The spindle pole body (SPB) localization of the fission yeast Schizosaccharomyces pombe TACC orthologue alp7p depends on the SPB protein csi1p. Compromised interaction between csi1p and alp7p delays bipolar spindle formation and leads to abnormal chromosome segregation.
Abstract
Accurate chromosome segregation requires timely bipolar spindle formation during mitosis. The transforming acidic coiled-coil (TACC) family proteins and the ch-TOG family proteins are key players in bipolar spindle formation. They form a complex to stabilize spindle microtubules, mainly dependent on their localization to the centrosome (the spindle pole body [SPB] in yeast). The molecular mechanism underlying the targeting of the TACC–ch-TOG complex to the centrosome remains unclear. Here we show that the fission yeast Schizosaccharomyces pombe TACC orthologue alp7p is recruited to the SPB by csi1p. The csi1p-interacting region lies within the conserved TACC domain of alp7p, and the carboxyl-terminal domain of csi1p is responsible for interacting with alp7p. Compromised interaction between csi1p and alp7p impairs the localization of alp7p to the SPB during mitosis, thus delaying bipolar spindle formation and leading to anaphase B lagging chromosomes. Hence our study establishes that csi1p serves as a linking molecule tethering spindle-stabilizing factors to the SPB for promoting bipolar spindle assembly.
INTRODUCTION
Timely bipolar spindle assembly facilitates proper kinetochore biorientation, thereby ensuring accurate chromosome segregation during mitosis (Walczak and Heald, 2008; Tanenbaum and Medema, 2010; Silkworth and Cimini, 2012). Spindle assembly takes place in prophase, with microtubule minus ends anchored at the spindle poles and microtubule plus ends interdigitating at the spindle midzone to form an antiparallel microtubule array. Spindle bipolarity is well established by metaphase as the interdigitating microtubules slide apart and the opposing forces within the spindle are balanced (Syrovatkina et al., 2013). Bipolar spindle formation requires synergistic coordination of microtubule-associated proteins (MAPs) and kinesin motors (Tanenbaum and Medema, 2010). In general, MAPs help organize spindle microtubules into an antiparallel microtubule array, and kinesins produce forces to elongate the array to separate two spindle poles (Fu et al., 2009; Peterman and Scholey, 2009; Syrovatkina et al., 2013). Intensive studies on bipolar spindle formation have focused on the conserved MAP PRC1/ase1p and the kinesin-5 Eg5/cut7p. PRC1/ase1p becomes more static upon cross-linking two antiparallel microtubules (Janson et al., 2007; Subramanian et al., 2013), whereas Eg5/cut7p is rotationally flexible (Kapitein et al., 2005). These structural features enable PRC1/ase1p and Eg5p/cut7 to function efficiently to maintain antiparallel spindle microtubules after spindle bipolarity has been established but nonefficiently sort near parallel spindle microtubules into an antiparallel microtubule array during early prophase. This implies that initial spindle microtubule organization may require another molecular mechanism.
One possible mechanism may involve the transforming acidic coiled-coil (TACC) family proteins, as cells lacking TACC proteins typically display abnormalities in initial bipolar spindle assembly (Peset and Vernos, 2008). Members of this family are characterized by the conserved TACC domain at their carboxyl termini and share a similar intracellular localization pattern and conserved functions in a wide range of organisms, including yeast (Sato et al., 2004), Caenorhabditis elegans (Bellanger and Gonczy, 2003; Srayko et al., 2003), Drosophila (Gergely et al., 2000b), and mammals (Gergely et al., 2003). During mitosis, TACC proteins mainly localize to spindle microtubules and the centrosome (the spindle pole body [SPB] in yeast; Sato et al., 2004; Peset and Vernos, 2008), where they form a complex with the conserved microtubule polymerase ch-TOG proteins via their TACC domains to stabilize kinetochore microtubules (Royle, 2012) and promote spindle formation (Peset and Vernos, 2008).
Clathrin, a key protein involved in membrane trafficking, has been reported to interact with TACC3 and subsequently form the clathrin–ch-TOG–TACC3 complex for localization to spindle microtubules (Fu et al., 2010; Lin et al., 2010; Booth et al., 2011; Hood et al., 2013), resulting in kinetochore fiber stabilization, an important process for spindle stabilization (Booth et al., 2011). Despite this, it remains unclear how TACC proteins are recruited to the centrosome/SPB, an equally important process required for spindle assembly and stabilization. Intriguingly, the conserved TACC domain is necessary for the localization of TACC proteins to the centrosome/SPB, independent of microtubules (Bellanger and Gonczy, 2003) and ch-TOG proteins (Gergely et al., 2000b; Sato et al., 2004), and the TACC domain does not interact with clathrin (Hood et al., 2013). These findings highlight the importance of the TACC domain in interacting with a centrosomal/SPB protein that remains to be determined.
The fission yeast centrosome/SPB protein csi1p is emerging as a key protein for ensuring faithful mitotic chromosome segregation (Hou et al., 2012). It is recruited to the SPB by the conserved SUN-domain protein sad1p (SUN1 in humans) for centromere clustering during interphase (Hou et al., 2012). Csi1p has also been implicated in spindle formation (Costa et al., 2013).
In this study, we show that csi1p promotes bipolar spindle assembly by recruiting the TACC orthologue alp7p to the SPB. The interaction domains in csi1p and alp7p lie at their carboxyl termini. When the interaction between csi1p and alp7p is compromised, alp7p and its binding protein alp14p/ch-TOG are absent from the SPBs, leading to transient monopolar spindle formation and subsequent anaphase B lagging chromosomes. Thus this work defines a new molecular mechanism regulating the SPB localization of the alp7p/TACC-alp14/ch-TOG complex and highlights the importance of this SPB complex in bipolar spindle formation and faithful chromosome segregation.
RESULTS
Csi1p interacts with alp7p
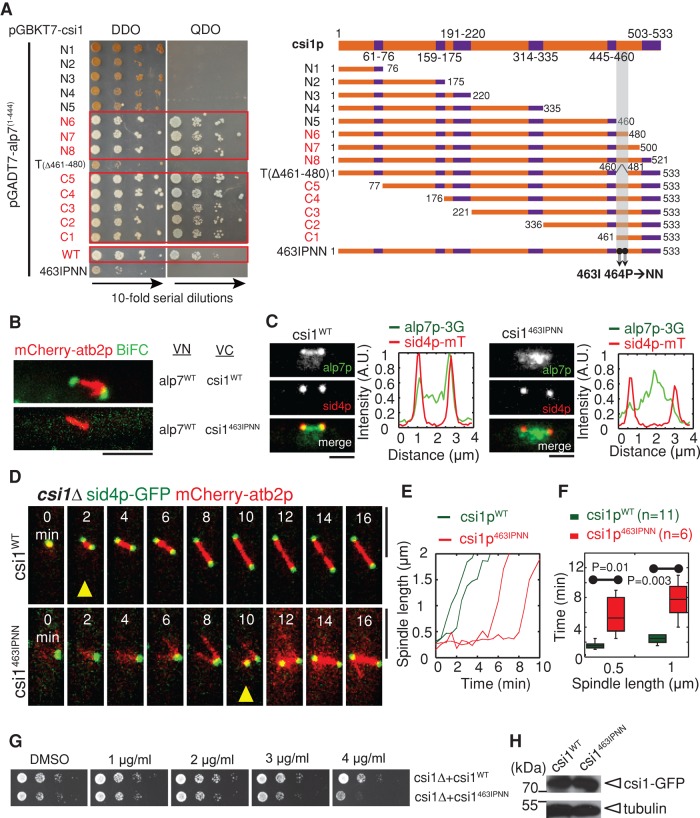
Csi1p has been shown to be an essential protein for centromere clustering during interphase (Hou et al., 2012). Our live-cell imaging screen identified csi1p as a key component for bipolar spindle formation (Costa et al., 2013; see later discussion of Figure 2A). To understand the role of csi1p in bipolar spindle formation, we carried out a yeast-two hybrid screen for csi1p-binding proteins. In this screen, an alp7p mutant lacking the last 31 residues at its carboxyl terminus was identified as a strong interacting protein of csi1p (Figure 1A), and the interaction between csi1p and alp7p was further confirmed by glutathione S-transferase (GST) pull-down assays, using full-length recombinant proteins histidine (His)-alp7p and GST-csi1p (Figure 1B). Therefore the carboxyl-terminal 31 residues of alp7p are not required for its interaction with csi1p, although these residues appear to affect the alp7p-csi1p interaction specifically in budding yeast in the yeast two-hybrid assays (see later discussion of Figure 5A).
FIGURE 2:
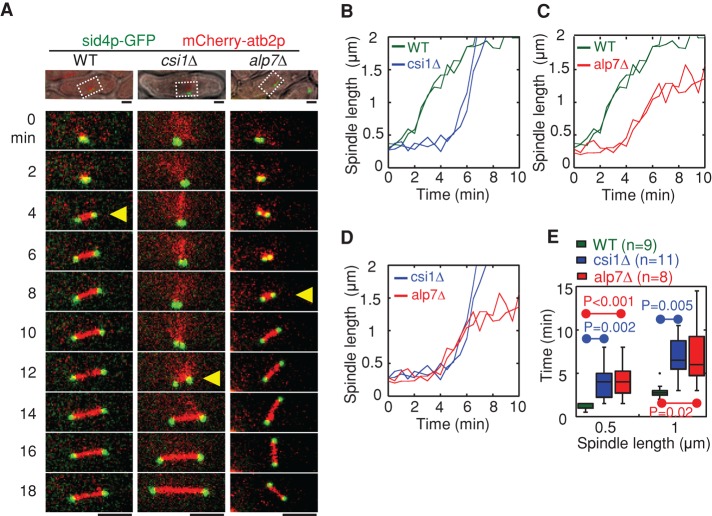
Csi1p is required for bipolar spindle assembly. (A) Maximum projection live-cell images of wild-type, csi1∆, and alp7∆ cells expressing sid4p-GFP (SPB marker) and mCherry-atb2p (α-tubulin). Yellow triangles mark bipolar spindles ∼1 μm in length. Highlighted regions in the differential interference contrast pictures. Scale bar, 2 μm. (B–D) Representative plots of spindle length against time for wild-type and csi1∆ cells (B), wild-type and alp7∆ cells (C), and csi1∆ and alp7∆ cells (D). (E) Box plots of time for assembly of bipolar spindles measuring 0.5 and 1 μm in length in wild-type, csi1∆, and alp7∆ cells. Student's t test was used to calculate p values. Cell numbers analyzed are indicated.
FIGURE 1:
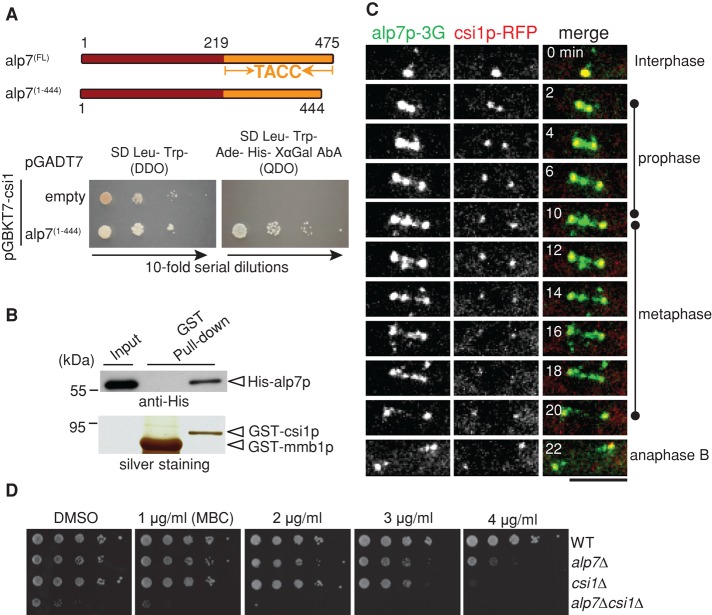
Csi1p interacts with alp7p. (A) Yeast two-hybrid assays testing the interaction between csi1p and alp7p. Y2HGold budding yeasts cotransformed with BD-csi1 and AD-alp7(1-444) or empty AD plasmids were subjected to 10-fold serial dilutions and spotted on SD/–Leu/–Trp (DDO) and SD/–Leu/–Trp/–Ade/–His (QDO) plus X-α-gal and Aureobasidin A (AbA) plates and incubated at 30°C for 4 d. (B) GST pull-down assays. Full-length recombinant proteins GST-csi1p and His-alp7p were produced in E. coli; the precipitation products were analyzed by Western blotting with anti-His antibody. Note that His-alp7p coprecipitated with GST-csi1p, not the control GST-mmb1p. (C) Maximum projection live-cell images of a cell expressing csi1p-TagRFP and alp7p-3GFP from their own promoters. Alp7p colocalized with csi17p at the SPBs throughout mitosis and appeared as distinct dots between the two SPBs during metaphase. Scale bar, 5 μm. (D) MBC sensitivity assays. Tenfold serial dilutions of wild-type (WT), alp7∆ (alp7p null), csi1∆ (csi1p null), and alp7∆csi1∆ (alp7p and csi1p null) cells were grown at 30°C for 4 d on YE5S plates containing dimethyl sulfoxide or the indicated concentrations of MBC. Note that cells lacking both alp7+ and csi1+ displayed an additive defect in cell growth.
FIGURE 5:
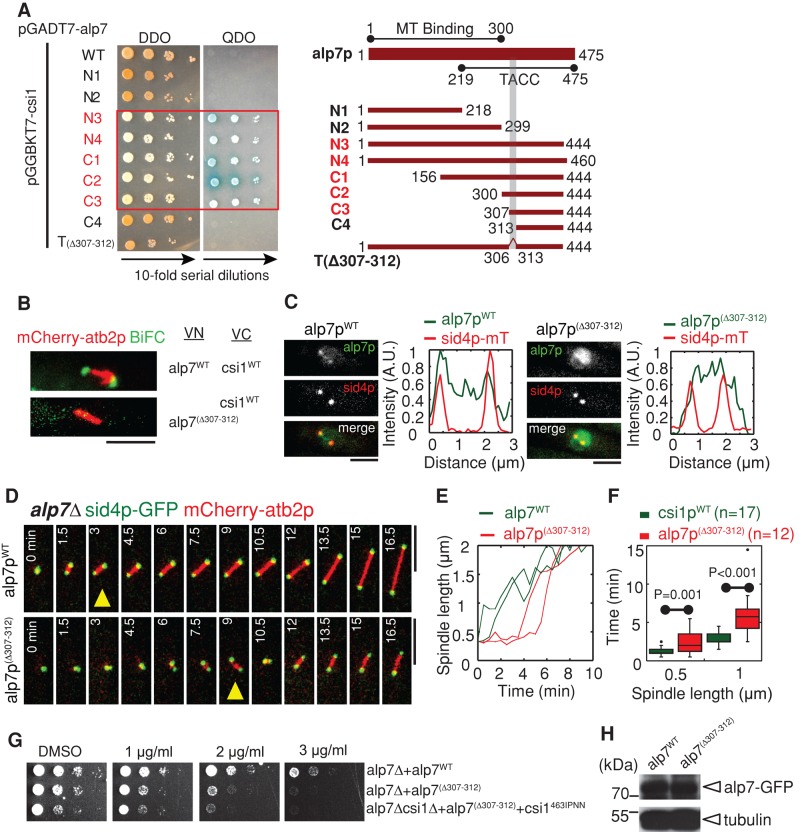
The alp7p TACC domain is responsible for interacting with csi1p. (A) Yeast two-hybrid assays for mapping the minimal csi1p interaction domain in csi1p. A series of alp7p deletion truncation mutants, as indicated in the schematic diagram, was used to test their interaction with csi1p, revealing that the domain 307–312 within the alp7p TACC domain is necessary for interacting with csi1p. (B) BiFC assays. Maximum projection images of cells expressing alp7pWT-VN or alp7p(∆307-312)-VN and csi1pWT-VC from the nmt1 promoter. Cells were cultured in EMM without thiamine for 14 h before imaging. Note that only the cell expressing wild-type alp7p gave BiFC signals. Scale bars, 5 μm. (C) Maximum projection images of alp7∆ cells expressing sid4p-mTomato and either wild-type alp7p (indicated as alp7pWT) or mutant alp7p(∆307-312) (indicated as alp7p(∆307-312)) from a alp7p promoter at the leu1-32 locus. Fluorescence intensity measurements were carried out to analyze alp7p signal profiles along the spindles. alp7p(∆307-312) no longer concentrated at the SPBs. Scale bars, 2 μm. (D) Maximum projection live-cell images of alp7pWT and alp7p(∆307-312) cells expressing sid4p-GFP and mCherry-atb2p. Yellow triangles mark bipolar spindles ∼1 μm in length. Bipolar spindle assembly in the alp7p(∆307-312) was impaired. Scale bars, 5 μm. (E) Representative plots of spindle length against time for alp7pWT and alp7p(∆307-312) cells. (F) Box plots of time for assembly of bipolar spindles measuring 0.5 and 1 μm in length in alp7pWT and alp7p(∆307-312) cells. Student's t test was used to calculate p values. Cell numbers analyzed are indicated. (G) MBC sensitivity assays for alp7WT, alp7(∆307-312), and csi1463IPNN and alp7(∆307-312) double-mutant cells. The cells were grown at 35°C for 4 d. Similar to alp7∆, alp7(∆307-312) cells were sensitive to MBC, and csi1463IPNN and alp7(∆307-312) displayed no obvious additive effect. (H) Western blot analysis of cells expressing alp7WT -GFP and alp7(∆307-312)-GFP.
We then used live-cell imaging to examine the colocalization of alp7p-3GFP (green fluorescent protein) and csi1p-TagRFP in wild-type cells. As shown in Figure 1C, alp7p displayed spindle localization, with strong preferential SPB localization, and colocalization with csi1p throughout mitosis (also see Supplemental Figure S1A). In addition, we observed alp7p signals between the two SPBs on the spindle, which may represent its kinetochore localization, as previously reported (Sato et al., 2004; Tang et al., 2013). These additional alp7p signals likely did not colocalize with csi1p, as even overexpressed csi1p from the nmt1 promoter did not localize to kinetochores (Supplemental Figure S1B). Intriguingly, csi1∆ and alp7∆ double-deletion mutant displayed additive sensitivity to the microtubule-depolymerizing drug methyl benzimidazole-2-yl-carbamate (MBC) in a dose-dependent manner (Figure 1D). Taken together, these findings suggest that csi1p physically and genetically interacts with alp7p.
Csi1p and alp7p are required for bipolar spindle assembly
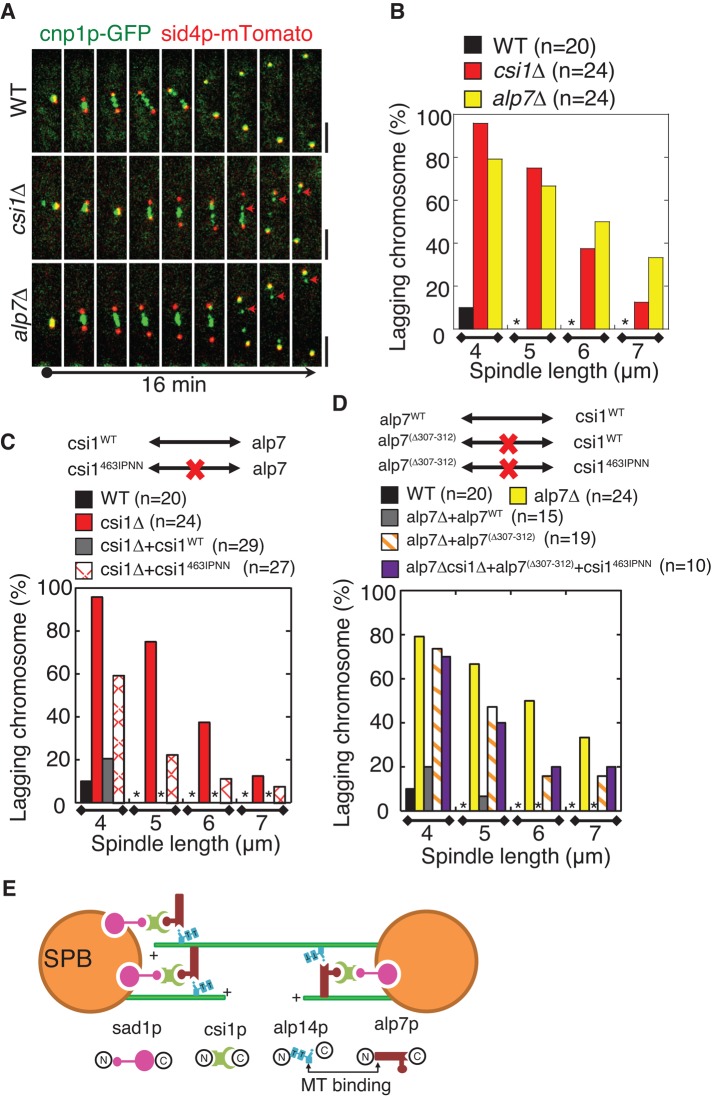
Both csi1p and alp7p are involved in regulating chromosome segregation (Supplemental Figure S2, A and B; Sato et al., 2004; Hou et al., 2012; Tang et al., 2013). Of importance, alp7p is required for microtubule organization (Thadani et al., 2009), mitotic spindle assembly (Sato et al., 2004; Sato and Toda, 2007), and proper attachment of spindle microtubules to kinetochores (Tang et al., 2013). Therefore, to assess the biological significance of the interaction between csi1p and alp7p, we first analyzed spindle dynamics, an important process for proper chromosome segregation. Live-cell imaging revealed that similar to alp7∆, csi1∆ cells displayed defective spindle dynamics during early mitosis, with the spindles initially forming a transient monopolar structure (Figure 2, A–D, and Supplemental Figure S2, E–G). In contrast, the wild-type spindles initially emerged as a dense, dot-like structure and quickly elongated to establish a bar-like bipolar structure 1 μm in length within 2–3 min (Figure 2, A and E). Further measurements of spindle dynamics in csi1∆ and alp7∆ mutant cells showed that a significantly longer time was required for these mutants to assemble a bipolar spindle of 1 μm in length compared with wild-type cells (7.0 ± 2.2 and 7.2 ± 3.9 min for csi1∆ and alp7∆ cells, respectively, vs. 3.0 ± 0.8 min for wild-type cells; Figure 2, D and E).
We then analyzed centromere clustering, another key process for proper chromosome segregation. In agreement with a previous report (Hou et al., 2012), 56% of the csi1∆ cells displayed centromere declustering (Supplemental Figure S1, C and D). However, no wild-type cells and only 2% of the alp7∆ cells displayed centromere declustering (Supplemental Figure S1, C and D). The high degree of similarity of spindle defect, not centromere declustering, caused by the absence of csi1p or alp7p suggests that the interaction between csi1p and alp7p is likely involved in bipolar spindle formation.
Csi1p recruits alp7p to the SPB during mitosis
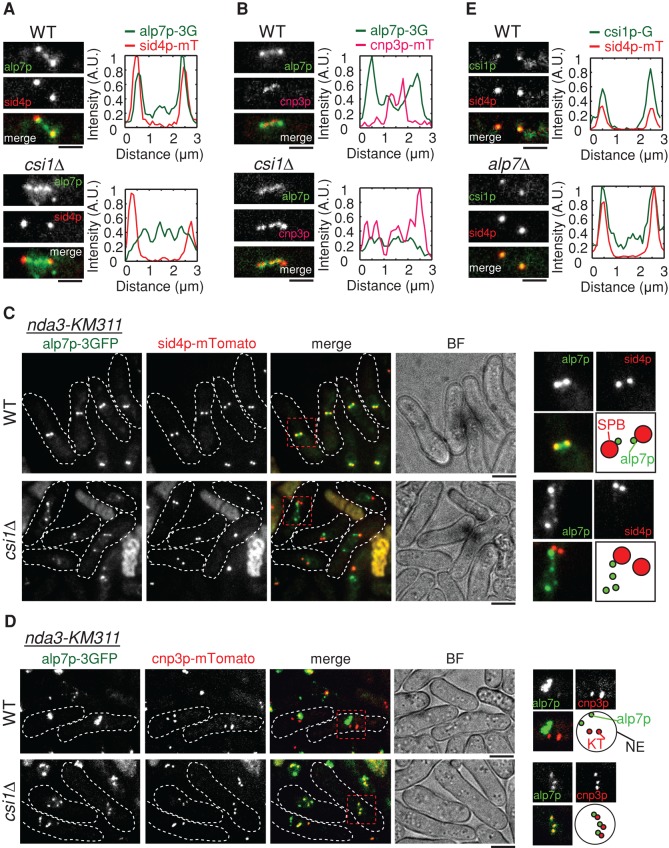
Next we analyzed the localization interdependence of alp7p and csi1p by confocal microscopy. Fluorescence intensity analysis of sum projection images showed that the majority of alp7p colocalized with csi1p at the SPBs in wild-type cells in early mitosis, whereas most alp7p signals appeared between the two SPBs as several distinct dots in csi1∆ cells (Figure 3A). Further fluorescence intensity analysis revealed that the alp7p dots between the two SPBs in csi1∆ cells colocalized with cnp3p (a kinetochore protein; CENP-C in humans) at the kinetochores (Figure 3B). To exclude the effect of spindle microtubules on alp7p localization, we then took advantage of the nda3-KM311 β-tubulin thermosensitive mutant strain (Hiraoka et al., 1984), which can be arrested in prophase by temperature shift to the restrictive temperature of 16ºC and has no spindle upon arrest. Similarly, we observed that at the restrictive temperature, the absence of csi1p caused delocalization of alp7p from the SPBs in prophase (Figure 3C), and the delocalized alp7p colocalized with cnp3p at the kinetochores (Figure 3D). Consistently, the absence of csi1p also caused delocalization of the alp7p-binding partner alp14p from the SPBs (Supplemental Figure S3). Moreover, we tested conversely whether alp7p is required for the SPB localization of csi1p. The fluorescence intensity analysis showed that the SPB localization of csi1p was not altered in the absence of alp7p (Figure 3E). Therefore we conclude that csi1p is required for the SPB localization of the alp7p-alp14p complex.
FIGURE 3:
Csi1p recruits alp7p to the SPBs. (A) Sum projection images of wild-type and csi1∆ cells expressing alp7p-3GFP and the SPB marker sid4p-mTomato (SPB marker) from their own promoters. Fluorescence intensity measurements were carried out with MetaMorph to analyze signal profiles of alp7p and sid4p along the spindle. Alp7p colocalized with sid4p at the SPBs in the wild-type cell but not in the csi1∆ cell; in the csi1∆ cell, alp7p appeared as distinct dots between the two SPBs. Scale bars, 2 μm. (B) Sum projection images of wild-type and csi1∆ cells expressing alp7p-3GFP and the kinetochore marker cnp3p-mTomato (kinetochore marker) from their own promoters. Fluorescence intensity measurements were carried out to analyze signal profiles of alp7p and cnp3p along the spindle. Alp7p colocalized with cnp3p at the kinetochores in the csi1∆ cell but not in the wild-type cell. Scale bars, 2 μm. (C) Sum projection images of nda3-KM311 wild-type and nda3-KM311 csi1∆ cells expressing alp7p-3GFP and sid4-mTomato. Before imaging, nda3-KM311 cells were cultured at their restrictive temperature 16°C for 6 h, a condition that can efficiently disassemble the spindles to arrest the cells at prophase/prometaphase. Magnified views on the right highlight the delocalization of alp7p from the SPBs in the absence of csi1p. Scale bars, 2 μm. (D) Sum projection images of nda3-KM311 wild-type and nda3-KM311 csi1∆ cells expressing alp7p-3GFP and cnp3p-mTomato. Before imaging, nda3-KM311 cells were cultured at their restrictive temperature 16°C for 6 h. Magnified views on the right highlight the colocalization of alp7p and cnp3p in the absence of csi1p. Scale bars, 2 μm. (E) Sum projection images of wild-type and alp7∆ cells expressing csi1p-GFP and sid43p-mTomato from their own promoters. Fluorescence intensity measurements were carried out to analyze signal profiles of csi1p and sid4p along the spindle. Csi1p colocalized with sid4p at the SPBs in both wild-type and alp7∆ cells. Scale bars, 2 μm.
The carboxyl terminus of csi1p is responsible for interacting with alp7p
In addition to centromere clustering, csi1p plays a role in bipolar spindle formation. It is likely that csi1p recruits alp7p to the SPBs for promoting bipolar spindle formation. To assess precisely the function of the csi1p-alp7p interaction, we sought to create a separation-of-function mutant for csi1p, whose interaction with alp7p is specifically inhibited. For this purpose, we used yeast two-hybrid assays to map the key csi1p residues responsible for interacting with alp7p. Multiple attempts revealed that Ile-463 and Pro-464 within the minimal domain (amino acids [aa] 461–480) lying at the carboxyl terminus of csi1p were indispensable for interacting with alp7p (Figure 4A).
FIGURE 4:
Csi1p carboxyl terminus is responsible for interacting with alp7p. (A) Yeast two-hybrid assays for mapping the minimal alp7p interaction domain in csi1p. A series of csi1p deletion truncation mutants, as indicated in the schematic diagram (coiled-coil domains indicated in purple), was used to test their interaction with alp71-444, revealing that domain 461–480 at the csi1p carboxyl terminus is important for interacting with alp71-444. Further, the two residues Ile-463 and Pro-464 within the minimal domain in csi1p are key residues responsible for the interaction with alp7p. (B) BiFC assays. Maximum projection images of cells expressing alp7pWT-VN and csi1pWT-VC or csi1p463IPNN-VC from the nmt1 promoter. Cells were cultured in EMM (Edinburgh minimal medium) without thiamine for 14 h before imaging. Note that only the cell expressing wild-type csi1p gave BiFC signals. Scale bars, 5 μm. (C) Maximum projection images of csi1∆ cells expressing alp7p-3GFP, sid4p-mTomato, and either wild-type csi1p (indicated as csi1WT) or mutant csi1p463IPNN (indicated as csi1463IPNN) from a csi1p promoter at the leu1-32 locus. Fluorescence intensity measurements were carried out to analyze alp7p signal profiles along the spindles. Alp7p no longer concentrated at the SPBs in the csi1p463IPNN cell. Scale bars, 2 μm. (D) Maximum projection live-cell images of csi1WT and csi1463IPNN cells expressing sid4p-GFP and mCherry-atb2p. Yellow triangles mark bipolar spindles ∼1 μm in length. Note that the csi1p463IPNN cell displayed transient monopolar spindle formation. Scale bars, 5 μm. (E) Representative plots of spindle length against time for csi1WT and csi1463IPNN cells. (F) Box plots of time for assembly of bipolar spindles measuring 0.5 and 1 μm in length in csi1WT and csi1463IPNN cells. Student's t test was used to calculate p values. Cell numbers analyzed are indicated. (G) MBC sensitivity assays for csi1WT and csi1463IPNN cells. The cells were grown at 30°C for 4 d. Similar to csi1∆, csi1463IPNN cells were sensitive to MBC. (H) Western blot analysis of cells expressing csi1WT-GFP and csi1463IPNN-GFP.
Csi1p is a low-abundance nuclear protein, with ∼500 molecules in a cell (Marguerat et al., 2012). Moreover, csi1p likely interacts with alp7p within a very short time window at prophase. These make it technically challenging to perform coimmunoprecipitation to test csi1p-alp7p interaction. Instead, we used the bimolecular fluorescence complementation (BiFC) assay, which has emerged as a key assay for examining protein–protein interaction in many model organisms, including yeast (Kodama and Hu, 2012). Generally, two complement peptide fragments of GFP (VN and VC in our study) are fused with proteins to be tested, and the interaction of the two fusion proteins can then bring the two complementary peptide fragments together to form a mature GFP molecule to give fluorescence signals. Consistently, BiFC assays confirmed that csi1p interacts with alp7p at the SPB (Figure 4B) and that the substitution of Ile-463 and Pro-464 for two asparagines in csi1p (designated as csi1p463IPNN) can effectively inhibit its interaction with alp7p in vivo (Figure 4B). Csi1p463IPNN localized to the SPB as wild-type csi1p (designated as csi1pWT; Supplemental Figure S4A), and their expression levels were comparable (Figure 4H). Further, csi1p463IPNN did not cause centromere declustering (Supplemental Figure S4, B and C). However, csi1p463IPNN caused delocalization of alp7p from the SPBs (Figure 4C), thus phenocopying the effect of the absence of csi1p on alp7p localization. Consistently, the delocalized alp7p colocalized with cnp3p at the kinetochores (Supplemental Figure S4E).
We then examined spindle dynamics in csi1∆ cells expressing csi1p463IPNN or csi1pWT. As expected, csi1pWT restored normal spindle dynamics, whereas csi1p463IPNN did not (Figure 4D). Further quantitative measurements also confirmed that csi1p463IPNN caused transient monopolar spindle formation (Figure 4E and Supplemental Figure S4D), and significantly longer time was required for csi1p463IPNN mutant cells to assemble a spindle 1 μm in length (i.e., 7.7 ± 2.6 and 2.4 ± 0.01 min for csi1p463IPNN and csi1pWT cells, respectively; Figure 4F). Moreover, similar to csi1∆, csi1p463IPNN cells were sensitive to MBC in a dose-dependent manner (Figure 4G). Hence csi1p463IPNN phenocopies the csi1∆ spindle defect, supporting the conclusion that csi1p recruits alp7p to the SPBs specifically for promoting bipolar spindle formation.
The alp7p TACC domain is responsible for interacting with csi1p
The TACC domain is necessary and sufficient for localizing TACC proteins to the centrosome/SPB (Gergely et al., 2000b; Bellanger and Gonczy, 2003; Sato et al., 2004). This prompted us to explore whether the TACC domain in alp7p is responsible for interacting with csi1p. Similarly, yeast two-hybrid assays were used to map the minimal csi1p-interacting domain in alp7p. Because the last 31 residues in alp7p appeared to affect its interaction with csi1p in budding yeast (Figure 5A; full-length alp7p displayed no interaction with csi1p), we chose to use a series of alp7p deletion truncation mutants lacking the 31 residues for the yeast two-hybrid assays. This attempt revealed that residues 307–312 within the TACC domain in alp7p were required for interacting with csi1p (Figure 5A). Further, alp7p lacking residues 307–312 (designated alp7p(∆307-312)) showed no BiFC signals when paired with csi1p in the BiFC assays (Figure 5B), suggesting that alp7p residues 307–312 are also important for interacting with csi1p in vivo. We then tested the expression levels of alp7pWT and alp7p(∆307-312) in vivo, showing that their expression levels were comparable (Figure 5H). Next live-cell imaging showed that alp7p(∆307-312) did not cause centromere declustering (Supplemental Figure S5, A and B), but its SPB localization was impaired (Figure 5C), with most alp7p(∆307-312) residing at the kinetochores (Supplemental Figure S5F). This led to transient monopolar spindle formation (Figure 5, D–F, and Supplemental Figure S5C). Moreover, similar to alp7∆, alp7p(∆307-312) cells were sensitive to MBC in a dose-dependent manner at 35ºC (Figure 5G). Of interest, csi1p463IPNN alp7p(∆307-312) double-mutant cells showed no obvious additive effect on MBC sensitivity (Figure 5G), confirming that csi1p463IPNN and alp7p(∆307-312) mutants likely operate in the same pathway.
Our further yeast two-hybrid assays showed that although the alp7p TACC domain also interacts with alp14p (Sato et al., 2004; Sato and Toda, 2007), the residues 461–467 at the extreme carboxyl terminus of alp7p are what is important for interacting with alp14p (Supplemental Figure S5D). Therefore the csi1p- and alp14p-interacting regions in the alp7p TACC domain do not overlap.
Proper interaction between csi1p and alp7p is required for faithful chromosome segregation
Timely bipolar spindle formation ensures faithful chromosome segregation (Walczak and Heald, 2008; Tanenbaum and Medema, 2010; Silkworth and Cimini, 2012). We then asked whether the transient monopolar spindle formation caused by the compromised interaction between csi1p and alp7p affects faithful chromosome segregation. To address this question, we first carried out live-cell imaging to examine kinetochore dynamics in wild-type, csi1∆, and alp7∆ cells. As shown in Figure 6A, both deletion mutants displayed remarkable anaphase B lagging chromosomes. To quantify this phenotype, we measured the percentage of anaphase B cells that displayed spindles 4, 5, 6, and 7 μm in length, respectively, and concomitantly showed lagging chromosomes. This analysis showed that a comparable large number of anaphase B csi1∆ and alp7∆ cells displayed lagging chromosomes, whereas no anaphase B wild-type cells with a spindle length >4 μm displayed lagging chromosomes (Figure 6B). Moreover, csi1pWT restored normal chromosome segregation in csi1∆ cells, whereas csi1p463IPNN, which cannot interact with alp7p, only partially restored normal chromosome segregation in csi1∆ cells (Figure 6C). Likewise, alp7pWT restored normal chromosome segregation in alp7∆ cells (Figure 6D); however, alp7p(∆307-312), which cannot interact with csi1p, partially rescued lagging chromosomes in alp7∆ cells (Figure 6D). Further, csi1p463IPNN alp7p(∆307-312) double-mutant cells showed a comparable degree of anaphase B chromosome lagging as single alp7p(∆307-312) mutant cells (Figure 6D), further confirming that csi1p463IPNN and alp7p(∆307-312) mutants operate in the same pathway. Taken together, these findings suggest that timely bipolar spindle formation mediated by the csi1p-alp7p complex at the SPB is required for faithful chromosome segregation.
FIGURE 6:
The coordination between csi1p and alp7p is required for accurate chromosome segregation. (A) Maximum projection live-cell images of wild-type, csi1∆, and alp7∆ cells expressing cnp1p-GFP and sid4p-mTomato from their own promoters. Anaphase B lagging chromosomes were detected in csi1∆ and alp7∆ cells (arrows). Scale bars, 2 μm. (B–D) Graphs of the percentage of wild-type, csi1∆, and alp7∆ cells (B), wild-type, csi1∆, csi1WT, and csi1463IPNN cells (C), and wild-type, alp7∆, alp7pWT, alp7p(∆307-312), and csi1463IPNN and alp7(∆307-312) double-mutant cells (D) displaying anaphase B lagging chromosomes. Quantification was categorized according to the spindle length (4, 5, 6, and 7 μm, respectively). Stars indicate no anaphase B lagging chromosomes. Note that csi1463IPNN and alp7(∆307-312) has no additive effect on anaphase B lagging chromosomes. Cell numbers analyzed are indicated. (E) A model for bipolar spindle assembly. Csi1p serves as a linking molecule between sad1p and alp7p to recruit the alp7p-alp14p complex to the SPBs for promoting bipolar spindle formation. The N-termini of sad1p and csi1p (Hou et al., 2012) and the C-termini of alp7p and alp14p (Sato et al., 2004) bind to each other, respectively. The microtubule-binding domains lie at the N-terminus (aa 1–300) of alp7p, outside of the TACC domain (Thadani et al., 2009), and the C-terminus of alp14p, adjacent to the dual TOG domains (indicated as T; Al-Bassam et al., 2012), respectively.Csi1p recruits alp7p to the SPBs through an interaction between csi1p C-terminus (aa 461–480) and a region (aa 307–312) in the TACC domain of alp7p (indicated as the protruding circular shape).
DISCUSSION
Spindle assembly and stabilization requires the Alp7p/TACC-alp14p/ch-TOG complex, which targets to kinetochore microtubules (Royle, 2012) and the spindle poles (Sato et al., 2004; Peset and Vernos, 2008; Royle, 2012). Although it is relatively clear that TACC and ch-TOG interact with the trimerized clathrin heavy chains (CHCs) to cross-bridge kinetochore microtubules for spindle stabilization (Fu et al., 2010; Lin et al., 2010; Hood et al., 2013), it remains unknown how the Alp7p/TACC-alp14p/ch-TOG complex is tethered to the spindle poles, a key process for timely bipolar spindle formation (Sato et al., 2004; Peset and Vernos, 2008). Here we demonstrate that alp7p and alp14p depend on csi1p for localization to the SPBs (Figure 3), and the SPB localization of alp7p and alp14p then promotes timely bipolar spindle formation (Figures 2, 4, and 5), thus ensuring faithful chromosome segregation (Figure 6). We propose that csi1p serves as a linking molecule, recruiting the microtubule-stabilizing factor alp7p-alp14p complex to the SPB, where this complex facilitates the lateral binding of adjacent microtubules for promoting bipolar spindle formation (Figure 6E).
In addition to localizing to spindle microtubules (Royle, 2012), the evolutionarily conserved alp7p/TACC-alp14p/ch-TOG complex shares another striking similarity in concentrating at the centrosomes in higher eukaryotic cells (Peset and Vernos, 2008) and the SPBs in yeast during mitosis (Usui et al., 2003; Sato et al., 2004. The targeting of the alp7p/TACC-alp14p/ch-TOG complex to the centrosome/SPB is a decisive step for mitotic spindle assembly in Drosophila (Gergely et al., 2000b) and C. elegans (Bellanger and Gonczy, 2003; Le Bot et al., 2003; Srayko et al., 2003) embryos, Xenopus egg extracts (O'Brien et al., 2005; Peset et al., 2005), human somatic cells (Gergely et al., 2000a, 2003), and yeasts (Usui et al., 2003; Sato et al., 2004). The mitotic kinase Aurora A has been shown to be essential for targeting TACC proteins to the centrosome (Giet et al., 2002; Bellanger and Gonczy, 2003; Le Bot et al., 2003; Peset et al., 2005). However, the sole yeast Aurora kinase, ark1, does not localize to the SPB (Petersen et al., 2001), and thus it is unlikely that it regulates the SPB localization of the yeast TACC alp7p. Moreover, alp7p contains no transmembrane domains, making it also unlikely for alp7p to localize to the SPB by inserting into the nuclear envelope. Intriguingly, the C. elegans TACC protein TAC-1 localizes to the centrosome independent of microtubules (Bellanger and Gonczy, 2003), and the conserved TACC domain is necessary and sufficient for localization to the centrosome in Drosophila embryos (Gergely et al., 2000b). In fission yeast, alp7p/TACC does not depend on alp14p/ch-TOG (Sato et al., 2004) or on microtubules (Figure 3C) for localizing to the SPB. All these findings raise the possibility that a conserved centrosomal/SPB protein or its functional homologues may be responsible for recruiting alp7p/TACC to the centrosome/SPB. Knowledge of such proteins is beginning to emerge. For example, NDEL1, a centrosomal protein involved in dynein function, is required for targeting TACC3 to the centrosome (Mori et al., 2007), and the centrosomin Cnn is required for proper localization of D-TACC to Drosophila embryonic centrosomes (Zhang and Megraw, 2007).
Csi1p has no amino acid sequence similarity to either NDEL1 or Cnn. However, the absence of csi1p causes nearly identical defects of spindle formation and chromosome segregation as the absence of alp7p (Figures 2 and 6, A and B), highlighting that the two proteins may function in the same pathway. Indeed, csi1p determines the SPB localization of alp7p, but the converse is not true (Figure 3). Csi1p is also required for the SPB localization of alp14p (Supplemental Figure S3). This may be via alp7p, as there is no direct interaction between csi1p and alp14p (Supplemental Figure S5E). The alp7p-alp14p complex is a key target of the Ran GTPase–dependent spindle assembly machinery (Sato and Toda, 2007). Our present work further extends this model, in which the Ran machinery is shown to target the alp7p-alp14p complex to the nucleus for accumulation. Thus our data suggest that upon entering the nucleus at mitosis onset, the alp7p-alp14p complex is tethered to the SPBs by csi1p for promoting bipolar spindle formation (Figure 3). The absence of csi1p, that is, losing the SPB docking site for alp7p, leads to alp7p mislocalization, with the majority of alp7p localizing to the kinetochores (Figure 3, B and D), consistent with the recent finding that alp7p can also dock at the kinetochores by interacting with the internal loop of the kinetochore protein Ndc80 (Tang et al., 2013). Of importance, in wild-type cells, alp7p does not display strong localization at the kinetochore in prophase and instead mainly localizes to the SPBs (Figure 3, B and C). Hence, by interacting with csi1p, alp7p is confined to the SPB region and thus is biased toward promoting bipolar spindle formation during early mitosis.
How does then the SPB localization of the alp7p-alp14p complex promote bipolar spindle formation? Csi1p is recruited to the SPBs by the SUN-domain protein sad1p through a physical interaction between their amino termini (Hou et al., 2012; Figure 6E). Our domain-mapping data further show that the extreme carboxyl terminus of csi1p (Figure 4A) interacts with a small region within the TACC domain of alp7p (Figure 5A) to recruit alp7p to the SPBs (Figure 6E). This interaction likely does not affect the interaction between alp7p and alp14p, given that the alp14p-interacting domain lies at the end of the alp7p carboxy terminus (Supplemental Figure S5D), not overlapping with the csi1p-interacting domain. These unique arrangements of domain structure enable alp7p not only to be tethered to the SPBs, but also simultaneously to form a complex with the microtubule-stabilizing factor/microtubule polymerase alp14p (Al-Bassam et al., 2012). Because alp7p and alp14p interact with each other through their extreme carboxyl termini (Sato et al., 2004; Supplemental Figure S5D), the two microtubule-binding domains, respectively from alp7p and alp14p as previously defined (Thadani et al., 2009; Al-Bassam et al., 2012), flank the csi1p-binding site in the alp7p-alp14 heterodimer, a configuration favorable for bundling microtubules. The absence of the alp7p-alp14 complex at the SPB results in monopolar spindles with protruding microtubules (Figures 2, 4D, and 5C), suggesting that the alp7p-alp14p complex at the SPBs can efficiently cross-bridge adjacent microtubules that just emerge from the two SPBs during prophase and thus allows for formation of the bar-like spindle in yeast. Hence csi1p recruits to the SPBs the microtubule-bundling and -stabilizing factor alp7p-alp14p complex to promote timely bipolar spindle formation.
Thus far csi1p has been shown to play roles in centromere clustering (Hou et al., 2012) and bipolar spindle formation (Figures 2 and 6E). Therefore, the lagging chromosomes caused by the absence of csi1p could be due to either centromere declustering or transient monopolar spindle formation. Given that the mutant csi1p463IPNN causes transient monopolar spindle formation (Figure 4D) but not centromere declustering (Supplemental Figure S4, B and C), it is an ideal separation-of-function mutant for assessing the individual contributions of centromere clustering and bipolar spindle formation to faithful chromosome segregation. Csi1p463IPNN still causes anaphase B lagging chromosomes but to a lesser degree than csi1∆ (Figure 6C and Supplemental Figure S6A), suggesting that not only bipolar spindle formation but also centromere clustering contributes to faithful chromosome segregation. Consistently, both alp7∆ and alp7p(∆307-312) mutants, which display normal centromere clustering (Supplemental Figures S2, C and D, and S5, A and B) but transient monopolar spindles (Figures 2 and 5D), show massive anaphase B lagging chromosomes (Figure 6, A and D, and Supplemental Figure S6B), reinforcing the conclusion that proper bipolar spindle formation is required for faithful chromosome segregation. Thus csi1p ensures faithful chromosome segregation not only by clustering centromeres at the SPBs (Hou et al., 2012), but also by promoting timely bipolar spindle formation.
Blast analysis shows no homologues of csi1p in other species, implying that csi1p may not be conserved through evolution or has evolved to functional homologues with a short conserved domain as alp7p/TACC (Peset and Vernos, 2008). Despite this, all csi1p binding proteins reported thus far, including sad1p, spc7p (Hou et al., 2012), and alp7p (Figure 1), are well conserved through evolution. In addition, the mechanism targeting alp7p/TACC to the centrosome/SPB is conserved. Therefore, to identify csi1p homologues in other species, it will be of great interest to explore the centrosomal proteins that are involved in bipolar spindle formation and have the ability to interact with the conserved TACC domain.
MATERIALS AND METHODS
Plasmids and yeast strains
Yeast genetics was carried out as previously described (Forsburg and Rhind, 2006), and yeast strains were created by either random spore digestion or tetrad dissection. Yeast culture media were purchased from ForMedium (Norfolk, UK). Mutagenesis was performed with the QuikChange II XL Site-Directed Mutagenesis Kit (Agilent Technologies, Santa Clara, CA). MBC sensitivity assays were carried out on YES5S (yeast extract medium supplemented with adenine, leucine, uracil, histidine, and lysine) plates supplied with the indicated concentration of the microtubule-depolymerizing drug MBC (Sigma-Aldrich, St. Louis, MO). Minichromosome loss assays were carried out as described in Niwa et al. (1989). All yeast strains and plasmids used in this study are listed in Supplemental Tables S1 and S2, respectively.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed using the Matchmaker Gold yeast two-hybrid system, along with Yeastmaker Yeast Transformation System 2 (Clontech, Mountain View, CA). Tenfold serial dilutions of Y2HGold cells containing bait and prey plasmids were cultured on double-dropout medium SD/–Leu/–Trp plates and quadruple-dropout medium SD/–Leu/–Trp/–Ade/–Ura plates containing 40 μg/ml X-α-Gal and 200 ng/ml Aureobasidin A at 30°C for 4 d.
Biochemistry
Recombinant proteins were purified from Escherichia coli using either glutathione Sepharose 4B resins (for GST-fused proteins; GE Healthcare, Pittsburgh, PA) or nickel resins (for His-tagged proteins; Qiagen, Valencia, CA). GST pull-down assays were then carried out by incubating GST-fused proteins bound to the glutathione resins with His-tagged proteins in Tris-buffered saline (TBS) plus 0.1% Triton X-100 at 4ºC for 4 h, followed by 5× intensive washing with the TBS plus 0.1% Triton X-100 buffer and 1× TBS buffer. The resulting pull-down products were analyzed by silver staining and Western blotting with anti-His antibody (GE Healthcare). For protein expression level analysis, yeast protein extract was prepared as previously described (Matsuo et al., 2006), followed by SDS–PAGE analysis and Western blotting with anti-GFP (Rockland Immunochemicals, Gilbertsville, PA) and anti-tubulin antibodies.
Microscopy and data analysis
Imaging was carried out as previously described (Tran et al., 2004). Briefly, a PerkinElmer spinning-disk confocal microscope equipped with a Zeiss PlanApo 100×/1.4 numerical aperture objective and the Photometrics electron-multiplying charge-coupled device camera Evolve 512 was used to carry out live-cell imaging at 26ºC in a temperature-controllable incubator. For maximum projection analysis, Z-stack images consisting of 11 planes (step size, 0.5 μm) were acquired every 30 s (for spindle dynamics analysis) or 1 min (for chromosome dynamics analysis); for sum projection analysis, Z-stack images consisting of 21 planes (step size, 0.25 μm) were acquired. Detailed imaging conditions are also described in the Supplemental Material. Spindle lengths were measured with MetaMorph (Molecular Devices, Sunnyvale, CA) and the MTrackJ plug-in in ImageJ (National Institutes of Health, Bethesda, MD) as previously described (Fu et al., 2009). Fluorescence intensity measurements were performed using the Linescan function in MetaMorph, and values were then normalized to the maximum fluorescence intensity in each comparison group. Student's t tests were determined using Excel. Box plots and graphs were generated with Kaleidagraph 4.5 (Synergy Software, Reading, PA).
Note added in proof. While our manuscript was in press, Takashi Toda (Cancer Research UK) and colleagues published a paper showing that pcp1p is also involved in the recruitment of alp7p to the SPB (Tang et al., 2014).
Supplementary Material
Acknowledgments
We thank Takashi Toda (Cancer Research UK, London, United Kingdom), Snezhana Oliferenko (King's College London, London, United Kingdom), Dannel McCollum (University of Massachusetts Medical School, Worcester, MA), Yoshinori Watanabe (University of Tokyo, Japan), Mitsuhiro Yanagida (University of Tokyo, Japan), and Fred Chang (Columbia University, New York, NY) for providing yeast strains. We also thank George Tsao and Jing Guo of the Faculty Core Facility at the University of Hong Kong for technical support. This work is supported by grants from the National Natural Science Foundation of China (31271439 and 31301109), the Hong Kong Research Grants Council, and the Hong Kong University Seed Funding Programme to C.F., the S.K. Yee Medical Research Fund 2011, and grants from the National Institutes of Health and the Agence Nationale de la Recherche to P.T.T.
Abbreviations used:
- EMM
Edinburgh minimal medium
- GST
glutathione S-transferase
- MAP
microtubule-associated protein
- MBC
methyl benzimidazole-2-yl-carbamate
- SPB
spindle pole body
- TACC
transforming acidic coiled-coil
- YE5S
yeast extract medium supplemented with adenine, leucine, uracil, histidine, and lysine
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-03-0786) on July 23, 2014.
Author contributions: F.Z., T.L., P.T.T., and C.F. designed and carried out experiments and analyzed data. F.Z., P.T.T., and C.F. wrote the article. All authors made comments.
REFERENCES
- Al-Bassam J, Kim H, Flor-Parra I, Lal N, Velji H, Chang F. Fission yeast Alp14 is a dose-dependent plus end-tracking microtubule polymerase. Mol Biol Cell. 2012;23:2878–2890. doi: 10.1091/mbc.E12-03-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellanger JM, Gonczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–1498. doi: 10.1016/s0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- Booth DG, Hood FE, Prior IA, Royle SJ. A TACC3/ch-TOG/clathrin complex stabilises kinetochore fibres by inter-microtubule bridging. EMBO J. 2011;30:906–919. doi: 10.1038/emboj.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa J, Fu C, Syrovatkina V, Tran PT. Imaging individual spindle microtubule dynamics in fission yeast. Methods Cell Biol. 2013;115:385–394. doi: 10.1016/B978-0-12-407757-7.00024-4. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C, Ward JJ, Loiodice I, Velve-Casquillas G, Nedelec FJ, Tran PT. Phospho-regulated interaction between kinesin-6 Klp9p and microtubule bundler Ase1p promotes spindle elongation. Dev Cell. 2009;17:257–267. doi: 10.1016/j.devcel.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, Tao W, Zheng P, Fu J, Bian M, Jiang Q, Clarke PR, Zhang C. Clathrin recruits phosphorylated TACC3 to spindle poles for bipolar spindle assembly and chromosome alignment. J Cell Sci. 2010;123:3645–3651. doi: 10.1242/jcs.075911. [DOI] [PubMed] [Google Scholar]
- Gergely F, Draviam VM, Raff JW. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 2003;17:336–341. doi: 10.1101/gad.245603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA. 2000a;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gergely F, Kidd D, Jeffers K, Wakefield JG, Raff JW. D-TACC: a novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000b;19:241–252. doi: 10.1093/emboj/19.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hood FE, Williams SJ, Burgess SG, Richards MW, Roth D, Straube A, Pfuhl M, Bayliss R, Royle SJ. Coordination of adjacent domains mediates TACC3-ch-TOG-clathrin assembly and mitotic spindle binding. J Cell Biol. 2013;202:463–478. doi: 10.1083/jcb.201211127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H, Zhou Z, Wang Y, Wang J, Kallgren SP, Kurchuk T, Miller EA, Chang F, Jia S. Csi1 links centromeres to the nuclear envelope for centromere clustering. J Cell Biol. 2012;199:735–744. doi: 10.1083/jcb.201208001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson ME, Loughlin R, Loiodice I, Fu C, Brunner D, Nedelec FJ, Tran PT. Crosslinkers and motors organize dynamic microtubules to form stable bipolar arrays in fission yeast. Cell. 2007;128:357–368. doi: 10.1016/j.cell.2006.12.030. [DOI] [PubMed] [Google Scholar]
- Kapitein LC, Peterman EJ, Kwok BH, Kim JH, Kapoor TM, Schmidt CF. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 2005;435:114–118. doi: 10.1038/nature03503. [DOI] [PubMed] [Google Scholar]
- Kodama Y, Hu CD. Bimolecular fluorescence complementation (BiFC): a 5-year update and future perspectives. BioTechniques. 2012;53:285–298. doi: 10.2144/000113943. [DOI] [PubMed] [Google Scholar]
- Le Bot N, Tsai MC, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr Biol. 2003;13:1499–1505. doi: 10.1016/s0960-9822(03)00577-3. [DOI] [PubMed] [Google Scholar]
- Lin CH, Hu CK, Shih HM. Clathrin heavy chain mediates TACC3 targeting to mitotic spindles to ensure spindle stability. J Cell Biol. 2010;189:1097–1105. doi: 10.1083/jcb.200911120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, Bahler J. Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell. 2012;151:671–683. doi: 10.1016/j.cell.2012.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuo Y, Asakawa K, Toda T, Katayama S. A rapid method for protein extraction from fission yeast. Biosci Biotechnol Biochem. 2006;70:1992–1994. doi: 10.1271/bbb.60087. [DOI] [PubMed] [Google Scholar]
- Mori D, Yano Y, Toyo-oka K, Yoshida N, Yamada M, Muramatsu M, Zhang D, Saya H, Toyoshima YY, Kinoshita K, et al. NDEL1 phosphorylation by Aurora-A kinase is essential for centrosomal maturation, separation, and TACC3 recruitment. Mol Cell Biol. 2007;27:352–367. doi: 10.1128/MCB.00878-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien LL, Albee AJ, Liu L, Tao W, Dobrzyn P, Lizarraga SB, Wiese C. The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Mol Biol Cell. 2005;16:2836–2847. doi: 10.1091/mbc.E04-10-0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Seiler J, Sardon T, Bejarano LA, Rybina S, Vernos I. Function and regulation of maskin, a TACC family protein, in microtubule growth during mitosis. J Cell Biol. 2005;170:1057–1066. doi: 10.1083/jcb.200504037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–388. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Peterman EJ, Scholey JM. Mitotic microtubule crosslinkers: insights from mechanistic studies. Curr Biol. 2009;19:R1089–1094. doi: 10.1016/j.cub.2009.10.047. [DOI] [PubMed] [Google Scholar]
- Petersen J, Paris J, Willer M, Philippe M, Hagan IM. The S. pombe Aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J Cell Sci. 2001;114:4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- Royle SJ. The role of clathrin in mitotic spindle organisation. J Cell Sci. 2012;125:19–28. doi: 10.1242/jcs.094607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Toda T. Alp7/TACC is a crucial target in Ran-GTPase-dependent spindle formation in fission yeast. Nature. 2007;447:334–337. doi: 10.1038/nature05773. [DOI] [PubMed] [Google Scholar]
- Sato M, Vardy L, Angel Garcia M, Koonrugsa N, Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silkworth WT, Cimini D. Transient defects of mitotic spindle geometry and chromosome segregation errors. Cell Div. 2012;7:19. doi: 10.1186/1747-1028-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr Biol. 2003;13:1506–1511. doi: 10.1016/s0960-9822(03)00597-9. [DOI] [PubMed] [Google Scholar]
- Subramanian R, Ti SC, Tan L, Darst SA, Kapoor TM. Marking and measuring single microtubules by PRC1 and kinesin-4. Cell. 2013;154:377–390. doi: 10.1016/j.cell.2013.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syrovatkina V, Fu C, Tran PT. Antagonistic spindle motors and MAPs regulate metaphase spindle length and chromosome segregation. Curr Biol. 2013;23:2423–2429. doi: 10.1016/j.cub.2013.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Medema RH. Mechanisms of centrosome separation and bipolar spindle assembly. Dev Cell. 2010;19:797–806. doi: 10.1016/j.devcel.2010.11.011. [DOI] [PubMed] [Google Scholar]
- Tang NH, Okada N, Fong CS, Arai K, Sato M, Toda T. Targeting Alp7/TACC to the spindle pole body is essential for mitotic spindle assembly in fission yeast. FEBS Lett. 2014 doi: 10.1016/j.febslet.2014.06.027. doi: 10.1016/j.febslet.2014.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang NH, Takada H, Hsu KS, Toda T. The internal loop of fission yeast Ndc80 binds Alp7/TACC-Alp14/TOG and ensures proper chromosome attachment. Mol Biol Cell. 2013;24:1122–1133. doi: 10.1091/mbc.E12-11-0817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thadani R, Ling YC, Oliferenko S. The fission yeast TACC protein Mia1p stabilizes microtubule arrays by length-independent crosslinking. Curr Biol. 2009;19:1861–1868. doi: 10.1016/j.cub.2009.09.063. [DOI] [PubMed] [Google Scholar]
- Tran PT, Paoletti A, Chang F. Imaging green fluorescent protein fusions in living fission yeast cells. Methods. 2004;33:220–225. doi: 10.1016/j.ymeth.2003.11.017. [DOI] [PubMed] [Google Scholar]
- Usui T, Maekawa H, Pereira G, Schiebel E. The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 2003;22:4779–4793. doi: 10.1093/emboj/cdg459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Zhang J, Megraw TL. Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires centrosomin motif 1. Mol Biol Cell. 2007;18:4037–4049. doi: 10.1091/mbc.E07-05-0474. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.