The actin nucleator Cordon Bleu (Cobl) is localized to the basal region of microvilli of epithelial cells, where it regulates microvilli length through its WH2 domains. The COBL domain recruits several BAR-containing proteins, including PACSIN 2 and ASAP1, suggesting a role in coordinating microvillar structure with membrane traffic.
Abstract
Cordon Bleu (Cobl) is a WH2-containing protein believed to act as an actin nucleator. We show that it has a very specific localization in epithelial cells at the basal region of microvilli, a localization unlikely to be involved in actin nucleation. The protein is localized by a central region between the N-terminal COBL domain and the three C-terminal WH2 domains. Ectopic expression of Cobl shortens apical microvilli, and this requires functional WH2 domains. Proteomic studies reveal that the COBL domain binds several BAR-containing proteins, including SNX9, PACSIN 2/syndapin 2, and ASAP1. ASAP1 is recruited to the base of microvilli by binding the COBL domain through its SH3. We propose that Cobl is localized to the basal region of microvilli both to participate in length regulation and to recruit BAR proteins that associate with the curved membrane found at the microvillar base.
INTRODUCTION
Polarized epithelial cells have provided an important model for understanding how cell polarity is established and maintained. These cells have apical and basolateral domains that are biochemically, functionally, morphologically, and biophysically distinct. The different protein and lipid compositions of the apical and basolateral domains are necessary for epithelial cell barrier and transporting functions. The apical domain is studded with microvilli to provide a large surface for the uptake of nutrients. This morphological specialization occurs despite the higher mechanical tension across the apical domain, as evidenced by the need for actin polymerization during endocytosis (for a recent review see Mooren et al., 2012). We have been interested in how microvilli are specifically assembled on the apical domain and therefore undertaken biochemical and cell biological analyses of microvilli to address this question. Our recent work has focused on the microvilli found on epithelial cells derived from cultured placental syncytiotrophoblasts, as these cells have abundant and easily imaged apical microvilli.
Microvilli have a central core bundle of actin filaments. The bundle is tethered laterally to the plasma membrane through ezrin, a member of the ezrin/radixin/moesin (ERM) family (Fehon et al., 2010). Also essential for microvilli is the scaffolding protein EBP50 (ERM-binding phosphoprotein of 50 kDa), which links directly from active ezrin to membrane-associated proteins through its two PDZ domains (Reczek and Bretscher, 1998; Saotome et al., 2004; Hanono et al., 2006; Garbett et al., 2010; Viswanatha et al., 2012).Microvilli from cultured cells are dynamic structures with a lifetime on the order of 7–15 min (Gorelik et al., 2003; Garbett and Bretscher, 2012), an essential part of which is certainly the assembly and disassembly of the F-actin core.
Even though the F-actin core of microvilli was first characterized >40 years ago and the polarity of the actin filaments established shortly thereafter (Tilney and Mooseker, 1971), there have been surprisingly few developments in understanding how its assembly and disassembly is regulated. F-actin cross-linking, bundling, and capping proteins important for microvilli morphology have been described in detail (Heintzelman and Mooseker, 1990; Bretscher, 1991), but the presumed nucleator of actin assembly necessary for microvillar biogenesis has remained elusive. Given the structure of F-actin in microvilli, the most likely nucleator is expected to be a member of the formin family of proteins. However, efforts to implicate a formin family member in microvilli formation have so far not been successful (our unpublished data).
Two recent proteomic analyses of brush borders isolated from epithelial cells suggested the presence of the actin nucleator Cordon Bleu (Cobl; McConnell et al., 2011; Revenu et al., 2012). Cobl is part of an emerging class of actin assembly nucleators containing wasp homology 2 (WH2) domains, modules that frequently bind monomeric actin. Cobl, as well as with Spire, leiomodin, and junction-mediating regulatory protein (JMY), can drive actin assembly using their WH2 domains (Qualmann and Kessels, 2009). Multiple WH2 domains are believed to increase the local concentration of actin monomers such that unbranched filament elongation can occur. Recent work also suggests that WH2 domains can be powerful severing agents (Husson et al., 2011; Jiao et al., 2014).
Cobl was first described in a gene trap expression screen in embryonic stem cells, named for its expression pattern in the notochord during mouse embryonic development (Gasca et al., 1995). Vertebrates have a single gene encoding Cobl and a second encoding a Cobl-like protein that shares sequence homology in the N-terminus, a region now termed the COBL domain (Carroll et al., 2003). Cobl has been shown to play roles in dendritogenesis in cultured neurons (Ahuja et al., 2007; Schwintzer et al., 2011) and primary cilia formation in zebrafish (Ravanelli and Klingensmith, 2011; Schüler et al., 2013). Here, we show that Cobl has a highly specific localization in epithelial cells, where it is localized to the basal region of microvilli and recruits additional factors and can regulate their length.
RESULTS
Cordon Bleu localizes to the basal region of microvilli
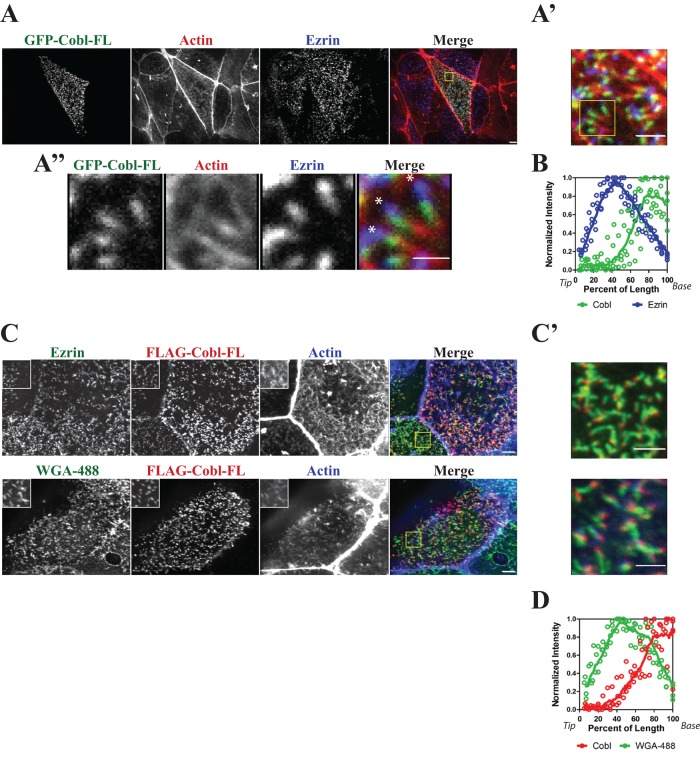
To determine the subcellular localization of Cobl in epithelial cells, we expressed a full-length version of green fluorescent protein (GFP)–tagged murine Cobl (GFP-Cobl-FL) in JEG-3 cells, a choriocarcinoma cell line with abundant microvilli. Maximum projection of confocal images through the cells showed a striking and highly specific punctate distribution on the apical surface. In cells stained for the microvillar protein ezrin, actin, and GFP-Cobl-FL, it was found to be specifically enriched at the basal region of microvilli (Figure 1, A and A’). In optimal images, an ezrin-rich region, GFP-Cobl-FL region, and actin-rich region could be discerned (Figure 1A”). Quantification of the normalized fluorescence intensity of GFP-Cobl-FL and endogenous ezrin along the length of the microvilli clearly revealed this relationship (Figure 1B).
FIGURE 1:
Cordon Bleu is localized to the basal region of microvilli. Maximum projections (XY) of confocal Z-stacks of JEG-3 cells. (A) Localization GFP-Cobl (green), F-actin (red), and endogenous ezrin (blue). Yellow box identifies area magnified in A’. (A”) Detailed image of area defined by yellow box in A’. Asterisks indicate the tips of microvilli. (B) Normalized intensity of ezrin (blue) and Cobl (green) of several microvilli (n > 5). Intensity was plotted (open circles) as a function of the percentage of the total microvillar length, with 0 and 100% representing tip and base, respectively. The data were fitted to a LOWESS function (solid line). (C) Localization of FLAG-Cobl, F-actin (blue), endogenous ezrin (green, top) or the membrane marker WGA-488 (green, bottom). Yellow box identifies magnified single-channel insets and is shown merged in C’. (D) Normalized intensity of WGA-488 (green) and Cobl (red) of several microvilli (n > 5). Intensity was plotted (open circles) as a function of the percentage of the total microvillar length, with 0 and 100% representing tip and base, respectively. The data were fitted to a LOWESS function (solid line). Scale bars, 5 μm (A, C), 2 μm (A’, C’), and 1 μm (A”).
The apparent localization of GFP-Cobl-FL at the basal region of microvilli could be complicated either by the GFP tag or possibly an effect on the distribution of ezrin. To address these issues, we expressed a FLAG-tagged construct (FLAG-Cobl-FL) and compared its localization with both ezrin and wheat germ agglutinin (WGA-488), a lectin that binds to cell surface proteins and serves as a plasma membrane marker. Once again, Cobl-FL was localized to the basal region of microvilli, as seen both by ezrin and plasma membrane staining (Figure 1, C and C’). FLAG-Cobl-FL localization at the base of microvilli shows little colocalization with WGA-488 (Figure 1D). This suggests that Cobl has a highly specific localization at the basal region of microvilli but not to actin filament minus ends that extend deeper into the cytoplasm.
Cobl is localized to microvilli by a region between the COBL and WH2 domains
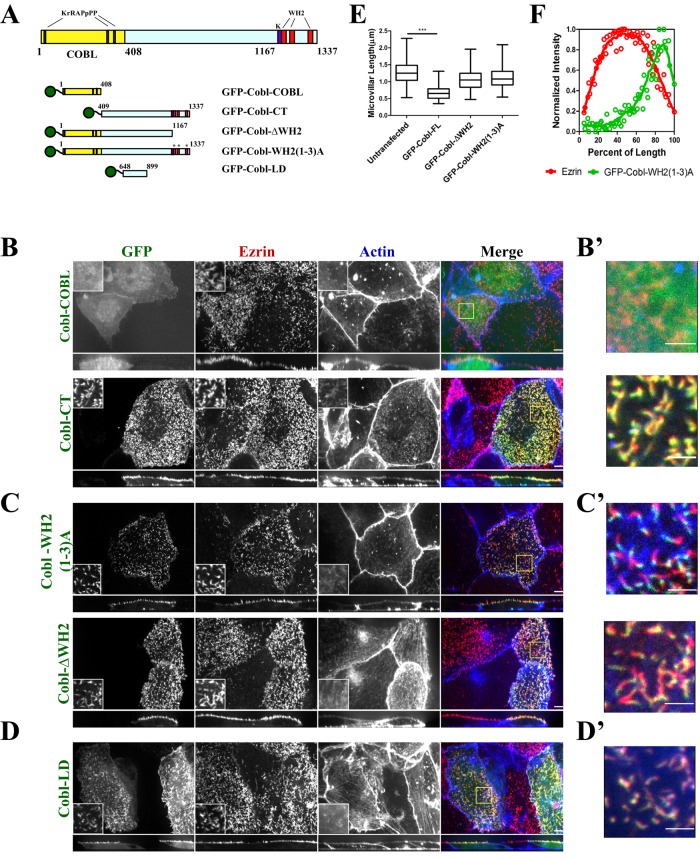
We next explored which region of Cobl is necessary for its localization to the base of microvilli. Cobl consists of an N-terminal COBL domain of 408 residues, with three WH2 domains in its C-terminal 170 residues (Figure 2A). We first made two complementary constructs, GFP-Cobl-COBL (amino acids [aa] 1–408) and GFP-Cobl-CT (aa 409–1337), each tagged with GFP on the N-terminal end (Figure 2B), to compare the localization of each in comparison with GFP-Cobl-FL (Figure 1A). Because the COBL domain has been shown to be essential for both protein–protein interactions and its localization to dendritic spines (Ahuja et al., 2007; Haag et al., 2012), we were surprised to find that GFP-Cobl-COBL was not enriched in microvilli but was cytoplasmic (Figure 2B, top). Conversely, expression of GFP-Cobl-CT without the COBL domain was still able to localize to microvilli (Figure 2B, bottom). Of interest, GFP-Cobl-CT is not as tightly restricted to the base as full-length Cobl. These data suggest that the COBL domain is not necessary for localization to microvilli.
FIGURE 2:
Cordon Bleu regulates microvilli length by its WH2 domains and is localized to microvilli independently of the COBL and WH2 domains. (A) Schematic of Cobl and the constructs used in this study. (B–D) Maximum projections (XY) of confocal Z-stacks and side views (XZ) stretched twofold of JEG-3 cells expressing the indicated constructs, then fixed and GFP (green), endogenous ezrin (red), and F-actin (blue) localized. Yellow boxes represent magnified single-channel insets and are shown merged in (B’–D’). Scale bars, 5 μm (B–D) and 2 μm (B’–D’). The contrast for F-actin was increased for clarity in C’, top. (E) Length of microvilli (micrometers) of untransfected JEG-3 cells or cells transfected to express GFP-Cobl-FL, GFP-Cobl-∆WH2, or GFP-Cobl-WH2(1–3)A. The box represents the 25th and 75th percentiles around the median, and the whiskers represent the maximum and minimum. ***p < 0.0001. (F) Normalized intensity of ezrin (red) and GPP-Cobl-WH2(1–3)A (green) of several microvilli (n > 5). Intensity was plotted (open circles) as a function of the percentage of the total microvillar length, with 0 and 100% representing tip and base, respectively. The data were fitted to a LOWESS function (solid line).
There are three WH2 domains in the C-terminal region of Cobl, each of which can bind an actin monomer (Ahuja et al., 2007). These WH2 domains are able to both nucleate the assembly of actin filaments and act as potent severing agents in vitro (Ahuja et al., 2007; Husson et al., 2011; Chen et al., 2013; Jiao et al., 2014). Adjacent to the first WH2 domain is a lysine-rich patch (Figure 2A) that is necessary for both nucleating and severing actin filaments (Husson et al., 2011; Jiao et al., 2014). To see whether functional WH2 domains were necessary for Cobl localization to the base of microvilli, we set out to generate mutants deficient in actin binding. Within each WH2 domain is an actin-binding consensus sequence, and mutations of these conserved residues can compromise this ability (Kelly et al., 2006; Zuchero et al., 2012). The WH2 domains of Cobl, Wiskott–Aldrich syndrome protein (WASP), suppressor of cyclical-AMP receptor (SCAR), and Spire, were aligned and conserved residues in the consensus sequence identified (Supplemental Figure S1B). We made point mutations in each individual WH2 domain (unpublished data), as well as in all three WH2 domains (GFP-Cobl-WH2(1–3)A). Several of these mutations have recently been used and verified to affect WH2 activity in Cobl (Chen et al., 2013). Expression of GFP-Cobl-WH2(1–3)A in JEG-3 cells still strongly localized to the base of microvilli (Figure 2C, top). Quantification of the normalized fluorescence intensity of GFP-Cobl-WH2(1–3)A and endogenous ezrin along the length of the microvilli clearly revealed this relationship (Figure 2F).
To further illustrate that the WH2 domains of Cobl are not essential for localization, we created a Cobl truncation mutant lacking the lysine-rich patch and all three WH2 domains (GFP-Cobl-∆WH2, aa 1–1167). GFP-Cobl-∆WH2 localized to the basal region of microvilli just like GFP-Cobl-WH2(1–3)A (Figure 2C, bottom). Further removal of the COBL domain from this construct (GFP-Cobl (409–1167)) still results in localization to microvilli (Supplemental Figure S1C), and expression of just the lysine-rich patch and the three WH2 domains (aa 1156–1338, Supplemental Figure S1D) is cytoplasmic. These data suggest that the WH2 domains of Cobl play no role in the localization to the basal region of microvilli.
With neither the COBL domain nor the WH2 domains playing an essential role in localization of Cobl to the basal region of microvilli, the large portion of the protein (residues 409–1167) between these, which is predicted to be unstructured, appears to harbor the localization domain. This region was further truncated into three separate 250-residue fragments, each of which was expressed individually in JEG-3 cells. One of these, comprising residues 648–899 localized to the basal region of microvilli, whereas the others did not (Figure 2D and Supplemental Figure S1E). To further demonstrate that this region is essential for the localization of Cobl, we made a construct in which this region was deleted in the context of the full-length protein (GFP-Cobl-∆LD, ∆648–899) and expressed it in JEG-3 cells. GFP-Cobl-∆LD is cytoplasmic, with no localization to microvilli (Supplemental Figure S1F). The 648–899 region of Cobl is therefore necessary and sufficient for the localization of Cobl to the basal region of microvilli, which is outside the COBL domain, a region believed to provide the protein's specific localization in other systems (Schwintzer et al., 2011; Haag et al., 2012).
The Cobl WH2 domains regulate the length of microvilli
Cells expressing GFP-Cobl-FL had visibly shorter microvilli. To quantify this effect, we measured the length of microvilli in three dimensions through confocal Z-stacks (Figure 2E). Using endogenous ezrin staining as our marker for microvillar length, we measured the length of microvilli in cells transfected with different Cobl constructs compared with adjacent untransfected cells in the same field. The median length of microvilli in untransfected cells was 1.3 μm, whereas in cells expressing GFP-Cobl-FL, microvilli were about half as long, with a median length of 0.6 μm (Figure 2E), using a bead standard to verify accuracy of the measurements (Supplemental Figure S2). To see whether this length regulation was mediated by the WH2 domains, we measured the lengths of microvilli in cells expressing GFP-Cobl-∆WH2 and GFP-Cobl-WH2(1–3)A. Expression of the constructs either lacking the WH2 domains or containing inactivated WH2 domains had no effect on microvillar length (Figure 2E). These data indicate that the WH2 domains of Cobl are necessary for the regulation of microvillar length. Because endogenous Cobl is present in Jeg-3 cells, we examined the effect of small interfering RNA (siRNA) knockdown or in Cas9/CRISPR genome-edited Cobl-knockout cells. Neither method of lowering Cobl expression affected microvilli length (Supplemental Figure S4)
The localization of Cobl to the basal region of microvilli is relatively stable
Components of microvilli can have very different dynamics in vivo, and these dynamics are regulated (Garbett and Bretscher, 2012). Even homologous proteins, such as the microvillar scaffolding proteins EBP50 and E3KARP (Na+/H+ exchanger type 3 kinase A regulatory protein), show vastly different dynamics even though they share similar localizations but presumably somewhat distinct functions (Garbett et al., 2013). We therefore examined the dynamics of Cobl to see whether it is a highly dynamic or relatively stable component at the basal region of microvilli.
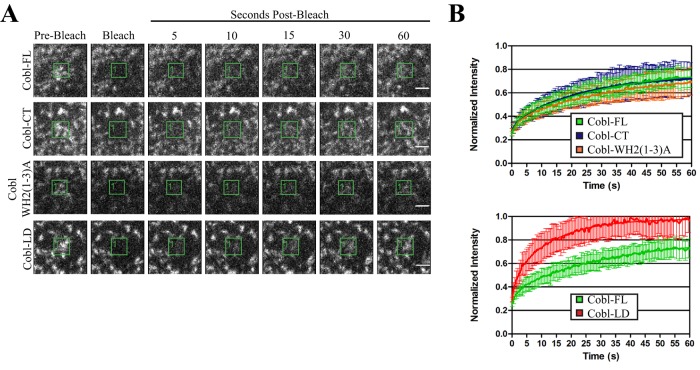
We explored the dynamics of Cobl using fluorescence recovery after photobleaching (FRAP) on JEG-3 cells expressing different GFP-Cobl constructs (Figure 3, A and B). Photobleaching of GFP-Cobl-FL exhibits relatively slow recovery and only recovers to ∼70% of the initial fluorescence intensity after photobleaching, just as for the relatively stable microvillar protein ezrin (Garbett and Bretscher, 2012). This suggests the Cobl undergoes a slow exchange rate between the cytosol and microvilli.
FIGURE 3:
Cordon Bleu is a stable component of microvilli. (A) Images of time points after photobleaching the boxed areas of cells expressing GFP-Cobl-FL, GFP-Cobl-CT, GFP-Cobl-WH2(1-3)A, and GFP-Cobl-LD. Scale bars, 2 μm. (B, top) Recovery curves after photobleaching the boxed areas of cells expressing GFP-Cobl FL (n = 12), GFP-Cobl-CT (n = 12), and GFP-Cobl-WH2(1–3)A (n = 12) and (B, bottom) GFP-Cobl-FL and GFP-Cobl-LD (n = 10). Recovery curves are normalized to the initial intensity in the boxed areas. Error bars, SD.
To determine whether the COBL domain or the WH2 domains contribute to this slow turnover, we used the constructs that lack the COBL domain (GFP-Cobl-CT) or contain inactive WH2 domains (GFP-Cobl-WH2(1–3)A). Expression of these constructs followed by photobleaching demonstrated that there is no significant difference in recovery curves compared with the full-length protein (Figure 3B, top). However, the dynamics of the localization domain alone, GFP-Cobl-LD, is much faster than that of the full-length protein (Figure 3B, bottom). Therefore regions outside of the localization domain and C-terminal to the COBL domain must also contribute to the stabilization of Cobl at the basal region of microvilli.
Identification of Cobl interaction partners
There are few known Cobl interaction partners, the best characterized of which are the PACSIN/syndapin family of proteins and the F-actin–binding protein Abp1 (Ahuja et al., 2007; Schwintzer et al., 2011; Haag et al., 2012). We sought to identify Cobl interaction partners to provide insight into the function of Cobl.
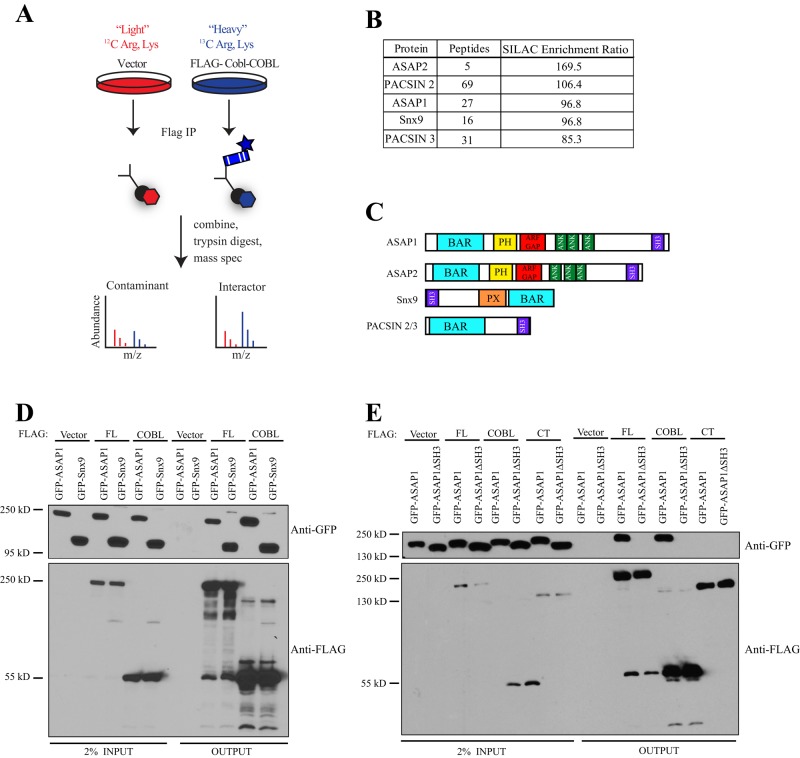
To identify potential interacting proteins, we used stable isotope labeling of amino acids in cell culture (SILAC) combined with quantitative mass spectrometry (Figure 4A). In outline, HEK293T cells stably expressing an empty vector control or 3x-FLAG-Cobl-COBL were used, as we have been unable to make a line stably expressing the full-length protein or the C-terminal region. FLAG immunoprecipitation was performed on both samples and the immunoprecipitates subsequently combined and trypsin digested. The digested samples were then subjected to quantitative mass spectrometry. Peptides identified as enriched in the control sample were considered background, and peptides enriched in the 3xFLAG-COBL sample were considered potential interactors of the COBL domain.
FIGURE 4:
Identification of Cordon Bleu interaction partners. (A) Schematic of SILAC experiment in HK293T cells. (B) Top hits showing total number of peptides identified for each candidate and the enrichment ratio of peptides in heavy:light. (C) Schematic showing domains of top SILAC hits. (D) 3x-FLAG-Cobl constructs were coexpressed in HEK293T cells with either GFP-ASAP1 or GFP-Snx9 and subjected to FLAG immunoprecipitation (IP) and then subsequently blotted for GFP and FLAG. (E) 3x-FLAG-Cobl constructs were coexpressed with either GFP-ASAP1 or GFP-ASAP1-∆SH3 and subjected to FLAG IP and subsequently blotted for GFP and FLAG.
The top candidates identified by our analysis are shown in Figure 4B, where the SILAC enrichment ratio is the quantitative measure of enrichment of peptides between heavy and light. Two are members of the PACSIN/syndapin family, and their interaction with Cobl has been described in detail (Ahuja et al., 2007; Haag et al., 2012). We also identified three novel Cobl-binding partners, including ArfGAP with Src homology 3 (SH3) domain, ankyrin repeat, and PH domain 1 (ASAP1), ArfGAP with SH3 domain, ankyrin repeat, and PH domain 2 (ASAP2), and sorting nexin 9 (SNX9). Of interest, all of our top candidates are Bin-amphiphysin-Rvs167 (BAR) proteins that also contain SH3 domains (Figure 4C).
To validate the results of the SILAC analysis, we coexpressed different FLAG-Cobl variants and GFP-ASAP1 or GFP-Snx9 in HEK293T cells and then immunoprecipitated the FLAG epitope and blotted for the GFP-tagged construct. Coimmunoprecipitations confirmed that both the full-length version of Cobl and the COBL domain alone were able to interact with ASAP1 and Snx9 (Figure 4D). To determine whether either of these proteins was relevant in the context of microvilli, we localized GFP-tagged versions of both ASAP1 and Snx9 in JEG-3 cells. GFP-ASAP1 was localized weakly to the base of microvilli (Figure 5A), whereas GFP-Snx9 was cytoplasmic (unpublished data).
FIGURE 5:
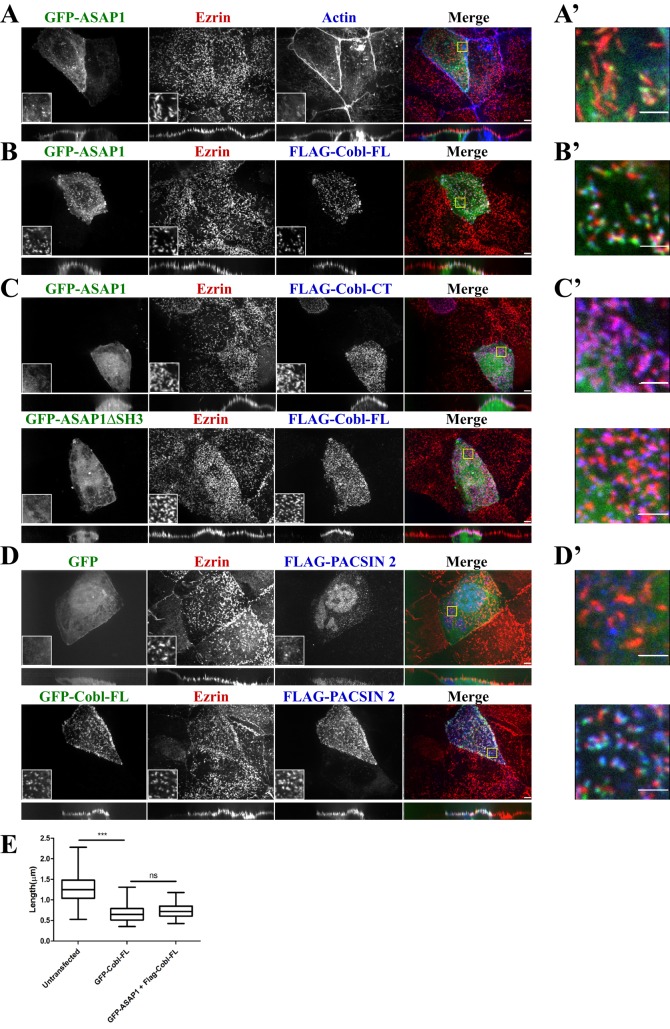
Cordon Bleu recruits ASAP1 and PACSIN 2 to microvilli. (A–D) Maximum projections (XY) of confocal Z-stacks and side views (XZ) stretched twofold of JEG-3 cells. Cells expressing the indicated constructs were fixed and stained for endogenous ezrin (red) and FLAG or F-actin (blue) as indicated. Yellow boxes identify magnified single-channel insets and are merged in A’–D’. Scale bars, 5 μm (A–D) and 2 μm (A’–D’). (E) Length of microvilli of untransfected JEG-3 cells or cells transfected to express GFP-Cobl-FL, GFP-ASAP1, or FLAG-Cobl-FL. The box represents the 25th and 75th percentiles around the median, and the whiskers represent the maximum and minimum. ***p < 0.0001.
We next sought to identify which region of ASAP1 interacts with the COBL domain. The interaction between Cobl and PACSIN1/2/3 is known to occur between the COBL domain and the SH3 domain of the PACSIN family (Schwintzer et al., 2011). Because ASAP1 harbors an SH3 domain in its C-terminal region, it is likely to mediate the interaction with Cobl. To test this, we constructed an expression construct of ASAP1 lacking the SH3 domain, GFP-ASAP1∆SH3, and coexpressed it with FLAG-tagged full-length Cobl, its COBL domain, or its C-terminal region. FLAG immunoprecipitates showed that the SH3 domain of ASAP1 is necessary for the recovery of GFP-ASAP1 by either 3xFLAG-Cobl-FL or 3xFLAG-Cobl-COBL (Figure 4E). Furthermore, the FLAG-Cobl-CT was not able to recover either GFP-ASAP1 or GFP-ASAP1∆SH3 from cell lysates (Figure 4E). These data document that the interaction between Cobl and ASAP1 occurs between the COBL domain of Cobl and the SH3 domain of ASAP1.
Cobl serves as a platform at the base of microvilli
GFP-ASAP1 is weakly localized to microvilli when expressed alone in JEG-3 cells (Figure 5, A and A’). However, upon coexpression with FLAG-Cobl-FL, GFP-ASAP1 is strongly enriched in microvilli (Figure 5, B and B’). This enrichment is dependent on the interaction between the SH3 domain of ASAP1 and the COBL domain, as GFP-ASAP1∆SH3, which is normally cytoplasmic (Supplemental Figure S3A), is not recruited with FLAG-Cobl-FL (Figure 5C, bottom). GFP-ASAP1 is also not recruited to microvilli with FLAG-Cobl-CT (Figure 5C, top) or FLAG-Cobl-COBL (Supplemental Figure S3B). This implies that Cobl recruits ASAP1 to the basal region of microvilli. Recruitment of ASAP1 does not further affect the length of microvilli, as the median length of microvilli is the same as the length of microvilli of cells expressing Cobl alone (Figure 5E).
Of interest, a similar effect is seen when FLAG-PACSIN 2 is expressed. On expression of FLAG-PACSIN2 alone, there is some enrichment to the apical domain but not specifically in microvilli (Figure 5, D and D’, top). Coexpression with GFP-Cobl-FL reveals significant enrichment of FLAG-PACSIN 2 in microvilli (Figure 5, D and D’, bottom), an effect not seen with coexpression of GFP-Cobl-LD (Supplemental Figure S3C). This suggests that Cobl influences multiple protein localization at the basal region of microvilli.
DISCUSSION
Many studies have been dedicated to microvilli dynamics (Garbett and Bretscher, 2012; Garbett et al., 2013; Yang et al., 2013) and regulation (Hanono et al., 2006; Zwaenepoel et al., 2012), most of which focus on proteins that localize and function along the entire length of microvilli. There are few examples of proteins that localize more locally to the basal region (Hanono et al., 2006; Hokanson and Bretscher, 2012; Garbett et al., 2013), and even less is understood about the functional consequences of this specific localization. In this study we demonstrated that the actin nucleator Cordon Bleu localizes to the basal region of microvilli but not to filament ends that extend deeper into the cytoplasm (Figure 1A”). We quantitatively demonstrate that Cobl is localized to the basal region of microvilli by comparing the normalized fluorescence intensity of endogenous ezrin to expressed GFP-Cobl-FL along the length of a microvillus. The peak fluorescence intensity for GFP-Cobl-FL is shifted more toward the basal region than for ezrin (Figure 1B). This shift in peak fluorescence intensity is more dramatically seen in cells expressing GFP-Cobl-WH2(1–3)A, as this construct does not shorten the overall length of microvilli but still has the ability to localize (Figure 2F). This localization brings into question whether Cobl acts as an actin nucleator in vivo, where one might expect the protein to be localized to the filament minus ends.
We also identified a novel localization domain (aa 648–899) outside of the canonical COBL domain (Figure 2D), which until this point has been believed to be essential for the specific localization of Cobl in other systems (Schwintzer et al., 2011; Haag et al., 2012). We attempted to determine how this region is so specifically localized by searching for interacting proteins, but so far without success. Another possibility is that it might have an affinity for lipids in a highly curved region, as is found at the base of microvilli.
In this study we also demonstrated that enhancing the concentration of Cobl is able to regulate the length of microvilli and that the WH2 domains are necessary for this function (Figure 2E). Because WH2 domains bind actin monomers, they are likely to elevate the local monomeric actin concentration, but it is difficult to envision a simple mechanism that would shorten microvilli. A more likely scenario is that the C-terminal region acts to locally sever actin filaments, as seen in vitro (Jiao et al., 2014), potentially allowing faster filament turnover, rather than plays a role as an actin filament nucleator. Loss of Cobl has also been shown to have a similar effect on the length of primary cilia in zebrafish (Ravanelli and Klingensmith, 2011), although primary cilia are composed of microtubules, not actin filaments, and so this is most likely an indirect consequence of loss of Cobl function.
Surprisingly, knocking down Cobl, either through siRNA transfection or with Cas9/CRISPR genome-edited cells, does not have a significant effect on the length of microvilli, at least not as measured by the techniques in this study. Our microvillar measurements were taken from confocal images with a 0.28-μm Z-step size, which means that the difference in length between knockdown and control cells would need to be greater than this length before being considered statistically different. We do not see a difference this large (Supplemental Figure S4). Thus reducing Cobl levels may in fact increase the length of microvilli, but we are unable to resolve such a difference at this time. There are other instances—namely, in cells from plastin-1– or desmoplakin–knockout mice, in which the change in microvilli length was <0.2 μm (Grimm-Günter et al., 2009; Sumigray and Lechler, 2012). More work will need to be done to determine whether this is the case in Cobl-knockdown cells. These results place Cobl on a growing list of microvilli length regulators, including desmoplakin, plastin-1, Drosophila Cad99c, and Eps8 (D'Alterio et al., 2005; Grimm-Günter et al., 2009; Sumigray and Lechler, 2012; Zwaenepoel et al., 2012), although in no case is the mechanism of regulation known.
Using FRAP, we demonstrated that Cobl is stably localized to the base of microvilli. Remarkably, we showed that the dynamics is not determined by either the COBL domain or the WH2 domains, as the dynamics of Cobl-CT and Cobl-WH2(1–3)A are not significantly different from that of full-length Cobl (Figure 3B). This is surprising, as we had suspected that a possible reason for the slow turnover of Cobl in microvilli might be protein–protein interactions mediated through the COBL domain. This suggests that there are regions outside of COBL and the localization domain that contribute to the stability of Cobl at the basal region of microvilli. Exploring which regions contribute to the dynamics of Cobl and how this influences function will be an interesting topic for future study.
Using SILAC, we identified two novel Cobl interacting partners, ASAP1 and SNX9 (Figure 4B). We further illustrated that the interaction between Cobl and ASAP1 occurs between the SH3 domain of ASAP1 and the COBL domain of Cobl (Figure 4E). Of interest, this interaction was also necessary for localization of ASAP1 to microvilli (Figure 5, A and B). We also observed a similar effect for the already established Cobl interaction partner PACSIN 2 (Figure 5D). These data are not in accord with the current model of Cobl recruitment. It has been suggested that in neurons the COBL domain is necessary for the specific localization of Cobl (Schwintzer et al., 2011; Haag et al., 2012) and that this localization is dependent on an interaction with a binding partner (Schwintzer et al., 2011; Haag et al., 2012). In the case of epithelial cells, the COBL domain is not necessary or sufficient for localization to the basal region of microvilli, and localization can occur independent of its ability to interact with its binding partners.
What might be the role of Cobl at the base of microvilli? It is difficult to envision how it could drive actin assembly from that location. Our finding that it serves as a platform for BAR-containing proteins suggests that it might be part of a specific structure associated with membrane curvature. The apical membrane of epithelial cells is known to be under tension, so one possibility is that it might associate with the curved membrane and transmit the tension across the base of the microvilli. Alternatively, it might represent nascent sites of endocytosis, as suggested by the Cobl-binding proteins. The strong enrichment of Cobl at the basal region of microvilli newly focuses attention on this region of the apical membrane.
MATERIALS AND METHODS
Antibodies and reagents
The mouse FLAG antibody and resin were from Sigma-Aldrich (St. Louis, MO), mouse GFP antibody was from Santa Cruz Biotechnology (Santa Cruz, CA), and the mouse E-cadherin antibody was from BD Biosciences (Mississauga, Canada). The rabbit antisera and affinity-purified antibodies against full-length human ezrin have been described (Bretscher, 1989). The anti-Cobl antibody was a kind gift from J. Klingensmith (Duke University, Durham, NC). Goat anti-mouse secondary antibodies conjugated to horseradish peroxidase were from Jackson ImmunoResearch Laboratories (West Grove, PA). Alexa Fluor 568– and Alexa Flour 660–conjugated donkey anti-rabbit antibodies and Alexa Fluor 660–conjugated phalloidin were from Invitrogen (Carlsbad, CA). IRDye 680– and 800–conjugated secondary antibodies were from LI-COR Biosciences (Lincoln, NE). WGA-488 was from Molecular Probes (Lafayette, CO).
DNA constructs and sequence alignments
Murine GFP-Cobl-FL was a kind gift from M. Kessels (Research Institute of the FSU Jena, Jena, Germany). Cobl truncations were cloned into pEGFP-C2 (Takara Bio, Shiga, Japan) as GFP-Cobl-COBL (1–408), GFP-Cobl-CT (409–1337), GFP-Cobl-LD (648–899), GFP Cobl (409–1167), GFP-Cobl-∆WH2 (1–1167), GFP-Cobl (409–658), and GFP-Cobl (895–1167). GFP-Cobl-FL, GFP-Cobl-COBL, and GFP-Cobl-CT were cloned into a modified PQCXIP (BD Biosciences) backbone that has an N-terminal 3xFLAG to create FLAG constructs. To generate GFP-Cobl-WH2(1–3)A, the WH2 domains of mouse Cobl were aligned with the WH2 domains of Drosophila Spire, human SCAR, human WASP, and human nWASP using ClustalW. The residues used in Kelly et al. (2006) to inactivate the WH2 domains were used as a reference to identify conserved lysines in Cobl. The following mutations were then introduced by site-directed mutagenesis of GFP-Cobl-FL using the QuikChange II XL kit (Agilent, Santa Clara, CA) to generate the GFP-Cobl-WH2(1–3)A mutant: L1189A, L1202A, L1229A, L1242A, L1317A, and L1330A. The internal deletion of the Cobl localization domain (∆648–899) was generated by two steps of overlapping PCR and inserted into pEGFP-C2. Murine GFP-ASAP1 in pEGFP-C1 was a kind gift from P. Randazzo (National Cancer Institute, Bethesda, MD). GFP-ASAP1∆SH3 (aa 1–1032) was cloned from GFP-ASAP1 and placed into pEGFP-C2. GFP-Snx9 and GFP-PACSIN2 were cloned into pEGFPC-2 from human ppSumo-Snx9 and human ppSumo-PACSIN2, which were a kind gift from H. Sondermann (Cornell University, Ithaca, NY). The siRNA targeting human Cordon Bleu (5′-AGCACGGCCUCACAACGUAtt-3′) was obtained from Ambion by Life Technologies (Grand Island, NY), and luciferase GL2 (5′-CGUACGCGGAAUACUUCGA-3′) was obtained from Thermo Fisher Scientific (Lafayette, CO) and Applied Biosystems (Grand Island, NY). The lentiCRISPR plasmid was obtained from Addgene (Cambridge, MA; plasmid 49535)
Cell culture and transfection
JEG-3 and HEK293T cells (American Type Culture Collection, Manassas, VA) were maintained in a 5% CO2 humidified atmosphere at 37°C. JEG-3 cells were cultured in MEM (Thermo Fisher Scientific) with 10% fetal bovine serum (FBS), and HEK293T in DMEM with 5% FBS. JEG-3 cells were transfected with polyethylenimine (Polysciences, Warrington, PA) and 1–2 μg of plasmid DNA, as described (Hanono et al., 2006). HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. For generation of stable HEK293T cell lines expressing 3xFLAG-COBL or 3xFLAG-empty vector, Phoenix-AMPHO cells were cotransfected with the foregoing constructs in pQCXIP in addition to a plasmid encoding vesicular stomatitis virus G using polyethylenimine. The infected HEK293T cells were then selected and maintained with 2 μg/ml puromycin (Sigma-Aldrich). For knocking down endogenous Cobl in Jeg-3 cells, cells were transfected with 10 nM siRNA using Lipofectamine RNAiMax, allowed to grow for 72 h, and then processed for immunofluorescence and microvilli measurements or Western blot analysis.
CRISPR genome-edited cell line generation
Creating stable genome-edited CRISPR Cobl cells, in which the endogenous Cobl gene was targeted for knockout, was performed as described previously (Shalem et al., 2014). The target guide sequence against the second exon of Cobl was placed into the LentiCRISPR backbone using the oligos 5′-CACCGCCAAGTTCTGCTGCGACCCG-3′ and 5′-AAACCGGGTCGCAGCAGAACTTGGC-3′. Jeg-3 cells expressing the CRISPR system were than selected for using 2 μg/ml puromycin in complete MEM for 15 d. Knockout was determined by Western blot analysis.
Immunoprecipitations and Western blotting
HEK293T cells transiently coexpressing 3xFLAG-Cobl and GFP-ASAP1 constructs for 24 h were lysed in lysis buffer (25 mM Tris, pH 7.4, 5% glycerol, 150 mM NaCl, 50 mM NaF, 0.1 mM Na3VO4, 10 mM β-glycerol phosphate, 8.7 mg/ml paranitrophenylphosphate, 0.3% Triton X-100, and protease inhibitor tablet [Roche, Indianapolis, IN]) and immunoprecipitated with M2 FLAG resin (Sigma-Aldrich) for 2 h. Immunoprecipitates were than washed four times in wash buffer (lysis buffer but with 0.2% Triton X-100 and no protease inhibitor tablet) and eluted from the FLAG resin with 200 μg/ml 3xFLAG peptide for 15 min at room temperature. Eluates were than denatured with Laemmli buffer, resolved by SDS-PAGE, transferred to Immobilon-FL (EMD Millipore, Billerica, MA), blotted with specific antibody, and visualized by enhanced chemiluminescence.
To determine percentage of Cobl knockdown, Western blots were probed with specific antibody and IRDye 680– or 800–conjugated secondary antibodies. The blot was than imaged using an Odyssey infrared imaging system (LI-COR Biosciences). Analysis to determine percentage Cobl knockdown was performed using Image Studio Lite 4.0 (LI-COR Biosciences). Samples were normalized to the E-cadherin loading control, and knockdown was determined as a percent of the control.
SILAC and mass spectrometry
For SILAC, HEK293T cells stably expressing either 3xFLAG-Empty Vector or 3xFLAG-Cobl-COBL were grown in MEM with 10% dialyzed FBS, 2 μg/ml puromycin (Invitrogen) and either [12C]arginine and lysine or [13C]arginine and lysine, respectively, for 3 wk to allow uniform labeling of all proteins. FLAG immunoprecipitations were performed as described, with modifications for mass spectrometry (Smolka et al., 2007; Viswanatha et al., 2012). Briefly, after immunoprecipitation, protein bound to FLAG resin was eluted in 50 mM Tris (pH 8.0) and 1% SDS and then precipitated with 50% ethanol, 49.9% acetone, and 0.1% acetic acid. Protein samples were then mixed, trypsin digested (Promega, Madison, WI) overnight at 37°C, and desalted in a C18 column (Waters, Milford, MA). The tryptic peptides were dehydrated in a speed vacuum and dissolved in 80% acetonitrile and 1% formic acid for fractionation by hydrophilic interaction chromatography. The resulting fractions were dried, dissolved in 0.1% trifluoroacetic acid, and injected into a mass spectrometer (Qexactive LC-MS/MS; Thermo Fisher Scientific). The data were analyzed using Proteome Discoverer (Thermo Fisher Scientific).
Immunofluorescence
JEG-3 cells were grown on coverslips and fixed in 3.7% formaldehyde at room temperature for 15 min. Coverslips were then washed three times in phosphate-buffered saline (PBS) and permeabilized with 0.2% Triton X-100 for 5 min at room temperature. Coverslips were than washed three more times with PBS and blocked in filtered 3% FBS in PBS for 10 min. Primary and secondary antibodies were made up in 3% FBS in PBS. Coverslips were washed three times in PBS between addition of primary and secondary antibodies. Alexa Fluor–conjugated phalloidin (Invitrogen) was added to the secondary when F-actin was visualized. Cells were imaged by time-lapse microscopy on a spinning disk (CSU-X; Yokogawa, Tokyo, Japan) with a spherical aberration correction device, a 100×/1.46 numerical aperture (NA) objective (Leica, Wetzlar, Germany) on an inverted microscope (DMI6000B; Leica), and an HQ2 CCD camera (Photometrics, Tucson, AZ). Maximum intensity projections were created using SlideBook (Intelligent Imaging Innovations, Denver, CO) and exported in Illustrator (Adobe, San Jose, CA).
Measurements of protein localization as a function of the percentage length of microvilli were performed in SlideBook. A line was drawn along the entire length of microvilli, and fluorescence intensity values were exported to Excel (Microsoft, Redmond, WA) for multiple microvilli. Data were normalized by plotting fluorescence intensity as a function of the percentage of the total length of microvilli, where 0 and 100% represents the tip and base, respectively. The resulting curve was fitted with a LOWESS function in Prism (GraphPad, La Jolla, CA).
Microvillus length measurements
The length of individual microvilli was measured using Volocity 3D image analysis software (PerkinElmer, Waltham, MA) to draw lines along the entire length of microvilli in three dimensions through several confocal Z-stacks. Confocal images were taken with 0.28-μm steps, and endogenous ezrin staining was used as a microvilli marker in all measurements. Lengths were exported to Prism and plotted as a whisker plot. Mann–Whitney tests were performed between adjacent untransfected and transfected cells to determine whether change in length was statistically significant. A bead calibration assay was performed to determine three-dimensional accuracy in which beads of known size were measured in the manner described, and the mean bead size was plotted against the actual size of the bead. The plot was fitted with a line of best fit, with the error bars representing the SD.
Live-cell imaging and FRAP
Transfected JEG-3 cells were grown in 35-mm glass-bottom dishes (MatTek, Ashland, MA), washed in PBS, and then maintained in low sodium bicarbonate phenol red–free MEM (Sigma-Aldrich) supplemented with 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.4) with 10% FBS and GlutaMAX (Invitrogen). Live cells were imaged by time-lapse microscopy on the spinning disk microscope at 37°C in an environmental chamber (Okolab, Ottaviano, Italy) controlled by SlideBook, version 5.5 (Intelligent Imaging Innovations). Regions selected for FRAP were illuminated either with a digital mirror illumination system (Mosaic; Andor Technology, Belfast, Northern Ireland) coupled to a 488-nm, 400-W argon laser or with a point-scanner galvanometer-based system (Vector; Intelligent Imaging Innovations) coupled to a 473-nm, 50-mW diode-pumped solid-state laser. A region independent of the region being bleached was monitored for the duration of each experiment to control for photobleaching. Movies were processed using SlideBook and analyzed using Excel and Prism. Normalized data from multiple biological replicates were fitted to a curve, and Prism was used to perform two-way analysis of variance.
Supplementary Material
Acknowledgments
We are very grateful to M. Kessels, P. Randazzo, and H. Sondermann for providing constructs and to J. Klingensmith for providing the anti-Cobl antibody. This work was supported by National Institutes of Health Grant GM39066.
Abbreviations used:
- Abp1
actin-binding protein 1
- ANK
ankyrin repeat
- ASAP1
Arf-GAP with SH3 domain, ANK repeat, and PH domain–containing protein 1
- ASAP2
Arf-GAP with SH3 domain, ANK repeat, and PH domain–containing protein 2
- BAR
Bin-amphiphysin-Rvs
- Cobl
Cordon Bleu
- E3KARP
NHE3 kinase A regulatory protein
- EBP50
ERM-binding protein of 50 kDa
- ERM
ezrin/radixin/moesin
- PACSIN
protein kinase C and casein kinase substrate in neurons protein
- PDZ
postsynaptic density 95/disks large/zona occludens-1
- PH
pleckstrin homology
- PX
Phox
- SCAR
suppressor of cyclical-AMP receptor
- SH3
Src homology 3
- SILAC
stable isotope labeling of amino acids in cell culture
- Snx9
sorting nexin 9
- WASP
Wiskott-Aldrich syndrome protein
- WH2
wasp homology 2 domain
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-06-1131) on July 16, 2014.
J.W. designed, performed, and analyzed all experiments. A.P.B. conceived, designed. and supervised the study. Both authors wrote the manuscript.
The authors declare that they have no conflict of interest.
REFERENCES
- Ahuja R, Pinyol R, Reichenbach N, Custer L, Klingensmith J, Kessels MM, Qualmann B. Cordon-bleu is an actin nucleation factor and controls neuronal morphology. Cell. 2007;131:337–350. doi: 10.1016/j.cell.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Rapid phosphorylation and reorganization of ezrin and spectrin accompany morphological changes induced in A-431 cells by epidermal growth factor. J Cell Biol. 1989;108:921–930. doi: 10.1083/jcb.108.3.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bretscher A. Microfilament structure and function in the cortical cytoskeleton. Annu Rev Cell Biol. 1991;7:337–374. doi: 10.1146/annurev.cb.07.110191.002005. [DOI] [PubMed] [Google Scholar]
- Carroll EA, Gerrelli D, Gasca S, Berg E, Beier DR, Copp AJ, Klingensmith J. Cordon-bleu is a conserved gene involved in neural tube formation. Dev Biol. 2003;262:16–31. doi: 10.1016/s0012-1606(03)00323-3. [DOI] [PubMed] [Google Scholar]
- Chen X, Ni F, Tian X, Kondrashkina E, Wang Q, Ma J. Structural basis of actin filament nucleation by tandem W domains. Cell Rep. 2013;3:1910–1920. doi: 10.1016/j.celrep.2013.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alterio C, Tran D, Yeung M, Hwang M, Li M, Arana C, Mulligan V, Kubesh M, Sharma P, Chase M, et al. Drosophila melanogaster Cad99C, the orthologue of human Usher cadherin PCDH15, regulates the length of microvilli. J Cell Biol. 2005;171:549–558. doi: 10.1083/jcb.200507072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehon RG, McClatchey AI, Bretscher A. Organizing the cell cortex: the role of ERM proteins. Nat Rev Mol Cell Biol. 2010;11:276–287. doi: 10.1038/nrm2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Bretscher A. PDZ interactions regulate rapid turnover of the scaffolding protein EBP50 in microvilli. J Cell Biol. 2012;198:195–203. doi: 10.1083/jcb.201204008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, LaLonde DP, Bretscher A. The scaffolding protein EBP50 regulates microvillar assembly in a phosphorylation-dependent manner. J Cell Biol. 2010;191:397–413. doi: 10.1083/jcb.201004115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garbett D, Sauvanet C, Viswanatha R, Bretscher A. The tails of apical scaffolding proteins EBP50 and E3KARP regulate their localization and dynamics. Mol Biol Cell. 2013;24:3381–3392. doi: 10.1091/mbc.E13-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasca S, Hill DP, Klingensmith J, Rossant J. Characterization of a gene trap insertion into a novel gene, cordon-bleu, expressed in axial structures of the gastrulating mouse embryo. Dev Genet. 1995;17:141–154. doi: 10.1002/dvg.1020170206. [DOI] [PubMed] [Google Scholar]
- Gorelik J, Shevchuk A, Frolenkov G, Diakonov I, Lab M, Kros C, Richardson G, Vodyanoy I, Edwards C, Klenerman D, et al. Dynamic assembly of surface structures in living cells. Proc Natl Acad Sci USA. 2003;100:5819–5822. doi: 10.1073/pnas.1030502100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm-Günter E-MS, Revenu C, Ramos S, Hurbain I, Smyth N, Ferrary E, Louvard D, Robine S, Rivero F. Plastin 1 binds to keratin and is required for terminal web assembly in the intestinal epithelium. Mol Biol Cell. 2009;20:2549–2562. doi: 10.1091/mbc.E08-10-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag N, Schwintzer L, Ahuja R, Koch N, Grimm J, Heuer H, Qualmann B, Kessels MM. The actin nucleator Cobl is crucial for Purkinje cell development and works in close conjunction with the F-actin binding protein Abp1. J Neurosci. 2012;32:17842–17856. doi: 10.1523/JNEUROSCI.0843-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanono A, Garbett D, Reczek D, Chambers DN, Bretscher A. EPI64 regulates microvillar subdomains and structure. J Cell Biol. 2006;175:803–813. doi: 10.1083/jcb.200604046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzelman MB, Mooseker MS. Assembly of the brush border cytoskeleton: changes in the distribution of microvillar core proteins during enterocyte differentiation in adult chicken intestine. Cell Motil. Cytoskeleton. 1990;15:12–22. doi: 10.1002/cm.970150104. [DOI] [PubMed] [Google Scholar]
- Hokanson DE, Bretscher AP. EPI64 interacts with Slp1/JFC1 to coordinate Rab8a and Arf6 membrane trafficking. Mol Biol Cell. 2012;23:701–715. doi: 10.1091/mbc.E11-06-0521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husson C, Renault L, Didry D, Pantaloni D, Carlier M-F. Cordon-Bleu uses WH2 domains as multifunctional dynamizers of actin filament assembly. Mol Cell. 2011;43:464–477. doi: 10.1016/j.molcel.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Walker M, Trinick J, Pernier J, Montaville P, Carlier M-F. Mutagenetic and electron microscopy analysis of actin filament severing by Cordon-Bleu, a WH2 domain protein. Cytoskeleton (Hoboken) 2014;71:170–183. doi: 10.1002/cm.21161. [DOI] [PubMed] [Google Scholar]
- Kelly AE, Kranitz H, Dötsch V, Mullins RD. Actin binding to the central domain of WASP/Scar proteins plays a critical role in the activation of the Arp2/3 complex. J Biol Chem. 2006;281:10589–10597. doi: 10.1074/jbc.M507470200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConnell RE, Benesh AE, Mao S, Tabb DL, Tyska MJ. Proteomic analysis of the enterocyte brush border. Am J Physiol Gastrointest Liver Physiol. 2011;300:G914–G926. doi: 10.1152/ajpgi.00005.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Galletta BJ, Cooper JA. Roles for actin assembly in endocytosis. Annu Rev Biochem. 2012;81:661–686. doi: 10.1146/annurev-biochem-060910-094416. [DOI] [PubMed] [Google Scholar]
- Qualmann B, Kessels MM. New players in actin polymerization–WH2-domain-containing actin nucleators. Trends Cell Biol. 2009;19:276–285. doi: 10.1016/j.tcb.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Ravanelli AM, Klingensmith J. The actin nucleator Cordon-bleu is required for development of motile cilia in zebrafish. Dev Biol. 2011;350:101–111. doi: 10.1016/j.ydbio.2010.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczek D, Bretscher A. The carboxyl-terminal region of EBP50 binds to a site in the amino-terminal domain of ezrin that is masked in the dormant molecule. J Biol Chem. 1998;273:18452–18458. doi: 10.1074/jbc.273.29.18452. [DOI] [PubMed] [Google Scholar]
- Revenu C, Ubelmann F, Hurbain I, El-Marjou F, Dingli F, Loew D, Delacour D, Gilet J, Brot-Laroche E, Rivero F, et al. A new role for the architecture of microvillar actin bundles in apical retention of membrane proteins. Mol Biol Cell. 2012;23:324–336. doi: 10.1091/mbc.E11-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saotome I, Curto M, McClatchey AI. Ezrin is essential for epithelial organization and villus morphogenesis in the developing intestine. Dev Cell. 2004;6:855–864. doi: 10.1016/j.devcel.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Schüler S, Hauptmann J, Perner B, Kessels MM, Englert C, Qualmann B. Ciliated sensory hair cell formation and function require the F-BAR protein syndapin I and the WH2 domain-based actin nucleator Cobl. J Cell Sci. 2013;126:196–208. doi: 10.1242/jcs.111674. [DOI] [PubMed] [Google Scholar]
- Schwintzer L, Koch N, Ahuja R, Grimm J, Kessels MM, Qualmann B. The functions of the actin nucleator Cobl in cellular morphogenesis critically depend on syndapin I. EMBO J. 2011;30:3147–3159. doi: 10.1038/emboj.2011.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalem O, Sanjana N, Hartenian E, Shi X, Scott D, Mikkelsen T, Heckl D, Ebert R, Root D, Doench J, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen S, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumigray KD, Lechler T. Desmoplakin controls microvilli length but not cell adhesion or keratin organization in the intestinal epithelium. Mol Biol Cell. 2012;23:792–799. doi: 10.1091/mbc.E11-11-0923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Mooseker M. Actin in the brush-border of epithelial cells of the chicken intestine. Proc Natl Acad Sci USA. 1971;68:2611–2615. doi: 10.1073/pnas.68.10.2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanatha R, Ohouo PY, Smolka MB, Bretscher A. Local phosphocycling mediated by LOK/SLK restricts ezrin function to the apical aspect of epithelial cells. J Cell Biol. 2012;199:969–984. doi: 10.1083/jcb.201207047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Singh V, Cha B, Chen T, Sarker R, Murtazina R, Jin S, Zachos N, Patterson G, Tse CM, et al. NHERF2 protein mobility rate is determined by a unique C-terminal domain that is also necessary for its regulation of NHE3 protein in OK cells. J Biol Chem. 2013;288:16960–16974. doi: 10.1074/jbc.M113.470799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuchero JB, Belin B, Mullins RD. Actin binding to WH2 domains regulates nuclear import of the multifunctional actin regulator JMY. Mol Biol Cell. 2012;23:853–863. doi: 10.1091/mbc.E11-12-0992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwaenepoel I, Naba A, Da Cunha MML, Del Maestro L, Formstecher E, Louvard D, Arpin M. Ezrin regulates microvillus morphogenesis by promoting distinct activities of Eps8 proteins. Mol Biol Cell. 2012;23:1080–1094. doi: 10.1091/mbc.E11-07-0588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.