
For germination tests, spores were incubated in 20 mM Tris buffer, pH 7.4, L-alanine (100 mM), L-lactate (50 mM) and NaHCO3 (50 mM) at 30°C. Spores were produced in CMB at different temperatures; 15°C, 20°C, 28°C, 30°C, 37°C. Spores were also produced in TY broth at 37°C and on RCM+SM plates at 37°C. Spores produced at 37°C in CMB and incubated in 20 mM Tris, pH 7.4, L-lactate (50 mM) and NaHCO3 (50 mM) only, were included as a negative control (WT Buffer). Spore germination was confirmed by phase contrast microscopy. Data labels (right) refer to percentage germination observed by phase contrast microscopy at the end of the experiment. Error bars represent the standard deviation of 3 independent experiments.
D-amino acids prevent L-amino acid induced germination
Unlike their L-isomers, D-alanine, D-cysteine, D-methionine, D-phenylalanine and D-serine all failed to trigger spore germination in either C. sporogenes or C. botulinum. Moreover, the D-amino acids prevented spore germination (defined as a <10% fall in OD600, equating to <1% germination observed microscopically) in C. botulinum and C. sporogenes induced by their equivalent L-amino acid (Table 1). To ascertain if specific D-amino acids could prevent germination by non-equivalent L-amino acids, D-serine was tested at a ten-fold excess of each of the five L-amino acids. Interestingly, D-serine prevented germination in C. sporogenes induced by L-cysteine, L-methionine, L-phenylalanine and L-serine, and to a lesser extent L-alanine. L-alanine and L-cysteine induced germination was only slightly affected by a ten-fold excess of D-serine in C. botulinum. However, when a 100-fold excess of D-alanine was added, spore germination in the presence of each of the five L-amino acids was prevented in C. botulinum. Finally, in order to assess whether the D-amino acid is acting specifically rather than being simply in excess of the L-amino acid germinant, we performed further tests using different germinants (L-alanine, 1 mM or L-serine, 20 mM) each combined with an excess (100 mM and 40 mM respectively) of the non-germinant L-valine. The addition of excess non-germinant in each experiment had no effect on final germination levels. However, the D-amino acids (in this case alanine or serine) continued to be inhibitory.
Table 1. Minimum D-amino acid concentration required to prevent* germination by its equivalent L-amino acid.
| D-amino acid concentration | ||
| L-amino acid/concentration | C. sporogenes | C. botulinum |
| L-alanine/1 mM | 10 mM | 100 mM |
| L-cysteine/10 mM | 100 mM | 100 mM |
| L-serine/10 mM | 20 mM | 100 mM |
| L-phenylalanine/10 mM | 20 mM | NT |
| L-methionine/10 mM | 100 mM | 100 mM |
Tests were conducted in 20 mM Tris buffer, pH 7.4, L-amino acid (1–10 mM), D-amino acid (10–100 mM) + L-lactate (50 mM) and NaHCO3 (50 mM) at 30°C. NT = D-phenylalanine was not inhibitory at 20 mM and could not be tested at a higher concentration due to low solubility.
*Defined as a <10% fall in OD600, equating to <1% germination observed microscopically.
Homologues of the C. botulinum nutrient germinant receptor operons in C. sporogenes
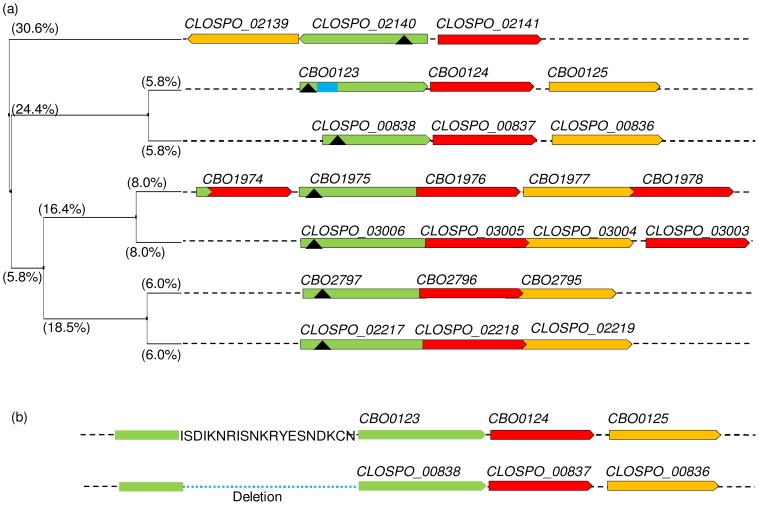
Homologues of Group I C. botulinum strain ATCC3502 GR sub-units (gerXA, gerXB, gerXC) were identified by BLASTp analyses against a draft un-assembled genome of C. sporogenes strain ATCC15579 (Figure 3). Analysis showed that the C. sporogenes strain contains three tricistronic GR operons and one tetracistronic GR operon. In comparison C. botulinum ATCC3502 has two tricistronic GR operons and one pentacistronic GR operon. Each strain has an additional orphan gerXB subunit homologue. Alignment using Clustal Omega and using Jalview to produce a tree showing the average distance based on amino acid sequence identity (%), revealed homology between the GR operons (Figure 3). Each operon in C. botulinum had a closely related operon in C. sporogenes (from 11.6–16.0% difference in identity for each gerXA), while the CLOSPO_02140 gerXA is most distant with regards to sequence % identity. Transmembrane helix (TMH) prediction analysis showed that the gerXA sub-units of C. botulinum and C. sporogenes have between three and five TMHs, and the gerXB subunits have ten TMHs. The gerXC subunits were predicted to be lipoproteins and encode a signal peptide. Interestingly, more detailed sequence analysis of the pentacistronic operon of C. botulinum reveals that although the first gene of the operon, CBO1974, is a full-length member of the gerXB family, the 5′ end of its coding region is overlapped by a small (162 bp) region of a gerXA gene, annotated as CBO1973A. Comparative analysis between C. botulinum and C. sporogenes ger homologues also revealed that the gerXA gene, CLOSPO_00838 lacks an uninterrupted region encoding 20 amino acid residues which map to residues 74–93 of the predicted translation for CBO0123 (Figure 3b). A database search shows that this deletion (with respect to C. botulinum strain ATCC3502) is not confined to C. sporogenes strain ATCC15579, but can be found in 10 of a total of 19 GerA peptide sequences from proteomes of C. sporogenes (2), Group I C. botulinum (4) and Group III C. botulinum (4) (data not shown). Similar deletions were not found in any Group II C. botulinum GerA peptides. The function of this apparently conserved region remains unknown.
Figure 3. Alignment of C. botulinum and C. sporogenes germinant receptor proteins.
Homologues of C. botulinum (strain ATCC3502) GRs were identified by BLASTp analyses using the draft un-assembled genome of C. sporogenes (strain ATCC15579). (a) Tree calculated (using Jalview [76]) from the pairwise sequence distances between GerXA only (determined from % sequence identities) of C. sporogenes (CLOSPO_number.) and C. botulinum (CBOnumber.) GRs, using the UPGMA algorithm [76]; average distances between GerXA (green) are shown on the branches. GerXB (red) and GerXC (yellow) are shown on the same tree (UPGMA produced identical-topology trees for each of the GerXB, GerXC proteins; distances not shown). (black triangle) Position of insertion sites of retargeted introns for mutations (in equivalent DNA sequence). Small green coloured region of CBO1974 represents a small protein fragment (CBO1973A), the coding sequence of which overlaps that of CBO1974, with homology to the C terminus of a GerXA protein. Blue square; 20 amino acid section that is deleted in its C. sporogenes homologue, CLOSPO_00838. (b) More detailed version of part of the above tree, showing the amino acid sequence encoded by a region in CBO0123 that is deleted from its C. sporogenes homologue, CLOSPO_00838.
Mutation of putative germinant receptors
To characterise the functionality of the putatively identified germination GRs, a series of single (C. botulinum and C. sporogenes), and triple (C. sporogenes) insertion mutants were constructed. Furthermore, a C. sporogenes quadruple insertional knockout GR mutant (gerXA 4−) was also generated. The current insertional knockout system does not allow multiple insertion selection, as following one insertion the single mutant is then erythromycin resistant. However, the mutant generation system was shown to be highly efficient, which negated the need for a different antibiotic selection in the multiple insertion mutants. All insertion events were tested by PCR which confirmed chromosomal integration of the intron (Figure S1). PCR using gene specific and intron specific primers confirmed insertion of the intron into the GR gene; this was further confirmed by PCR with gene specific primers flanking the insertion site producing a ∼2 kb product (Figure S1). Insertion events were verified by Southern hybridization using an intron specific probe which confirmed the correct number of insertion events in all the constructed mutants (Figure S1c).
C. botulinum requires two tri-cistronic receptors for germination
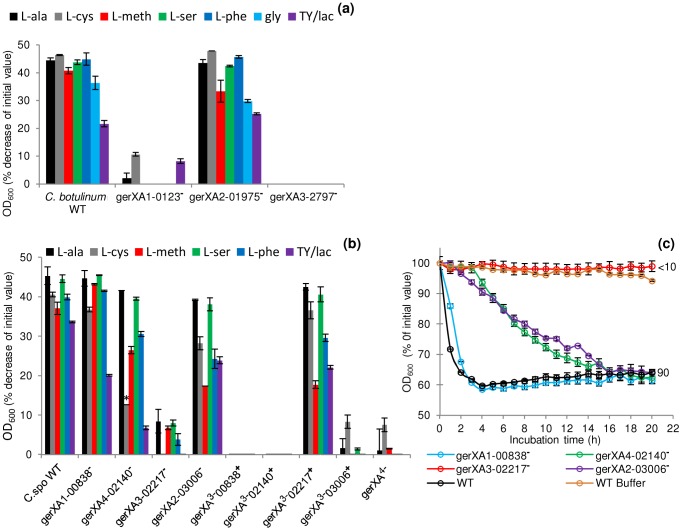
To characterise the GRs and to identify their cognate germinants, single insertional knockout mutants (gerXA1-0123−, gerXA2-1975−, gerXA3-2797−) were created for each of the identified GR operons in C. botulinum. Spores generated from these mutants were then analysed for amino acid stimulated germination using L-alanine, L-cysteine, L-methionine, L-serine, L-phenylalanine or glycine, all in the presence of L-lactate (Figure 4a). The OD600 of wild-type spores decreased (∼40%) indicating efficient and complete spore germination in the presence of each amino acid. There was a ∼20% decrease in OD600 with TY medium + L-lactate. In contrast, the gerXA3-2797− mutant failed to germinate with any of the amino acids tested even after 20 h of exposure (<1% germination observed microscopically). Moreover, germination was not observed with a nutrient rich broth (TY + L-lactate) suggesting that CBO2797 is essential for amino acid stimulated germination. The mutant gerXA1-0123− also failed to germinate to the same extent as the wild type, with a small decrease (<10%) in OD600 observed with L-cysteine and with nutrient rich broth (TY + L-lactate). In neither case could spore germination be observed microscopically. These results suggest that CBO0123 is also essential for amino acid induced germination. Interestingly, the gerXA2-1975− mutant showed similar germination patterns to those of WT spores (Figure 4a). Complementation was performed by two different approaches; using plasmid pMTL83151esp, which relies on the putative native promoter of the gene, or using pMTL83151fdx which includes the powerful promoter Pfdx of the ferredoxin gene (fdx) from C. sporogenes. Complementation was successful for one of the two GerXAs observed to be important for nutrient-induced germination. Introduction of the receptor CBO0123-CBO0124-CBO0125 complementation vector (pMTL83151esp or pMTL83151fdx) successfully restored germination to the mutant gerXA1-0123−, albeit at a different rate compared to that of the wild type (Figure 5a). Introduction of the GR CBO2797-CBO2796-CBO2795 complementation vector (pMTL83151esp or pMTL83151fdx) drastically reduced sporulation efficiency, giving insufficient spores to allow assessment of the germination phenotype.
Figure 4. Mutation of specific receptors precludes amino acid stimulated germination.
Spores were incubated in 20 mM Tris buffer, pH 7.4, amino acid (100 mM) + L-lactate (50 mM) and NaHCO3 (50 mM) at 30°C for 20 hours with L-alanine, L-cysteine, L-methionine, L-serine, L-phenylalanine, glycine (C. botulinum only) or in TY + L-lactate (50 mM). (a) C. botulinum single insertional knockout mutants and wild type spore germination. (b) C. sporogenes single insertional knockout mutants, triple insertional knockout mutants, quadruple insertional knockout GR mutant and wild type spore germination. * L-cysteine is a relatively insoluble amino acid and precipitates out of solution after 2 hours. Due to the 4 hour delay in germination of the mutant gerXA4-02140− cysteine precipitation caused OD600 readings to be unrepresentative and therefore germination was confirmed by microscopy. (c) Alanine induced germination rates of single insertional knockout GR mutants for C. sporogenes were determined using spores generated from the wild type (C. sporogenes ATCC15579) and mutants gerXA1-00838−, gerXA4-02140−, gerXA3-02217−, gerXA2-03006−. WT spores incubated in buffer only (see above), were included as a negative control (WT Buffer). Data labels (right) refer to percentage germination observed by phase contrast microscopy at the end of the experiment. Error bars in (a–c) represent the standard deviation of 3 independent experiments. Spore germination was confirmed by phase contrast microscopy.
Figure 5. Germination rates of complemented GR mutants for C. botulinum and C. sporogenes.
Spores were incubated in 20 mM Tris buffer, pH 7.4, amino acid (100 mM) + L-lactate (50 mM) + NaHCO3 (50 mM) at 30°C with L-cysteine (C. botulinum), L-alanine (C. sporogenes). (a) C. botulinum mutant gerXA1-0123− complemented with plasmid pMTL8315esp (gerXA1-0123−esp) or plasmid pMTL8315fdx (gerXA1-0123−fdx). (b) C. sporogenes mutant gerXA3-02217− complemented with plasmid pMTL8315esp (gerXA3-02217−esp) or plasmid pMTL8315fdx (gerXA3-02217−fdx). There were two negative controls. Firstly, the uncomplemented mutant (gerXA1-0123− or gerXA3-02217−), secondly WT spores incubated in 20 mM Tris buffer (pH 7.4) + L-lactate (50 mM) + NaHCO3 (50 mM) only (WT Buffer). Spore germination was confirmed by phase contrast microscopy. Error bars represent the standard deviation of 3 independent experiments. Data labels (right) refer to percentage germination observed by phase contrast microscopy at the end of the experiment.
Finally, these results were further supported by examining the number of colonies formed on TY plates after incubation for 2 days at 37°C. All spore crops were adjusted to a final concentration of ∼1×108 spores/ml, serially diluted, and plated on to TY agar. Single mutant gerXA2-1975− showed comparable numbers of colonies to the wild-type. In contrast, mutants gerXA1-0123− and gerXA3-2797− exhibited a greatly reduced colony forming efficiency (Figure 6).
Figure 6. Capacity of WT or mutant spores to germinate and form colonies.
C. botulinum and C. sporogenes wild type and mutant spore suspensions were enumerated using a haemocytometer and adjusted to a final concentration of ∼1×108 spores/ml. Spores were heat activated (80°C, 15 min), serially diluted in 0.85% saline, and plated in triplicate on to TY agar before anaerobic incubation (37°C, 48 hrs), after which colonies were enumerated. Data presented represent the mean log10 colony-forming units/ml from triplicate plates, with error bars representing the standard deviation of the mean.
A single tri-cistronic receptor in C. sporogenes is essential for germination
To characterise the C. sporogenes GRs and to identify their cognate germinants, single insertional knockout mutants (gerXA1-00838−, gerXA2-03006−, gerXA3-02217−, gerXA4-02140−) were created for each of the identified GR operons. The OD600 of wild-type spores decreased (∼40%) indicating efficient and complete spore germination (Figure 4b). Mutant gerXA3-02217− failed to germinate (<10% fall in OD600; spore germination not observed microscopically) with any of the amino acids tested after 20 h of exposure (Figure 4b & 4c). Spore germination was not observed with a nutrient rich broth (TY + L-lactate) demonstrating that CLOSPO_02217 is required for amino acid stimulated germination. Germination of mutant gerXA2-03006− showed an initial delay of one hour compared to the wild type with all the amino acids tested (e.g. Figure 4c). However, following this interval, germination proceeded to the same extent, albeit at a slower rate compared to the wild type (Figure 4b & 4c). Similarly, gerXA4-02140− had an initial germination postponement of four hours and then proceeded to germinate fully, but also at a slower rate. No discernible phenotype was observed following insertional inactivation of the gerXA1 (mutant gerXA1-00838−) GR compared to the WT (Figure 4b & 4c).
To further understand the function of the individual GRs in germination, triple insertional knockout GerXA mutants (gerXA 3−00838+, gerXA 3−02140+, gerXA 3−02217+, gerXA 3−03006+) and a quadruple insertional knockout GerXA mutant gerXA 4− were created. Mutation of three GerXAs resulted in one remaining potentially functional GerXA so any possible interaction between GerXAs is excluded and so enables dissection of the specific germinant recognised. As anticipated, spores from the quadruple insertional knockout mutant gerXA 4− failed to germinate (<10% fall in OD600, germination not observed microscopically) with any of the amino acids or in nutrient rich broth (TY + L-lactate) (Figure 4b). Furthermore, the three triple mutants gerXA 3−00838+, gerXA 3−02140+, and gerXA 3−03006+ also failed to germinate with the amino acid systems or in nutrient rich broth (TY + L-lactate). Mutant gerXA 3−02217+ which has only a single active GerXA present (CLOSPO_02217), displayed comparable germination rates to the wild type with all the amino acids tested and the nutrient rich broth (TY + L-lactate) (Figure 4b). Complementation of all the GerXAs further confirmed these findings. Particularly important was the introduction of the GR CLOSPO_02217- CLOSPO_02218- CLOSPO_02219 complementation vectors (pMTL83151esp or pMTL83151fdx), as these fully restored germination to the mutant gerXA3-02217− to WT levels (Figure 5b).
Finally, the number of colonies formed on TY plates after incubation for 2 days at 37°C was determined. All spore crops were adjusted to a final concentration of ∼1×108 spores/ml, serially diluted, and plated on to TY agar. The wild-type and single insertion mutants gerXA1-00838−, gerXA4-02140− and gerXA2-03006− formed comparable numbers of colonies. In contrast, mutant gerXA3-02217− showed a greatly reduced colony forming efficiency (Figure 6). Triple mutants gerXA 3−00838+, gerXA 3−02140+, gerXA 3−03006+ and the quadruple insertional knockout mutant gerXA 4− exhibited a significantly reduced colony forming efficiency compared to the WT. Importantly, the triple mutant gerXA 3−02217+, revealed comparable numbers of colonies to the wild type (Figure 6).
Discussion
One important feature that has contributed to the success of botulinum neurotoxin-forming clostridia, and all other clostridia, is their ability to form highly resistant endospores. Under suitable conditions the spores germinate with associated loss in resistance properties, and cell multiplication recommences. Spore germination occurs through a number of steps that are poorly understood in clostridia. The present study has identified which nutrient germinants are able to stimulate spore germination in Group I C. botulinum ATCC3502 and in C. sporogenes ATCC15579, has for the first time identified which of the individual GRs are responding to these nutrient germinants. A survey of the available genomes of Group I C. botulinum and its close relative, C. sporogenes reveals that although the general trend is to possess three or four operons encoding spore GR proteins, the fine detail of this organisation varies. The gerXC subunits were predicted to be lipoproteins and encode a signal peptide. In Bacillus, a lipobox consensus sequence (GCX) has been recognised within the first 30 residues of the GRs C subunits, where the cysteine in this motif is diacylglycerylated to facilitate cleavage of the signal peptide. In C. botulinum this lipobox was observed in gerXC gene CBO0125 and in two C. sporogenes gerXC genes CLOSPO_00836 and CLOSPO_02139. However, diacylglycerol addition does not appear to be essential for function [29]. In addition to the existence of orphan ger genes (such as CBO2300 in Group I C. botulinum ATCC3502), there is evidence of genetic recombination at these loci, most obviously the presence of multiple gerXB genes in the pentacistronic operon of Group I C. botulinum ATCC3502 (CBO1974-1978), and in its tetracistronic equivalent in C. sporogenes ATCC15579 (CLOSPO_03006-03003). Less obvious evidence includes the small fragment of a gerXA gene apparently inserted into the beginning of the first ‘extra’ gerXB gene of the Group I C. botulinum ATCC3502 pentacistronic operon, and the extra (or deleted) 20 codons discovered by comparison of the coding sequences of CBO0123 and CLOSPO_00838. Site-directed mutagenesis studies will be required to determine the functional status of these genetic differences.
Spore germination in Group I C. botulinum ATCC3502 and C. sporogenes ATCC15579 was triggered by a variety of amino acids, often in combination with L-lactate. L-phenylalanine + L-lactate and L-cysteine + L-lactate were the most effective germinants for C. botulinum, while L-cysteine + L-lactate were the most effective germinant for C. sporogenes. L-lactate had no discernible effect on spore germination in the presence of L-alanine or L-cysteine, but was essential for germination induced by L-methionine, L-serine or L-phenylalanine. Previous studies have reported a variable effect of L-lactate on amino acid induced spore germination in Group I C. botulinum and C. sporogenes [1], [44], [46]. Germination of Group I C. botulinum and C. sporogenes with L-serine and glycine has been reported previously [44], [49], while germination induced by L-methionine + L-lactate in Group I C. botulinum appears to be a novel finding. L-methionine triggered germination has been previously reported in C. sporogenes [49], B. anthracis [50] and C. tetani [51]. Germination in C. botulinum and C. sporogenes was also induced by L-phenylalanine + L-lactate, as reported previously in C. bifermentans [52] and C. sordellii [30], whereas the GR GerI in B. cereus interacts with L-phenylalanine in combination with inosine [53]. Moreover, L-phenylalanine stimulation of C. botulinum germination was more effective than that obtained with L-alanine. It may be that L-phenylalanine and L-lactate interact before interacting with the GR or that L-lactate and L-phenylalanine may directly affect the GR together or sequentially. Any effect is unlikely to be due to the hydrophobic nature of L-phenylalanine, as L-alanine and L-cysteine also have polar side chains and induce germination efficiently. In the present study, germination was more rapid when induced by single amino acids with L-lactate than in the nutrient rich medium TY; a similar observation has been made for Group II C. botulinum [54]. However, the addition of L-lactate to TY increased the germination rate significantly.
The production of spore crops is usually performed under conditions that maximise the spore yield [55], [56]. However, mounting evidence now suggests that sporulation conditions may have a direct effect on germination efficiency in Bacillus [55]–[57]. In the present study, a greater yield of C. sporogenes spores was achieved on RCM+SM plates compared with CMB and TY broths. However, the germination rate was initially more rapid with spores produced in CMB, albeit all spore crops achieved a similar extent of germination after 16 h. B. subtilis spores produced in a liquid medium germinated more readily than spores produced on plates [58]. Similarly, germination of B. subtilis spores with dodecylamine was also highly dependent on the method used for spore preparation [59]. Although the present study shows that media composition for sporulation does have an impact on germination, the reasons for these findings remain unclear. The effect of sporulation temperature in CMB on the yield of spores and their subsequent germination was also assessed. A greater number of spores were formed at 37°C than at lower temperatures, and they also germinated more readily. Thus, on this occasion spore yield and spore germination were positively correlated. For Group II C. botulinum, sporulation temperature affected spore yield and fatty acid content, but not heat resistance or germination [60]. For B. subtilis, sporulation temperature affected resistance to wet heat and spore coat protein levels [61]. Certainly, for C. sporogenes it is apparent that sporulation conditions have a direct effect on subsequent germination with the selected amino acids. It remains to be established whether this effect is due to the number and/or state of GRs, or to as yet unknown proteins that are involved in the germination pathway. However, it is clear from these results that sporulation conditions should be considered, especially when Clostridium studies in the food industry are to be performed to evaluate processing strategies, as these typically use spores produced under conditions where the yield has been maximised.
In the present study, D-amino acids failed to trigger spore germination and also prevented germination induced by their respective L-amino acid, as reported previously for other strains of Group I C. botulinum and C. sporogenes [44], [47], [48], [62]–[64]. It is noted that D-alanine was previously reported to be a competitive inhibitor of L-alanine induced germination in C. sporogenes [48]. However, D-alanine did not prevent germination of spores of Group II C. botulinum types B, E and F in L-alanine + l-lactate + NaHCO3 (pH 7·0) when added at ten-times the concentration of l-alanine [54]. Interestingly, in the present study, D-serine prevented germination induced by L-amino acids in C. sporogenes, and D-alanine prevented germination induced by L-amino acids in C. botulinum. These observations are consistent with those made by Montville et al. (1985), who reported that L-cysteine triggered germination was inhibited by D-alanine as well as by D-cysteine, and that L-alanine-triggered germination was inhibited by D-cysteine as well as by D-alanine in Group I C. botulinum strains B-aphis and Ba410 [64]. Montville et al. suggested that alanine and cysteine shared a common germinant binding site in spores of these two strains [64]. However, kinetic studies (e.g. [43]) are required to establish if the position is the same for Group I C. botulinum ATCC3502 and C. sporogenes ATCC15579. Studies with B. megaterium and B. subtilis suggest that the B-protein subunit of the GR presents the site for the receptor-ligand binding [65], [66], and although no evidence is presently available, the position may be similar in C. botulinum and C. sporogenes. Furthermore, C. botulinum (ATCC3502) and C. sporogenes (ATCC15579) both contain five putative alanine racemase genes. Alanine racemase is able to convert the germinant L-alanine into inhibitory D-alanine in B. cereus [67]. However, despite these clostridia containing five putative racemase genes, germination of Group I botulinum spores appeared not to be influenced by l-alanine racemase activity [44].
Molecular dissection of spore germination in Group I C. botulinum strain ATCC3502 demonstrated that two GerXAs were required for amino acid stimulated germination. The interruption of either gene CBO0123 or CBO2797 (mutants gerXA1-0123− and gerXA3-2797−) resulted in no observable germination. Thus, it has been shown that for amino acid stimulated germination there is a minimum requirement for the GerXAs produced by these two GRs, and while the product of gene CBO1975 appears to be inactive, it cannot be ruled out that the other products (GerXB and GerXC) of this operon are functional. The requirement for two GRs for germination has been previously reported in B. subtilis [68]. In this bacterium the GRs GerB and GerK interact and responded to a cocktail of L-asparagine, D-glucose, D-fructose, and K+ (AGFK) [68]. The B. anthracis GRs GerK and GerL also act cooperatively with alanine to stimulate the germination pathway [69]. Importantly, they can also act individually and initiate germination with proline and methionine (GerK) or serine and valine (GerL) as cogerminants in conjunction with inosine [69]. It is presently unclear how pairs of GRs come together to induce germination; potential hypotheses include: (i) one GR of the pair is involved in the binding of the germinant and the second GR is involved in a signalling capacity; (ii) both GRs together may be required to form the germinant binding site; (iii) one GR may physically stabilise the receptor that receives the germinant. What is clear is that more evidence is required to characterise the individual role of each GR in germinant recognition. In the present study, single C. botulinum GRs failed to induce germination, either with single or with combinations of amino acids, or with components of a rich growth medium. Complementation restored wild type levels of germination to the mutant gerXA1-0123−, albeit at a slower rate compared to that of the wild type. However restoration of germination efficiency could not be assessed for the complementation mutant gerXA3-2797− due to its poor sporulation efficiency. The failure of plasmid complemented mutants to regain wild type sporulation levels has been reported previously in C. perfringens [70]. Moreover, the use of multicopy plasmids can fail to restore the phenotype to wild type levels in clostridia [71]. It may be that in this instance inappropriately elevated levels of the GR proteins in the complementation mutant gerXA3-2797− diminished sporulation efficiency. Undoubtedly, until techniques become available for stable integration of a single chromosomal copy of the complementing DNA, complementation studies in clostridia will remain challenging. Group I C. botulinum ATCC3502 also has a pentacistronic putative GR. This third GR, CBO1975-1977 (gerXA2-XB2-XC2), is unusual as it is flanked by two additional gerXB genes (CBO1974 and CBO1978) and is closely related to a GR gene cluster characterized in C. sporogenes ATCC15579 (this work). Furthermore, upstream of CBO1974 there is a pseudogene, CBO1973A with a disrupted ORF which would encode the C-terminal region of a GerA protein. This pseudogene overlaps the putative start of CBO1974 (gerXB). Insertion mutant (gerXA2-1975−) showed no attributable phenotype, with a similar germination pattern to the wild type strain. The pseudogene CBO1973A may hint at the possibility of recombinational events that have occurred at this locus that may have disrupted the normal control regions for correct GR expression. It cannot be ruled out that germination may be stimulated by suitable environmental stimuli that are not found in the nutrient rich medium, TY broth or any of the specific germinants tested in this work.
C. sporogenes is often regarded as the non-toxigenic equivalent of Group I C. botulinum [2], [19]. Comparisons of C. botulinum and C. sporogenes are important if C. sporogenes is to be used as a valid surrogate model in spore germination and other studies. There are four genes encoding GerXAs in C. sporogenes ATCC15579, and only one of these (CLOSPO_02217) was essential for amino acid induced germination. Two other GerXA proteins (products of CLOSPO_02140 and CLOSPO_03006) increased the rate of germination, providing that the product of CLOSPO_02217 was also present. Therefore, one GR (CLOSPO_02217-02219; and at least CLOSPO_02217) is required for amino acid stimulated spore germination. The remaining three GerXAs were not essential, but it cannot be ruled out that other products (GerXB and GerXC) of these operons may be functional and act synergistically with the CLOSPO_02217-02219 GR. Indeed, preliminary proteomics data from purified spores reveals that the gerXC gene CLOSPO_00836 is translated into a protein (data not shown). Although in Bacillus all subunits of GRs are required for a response to amino acids, the function of each individual subunit still remains to be elucidated [72]. The finding that, in C. sporogenes ATCC15579, some or all of the proteins from two GRs (CLOSPO_02139-02141 and CLOSPO_03003-03006) contribute to increase the rate of spore germination induced by a third GR (CLOSPO_02217-02219; or at least CLOSPO_02217) implies a close interaction between GR proteins. Different GRs may interact directly (and/or compete) with each other [73] or possibly one GR may facilitate access of the germinant to another GR. Although these two GRs were not stimulated by individual or a combination of amino acids, or by components of a rich microbiological growth medium, it is possible that they may individually respond to some other as yet unknown germinant. Interestingly, when the wild-type and various mutants containing an active CLOSPO_02217 were plated out on a rich growth medium recovery was complete (∼107 CFU/ml), while in mutants where CLOSPO_02217 was insertionally inactivated (including in the quadruple gerXA 4− mutant) the number of colonies recovered was significantly lower (∼103–104 CFU/ml). The recovery, at a very low frequency, of any spores in the absence of CLOSPO_02217 (including in the quadruple gerXA 4− mutant) may be due to an alternative low efficiency receptor system distinct from the ger family, or perhaps to stochastic effects (e.g. cortex-lytic enzymes or ion/water channels). Similar observations have been made in Bacillus subtilis [74]. It is perhaps not surprising that this GR (CLOSPO_02217-02219) was involved in germination with the selected amino acids as it shares >85% homology with the functional and now characterised C. botulinum GR (CBO2795-2797). It is interesting that this GerXA can operate independently of any other GerXA, unlike in C. botulinum. One hypothesis is that the short deletion in gerXA of CLOSPO_00838 (when compared to its active C. botulinum homologue, CBO0123) is associated with loss of function. This mutational event in C. sporogenes may have brought about an evolutionary pressure which has allowed adaption of this organism to survive with only a single functional GR operon (or at least a single functional GerXA). Certainly the germination mechanism of C. sporogenes ATCC15579 is different to that of C. botulinum ATCC3502. Alignment of putative GRs with known functioning GRs appears to be problematic, as at present this is done over the whole protein in the absence of detailed knowledge of the functionality of the putative GR. However, based on comparison of the whole proteins, the GerXA of homologous GR proteins CBO2795-2797/CLOSPO_02217-02219 are functionally active, while no discernible function could be identified for the GerXA of GR proteins CBO1974-1978/CLOSPO_03003-03006. The GerXAs of other GR proteins were either essential (in C. botulinum ATCC3502) or contribute to the rate (in C. sporogenes ATCC15579). The results also call into the question the use of C. sporogenes as a suitable substitute for C. botulinum with regards to germination rates and germination substrates. More work is required to fully understand the role of each GR in clostridia and indeed why some species contain multiple GR operons and others can function with just one. The testing of additional strains would seem to be appropriate.
The long term aim is that as more is understood of the complex germination systems in clostridia, it may be possible to devise specific strategies to disrupt this process. This would be of great benefit to help control pathogenic clostridia, for example in the food industry, and might also help to control spore-disseminated nosocomial infections such as those caused by C. difficile.
Materials and Methods
Receptor identification and alignment
Homologues of C. botulinum ATCC3502 GR sub-units (gerXA, gerXB, gerXC) were identified by BLASTp analyses against a draft un-assembled genome of C. sporogenes strain ATCC15579. Alignment of C. sporogenes receptors with C. botulinum was performed using Clustal Omega [75] and Jalview [76] was utilised to produce a tree showing the average distance using % identity. Protein domain analysis was performed using Pfam [77]. Transmembrane helix prediction analysis of the GR sub-units was implemented using TMHMM [78].
Bacterial strains and growth conditions
Proteolytic C. botulinum strain ATCC3502 (neurotoxin subtype A1) and C. sporogenes ATCC15579 were grown anaerobically at 37°C in tryptone-yeast medium (TY). The Escherichia coli strain Top10 (Invitrogen) was used for plasmid maintenance and the E. coli strain CA434 [79] was used for conjugal transfer. Both strains of E. coli were grown aerobically in Luria-Bertani medium (LB) at 37°C. Where appropriate, growth medium was supplemented with antibiotics at the following final concentrations; ampicillin 100 µg/ml, chloramphenicol 25 µg/ml, cycloserine 250 µg/ml, thiamphenicol 15 µg/ml, erythromycin 500 µg/ml (E. coli), 20 µg/ml (C. botulinum) 2.5 µg/ml (C. sporogenes), and the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal) 80 µg/ml. All bacterial media supplements were purchased from Sigma. Spores of C. botulinum and C. sporogenes strains were prepared in TY, Reinforced Clostridial Medium plus skimmed milk (RCM+SM) or Robertson's cooked meat broth (CMB) (Southern Group Laboratories) and incubated at 15, 20, 28, 30, 37 or 42°C for a period of 10 days. Spores were cleaned and stored as described previously [54].
PCR, cloning and Southern hybridisation
Constructed mutants and plasmids utilised in this study are presented in Table 2. Primers used for verification of successful insertion events and the Southern blot probes are listed in Table S2 (supplementary material). PCR experiments were carried out using Phusion High-Fidelity PCR Master Mix with GC Buffer kit (Thermo Fisher). Plasmid isolation and PCR purification was performed using the Wizard Plus SV Minipreps DNA Purification System and the Wizard SV Gel and PCR Clean-Up System (Promega) respectively, as described in the provided Technical Manual. Chromosomal DNA isolation from suspected mutants were prepared as previously described [16]. Restriction endonucleases and T4 DNA ligase were purchased from New England BioLabs and used according to the manufacturer's instructions. Southern hybridisation was performed to confirm the correct number of insertion events had occurred. The hybridisation probe was constructed by PCR to target the inserted intron using the primers Erm-F and Erm-R (Table S2). Genomic DNA (1 µg) was digested overnight with HindIII restriction enzyme and the fragments separated on a 1% agarose gel. Southern blot analysis was performed with ECL detection using a commercial kit (Amersham ECL Direct Nucleic Acid Labelling and Detection System) according to the manufacturer's instructions.
Table 2. Constructed mutants and plasmids utilised in this study.
| Strain | CDS annotation | Locus | Insertion site | Mutant | Description |
| C. botulinum (ATCC3502) | gerXA1 | CBO0123 | 42s | gerXA1-0123− | Single insertional knockout |
| gerXA2 | CBO1975 | 56s | gerXA2-1975− | Single insertional knockout | |
| gerXA3 | CBO2797 | 1344a | gerXA3-2797− | Single insertional knockout | |
| C. sporogenes (ATCC15579) | gerXA1 | CLOSPO_00838 | 175s | gerXA1-00838− | Single insertional knockout |
| gerXA2 | CLOSPO_03006 | 795s | gerXA2-03006− | Single insertional knockout | |
| gerXA3 | CLOSPO_02217 | 61s | gerXA3-02217− | Single insertional knockout | |
| gerXA4 | CLOSPO_02140 | 601s | gerXA4-02140− | Single insertional knockout | |
| gerXA1, | CLOSPO_00838, | 175s, | gerXA 3−02140+ | Triple insertional knockout leaving one functional GerXA gerXA4 | |
| gerXA2, | CLOSPO_03006, | 795s, | |||
| gerXA3 | CLOSPO_02217 | 61s | |||
| gerXA1, | CLOSPO_00838, | 175s, | gerXA 3−02217+ | Triple insertional knockout leaving one functional GerXA gerXA3 | |
| gerXA2, | CLOSPO_03006, | 795s, | |||
| gerXA4 | CLOSPO_02140 | 601s | |||
| gerXA1, | CLOSPO_00838, | 175s, | gerXA 3−03006+ | Triple insertional knockout leaving one functional GerXA gerXA2 | |
| gerXA3, | CLOSPO_02217, | 61s, | |||
| gerXA4 | CLOSPO_02140 | 601s | |||
| gerXA2, | CLOSPO_03006, | 795s, | gerXA 3−00838+ | Triple insertional knockout leaving one functional GerXA gerXA1 | |
| gerXA3, | CLOSPO_02217, | 61s, | |||
| gerXA4 | CLOSPO_02140 | 601s | |||
| gerXA1, | CLOSPO_00838, | 175s, | gerXA 4− | Quadruple insertional knockout. Insertion in all known gerXA sub-units | |
| gerXA2, | CLOSPO_03006, | 795s, | |||
| gerXA3, | CLOSPO_02217, | 61s, | |||
| gerXA4 | CLOSPO_02140 | 601s | |||
| Plasmids | pMTL007C-E2 | Clostridial mutagenesis plasmid | |||
| pMTL8315esp | GR expression plasmid with no fdx promoter. Relies on native promoter of inserted genes | ||||
| pMTL8315fdx | GR expression plasmid with fdx promoter |
− indicates the gene is absent; + indicates that only that gene is functional; no− describes the number of genes knocked out. a = antisense orientation insertion site; s = sense orientation insertion site. The designation of “X” before the letter A is used in clostridia as unlike Bacillus, GRs in these species have not yet been attributed to specific germinants.
Germinants and spore germination
The potential germinants, including; L-alanine, L-serine, L-cysteine, L-methionine, L-phenylalanine, glycine (0.5 mM–100 mM) and antagonists d-alanine, D-cysteine, D-methionine, D-phenylalanine, D-serine (10 mM–200 mM depending on solubility) (Sigma) were all prepared in Tris-HCl buffer (20 mM, pH 7.4) with NaHCO3 (50 mM), with or without L-lactate (50 mM). NaHCO3 was a non-essential component that increases the rate and overall extent of germination by approximately 10% [54]. Germinant solutions were prepared and filter sterilised (0.45-µm syringe filter, Millipore, Bedford, MA). The pH of the germinant solutions was adjusted to evaluate the effect of pH on spore germination at pH 3 to pH 10. Prior to the addition of germinants all spore suspensions were heat activated at 80°C for 10 min. Germination of spores at 30°C was measured by a decrease in optical density (OD) at 600 nm every 5 min using a Bioscreen C analyser system (Labsystems, Basingstoke, UK) under aerobic conditions. Germination was expressed in terms of measured OD600 as a percentage of the initial OD600. To validate the OD600 measurements, at the completion of each test the proportion of germinated spores was visualised by the assessment of 200 spores in at least ten fields using phase-contrast microscopy. Typically, full germination was indicated when the OD600 fell to ∼40% of its initial value. In some tests a small fall in OD600 was observed (<10% of initial value). This was attributed to settling of spores in the Bioscreen wells, and was not accompanied by microscopic observation of spore germination. Finally, the capacity of spores to germinate and form colonies was assessed. Spore suspensions were enumerated using a haemocytometer and adjusted to a final concentration of ∼1×108 spores/ml. Spores were then heat activated (80°C, 15 min), serially diluted in 0.85% saline, and plated in triplicate on to TY agar before incubation anaerobically (37°C, 48 hrs).
Mutant generation
Clostridium mutants were generated using the Clostron system, which inserts an erythromycin resistance cassette into the targeted gene of interest. Target sites were identified using the Pertuka method [80] and mutants were generated (Table 2) as described by Heap et al. [81]. gerXA GR subunits were targeted in all germination operons. Re-targeted introns were ligated into the pMTL007C-E2 vector following restriction digest with HindIII/BsrGI. All retargeted introns were sequence verified before transformation into E. coli CA434. Confirmed sequenced plasmids were then conjugated into their respective hosts. Finally, primers were designed and used to confirm that the intron was present and in the correct orientation in the target gene/genes of interest (Table S2). The current insertional knockout system does not allow selective isolation of clones containing a second intron insertion as following one insertion the mutant strain is then erythromycin resistant. To create the double, triple and quadruple insertional knockout mutants an alternative approach was taken in which clones containing intron insertions were identified by screening large numbers of colonies rather than by antibiotic selection. Plasmid re-targeting was carried out as above and transferred in to C. botulinum or C. sporogenes using E. coli CA434. Confirmation of successful trans-conjugation events were screened on TY agar plates containing cycloserine (250 µg/ml) and thiamphenicol (15 µg/ml). To create the multiple insertional knockout mutants the process was repeated using successive rounds of plasmid targeting and gene insertion. Successful integration of introns into the target genes was confirmed by PCR using primers flanking the target sites (Table S2). Furthermore, to confirm that the required number of insertion events had occurred, genomic DNA from the mutants was digested with HindIII and subjected to Southern analysis using an intron specific probe.
Complementation
For complementation, two plasmids, pMTL8315esp and pMTL8315fdx were constructed (Figure S2). Primers, with Esp3I restriction sites were designed to amplify two GR fragments; one included the 5′ noncoding region encompassing the putative promoter and was ligated into pMTL8315esp; the second fragment contained the coding operon only and was ligated into the plasmid pMTL8315-fdx which contains the strong promoter Pfdx of the ferredoxin gene (fdx) from C. sporogenes NCIMB 10696. Following confirmation by sequencing, GR plasmids were then transconjugated into their respective mutants using E. coli CA434 as described earlier. Spore crops were produced as above, except with the addition of thiamphenicol (15 µg/ml) to the media to maintain the plasmid.
Supporting Information
Confirmation of successful insertional mutagenesis. A, PCR screens of C. botulinum ger gene mutants confirms that the intron has successfully inserted into the target gene. Primer set 1 anneals to the ErmRAM and confirms that the RAM is spliced and therefore integrated (expected band size of mutants 900 bp). Primer set 2, intron binding EBS universal primer plus gene specific forward primer amplified sequence across the intron-exon junction to confirm intron insertion into the desired gene (expected band size of mutants 250 bp). Primer set 3 anneals to each end of the target gene and confirms the intron is present in the target gene (expected band size of mutants ∼2 kb). Lanes labelled WT show the PCR products (primer set 3) generated for each ger A gene when wild type DNA is used as template. B, Diagrammatic representation of results shown in A revealing primer binding sites and length of pcr products. C, Southern hybridisation was performed to confirm the correct number of insertion events had occurred. The hybridisation probe was designed to target the chromosomally inserted ErmRAM sequence. Genomic DNA of all strains was digested overnight with HindIII. Expected band sizes were as follows; gerXA1-0123− 2.9 kb, gerXA2-1975− 6 kb, gerXA3-2797− 4 kb. The negative control, WT genomic DNA, generated no signal for the probe.
(PDF)
Constructed plasmids used for expression of Clostridium receptors in complementation experiments. Complementation plasmid components; catP (chloramphenicol acetyltransferase) conferring thiamphenicol resistance in E.coli and chloramphenicol resistance in Clostridium. repH (pCB102); Gram-positive replicon. ColE1; Gram-negative replicon. traJ; plasmid conjugative transfer protein. Esp3I; restriction enzyme site used for insertion of GR operons. Plasmid pMTL8315fdx contains the strong promoter Pfdx of the ferredoxin gene (fdx) from C. sporogenes NCIMB 10696. Plasmid pMTL8315esp contains no additional promoter and relies on a wild type copy of the inserted gene under the control of its native promoter (assumed to be located in the intergenic region upstream of the first CDS of the operon). Both plasmids are adaptations of the pMTL80000 modular plasmid system of Heap et al., 2009 [82].
(PDF)
Effect of amino acid concentrations on clostridial spore germination. * C. botulinum only, no germination detected in C. sporogenes at 100 mM. A selection of amino acids, were assessed for their individual effect on germination of spores of C. botulinum and C. sporogenes in the presence of Tris-HCl buffer (pH 7.4), L-lactate (50 mM) and NaHCO3 (50 mM). Germination was determined by a decrease in optical density (OD) at 600 nm using a Bioscreen C analyser and validated by phase-contrast microscopy.
(PDF)
Primers used in this study. + denotes resulting product contains >500 bp upstream of the named gene to be certain that a native promoter is included.
(PDF)
Acknowledgments
The authors would like to thank Dr. Fran Mulholland for his advice on proteomics data.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This research was funded by the BBSRC Institute Strategic Programme on Gut Health and Food Safety (BB/J004529/1). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Peck MW (2009) Biology and genomic analysis of Clostridium botulinum. In: Poole RK, editor. Advances in Microbial Physiology: Academic Press. pp. 183–265, 320. [DOI] [PubMed] [Google Scholar]
- 2. Carter AT, Paul CJ, Mason DR, Twine SM, Alston MJ, et al. (2009) Independent evolution of neurotoxin and flagellar genetic loci in proteolytic Clostridium botulinum . BMC Genomics 10: 115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peck MW, Stringer SC, Carter AT (2011) Clostridium botulinum in the post-genomic era. Food Microbiol 28: 183–191. [DOI] [PubMed] [Google Scholar]
- 4. Hill KK, Smith TJ (2013) Genetic diversity within Clostridium botulinum serotypes, botulinum neurotoxin gene clusters and toxin subtypes. Curr Top Microbiol Immunol 364: 1–20. [DOI] [PubMed] [Google Scholar]
- 5. Barash JR, Arnon SS (2014) A novel strain of Clostridium botulinum that produces type B and type H botulinum toxins. J Infect Dis 209: 183–191. [DOI] [PubMed] [Google Scholar]
- 6. Poulain B, Popoff MR, Molgo J (2008) How do the Botulinum Neurotoxins block neurotransmitter release: from botulism to the molecular mechanism of action. The Botulinum J 1: 14. [Google Scholar]
- 7.Bruggemann H, Wollherr A, Mazuet C, Popoff M (2011) Clostridium botulinum. In: Fratamico P, Liu Y, Kathariou S, editors. Genomes of Foodborne and Waterborne Pathogens: ASM Press. pp. 185–212. [Google Scholar]
- 8.Lindström M, Fredriksson-Ahomaa M, and Korkeala H (2009) Molecular epidemiology of group I and group II Clostridium botulinum In: Brüggemann H, Gottschalk, G., editor. Clostridia, molecular biology in the post-genomic era. Caister Academic Press, Norfolk, United Kingdom. pp. 103–130. [Google Scholar]
- 9. Hackett R, Kam PC (2007) Botulinum toxin: pharmacology and clinical developments: a literature review. Med Chem 3: 333–345. [DOI] [PubMed] [Google Scholar]
- 10.Hatheway C (1988) Botulism. In: Balows A, Hausler WJ, Ohashi M, Turano A, Lennete EH, editors. Laboratory Diagnosis of Infectious Diseases: Springer New York. pp. 111–133. [Google Scholar]
- 11.Johnson EA (2007) Food microbiology: fundamentals and frontiers. In: Doyle MP, Beuchat, L. R., editor. Clostridium botulinum. 3rd ed. Food microbiology: fundamentals and frontiers: ASM Press. pp. 401–421. [Google Scholar]
- 12. Johnson EA (1999) Clostridial toxins as therapeutic agents: benefits of nature's most toxic proteins. Annu Rev Microbiol 53: 551–575. [DOI] [PubMed] [Google Scholar]
- 13. Raphael BH, Luquez C, McCroskey LM, Joseph LA, Jacobson MJ, et al. (2008) Genetic homogeneity of Clostridium botulinum type A1 strains with unique toxin gene clusters. Appl Environ Microbiol 74: 4390–4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fang PK, Raphael BH, Maslanka SE, Cai S, Singh BR (2010) Analysis of genomic differences among Clostridium botulinum type A1 strains. BMC Genomics 11: 725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raphael BH, Joseph LA, McCroskey LM, Luquez C, Maslanka SE (2010) Detection and differentiation of Clostridium botulinum type A strains using a focused DNA microarray. Mol Cell Probe 24: 146–153. [DOI] [PubMed] [Google Scholar]
- 16. Sebaihia M, Peck MW, Minton NP, Thomson NR, Holden MT, et al. (2007) Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res 17: 1082–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inkster T, Cordina C, Siegmeth A (2011) Septic arthritis following anterior cruciate ligament reconstruction secondary to Clostridium sporogenes; a rare clinical pathogen. J Clin Pathol 64: 820–821. [DOI] [PubMed] [Google Scholar]
- 18.McClure PJ (2006) Spore-forming bacteria. In: Blackburn CdW, editor. Food spoilage microorganisms. No. 122 ed: Woodhead Publishing limited pp. 579–623. [Google Scholar]
- 19. Brown JL, Tran-Dinh N, Chapman B (2012) Clostridium sporogenes PA 3679 and its uses in the derivation of thermal processing schedules for low-acid shelf-stable foods and as a research model for proteolytic Clostridium botulinum . J Food Prot 75: 779–792. [DOI] [PubMed] [Google Scholar]
- 20. Taylor RH, Dunn ML, Ogden LV, Jefferies LK, Eggett DL, et al. (2013) Conditions associated with Clostridium sporogenes growth as a surrogate for Clostridium botulinum in nonthermally processed canned butter. J Dairy Sci 96: 2754–2764. [DOI] [PubMed] [Google Scholar]
- 21. Collins MD, Lawson PA, Willems A, Cordoba JJ, Fernandez-Garayzabal J, et al. (1994) The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol 44: 812–826. [DOI] [PubMed] [Google Scholar]
- 22. Jacobson MJ, Lin G, Whittam TS, Johnson EA (2008) Phylogenetic analysis of Clostridium botulinum type A by multi-locus sequence typing. Microbiology 154: 2408–2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bradbury M, Greenfield P, Midgley D, Li D, Tran-Dinh N, et al. (2012) Draft genome sequence of Clostridium sporogenes PA 3679, the common nontoxigenic surrogate for proteolytic Clostridium botulinum . J Bacteriol 194: 1631–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Setlow P (2014) Germination of Spores of Bacillus Species: What We Know and Do Not Know. J Bacteriol 196: 1297–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stringer SC, Webb MD, Peck MW (2009) Contrasting effects of heat treatment and incubation temperature on germination and outgrowth of individual spores of nonproteolytic Clostridium botulinum bacteria. Appl Environ Microbiol 75: 2712–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stringer SC, Webb MD, Peck MW (2011) Lag time variability in individual spores of Clostridium botulinum . Food Microbiol 28: 228–235. [DOI] [PubMed] [Google Scholar]
- 27. Webb MD, Stringer SC, Le Marc Y, Baranyi J, Peck MW (2012) Does proximity to neighbours affect germination of spores of non-proteolytic Clostridium botulinum? Food Microbiol 32: 104–109. [DOI] [PubMed] [Google Scholar]
- 28.Christie G (2012) Initiation of germination in Bacillus and Clostridium spores. In: Bacterial Spores: Current Research and Applications. pp 89–106. [Google Scholar]
- 29. Paredes-Sabja D, Setlow P, Sarker MR (2011) Germination of spores of Bacillales and Clostridiales species: mechanisms and proteins involved. Trends Microbiol 19: 85–94. [DOI] [PubMed] [Google Scholar]
- 30. Ramirez N, Abel-Santos E (2010) Requirements for germination of Clostridium sordellii spores in vitro. J Bacteriol 192: 418–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liggins M, Ramirez N, Magnuson N, Abel-Santos E (2011) Progesterone Analogs Influence Germination of Clostridium sordellii and Clostridium difficile Spores In Vitro. J Bacteriol 193: 2776–2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adam KH, Brunt J, Brightwell G, Flint SH, Peck MW (2011) Spore germination of the psychrotolerant, red meat spoiler, Clostridium frigidicarnis . Lett Appl Microbiol 53: 92–97. [DOI] [PubMed] [Google Scholar]
- 33. Moore P, Kyne L, Martin A, Solomon K (2013) Germination efficiency of clinical Clostridium difficile spores and correlation with ribotype, disease severity and therapy failure. J Med Microbiol 62: 1405–1413. [DOI] [PubMed] [Google Scholar]
- 34. Paredes-Sabja D, Shen A, Sorg JA (2014) Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends Microbiol 22: 406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Banawas S, Paredes-Sabja D, Korza G, Li Y, Hao B, et al. (2013) The Clostridium perfringens germinant receptor protein GerKC is located in the spore inner membrane and is crucial for spore germination. J Bacteriol 195: 5084–5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Francis MB, Allen CA, Shrestha R, Sorg JA (2013) Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog 9: e1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Heeg D, Burns DA, Cartman ST, Minton NP (2012) Spores of Clostridium difficile clinical isolates display a diverse germination response to bile salts. PLoS One 7: e32381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Howerton A, Ramirez N, Abel-Santos E (2011) Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol 193: 274–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Paredes-Sabja D, Setlow P, Sarker MR (2009) The protease CspB is essential for initiation of cortex hydrolysis and dipicolinic acid (DPA) release during germination of spores of Clostridium perfringens type A food poisoning isolates. Microbiology 155: 3464–3472. [DOI] [PubMed] [Google Scholar]
- 40. Ramirez N, Liggins M, Abel-Santos E (2010) Kinetic evidence for the presence of putative germination receptors in Clostridium difficile spores. J Bacteriol 192: 4215–4222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sarker MR, Paredes-Sabja D (2012) Molecular basis of early stages of Clostridium difficile infection: germination and colonization. Future Microbiol 7: 933–943. [DOI] [PubMed] [Google Scholar]
- 42. Sorg JA, Sonenshein AL (2009) Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol 191: 1115–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sorg JA, Sonenshein AL (2010) Inhibiting the initiation of Clostridium difficile spore germination using analogs of chenodeoxycholic acid, a bile acid. J Bacteriol 192: 4983–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alberto F, Broussolle V, Mason DR, Carlin F, Peck MW (2003) Variability in spore germination response by strains of proteolytic Clostridium botulinum types A, B and F. Lett Appl Microbiol 36: 41–45. [DOI] [PubMed] [Google Scholar]
- 45. Xiao Y, Francke C, Abee T, Wells-Bennik MH (2011) Clostridial spore germination versus bacilli: genome mining and current insights. Food Microbiol 28: 266–274. [DOI] [PubMed] [Google Scholar]
- 46. Broussolle V, Alberto F, Shearman CA, Mason DR, Botella L, et al. (2002) Molecular and physiological characterisation of spore germination in Clostridium botulinum and C-sporogenes . Anaerobe 8: 89–100. [Google Scholar]
- 47. Ando Y (1973) Studies on germination of spores of clostridial species capable of causing food poisoning (I) Factors affecting the germination of spores of Clostridium botulinum type A in a chemically defined medium. J Food Hyg Soc Jpn 14: 457–461. [Google Scholar]
- 48. Uehara M, Frank HA (1965) Factors affecting alanine-induced germination of clostridial spores. Spores III: American Society for Microbiology 38–46. [Google Scholar]
- 49. Ishimori T, Takahashi K, Goto M, Nakagawa S, Kasai Y, et al. (2012) Synergistic effects of high hydrostatic pressure, mild heating, and amino acids on germination and inactivation of Clostridium sporogenes spores. Appl Environ Microbiol 78: 8202–8207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ross C, Abel-Santos E (2010) The Ger receptor family from sporulating bacteria. Curr Issues Mol Biol 12: 147–158. [PMC free article] [PubMed] [Google Scholar]
- 51. Shoesmith JG, Holland KT (1972) The germination of spores of Clostridium tetani . J Gen Microbiol 70: 253–261. [DOI] [PubMed] [Google Scholar]
- 52. Waites WM, Wyatt LR (1971) Germination of spores of Clostridium bifermentans by certain amino acids, lactate and pyruvate in the presence of sodium or potassium ions. J Gen Microbiol 67: 215–222. [DOI] [PubMed] [Google Scholar]
- 53. Hornstra LM, de Vries YP, Wells-Bennik MH, de Vos WM, Abee T (2006) Characterization of germination receptors of Bacillus cereus ATCC 14579. Appl Environ Microbiol 72: 44–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Plowman J, Peck MW (2002) Use of a novel method to characterize the response of spores of non-proteolytic Clostridium botulinum types B, E and F to a wide range of germinants and conditions. J Appl Microbiol 92: 681–694. [DOI] [PubMed] [Google Scholar]
- 55. Hornstra LM, de Vries YP, de Vos WM, Abee T (2006) Influence of sporulation medium composition on transcription of ger operons and the germination response of spores of Bacillus cereus ATCC 14579. Appl Environ Microbiol 72: 3746–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ramirez-Peralta A, Zhang P, Li YQ, Setlow P (2012) Effects of sporulation conditions on the germination and germination protein levels of Bacillus subtilis spores. Appl Environ Microbiol 78: 2689–2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nguyen Thi Minh H, Durand A, Loison P, Perrier-Cornet JM, Gervais P (2011) Effect of sporulation conditions on the resistance of Bacillus subtilis spores to heat and high pressure. Appl Microbiol Biotechnol 90: 1409–1417. [DOI] [PubMed] [Google Scholar]
- 58. Rose R, Setlow B, Monroe A, Mallozzi M, Driks A, et al. (2007) Comparison of the properties of Bacillus subtilis spores made in liquid or on agar plates. J Appl Microbiol 103: 691–699. [DOI] [PubMed] [Google Scholar]
- 59. Setlow B, Cowan AE, Setlow P (2003) Germination of spores of Bacillus subtilis with dodecylamine. J Appl Microbiol 95: 637–648. [DOI] [PubMed] [Google Scholar]
- 60. Peck MW, Evans RI, Fairbairn DA, Hartley MG, Russell NJ (1995) Effect of sporulation temperature on some properties of spores of non-proteolytic Clostridium botulinum . Int J Food Microbiol 28: 289–297. [DOI] [PubMed] [Google Scholar]
- 61. Melly E, Genest PC, Gilmore ME, Little S, Popham DL, et al. (2002) Analysis of the properties of spores of Bacillus subtilis prepared at different temperatures. J Appl Microbiol 92: 1105–1115. [DOI] [PubMed] [Google Scholar]
- 62. Johnstone K (1994) The trigger mechanism of spore germination: current concepts. J Appl Bacteriol 76: 17S–24S. [DOI] [PubMed] [Google Scholar]
- 63. Rowley DB, Feeherry F (1970) Conditions affecting germination of Clostridium botulinum 62A spores in a chemically defined medium. J Bacteriol 104: 1151–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Montville TJ, Jones SB, Conway LK, Sapers GM (1985) Germination of spores from Clostridium botulinum B-aphis and Ba410. Appl Environ Microbiol 50: 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Christie G, Lowe CR (2007) Role of chromosomal and plasmid-borne receptor homologues in the response of Bacillus megaterium QM B1551 spores to germinants. J Bacteriol 189: 4375–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Moir A, Corfe BM, Behravan J (2002) Spore germination. Cell Mol Life Sci 59: 403–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Dodatko T, Akoachere M, Muehlbauer SM, Helfrich F, Howerton A, et al. (2009) Bacillus cereus spores release alanine that synergizes with inosine to promote germination. PLoS ONE 4: e6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yi X, Liu J, Faeder JR, Setlow P (2011) Synergism between different germinant receptors in the germination of Bacillus subtilis spores. J Bacteriol 193: 4664–4671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Fisher N, Hanna P (2005) Characterization of Bacillus anthracis germinant receptors in vitro. J Bacteriol 187: 8055–8062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Li J, Chen J, Vidal JE, McClane BA (2011) The Agr-Like quorum-sensing system regulates sporulation and production of enterotoxin and Beta2 toxin by Clostridium perfringens type A non-food-borne human gastrointestinal disease strain F5603. Infection and Immunity 79: 2451–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ng YK, Ehsaan M, Philip S, Collery MM, Janoir C, et al. (2013) Expanding the repertoire of gene tools for precise manipulation of the Clostridium difficile genome: allelic exchange using pyrE alleles. PLoS ONE 8: e56051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P (2011) Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Mol Microbiol 81: 1061–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Cabrera-Martinez RM, Tovar-Rojo F, Vepachedu VR, Setlow P (2003) Effects of overexpression of nutrient receptors on germination of spores of Bacillus subtilis . J Bacteriol 185: 2457–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Paidhungat M, Setlow P (2000) Role of ger proteins in nutrient and nonnutrient triggering of spore germination in Bacillus subtilis . J Bacteriol 182: 2513–2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, et al. (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7: 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ (2009) Jalview Version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Punta M, Coggill PC, Eberhardt RY, Mistry J, Tate J, et al. (2012) The Pfam protein families database. Nucleic Acids Res 40: D290–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Krogh A, Larsson B, von Heijne G, Sonnhammer EL (2001) Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 79. Purdy D, O'Keeffe TAT, Elmore M, Herbert M, McLeod A, et al. (2002) Conjugative transfer of clostridial shuttle vectors from Escherichia coli to Clostridium difficile through circumvention of the restriction barrier. Mol Microbiol 46: 439–452. [DOI] [PubMed] [Google Scholar]
- 80. Perutka J, Wang WJ, Goerlitz D, Lambowitz AM (2004) Use of computer-designed group II introns to disrupt Escherichia coli DExH/D-box protein and DNA helicase genes. J Mol Biol 336: 421–439. [DOI] [PubMed] [Google Scholar]
- 81. Heap JT, Kuehne SA, Ehsaan M, Cartman ST, Cooksley CM, et al. (2010) The ClosTron: Mutagenesis in Clostridium refined and streamlined. J Microbiol Methods 80: 49–55. [DOI] [PubMed] [Google Scholar]
- 82. Heap JT, Pennington OJ, Cartman ST, Minton NP (2009) A modular system for Clostridium shuttle plasmids. J Microbiol Methods 78: 79–85. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Confirmation of successful insertional mutagenesis. A, PCR screens of C. botulinum ger gene mutants confirms that the intron has successfully inserted into the target gene. Primer set 1 anneals to the ErmRAM and confirms that the RAM is spliced and therefore integrated (expected band size of mutants 900 bp). Primer set 2, intron binding EBS universal primer plus gene specific forward primer amplified sequence across the intron-exon junction to confirm intron insertion into the desired gene (expected band size of mutants 250 bp). Primer set 3 anneals to each end of the target gene and confirms the intron is present in the target gene (expected band size of mutants ∼2 kb). Lanes labelled WT show the PCR products (primer set 3) generated for each ger A gene when wild type DNA is used as template. B, Diagrammatic representation of results shown in A revealing primer binding sites and length of pcr products. C, Southern hybridisation was performed to confirm the correct number of insertion events had occurred. The hybridisation probe was designed to target the chromosomally inserted ErmRAM sequence. Genomic DNA of all strains was digested overnight with HindIII. Expected band sizes were as follows; gerXA1-0123− 2.9 kb, gerXA2-1975− 6 kb, gerXA3-2797− 4 kb. The negative control, WT genomic DNA, generated no signal for the probe.
(PDF)
Constructed plasmids used for expression of Clostridium receptors in complementation experiments. Complementation plasmid components; catP (chloramphenicol acetyltransferase) conferring thiamphenicol resistance in E.coli and chloramphenicol resistance in Clostridium. repH (pCB102); Gram-positive replicon. ColE1; Gram-negative replicon. traJ; plasmid conjugative transfer protein. Esp3I; restriction enzyme site used for insertion of GR operons. Plasmid pMTL8315fdx contains the strong promoter Pfdx of the ferredoxin gene (fdx) from C. sporogenes NCIMB 10696. Plasmid pMTL8315esp contains no additional promoter and relies on a wild type copy of the inserted gene under the control of its native promoter (assumed to be located in the intergenic region upstream of the first CDS of the operon). Both plasmids are adaptations of the pMTL80000 modular plasmid system of Heap et al., 2009 [82].
(PDF)
Effect of amino acid concentrations on clostridial spore germination. * C. botulinum only, no germination detected in C. sporogenes at 100 mM. A selection of amino acids, were assessed for their individual effect on germination of spores of C. botulinum and C. sporogenes in the presence of Tris-HCl buffer (pH 7.4), L-lactate (50 mM) and NaHCO3 (50 mM). Germination was determined by a decrease in optical density (OD) at 600 nm using a Bioscreen C analyser and validated by phase-contrast microscopy.
(PDF)
Primers used in this study. + denotes resulting product contains >500 bp upstream of the named gene to be certain that a native promoter is included.
(PDF)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.