The subcellular distribution of yeast β-importins inverts upon acute glucose deprivation, likely due to collapse of the RanGTP nuclear–cytoplasmic gradient. This redistribution of β-importins likely results in rapid widespread alterations of the traffic of macromolecules between the nucleus and cytoplasm in response to glucose limitation.
Abstract
The importin-β family members (karyopherins) mediate the majority of nucleocytoplasmic transport. Msn5 and Los1, members of the importin-β family, function in tRNA nuclear export. tRNAs move bidirectionally between the nucleus and the cytoplasm. Nuclear tRNA accumulation occurs upon amino acid (aa) or glucose deprivation. To understand the mechanisms regulating tRNA subcellular trafficking, we investigated whether Msn5 and Los1 are regulated in response to nutrient availability. We provide evidence that tRNA subcellular trafficking is regulated by distinct aa-sensitive and glucose-sensitive mechanisms. Subcellular distributions of Msn5 and Los1 are altered upon glucose deprivation but not aa deprivation. Redistribution of tRNA exportins from the nucleus to the cytoplasm likely provides one mechanism for tRNA nuclear distribution upon glucose deprivation. We extended our studies to other members of the importin-β family and found that all tested karyopherins invert their subcellular distributions upon glucose deprivation but not aa deprivation. Glucose availability regulates the subcellular distributions of karyopherins likely due to alteration of the RanGTP gradient since glucose deprivation causes redistribution of Ran. Thus nuclear–cytoplasmic distribution of macromolecules is likely generally altered upon glucose deprivation due to collapse of the RanGTP gradient and redistribution of karyopherins between the nucleus and the cytoplasm.
INTRODUCTION
In eukaryotes, macromolecules must be transported selectively and efficiently between the cytoplasm and the nucleus. Coordination of nucleocytoplasmic transport is essential for diverse cellular processes, such as basal replication, transcription, and translation, as well as for regulation of the cell cycle, transcriptional activation and repression, and other cellular processes (Macara, 2001). To ensure proper subcellular distribution of macromolecules, their transport into and out of the nucleus is tightly controlled. Proteins of the importin-β (also known as karyopherin-β) family mediate the majority of macromolecular nucleocytoplasmic transport. Members of this family shuttle between the nucleus and the cytoplasm through nuclear pores. Cells encode numerous importin-β family members; exportins are the family members dedicated to the nuclear export process, whereas importins transport cargo from the cytoplasm to the nucleus. The different family members vary greatly in their substrate specificities.
Nucleocytoplasmic transport is controlled by small GTPase Ran, which binds to the karyopherins to regulate association or dissociation of the transport complexes (Nachury and Weis, 1999; Strom and Weis, 2001; Fried and Kutay, 2003; Madrid and Weis, 2006). Ran exists primarily in a GTP-bound form in the nucleus and in a GDP-bound form in the cytoplasm. This asymmetric distribution results from the distinct subcellular localizations of the Ran cycle regulators, the nuclear guanine nucleotide exchange factor (RanGEF, RCC1 in vertebrates, Prp20 in yeast) and the cytoplasmic GTPase-activating protein (RanGAP in vertebrates, Rna1 in yeast; Ohtsubo et al., 1989; Hopper et al., 1990; Bischoff and Ponstingl, 1991; Bischoff et al., 1994). Exportins bind cargo, directly or indirectly, and RanGTP in the nucleus and then move to the cytoplasm. RanGAP stimulates GTP hydrolysis of RanGTP to dissociate the export complexes and thereby release cargo in the cytoplasm. In contrast, importins bind cargo, directly or via an adaptor, in the cytoplasm. The importin–cargo complex moves to the nucleus, where it encounters RanGTP, leading to release of the cargo in the nucleus. Then exportins/importins are recycled for the next round of transport (for review see Chook and Suel, 2011).
Transport through nuclear pores is intimately linked to cell physiology. Changes in the cellular environment, by externally applied stress, aging, or disease, alter nucleocytoplasmic traffic (for review see Kodiha et al., 2010). tRNA subcellular movement is one of the examples that exhibits altered subcellular distribution upon nutrient deprivation. It was believed that pre-tRNAs transcribed in the nucleus are only exported from the nucleus to the cytoplasm, where the translation machinery is located. However, it is now known that tRNAs also move constitutively from the cytoplasm to the nucleus via retrograde nuclear import and can again access the cytoplasm via tRNA reexport (Shaheen and Hopper, 2005; Takano et al., 2005; Whitney et al., 2007). The tRNA retrograde pathway is regulated in response to loss of nutrients, as cells exhibit an even distribution of tRNA throughout the nucleus and the cytoplasm under nutrient-replete conditions, whereas tRNA nuclear accumulation occurs upon amino acid (aa), glucose, or Pi deprivation (Grosshans et al., 2000; Shaheen and Hopper, 2005; Hurto et al., 2007; Whitney et al., 2007).
Movement of tRNAs between the nucleus and the cytoplasm proceeds via association of importin-β family members. In vertebrates, importin-β family member exportin-t functions in tRNA nuclear export (Arts et al., 1998; Kutay et al., 1998). Exportin-t preferentially binds to the appropriately structured tRNA backbone with mature tRNA 5′ and 3′ ends, but it has no preference for intron-containing or intron-less tRNAs (Arts et al., 1998; Lund and Dahlberg, 1998; Lipowsky et al., 1999; Cook et al., 2009). The Saccharomyces cerevisiae exportin-t homologue is Los1 (Hopper et al., 1980; Hellmuth et al., 1998; Sarkar and Hopper, 1998). End-processed, intron-containing pre-tRNAs accumulate in los1Δ cells due to defects in primary nuclear export of pre-tRNAs to the cytoplasm, where the splicing machinery resides (Sarkar and Hopper, 1998; Yoshihisa et al., 2003, 2007; Murthi et al., 2010).
The vertebrate importin-β family member exportin-5 (Exp-5) has also been implicated in the nuclear export of tRNA; however, it is believed to play only a minor role in tRNA nuclear export, and instead it primarily functions in the nuclear export of microRNAs (Bohnsack et al., 2002; Calado et al., 2002; Lund et al., 2004; Shibata et al., 2006; Mingot et al., 2013; for review see Leisegang et al., 2012). The yeast homologue Msn5 is well known to export particular phosphorylated transcription factors to the cytoplasm (for review see Hopper, 1999). Msn5 is likely also involved in tRNA nuclear export (Takano et al., 2005; Shibata et al., 2006; Murthi et al., 2010). However, in contrast to los1Δ cells, deletion of MSN5 does not result in the accumulation of end-processed, intron-containing pre-tRNAs (Murthi et al., 2010). Thus the current working model is that Msn5 participates in tRNA nuclear reexport but not primary tRNA nuclear export for tRNAs that are encoded by intron-containing genes, whereas Los1 likely functions in both primary nuclear export and the reexport step for this class of tRNAs (Murthi et al., 2010).
The mechanisms regulating tRNA nuclear–cytoplasmic distribution remain unclear. There are multiple possible mechanisms by which tRNA subcellular distribution could be regulated, including nutrient-regulated tethering of tRNAs and regulation of karyopherins in response to nutrient conditions. Previous studies showed that the subcellular distributions of the nuclear–cytoplasmic shuttling proteins, Los1 and Msn5, are affected by carbon source and other stress conditions (Ghavidel et al., 2007; Karkusiewicz et al., 2011; Pierce et al., 2013). In cells grown with a fermentable sugar as the carbon source, Los1 is located primarily in the nucleus. In contrast, in cells grown in a nonfermentable carbon source, under glucose deprivation, or when the cells are exposed to DNA damaging agents, Los1 is primarily cytoplasmic, separated from the tRNAs awaiting nuclear export (Ghavidel et al., 2007; Karkusiewicz et al., 2011; Pierce et al., 2014). The subcellular distribution of Msn5 is also affected by stress. Msn5 is concentrated in nuclei of unstressed cells, but it is located in the cytoplasm upon exposure to ethanol, heat, starvation, or severe oxidative stress (Pierce et al., 2014; Quan et al., 2006, 2007).
To test the hypothesis that tRNA exportins are affected by nutrient status to regulate tRNA subcellular dynamics, we investigated the regulation of endogenous levels of Los1 and Msn5 under nutrient deprivation conditions known to affect tRNA subcellular distribution. By employing yeast, S. cerevisiae, as a model organism, we investigate the possible mechanisms regulating nucleocytoplasmic transport in response to nutrient availability.
RESULTS
tRNA reexport is regulated by nutrient availability
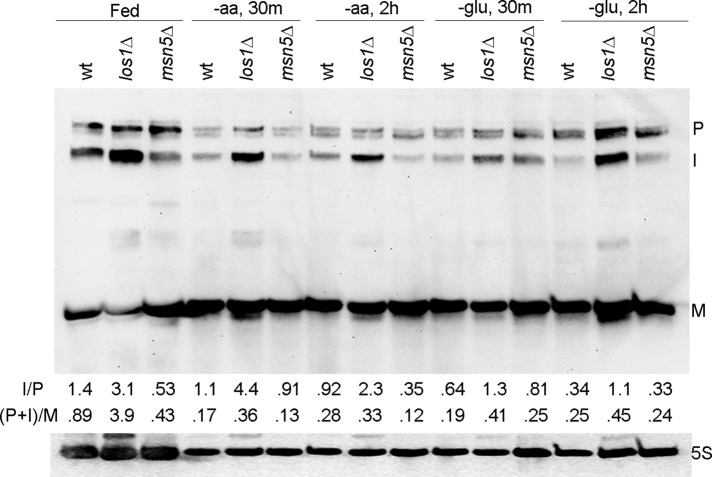
The subcellular distribution of tRNAs between the nucleus and the cytoplasm is a result of the balance among the rates of primary export, nuclear import, and reexport. Because retrograde tRNA nuclear import is constitutive (Murthi et al., 2010), we previously proposed that the tRNA reexport step is regulated by nutrient availability. To investigate this proposal, we performed Northern analyses to examine tRNA species in cells that are fed or deprived of aa or glucose. Because the yeast splicing machinery is located on the cytoplasmic surface of mitochondria (Yoshihisa et al., 2003, 2007), defects in primary nuclear export result in the accumulation of end-processed, intron-containing pre-tRNAs. Thus, if the response to nutrient availability modulates the primary tRNA nuclear export step, end-processed, intron-containing pre-tRNAs should accumulate upon nutrient deprivation, whereas if there are defects in the reexport step, spliced tRNAs should accumulate. RNAs were isolated from fed and nutrient-deprived cells. Under nutrient-replete conditions, cells deleted for LOS1 accumulated elevated levels of unspliced tRNA, whereas wild-type (wt) cells and cells deleted for MSN5 did not (Figure 1, ratio I/P), consistent with previous studies that primary tRNA nuclear export is defective in los1Δ cells. Northern analysis of tRNA in wt cells deprived of aa or glucose for 30 min or 2 h showed that the levels of primary and end-processed, intron-containing pre-tRNAs decreased compared with fed conditions (Figure 1, ratio (P + I)/M); this decrease likely results from decreased transcription (Upadhya et al., 2002; Ciesla et al., 2007). Unlike los1Δ cells, which are defective in primary tRNA export, the ratio of end-processed, intron-containing pre-tRNAs to primary transcript did not increase when wt or msn5Δ cells were deprived of nutrients (Figure 1, ratio I/P). Although wt cells deprived of aa or glucose rapidly accumulate tRNA in the nucleus (Whitney et al., 2007), we did not observe defects in primary tRNA nuclear export under these conditions. The data provide further support for the proposal that tRNA nuclear reexport, rather than primary export, is regulated by nutrient availability.
FIGURE 1:
The tRNA nuclear reexport step is likely regulated in response to nutrient availability. Northern analysis of pre-tRNAIle and mature tRNAIle in wt and mutant cells under fed and nutrient-deprived conditions. I, end-processed, intron-containing pre-tRNA; M, mature tRNA; P, primary tRNA transcript. Ratio I/P, end-processed, intron-containing pre-tRNA to primary transcript. Ratio (P + I)/M, both precursor tRNAs to mature tRNA. 5S rRNA serves as loading control.
Subcellular distributions of Msn5 and Los1 are not altered upon aa deprivation
The mechanisms regulating tRNA reexport in response to nutrient availability remain unclear (for review see Hopper, 2013). We investigated one possible mechanism by which the tRNA exportins are affected by nutrient status. We examined steady-state protein levels, posttranslational modifications, and subcellular distributions of Los1 and Msn5 under nutrient-replete and depletion conditions. Endogenously tagged Msn5 and Los1 were generated by Msn5–green fluorescent protein (GFP) and Los1-GFP gene replacements (Huh et al., 2003). To test whether GFP-tagged tRNA exportins are functional, we performed fluorescence in situ hybridization (FISH). The studies reveal that cells with Msn5-GFP or Los1-GFP gene replacements evidence a distribution of tRNAs between the nucleus and the cytoplasm that is indistinguishable from the distribution in wt cells and unlike cells that possess mutations of either LOS1 or MSN5 (Supplemental Figure S1). Thus GFP-tagged versions of these proteins maintain their functions in tRNA nuclear export.
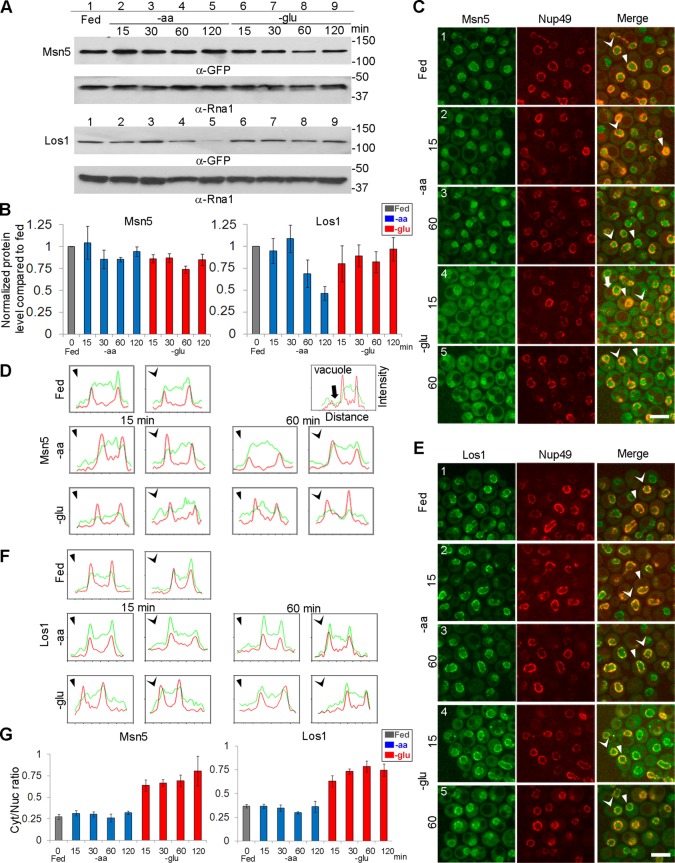
We determined the steady-state expression levels of the functional tagged tRNA exportins in nutrient-replete and depletion conditions by Western blotting, as normalized to the control protein, Rna1. Cultures were deprived of all aa for various times (15, 30, 60, or 120 min). Comparing the normalized protein levels from cells that were aa deprived to the levels under fed conditions (i.e., Msn5/Rna1 under experimental conditions to Msn5/Rna1 under fed conditions), we found that Msn5-GFP levels were unaltered throughout the time course (Figure 2, A, top, lanes 1–5, and B). In contrast, Los1-GFP levels decreased after 1 h of aa deprivation, reaching ∼46% of fed levels by 2 h of aa deprivation (Figure 2, A, bottom, lanes 1–5, and B). Thus aa deprivation does not markedly affect Msn5-GFP steady-state levels but does affect Los1-GFP levels after extended periods of aa deprivation.
FIGURE 2:
Redistribution of Msn5 and Los1 pools from the nucleus to the cytoplasm upon glucose but not aa deprivation. (A) Western blot analysis showing Msn5-GFP and Los1-GFP expression. The time points of aa and glucose deprivation are indicated. Rna1 is the loading control. (B) Quantification of Msn5-GFP and Los1-GFP expression by Western blot analysis from three independent experiments. Relative protein levels were obtained by determining the amount of GFP-tagged protein normalized to Rna1 for each condition compared with that in fed conditions. Gray bars, fed conditions; blue bars, aa removal for indicated time points; red bars, glucose removal for indicated time points. (C) Confocal images of the subcellular distribution of endogenously expressed Msn5-GFP after the indicated times of removal of aa or glucose. (D) Intensity plot profiles of the subcellular distribution of Msn5-GFP. Intensity plot profiles of single sections are shown for two independent cells for each condition. The cells in C that are scanned and plotted in D are indicated with arrowheads of the same shape. Red lines, Nup49-mCherry intensities; green lines, Msn5-GFP intensities. (E) Subcellular distribution of Los1-GFP after the indicated times of removal of aa or glucose. (F) Intensity plot profiles of the subcellular distribution of Los1-GFP. The cells in E that are scanned and plotted in F are indicated with arrowheads of the same shape. Red lines, Nup49-mCherry intensities; green lines, Los1-GFP intensities. Bar, 5 μm. (G) Intensity bar charts of cellular fluorescence intensity of Msn5-GFP or Los1-GFP. Ratios for each condition were obtained by comparing mean fluorescence intensity in the cytoplasm to mean fluorescence intensity in the nucleus (n = 150 cells). Gray bars, fed conditions; blue bars, aa removal for indicated time points; red bars, glucose removal for indicated time points.
Because tRNA nuclear accumulation occurs within 10 min of aa removal (Whitney et al., 2007) and at this time point the expression levels of both Msn5 and Los1 are unchanged, we investigated whether these proteins are posttranslationally modified in response to nutrient status. To identify modifications, we purified the tagged versions of Msn5 and Los1 from cells grown under fed or 1-h aa-deprived conditions and then analyzed the resulting protein modifications by mass spectrometry. None of the peptides had detectable levels of phosphate or ubiquitin modifications (~70% coverage for Msn5 and ~60% coverage for Los1). Although acetylated or methylated peptides were found, the relative levels of these peptides from fed and aa-starved conditions were unaltered for Msn5 and Los1 (Supplemental Table S1). Consistently, there are no identified modifications of Msn5 and Los1 in the Phosida database (www.phosida.com/), which integrates thousands of high-confidence in vivo phosphosites identified by mass spectrometry–based proteomics in various species.
Because neither steady-state protein levels nor assessable protein modifications of Los1 or Msn5 appears to regulate tRNA nuclear accumulation upon acute aa deprivation, we determined whether nutrient availability influences the steady-state subcellular distributions of Msn5 and Los1. To address this possibility, we used confocal imaging of live cells harboring endogenous Msn5-GFP and Los1-GFP and the exogenously expressed nuclear pore protein Nup49-mCherry to demarcate the nuclear boundary. Msn5-GFP was predominantly nuclear under fed conditions. When wt cells were deprived of aa for 0–2 h, Msn5-GFP maintained its predominately nuclear location (Figure 2C, compare 1–3). To obtain a graphic display of the subcellular distribution of Msn5 in the different conditions, we generated intensity plot profiles of single confocal sections of individual cells (Figure 2D). We also measured the cellular fluorescence intensity in the cytoplasm and nucleus using a sum intensity projection. Because Msn5-GFP is not in the vacuole (Figure 2D, white arrow), the intensity analyses avoid the area of vacuole (Figure 2, C, white arrow, and D). Figure 2C depicts the images of single confocal sections upon nutrient deprivation for 15 or 60 min, whereas Figure 2G provides the cytoplasmic/nuclear ratios of fluorescence intensity for all examined time points. As supported by the intensity plot profiles and relative ratios (Figure 2, D and G), the subcellular distribution of Msn5 is insensitive to aa availability.
The subcellular distribution of Los1 was also insensitive to aa availability, as Los1-GFP localized to the nuclear periphery and the nucleoplasm under fed conditions and throughout the time course, from 0 to 2 h, of aa deprivation (Figure 2, E–G). Thus the subcellular distributions of Msn5 and Los1 are unaltered upon acute aa deprivation.
Taken together, the results indicate that alterations of steady-state protein levels, assessable protein modifications, and subcellular distributions of Los1 or Msn5 do not account for tRNA nuclear accumulation upon acute aa deprivation. In the following, we discuss other possible mechanisms.
Steady-state subcellular distributions of Msn5 and Los1 are altered upon glucose deprivation
To investigate whether Msn5 and Los1 are regulated in response to glucose availability, we examined their steady-state levels upon glucose deprivation. Comparing normalized protein levels from cells that were glucose deprived to the levels under fed conditions, we find that Msn5-GFP maintained at least 72% of protein levels (Figure 2, A, top, lanes 1 and 5–9, and B). Los1-GFP levels were maintained at ≥80% upon glucose deprivation (Figure 2, A, bottom, lanes 1 and 5–9, and B). In addition, the relative levels of assessable modified peptides for fed and glucose-deprived conditions were unchanged for both purified Msn5 and Los1 (Supplemental Table S1). Thus glucose deprivation does not dramatically alter the steady-state levels or posttranslational modifications of Msn5 or Los1.
In contrast to the unaltered nuclear–cytoplasmic distributions of Msn5-GFP and Los1-GFP upon aa deprivation, both proteins are redistributed in the cells upon glucose deprivation. Cells were deprived of glucose for various times (15, 30, 60, or 120 min). The steady-state subcellular distribution of Msn5-GFP changed from primarily nucleoplasmic to cytoplasmic by 15 min of glucose deprivation and remained predominately cytoplasmic throughout the time course (Figure 2C, compare 1, 4, and 5). Consistent with the visual images, the intensity plot profiles showed that under fed conditions Msn5-GFP (green) was distributed between the two Nup49-mCherry peaks (red), which mark the nuclear boundaries (Figure 2D). By 15 min of glucose deprivation, the Msn5-GFP signal redistributed to primarily cytoplasmic (Figure 2D), and the cytoplasmic/nuclear ratio increased about threefold compared with fed conditions (Figure 2G).
Under fed conditions, Los1-GFP localized to the nuclear periphery and the nucleoplasm. However, when cells were deprived of glucose for as briefly as 15 min, part of the Los1-GFP pool was localized to the cytoplasm, whereas a portion of the pool remained at the nuclear periphery (Figure 2E, compare 1, 4, and 5). As examined by pixel intensity profile, Los1-GFP levels increased in the cytoplasm and decreased in the nucleoplasm throughout the time course of glucose deprivation (Figure 2F). By 15 min of glucose removal, the cytoplasmic/nuclear ratio of Los1 increased about twofold compared with fed conditions (Figure 2G). Although part of the Los1-GFP pool remained at the nuclear periphery, we were unable to distinguish by confocal imaging between the outer and inner nuclear periphery. Whereas the subcellular distribution of Los1-GFP was altered by glucose availability, the change was somewhat more subtle than that evidenced for Msn5-GFP. Thus the subcellular distributions of these two endogenously expressed karyopherins between the nucleus and the cytoplasm were altered upon glucose deprivation.
The previously published localization studies of Msn5 and Los1 were performed using fixed cells and, in some cases, high-copy protein expression levels (Quan et al., 2006, 2007; Ghavidel et al., 2007; Karkusiewicz et al., 2011; Pierce et al., 2014); here we provide confocal live cell imaging of endogenously expressed functional Msn5 and Los1. To compare our results using endogenous Msn5-GFP and Los1-GFP to previous studies, we also performed indirect immunofluorescence (IF) using multicopy functional Myc-tagged Msn5-Myc and Los1-Myc. The results for Msn5 were very similar by the two methods. However, overexpressed Los1 had a larger nucleoplasmic pool under fed conditions than for endogenous Los1 and a larger cytoplasmic pool upon glucose deprivation (Supplemental Figure S2). Taken together, the data show that the steady-state distributions of Msn5 and Los1 between the nucleus and cytoplasm are altered upon glucose deprivation but not upon aa deprivation.
Altered subcellular distribution of tRNA exportins upon glucose deprivation is rapid and reversible
Nuclear accumulation of tRNA in response to acute glucose starvation is rapid and reversible (Whitney et al., 2007). One way this could occur is via the redistribution of the steady-state pools of tRNA exportins, Los1 and Msn5, from the nucleus to the cytoplasm upon removal of glucose. If tRNA nuclear accumulation is, in fact, due to the altered subcellular distribution of the exportins, then one would expect that the subcellular dynamics of the exportins should be fast, preceding tRNA distribution changes, and reversible. To test this prediction, we conducted live-cell imaging using microfluidics. Microfluidics permitted appropriate aeration and immediate media exchange, thus allowing the tracking living cells in various growth conditions. We first located Msn5-GFP under fed conditions and then followed the response of Msn5-GFP upon glucose deprivation. Under fed conditions, the steady-state distribution of Msn5-GFP was predominantly nuclear, as expected (Figure 3A). The steady-state distribution of Msn5-GFP became more cytoplasmic after 5 min of glucose removal (Figure 3A). To test reversibility, we reintroduced glucose to cells that had been deprived of glucose for 10 min. Within 10 min of refeeding, the majority of the Msn5 pool was again nuclear (Figure 3A). Quantification of subcellular dynamics of Msn5-GFP supported that the cytoplasmic pools of Msn5-GFP increased within 10 min of glucose removal and then the cytoplasmic/nuclear ratios decreased after refeeding (Figure 3C). Similarly, part of the Los1-GFP pool moved from the nuclear periphery to the cytoplasm after 5 min of glucose removal (Figure 3, B and C). The subcellular dynamics of Los1-GFP was also reversible, even though the redistribution of Los1-GFP was more subtle than that of Msn5-GFP (Figure 3, B and C). Thus redistribution of tRNA exportins in response to acute glucose starvation is rapid and reversible, and their subcellular dynamics is consistent with tRNA nuclear accumulation. Because yeast produces ∼3 million tRNAs/generation (Waldron and Lacroute, 1975), and Los1-mediated tRNA nuclear export is limiting (Ghavidel et al., 2007), redistribution of Los1 and Msn5 likely accounts for altered tRNA location upon glucose deprivation.
FIGURE 3:
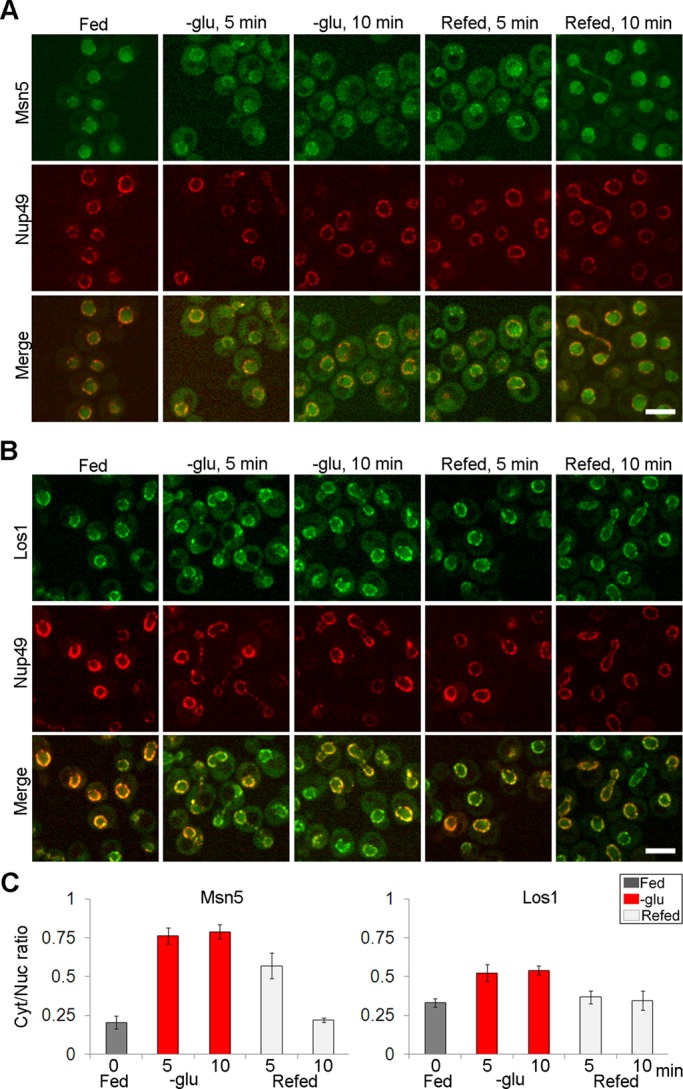
Redistribution of tRNA exportins upon glucose deprivation is rapid and reversible. (A) Confocal imaging of endogenously tagged Msn5-GFP and (B) Los1-GFP after the indicated times of removal or reintroduction of glucose by microfluidics. Bar, 5 μm. (C) Ratios for each condition were obtained by comparing mean fluorescence intensity in the cytoplasm to mean fluorescence intensity in the nucleus (n = 120 cells). Gray bars, fed conditions; red bars, glucose removal for indicated time points; light gray bars, refed conditions.
Steady-state subcellular distributions of other importin-β family members are altered upon glucose deprivation
To learn whether redistribution of Los1 and Msn5 upon glucose deprivation is a general phenomenon that also occurs for other members of the importin-β family, we expanded the analyses to study the subcellular distributions of other importin-β family members upon nutrient deprivation. We studied one additional exportin, Crm1, and two importins, Mtr10 and Kap95.
To investigate whether these family members were modulated by nutrient availability, we employed endogenously GFP-tagged constructs generated by gene replacement (Huh et al., 2003). Because CRM1 and KAP95 are essential genes, the fact that the yeast strains with endogenously tagged Crm1-GFP and Kap95-GFP are viable and grow similarly to wt cells provides evidence that these tagged versions are active. MTR10 is not essential, but mtr10Δ cells exhibit altered cellular morphology (Murthi et al., 2010). Because cells harboring the Mtr10-GFP replacement possess normal cellular morphology, this tagged version of Mtr10 is also functional.
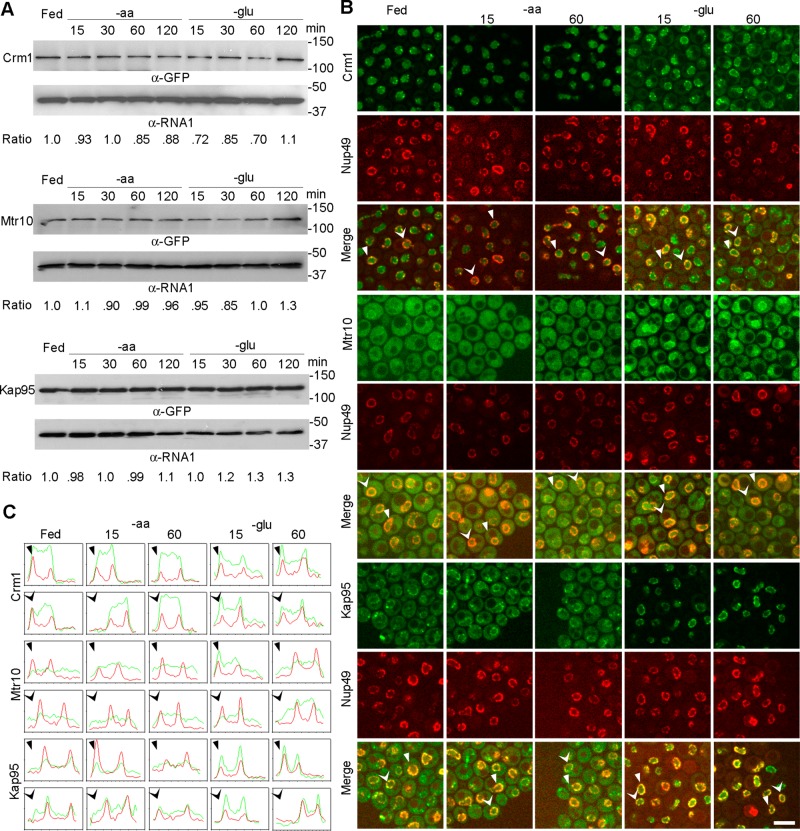
We first determined the levels of each of these proteins by Western blotting, comparing the level of the karyopherins to Rna1. Cultures were harvested at four time points (15, 30, 60, and 120 min) after aa or glucose deprivation. The steady-state levels of Crm1-GFP, Mtr10-GFP, and Kap95-GFP appeared to be relatively constant (Figure 4A).
FIGURE 4:
Crm1, Mtr10, and Kap95 exhibit inverted subcellular distributions upon glucose but not aa deprivation. (A) Western blot analysis showing Crm1-GFP, Mtr10-GFP, and Kap95-GFP steady state levels under fed conditions and aa or glucose depletion conditions for the indicated times. Rna1 is the loading control. Ratio, relative protein level normalized to Rna1 in each condition compared with that in fed condition (B) Confocal images of the subcellular distributions of endogenously tagged Crm1-GFP, Mtr10-GFP, and Kap95-GFP under fed conditions and aa or glucose depletion conditions for the indicated times. Bar, 5 μm. (C) Intensity plot profiles of the subcellular distribution of karyopherin-GFP. The cells in B that are scanned and plotted in C are indicated with arrowheads of the same shape. Red lines indicate Nup49-mCherry intensities; green lines indicate karyopherin-GFP intensities.
Crm1 functions in the nuclear export of proteins, RNA, and ribosome subunits (Maurer et al., 2001; Johnson et al., 2002; Verheggen and Bertrand, 2012). To address whether nutrient deprivation regulates the subcellular distribution of Crm1-GFP, we observed the location of Crm1-GFP in cells deprived of aa or glucose for 0, 15, and 60 min. As determined by confocal imaging, Crm1-GFP was predominantly nuclear under nutrient-replete conditions. When cells were deprived of aa for 15 or 60 min, Crm1 retained its primarily nuclear location (Figure 4B). However, upon glucose deprivation for 15 min, part of the Crm1-GFP signal was located in the cytoplasm; after longer periods of glucose deprivation, the Crm1-GFP pool gradually became less nucleoplasmic and more cytoplasmic, although some proteins were retained at the nuclear periphery (Figure 4B). Assessment of Crm1-GFP distribution by intensity plot profiles confirmed the visual assessment (Figure 4C). Thus, similar to Msn5 and Los1, glucose availability, but not aa availability, affects the steady-state subcellular distribution of Crm1-GFP.
Mtr10 mediates nuclear import of mRNA-binding protein Npl3 (Kadowaki et al., 1994; Pemberton et al., 1997; Senger et al., 1998). In addition, cells bearing a deletion of MTR10 fail to accumulate the RNA subunit of telomerase or tRNA nuclear pools upon aa deprivation, presumably because Mtr10 is required, directly or indirectly, for their import from the cytoplasm to the nucleus (Ferrezuelo et al., 2002; Shaheen and Hopper, 2005; Murthi et al., 2010). Mtr10 redistributes from the cytoplasm to the nuclear periphery upon DNA replication stress (Tkach et al., 2012). Examination of Mtr10-GFP subcellular distribution showed that the majority of the endogenous Mtr10-GFP pool was evenly distributed in the cytoplasm of cells that received all nutrients or were deprived of aa for 15 or 60 min (Figure 4B). In contrast, in cells deprived of glucose for 15 min, Mtr10-GFP was primarily located at the nuclear rim and in the nucleoplasm with residual cytoplasmic pools (Figure 4B); these distributions were supported by intensity plot profiles (Figure 4C).
The well-characterized Kap95 functions in nuclear import of nuclear localization signal (NLS)-containing proteins (Enenkel et al., 1995; Gilchrist and Rexach, 2003). Consistent with earlier studies, we found endogenous Kap95-GFP to be primarily cytoplasmic with some nuclear rim signal under nutrient-replete conditions. After 15 or 60 min of aa deprivation, Kap95 maintained its steady-state cytoplasmic pools (Figure 4B). However, upon 15 min of glucose deprivation, the Kap95-GFP pool was concentrated at the nuclear rim, colocalizing with the nuclear pore protein Nup49 (Figure 4B). The cytoplasmic Kap95-GFP pool dramatically decreased upon prolonged glucose deprivation compared with fed and aa deprivation conditions (Figure 4B). Intensity plot profiles of confocal imaging supported the visual results (Figure 4C). Thus, like Msn5 and Los1, all tested members of importin-β family, both importins and exportins, invert their steady-state subcellular distributions upon glucose deprivation but not upon aa deprivation.
Collapse of the RanGTP gradient affects the steady-state subcellular distributions of importin-β family members
Because we observed that all tested importin-β family members invert their steady-state subcellular distributions upon glucose availability, we investigated the possible mechanisms. Assembly and disassembly of karyopherin-cargo transport complexes are regulated by the small GTPase Ran (Strom and Weis, 2001; Madrid and Weis, 2006; Fried and Kutay, 2003). Under normal growth conditions, Ran is predominantly nuclear. However, when cells are exposed to starvation, heat, ethanol, or hydrogen peroxide, Ran equilibrates between nucleus and cytoplasm (Stochaj et al., 2000). In addition, removal of glucose from cells results in a rapid decrease in ATP level and an immediate increase in AMP/ATP ratio (Wilson et al., 1996; Ashe et al., 2000). Levels of GTP are intrinsically linked to ATP levels because GDP is converted to GTP by nucleoside diphosphate kinase, which uses ATP as the phosphate donor (Parks and Agarwal, 1973; Kikkawa et al., 1990). Shortage of GTP-bound Ran upon ATP depletion results in inhibition of Ran-dependent nuclear transport (Schwoebel et al., 2002). Because the subcellular distributions of all tested karyopherins were altered in response to loss of glucose, we tested the possibility that this redistribution is coordinated with alterations of the RanGTP gradient.
Yeast contains two paralogues of the mammalian Ran, encoded by the GSP1 and GSP2 genes. We examined the subcellular distribution of endogenously tagged Gsp2-GFP under fed and nutrient deprivation conditions. As determined by confocal imaging, Gsp2-GFP was predominantly nuclear when cells were in nutrient-replete conditions. In cells deprived of aa for 15 or 60 min, Gsp2-GFP retained its primarily nuclear location (Figure 5A). However, upon glucose deprivation for 15 or 60 min, the steady-state Gsp2-GFP pool was primarily cytoplasmic (Figure 5A). Redistribution of Ran likely causes alteration of the nuclear–cytoplasmic RanGTP gradient.
FIGURE 5:
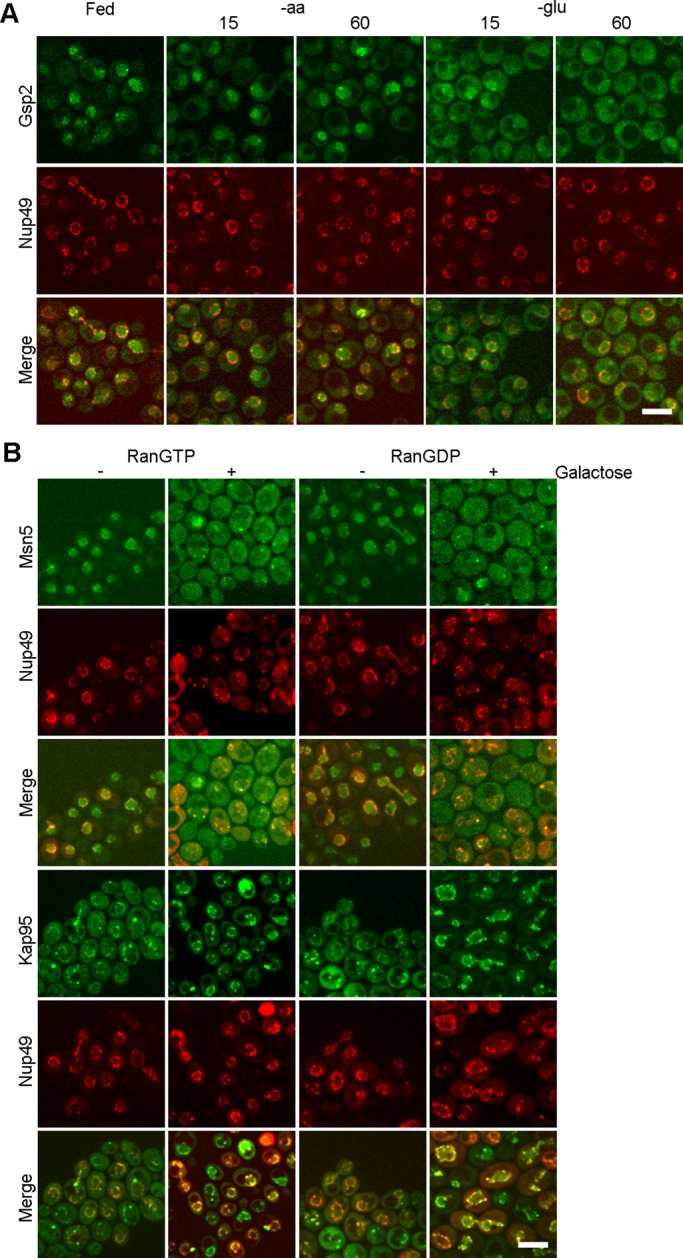
Redistribution of karyopherins upon glucose deprivation is coordinated with the RanGTP gradient. (A) Microfluidics to study the subcellular dynamics of endogenously tagged Gsp2-GFP under fed and nutrient deprivation conditions for the indicated times. (B) Confocal imaging of Msn5-GFP and Kap95-GFP in cells with galactose-inducible Ran locked in the GTP- or GDP-bound state. –, locations of proteins before addition of galactose; +, locations of proteins after addition of galactose for 2 h. Bar, 5 μm.
To test whether collapse of the RanGTP gradient per se causes redistribution of karyopherins, we used yeast strains with an inducible Ran locked in the GTP- or GDP-bound state. Gsp1 with G21V mutation would bind to, but not hydrolyze, GTP, thus stabilizing the GTP-bound form of Ran. The T24N mutation of Gsp1 is defective in GTP binding, and thus it remains predominantly in the GDP-bound form (Kornbluth et al., 1994). Because expression of Gsp1-G21V and Gsp1-T24N in yeast cells causes dominant lethality (Schlenstedt et al., 1995), we constructed galactose-inducible Gsp1-G21V or Gsp1-T24N to induce their expression and hence disrupt the RanGTP gradient in a regulated manner. These plasmids encoding Ran mutant proteins were introduced into yeast strains harboring endogenous Msn5-GFP and Kap95-GFP.
As assessed by confocal live-cell imaging, without galactose induction of the RanGTP- or RanGDP-locked mutant proteins, Msn5-GFP was predominantly nuclear, as expected (Figure 5B). In contrast, after 2 h of galactose (Gal) induction, Msn5-GFP was primarily cytoplasmic, with inducible Ran locked in either the GTP- or the GDP-bound form (Figure 5B). The subcellular distribution of Los1-GFP was also altered after galactose induction of RanGTP- or RanGDP-locked mutants (Supplemental Figure S3). Assessment of Kap95-GFP subcellular distribution showed that Kap95 was predominantly cytoplasmic, and part of the signal associated at the nuclear rim before galactose induction. However, after 2-h induction of Ran mutant proteins, the Kap95-GFP pool markedly increased to the nuclear periphery and the cytoplasmic pool decreased in cells with RanGTP-locked mutant, whereas Kap95 was concentrated at the nuclear rim in cells with RanGDP-locked mutant (Figure 5B). Thus the distributions of both an exportin and an importin between the nucleus and the cytoplasm were inverted in cells with either RanGTP- or RanGDP-locked mutants. The results show that defects in the RanGTP gradient by introducing either RanGTP- or RanGDP-locked forms affect the subcellular distributions of karyopherins.
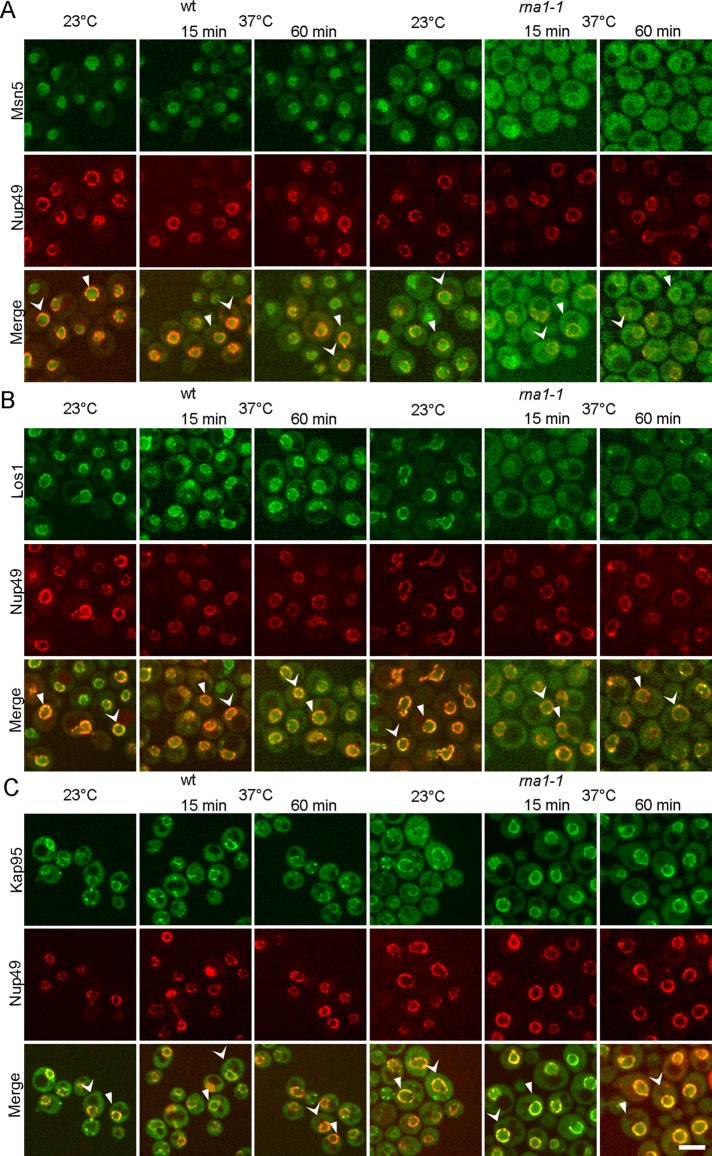
In the foregoing experiments, we observed nuclear–cytoplasmic redistribution of the karyopherins after 2 h, but not within 15 min, the time at which cells respond to glucose deprivation. The slowed kinetics is likely due to the facts that by necessity cells contain endogenous wt Ran and that production of the mutant proteins requires galactose induction. To address this issue, we altered RanGAP using the rna1-1 strain, which encodes a temperature-sensitive (ts) RanGAP (Hopper et al., 1990). Rna1 is necessary for GTP hydrolysis of RanGTP to RanGDP in the cytoplasm (Hopper et al., 1990; Becker et al., 1995; Bischoff et al., 1995; Corbett et al., 1995). Endogenously expressed GFP-tagged karyopherins, including Msn5, Los1, and Kap95, were generated in the rna1-1 strain by homologous recombination.
In both wt and rna1-1 cells, Msn5 was predominantly nuclear at the permissive temperature (23°C); however, there was a cytoplasmic pool in rna1-1 cells, perhaps due to a defect even at 23°C (Figure 6A). After 15 min at nonpermissive temperature (37°C), in wt cells Msn5-GFP remained primarily nuclear, although some cytoplasmic signal could be detected. In stark contrast, after 15 min at 37°C, Msn5-GFP in rna1-1 cells was primarily cytoplasmic (Figure 6A). Intensity plot profiles confirmed that there was much less nuclear signal and more cytoplasmic signal in rna1-1 cells at 37°C than for wt cells (Supplemental Figure S4A).
FIGURE 6:
Collapse of the RanGTP gradient affects the subcellular distributions of karyopherins. The RanGTP gradient was altered by inactivation of RanGAP in the rna1-1 strain. Microfluidics to study the subcellular dynamics of endogenously tagged (A) Msn5-GFP, (B) Los1-GFP, and (C) Kap95-GFP in wt and rna1-1 ts mutant cells at permissive (23°C) and nonpermissive (37°C) temperatures for the indicated times. Cells that are scanned and plotted in Supplemental Figure S4 are indicated with arrowheads of the same shape. Bar, 5 μm.
Los1-GFP in both wt and rna1-1 cells was located at the nuclear periphery at 23°C. After 15 min at 37°C, Los1-GFP in wt cells retained its primarily nuclear periphery distribution, although some cytoplasmic signal was detected (Figure 6B). However, in rna1-1 cells, within 15 min at 37°C, Los1-GFP was primarily cytoplasmic, with significantly decreased nuclear rim signal (Figure 6B), supported by intensity plot profiles (Supplemental Figure S4B).
In both wt and rna1-1 strains at 23°C, Kap95-GFP was predominantly cytoplasmic, and part of the signal was associated with the nuclear rim (Figure 6C). After shifting wt cells to 37°C for 15 or 60 min, Kap95-GFP remained predominantly cytoplasmic. In contrast, the Kap95-GFP signal remarkably increased at the nuclear periphery and decreased in the cytoplasm in rna1-1 cells after 15 min at 37°C (Figure 6C), confirmed by intensity plot profiles (Supplemental Figure S4C).
Previous studies showed that intron-containing tRNAs accumulate when cells possess mutations of the regulation of the Ran pathway (RanGAP-rna1-1 and RanGEF-prp20; Hopper et al., 1978; Kadowaki et al., 1993), and tRNAs accumulate in the nucleus in rna1-1 cells (Sarkar and Hopper, 1998; Shaheen and Hopper, 2005; Gu et al., 2005; McGuire and Mangroo, 2012). Our studies provide additional insight into these tRNA export defects, as tRNAs or pre-tRNAs to be exported and tRNA exportins are primarily in two separate subcellular compartments in rna1-1 cells at the nonpermissive temperature. Consistent with in vivo labeling studies in rna1-1 cells (Hopper et al., 1980), by using Northern analyses, we show that pre-tRNAs accumulate in rna1-1 cells after shifting to the nonpermissive temperature for 10 min (Supplemental Figure S5). The data support that pre-tRNA accumulation in the rna1-1 strain is coordinated to redistribution of tRNA exportin in this mutant.
Taken together, the results indicate that collapse of the RanGTP gradient causes marked inverse subcellular distributions of karyopherins between the nucleus and the cytoplasm. Increased cellular RanGTP or RanGDP levels results in similar consequences. In rna1-1 cells at 37°C, nuclear–cytoplasmic distributions of the karyopherins altered within 15 min, the time at which cells respond to glucose deprivation. Thus collapse of the RanGTP gradient results in similar kinetics as glucose deprivation for redistributed karyopherins, and our data support the notion that redistribution of karyopherins upon glucose deprivation is likely due to collapse of the RanGTP gradient.
DISCUSSION
We provide evidence that tRNA nuclear reexport is subject to complex regulation as cells use distinct mechanisms to respond to aa versus glucose availability. Within 10 min of aa removal, tRNA nuclear accumulation occurs (Whitney et al., 2007); however, at this time point the steady-state protein levels, assayable protein modifications, and subcellular distributions of Los1 and Msn5 resemble those in fed conditions. Mechanisms independent of Los1’s and/or Msn5’s subcellular distribution or activity likely account for how tRNA reexport responds to aa depletion. Because tRNAs can be aminoacylated in the nucleus by the nuclear pool of aminoacyl-tRNA synthetases, uncharged tRNA may accumulate in the nucleus when tRNA charging is defective or when cells are deprived of aa that they are unable to synthesize (Lund and Dahlberg, 1998; Sarkar et al., 1999; Grosshans et al., 2000; Azad et al., 2001; Feng and Hopper, 2002; Whitney et al., 2007; Murthi et al., 2010). Charging defects in methionyl-tRNA synthetase caused defective nuclear export of cognate tRNAMet; however, nuclear export of noncognate tRNAIle and tRNATyr was not affected (Sarkar et al., 1999). Similarly, depletion of yeast THG1 results in uncharged tRNAHis (Gu et al., 2005). thg1Δ cells accumulate tRNAHis but not tRNATyr in the nucleus (Gu et al., 2005). Thus nuclear aminoacylation status of tRNA could provide one regulatory mechanism for cognate tRNA nuclear reexport.
tRNATyr, tRNALeu, tRNAIle, tRNAMet, and tRNAHis upon aa deprivation accumulate in nuclei of BY4741 cells (Grosshans et al., 2000; Shaheen and Hopper, 2005; Hurto et al., 2007; Whitney et al., 2007; Murthi et al., 2010). Yeasts are able to produce all 20 aa unless they harbor mutations of particular aa biosynthesis pathways. After aa deprivation, BY4741 cells are able to produce prototropic amino acids, such as Tyr and Ile, and thus tRNATyr and tRNAIle are charged (Whitney et al., 2007). However, these charged tRNAs accumulate in the nucleus when cells are depleted of all aa. The data suggest that, besides tRNA charging status, a separate aa-sensitive mechanism accounts for nuclear accumulation of these tRNAs.
Glucose availability regulates the subcellular distributions of Los1 and Msn5 between the nucleus and the cytoplasm. Under fed conditions, Los1 and Msn5 are primarily nuclear, where they are able to interact with tRNAs and then export them to the cytoplasm. After glucose deprivation, Msn5, with kinetics identical to that of tRNA nuclear accumulation, becomes primarily cytoplasmic, where it is unable to access nuclear tRNAs to deliver them to the cytoplasm; for this condition, part of the Los1 pool redistributes to the cytoplasm and part of the Los1 pool remains at the nuclear periphery. After glucose deprivation, partial maintenance of Los1 at the nuclear periphery may be sufficient for exporting the low levels of intron-containing pre-tRNAs that are transcribed under these conditions, and thereby primary tRNA nuclear export is not defective and end-processed, intron-containing pre-tRNAs do not accumulate. Redistribution of tRNA exportins upon glucose removal provides an explanation for nuclear accumulation of imported cytoplasmic tRNAs. Thus subcellular distributions of tRNA exportins likely regulate tRNA nuclear reexport in response to glucose availability.
Here we report that Ran redistributes to the cytoplasm upon glucose removal. Because RanGEF is in the nucleus and RanGAP is in the cytoplasm (Ohtsubo et al., 1989; Hopper et al., 1990; Corbett et al., 1995), under fed conditions Ran is primarily in its GTP-bound state in the nucleus (Figure 7A), whereas upon glucose deprivation Ran is likely predominantly in its GDP-bound state in the cytoplasm (Figure 7B). Moreover, purposeful collapse of the RanGTP gradient via RanGTP- or RanGDP-locked mutants or inactivation of RanGAP by the rna1-1 mutation caused inverted subcellular distributions of karyopherins. Thus redistribution of karyopherins upon glucose deprivation is likely due to collapse of the RanGTP gradient. Whether this disruption is upstream or downstream of other nutrient responsive pathway is unknown. tRNA retrograde traffic is independent of Gcn2 but dependent on the PKA pathway (Whitney et al., 2007; Pierce et al., 2014). In cells with constitutive PKA activity (tpk1w1 tpk2Δ tpk3Δ bcy1Δ), tRNAs fail to accumulate in nuclei, and Los1 and Msn5 remain nuclear upon glucose deprivation (Whitney et al., 2007; Pierce et al., 2014). Owing to the nature of the deletion strains, the kinetics of responses to the PKA pathway have not been explored. There is also a connection between P-body formation and tRNA nuclear–cytoplasmic trafficking, as both have similar kinetics in response to glucose deprivation and both depend on Dhh1 and Pat1 (Hurto and Hopper, 2011; Parker, 2012); however, the distributions of Msn5 and Los1 in dhh1Δ pat1Δ cells or cells with overexpressed Dhh1 or Pat1 have not been explored. Thus the roles of PKA and P-body formation pathways in Msn5 and Los1 subcellular distribution and their connections to the Ran pathway require further study.
FIGURE 7:
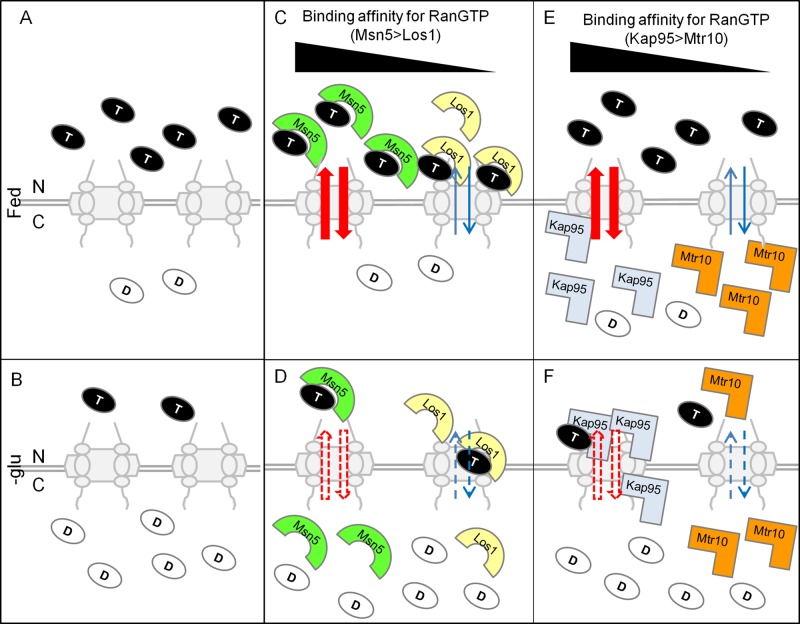
Working model for the mechanism of redistribution of karyopherins by collapse of the RanGTP gradient upon glucose deprivation. (A) RanGTP gradient under fed condition. C, cytoplasm; D, RanGDP; N, nucleus; T, RanGTP. (B) After glucose deprivation, redistributed Ran may cause collapse of the RanGTP gradient. (C) Exportins are primarily nuclear under fed condition. (D) Exportins are redistributed to the cytoplasm upon glucose deprivation. (E) Importins are primarily cytoplasmic under fed condition. (F) Importins are redistributed upon glucose deprivation. Black triangle indicates the RanGTP binding affinity of karyopherins from strong (left) to weak (right). Red arrows indicate greater shuttling of karyopherins than shuttling of other karyopherins, depicted by blue arrows. Solid arrows indicate normal shuttling under fed condition, and dotted arrows indicate aberrant shuttling when cells are deprived of glucose.
Although we were initially motivated to study the regulation of tRNA subcellular dynamics, we extended our analyses to other karyopherins, both importins and exportins. All studied karyopherins show inverted distribution between the nucleus and the cytoplasm upon glucose deprivation but not aa deprivation. Individual karyopherins demonstrated somewhat different patterns in subcellular redistribution upon glucose deprivation. How do cells differentially regulate the subcellular distribution of individual karyopherins upon glucose deprivation? One possible explanation could relate to the distinct binding affinities of individual karyopherins for RanGTP, as the affinities of karyopherins for RanGTP differ significantly (Hahn and Schlenstedt, 2011). In yeast, binding of Crm1 and Los1 to RanGTP is undetectable in the absence of their cargo in vitro (Hellmuth et al., 1998; Maurer et al., 2001; Hahn and Schlenstedt, 2011), and thus the RanGTP binding affinity for Msn5 is higher than for Los1 and Crm1. Because Msn5 displays more dramatic redistribution upon glucose removal than Los1 or Crm1 (Figure 7, C and D), the differential redistribution of karyopherins upon glucose deprivation correlates with their RanGTP binding affinities. Consistently, Kap95 has strong affinity for RanGTP, and its subcellular distribution is dramatically altered upon glucose deprivation (Figure 7, E and F). In contrast, Mtr10 has weaker affinity for RanGTP, and it exhibits only a subtle redistribution upon glucose removal (Figure 7, E and F). Therefore divergent binding affinities of karyopherins for RanGTP may provide a mechanism for their individual responses.
Glucose availability appears to regulate global transport of macromolecules between the nucleus and the cytoplasm. Redistribution of karyopherins upon glucose deprivation is likely due to collapse of the RanGTP gradient, and the individual redistribution of karyopherins appears to be correlated with their RanGTP binding affinities (Figure 7). Consistently, nuclear import of several proteins is defective in rna1-1 cells, in which the RanGTP gradient is affected (Corbett et al., 1995). Whether the subcellular distributions of karypherins are the major determinants that control nucleocytoplasmic transport for their cargoes remains unclear. Several levels of control could add complexity in regulating subcellular dynamics of individual cargo, including competition between different cargoes for binding to karyopherins (Quan et al., 2006), posttranslational modifications of proteins that interact with karyopherins (Hopper, 1999), and balance among the rates of import and export. Therefore, whereas glucose deprivation globally regulates the subcellular distribution of karyopherins and likely a significant portion of nuclear–cytoplasmic trafficking of macromolecules, individual members and their cargo may respond differently. In sum, cells are able to sense distinct nutrient status and use separate mechanisms to regulate nucleocytoplasmic transport in response to different stresses.
MATERIALS AND METHODS
Yeast strains and plasmids
Most experiments used yeast strain BY4741 (MATa his3Δ leu2Δ met15Δ ura3Δ) and its derivatives. BY4741 is the parent for the collection of yeast strains that endogenously express GFP-tagged proteins (Huh et al., 2003; Invitrogen–Life Technologies, Grand Island, NY). The strain containing the rna1-1 mutation is derived from BY4741 (Li et al., 2011). Yeast strains were maintained in synthetic defined media (SC) lacking the appropriate nutritional ingredients for plasmid selection. Genomic GFP-tagged Msn5-GFP, Los1-GFP, or Kap95-GFP ts strains containing the rna1-1 mutation were generated by introducing DNA fragments, which contain homologous sequences with GFP sequences and a HIS3 selection marker (Longtine et al., 1998), to rna1-1.
Plasmid Nup49-mCherry with a LEU2 selection marker was constructed as described (Lai et al., 2009). To construct a plasmid harboring Nup49-mCherry with a URA3 selection marker (pIVY48), the SacI/NaeI fragment of plasmid Nup49-mCherry was subcloned into plasmid pRS426. Plasmid with galactose-inducible Gsp1-G21V and Gsp1-T26N were constructed by site-directed mutagenesis as described (Kornbluth et al., 1994).
Oligonucleotides
The sequences of the oligonucleotides used are provided in the Supplemental Materials and Methods.
Northern analysis
Small RNAs were isolated and separated by electrophoresis and transferred onto a Hybond N+ membrane (Amersham–GE Healthcare, Pittsburgh, PA) as described (Murthi et al., 2010). tRNAs were detected with a digoxigenin-labeled probe complementary to tRNAIle as described (Wu et al., 2013). The relative levels of precursor and mature tRNAs were determined using ImageJ (National Institutes of Health, Bethesda, MD).
In situ hybridization
FISH was performed as previously described (Sarkar and Hopper, 1998), with the modifications detailed in Stanford et al. (2004).
Western analysis
GFP fusion protein expression was assessed using chemiluminescence-based Western blot analysis following standard protocols as described (Chu and Hopper, 2013). The membrane was probed with primary antibodies: 1:1000 dilution anti-GFP (clone CBP-KK1; Roche, Indianapolis, IN) and anti-Rna1 at 1:10,000 dilution (Hopper et al., 1990). Secondary antibodies were horseradish peroxidase–conjugated anti-mouse and anti-rabbit immunoglobulin G (IgG; GE Healthcare) at 1:3000 dilution. Protein signals were quantified using ImageJ.
Posttranslational modification analysis
Protein purification and mass spectrometry analysis of posttranslational modifications are described in the Supplemental Materials and Methods.
Immunofluorescence
Immunofluorescence was conducted as previously described (Tolerico et al., 1999). The location of multicopy Msn5-Myc and Los1-Myc was determined by using mouse monoclonal anti-Myc (12CA5; Roche) at 1:100 dilution. Fluorescein isothiocyanate–conjugated goat anti-mouse IgG (Jackson ImmunoResearch Labs, West Grove, PA) was used at 1:400 dilution to locate the primary antibody. Cells were counterstained with 4′,6-diamidino-2-phenylindole (0.1 μg/ml) to locate DNA.
Microfluidics
To study the intracellular dynamics of GFP-tagged proteins in wt and ts strains, a microfluidics system (Cell ASIC; EMD Millipore, Darmstadt, Germany) was used. The temperature was controlled by a microfluidic control system (Cell ASIC). Changes in the media and incubation times were programmed using ONIX FG software (Cell ASIC) according to the manufacturer's protocol.
Microscopy and imaging
To view live cells by confocal microscopy, cells were placed on a slide containing a thin layer of appropriate medium with 20% gelatin and 0.1 mM n-propylgallate as previously described (Wu et al., 2006). Alternatively, cells were analyzed using the microfluidics system. Monitoring of live cells was performed using a Nikon microscope (Nikon, Melville, NY) equipped with a spinning disk confocal apparatus (UltraView; PerkinElmer Life and Analytical Science, Waltham, MA) and a cooled charge-coupled device (CCD) camera (ORCA-AG; Hamamatsu, Bridgewater, NJ). Cells were visualized using 488-nm (green) and 568-nm (red) argon ion lasers and a 100×/1.4 numerical aperture objective lens. Maximum intensity projections of images were created using UltraView ERS software, and image analyses of single 0.4-μm optical sections were performed using ImageJ. To measure the subcellular fluorescence intensity in the nucleus and cytoplasm, we used sum intensity projections and drew a square measuring area in ImageJ as described (Wu and Pollard, 2005). Epifluorescence imaging was accomplished using a Nikon Microscope Eclipse 90i equipped with a CoolSNAP HQ2 CCD camera (Photometrics, Tucson, AZ) and Nis-Elements software (3.1; Nikon). Photoshop (Adobe, San Jose, CA) was used for image assembly.
Supplementary Material
Acknowledgments
We thank members of A.K.H.’s lab for scientific discussions and S. Osmani for comments on the manuscript. We thank C. Boone for the ts strain collection, J.-Q. Wu and his lab members for assistance with confocal microscopy and data analysis, and V. Wysocki and L. Zhang for mass spectrometry analysis and discussion. This work was supported by National Institutes of Health Grant GM27930 to A.K.H. and Taiwan Merit Scholarship NSC-095-SAF-I-564-049-TMS to H.-Y.H.
Abbreviations used:
- aa
amino acid
- FISH
fluorescence in situ hybridization
- GFP
green fluorescent protein
- IF
immunofluorescence
- ts
temperature-sensitive
- wt
wild type
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-04-0948) on July 23, 2014.
REFERENCES
- Arts GJ, Kuersten S, Romby P, Ehresmann B, Mattaj IW. The role of exportin-t in selective nuclear export of mature tRNAs. EMBO J. 1998;17:7430–7441. doi: 10.1093/emboj/17.24.7430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe MP, De Long SK, Sachs AB. Glucose depletion rapidly inhibits translation initiation in yeast. Mol Biol Cell. 2000;11:833–848. doi: 10.1091/mbc.11.3.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azad AK, Stanford DR, Sarkar S, Hopper AK. Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol Biol Cell. 2001;12:1381–1392. doi: 10.1091/mbc.12.5.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker J, Melchior F, Gerke V, Bischoff FR, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- Bischoff FR, Klebe C, Kretschmer J, Wittinghofer A, Ponstingl H. RanGAP1 induces GTPase activity of nuclear Ras-related Ran. Proc Natl Acad Sci USA. 1994;91:2587–2591. doi: 10.1073/pnas.91.7.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Krebber H, Kempf T, Hermes I, Ponstingl H. Human RanGTPase-activating protein RanGAP1 is a homologue of yeast Rna1p involved in mRNA processing and transport. Proc Natl Acad Sci USA. 1995;92:1749–1753. doi: 10.1073/pnas.92.5.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- Bohnsack MT, Regener K, Schwappach B, Saffrich R, Paraskeva E, Hartmann E, Gorlich D. Exp5 exports eEF1A via tRNA from nuclei and synergizes with other transport pathways to confine translation to the cytoplasm. EMBO J. 2002;21:6205–6215. doi: 10.1093/emboj/cdf613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Treichel N, Muller EC, Otto A, Kutay U. Exportin-5-mediated nuclear export of eukaryotic elongation factor 1A and tRNA. EMBO J. 2002;21:6216–6224. doi: 10.1093/emboj/cdf620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chook YM, Suel KE. Nuclear import by karyopherin-betas: recognition and inhibition. Biochim Biophys Acta. 2011;1813:1593–1606. doi: 10.1016/j.bbamcr.2010.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu HY, Hopper AK. Genome-wide investigation of the role of the tRNA nucleus-cytoplasm trafficking pathway in regulation of the yeast S. cerevisiae transcriptome and proteome. Mol Cell Biol. 2013;26 doi: 10.1128/MCB.00785-13. 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla M, Towpik J, Graczyk D, Oficjalska-Pham D, Harismendy O, Suleau A, Balicki K, Conesa C, Lefebvre O, Boguta M. Maf1 is involved in coupling carbon metabolism to RNA polymerase III transcription. Mol Cell Biol. 2007;27:7693–7702. doi: 10.1128/MCB.01051-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- Corbett AH, Koepp DM, Schlenstedt G, Lee MS, Hopper AK, Silver PA. Rna1p, a Ran/TC4 GTPase activating protein, is required for nuclear import. J Cell Biol. 1995;130:1017–1026. doi: 10.1083/jcb.130.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enenkel C, Blobel G, Rexach M. Identification of a yeast karyopherin heterodimer that targets import substrate to mammalian nuclear pore complexes. J Biol Chem. 1995;270:16499–16502. doi: 10.1074/jbc.270.28.16499. [DOI] [PubMed] [Google Scholar]
- Feng W, Hopper AK. A Los1p-independent pathway for nuclear export of intronless tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2002;99:5412–5417. doi: 10.1073/pnas.082682699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrezuelo F, Steiner B, Aldea M, Futcher B. Biogenesis of yeast telomerase depends on the importin mtr10. Mol Cell Biol. 2002;22:6046–6055. doi: 10.1128/MCB.22.17.6046-6055.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fried H, Kutay U. Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci. 2003;60:1659–1688. doi: 10.1007/s00018-003-3070-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavidel A, Kislinger T, Pogoutse O, Sopko R, Jurisica I, Emili A. Impaired tRNA nuclear export links DNA damage and cell-cycle checkpoint. Cell. 2007;131:915–926. doi: 10.1016/j.cell.2007.09.042. [DOI] [PubMed] [Google Scholar]
- Gilchrist D, Rexach M. Molecular basis for the rapid dissociation of nuclear localization signals from karyopherin alpha in the nucleoplasm. J Biol Chem. 2003;278:51937–51949. doi: 10.1074/jbc.M307371200. [DOI] [PubMed] [Google Scholar]
- Grosshans H, Hurt E, Simos G. An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev. 2000;14:830–840. [PMC free article] [PubMed] [Google Scholar]
- Gu W, Hurto RL, Hopper AK, Grayhack EJ, Phizicky EM. Depletion of Saccharomyces cerevisiae tRNA(His) guanylyltransferase Thg1p leads to uncharged tRNAHis with additional m(5)C. Mol Cell Biol. 2005;25:8191–8201. doi: 10.1128/MCB.25.18.8191-8201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S, Schlenstedt G. Importin beta-type nuclear transport receptors have distinct binding affinities for Ran-GTP. Biochem Biophys Res Commun. 2011;406:383–388. doi: 10.1016/j.bbrc.2011.02.051. [DOI] [PubMed] [Google Scholar]
- Hellmuth K, Lau DM, Bischoff FR, Kunzler M, Hurt E, Simos G. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol Cell Biol. 1998;18:6374–6386. doi: 10.1128/mcb.18.11.6374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK. Nucleocytoplasmic transport: inside out regulation. Curr Biol. 1999;9:R803–R806. doi: 10.1016/s0960-9822(99)80494-1. [DOI] [PubMed] [Google Scholar]
- Hopper AK. Transfer RNA post-transcriptional processing, turnover, subcellular dynamics in the yeast Saccharomyces cerevisiae. Genetics. 2013;194:43–67. doi: 10.1534/genetics.112.147470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper AK, Banks F, Evangelides V. A yeast mutant which accumulates precursor tRNAs. Cell. 1978;14:211–219. doi: 10.1016/0092-8674(78)90108-3. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Schultz LD, Shapiro RA. Processing of intervening sequences: a new yeast mutant which fails to excise intervening sequences from precursor tRNAs. Cell. 1980;19:741–751. doi: 10.1016/s0092-8674(80)80050-x. [DOI] [PubMed] [Google Scholar]
- Hopper AK, Traglia HM, Dunst RW. The yeast RNA1 gene product necessary for RNA processing is located in the cytosol and apparently excluded from the nucleus. J Cell Biol. 1990;111:309–321. doi: 10.1083/jcb.111.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- Hurto RL, Hopper AK. P-body components, Dhh1 and Pat1, are involved in tRNA nuclear-cytoplasmic dynamics. RNA. 2011;17:912–924. doi: 10.1261/rna.2558511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurto RL, Tong AH, Boone C, Hopper AK. Inorganic phosphate deprivation causes tRNA nuclear accumulation via retrograde transport in Saccharomyces cerevisiae. Genetics. 2007;176:841–852. doi: 10.1534/genetics.106.069732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson AW, Lund E, Dahlberg J. Nuclear export of ribosomal subunits. Trends Biochem Sci. 2002;27:580–585. doi: 10.1016/s0968-0004(02)02208-9. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Chen S, Hitomi M, Jacobs E, Kumagai C, Liang S, Schneiter R, Singleton D, Wisniewska J, Tartakoff AM. Isolation and characterization of Saccharomyces cerevisiae mRNA transport-defective (mtr) mutants. J Cell Biol. 1994;126:649–659. doi: 10.1083/jcb.126.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadowaki T, Goldfarb D, Spitz LM, Tartakoff AM, Ohno M. Regulation of RNA processing and transport by a nuclear guanine nucleotide release protein and members of the Ras superfamily. EMBO J. 1993;12:2929–2937. doi: 10.1002/j.1460-2075.1993.tb05955.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karkusiewicz I, Turowski TW, Graczyk D, Towpik J, Dhungel N, Hopper AK, Boguta M. Maf1 protein, repressor of RNA polymerase III, indirectly affects tRNA processing. J Biol Chem. 2011;286:39478–39488. doi: 10.1074/jbc.M111.253310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikkawa S, Takahashi K, Shimada N, Ui M, Kimura N, Katada T. Conversion of GDP into GTP by nucleoside diphosphate kinase on the GTP-binding proteins. J Biol Chem. 1990;265:21536–21540. [PubMed] [Google Scholar]
- Kodiha M, Crampton N, Shrivastava S, Umar R, Stochaj U. Traffic control at the nuclear pore. Nucleus. 2010;1:237–244. doi: 10.4161/nucl.1.3.11444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornbluth S, Dasso M, Newport J. Evidence for a dual role for TC4 protein in regulating nuclear structure and cell cycle progression. J Cell Biol. 1994;125:705–719. doi: 10.1083/jcb.125.4.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Lipowsky G, Izaurralde E, Bischoff FR, Schwarzmaier P, Hartmann E, Gorlich D. Identification of a tRNA-specific nuclear export receptor. Mol Cell. 1998;1:359–369. doi: 10.1016/s1097-2765(00)80036-2. [DOI] [PubMed] [Google Scholar]
- Lai TP, Stauffer KA, Murthi A, Shaheen HH, Peng G, Martin NC, Hopper AK. Mechanism and a peptide motif for targeting peripheral proteins to the yeast inner nuclear membrane. Traffic. 2009;10:1243–1256. doi: 10.1111/j.1600-0854.2009.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leisegang MS, Martin R, Ramirez AS, Bohnsack MT. Exportin t and Exportin 5: tRNA and miRNA biogenesis—and beyond. Biol Chem. 2012;393:599–604. doi: 10.1515/hsz-2012-0146. [DOI] [PubMed] [Google Scholar]
- Li Z, Vizeacoumar FJ, Bahr S, Li J, Warringer J, Vizeacoumar FS, Min R, Vandersluis B, Bellay J, Devit M, et al. Systematic exploration of essential yeast gene function with temperature-sensitive mutants. Nat Biotechnol. 2011;29:361–367. doi: 10.1038/nbt.1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipowsky G, Bischoff FR, Izaurralde E, Kutay U, Schafer S, Gross HJ, Beier H, Gorlich D. Coordination of tRNA nuclear export with processing of tRNA. Rna. 1999;5:539–549. doi: 10.1017/s1355838299982134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE. Proofreading and aminoacylation of tRNAs before export from the nucleus. Science. 1998;282:2082–2085. doi: 10.1126/science.282.5396.2082. [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science. 2004;303:95–98. doi: 10.1126/science.1090599. [DOI] [PubMed] [Google Scholar]
- Macara IG. Transport into and out of the nucleus. Microbiol Mol Biol Rev. 2001;65:570–594. doi: 10.1128/MMBR.65.4.570-594.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madrid AS, Weis K. Nuclear transport is becoming crystal clear. Chromosoma. 2006;115:98–109. doi: 10.1007/s00412-005-0043-3. [DOI] [PubMed] [Google Scholar]
- Maurer P, Redd M, Solsbacher J, Bischoff FR, Greiner M, Podtelejnikov AV, Mann M, Stade K, Weis K, Schlenstedt G. The nuclear export receptor Xpo1p forms distinct complexes with NES transport substrates and the yeast Ran binding protein 1 (Yrb1p) Mol Biol Cell. 2001;12:539–549. doi: 10.1091/mbc.12.3.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuire AT, Mangroo D. Cex1p facilitates Rna1p-mediated dissociation of the Los1p-tRNA-Gsp1p-GTP export complex. Traffic. 2012;13:234–256. doi: 10.1111/j.1600-0854.2011.01304.x. [DOI] [PubMed] [Google Scholar]
- Mingot JM, Vega S, Cano A, Portillo F, Nieto MA. eEF1A mediates the nuclear export of SNAG-containing proteins via the Exportin5-aminoacyl-tRNA complex. Cell Rep. 2013;5:727–737. doi: 10.1016/j.celrep.2013.09.030. [DOI] [PubMed] [Google Scholar]
- Murthi A, Shaheen HH, Huang HY, Preston MA, Lai TP, Phizicky EM, Hopper AK. Regulation of tRNA bidirectional nuclear-cytoplasmic trafficking in Saccharomyces cerevisiae. Mol Biol Cell. 2010;21:639–649. doi: 10.1091/mbc.E09-07-0551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsubo M, Okazaki H, Nishimoto T. The RCC1 protein, a regulator for the onset of chromosome condensation locates in the nucleus and binds to DNA. J Cell Biol. 1989;109:1389–1397. doi: 10.1083/jcb.109.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R. RNA degradation in Saccharomyces cerevisiae. Genetics. 2012;191:671–702. doi: 10.1534/genetics.111.137265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks REJ., Agarwal RP. Nucleoside diphosphokinases. In: Boyer ed. PD., editor. In: The Enzymes. 3rd ed. New York: Academic Press; 1973. pp. 307–333. [Google Scholar]
- Pemberton LF, Rosenblum JS, Blobel G. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J Cell Biol. 1997;139:1645–1653. doi: 10.1083/jcb.139.7.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce JB, van der Merwe G, Mangroo D. Protein kinase A is part of a mechanism that regulates nuclear reimport of the nuclear tRNA export receptors Los1p and Msn5p. Eukaryot Cell. 2014;13:209–230. doi: 10.1128/EC.00214-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quan X, Tsoulos P, Kuritzky A, Zhang R, Stochaj U. The carrier Msn5p/Kap142p promotes nuclear export of the hsp70 Ssa4p and relocates in response to stress. Mol Microbiol. 2006;62:592–609. doi: 10.1111/j.1365-2958.2006.05395.x. [DOI] [PubMed] [Google Scholar]
- Quan X, Yu J, Bussey H, Stochaj U. The localization of nuclear exporters of the importin-beta family is regulated by Snf1 kinase, nutrient supply and stress. Biochim Biophys Acta. 2007;1773:1052–1061. doi: 10.1016/j.bbamcr.2007.04.014. [DOI] [PubMed] [Google Scholar]
- Sarkar S, Azad AK, Hopper AK. Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1999;96:14366–14371. doi: 10.1073/pnas.96.25.14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Hopper AK. tRNA nuclear export in Saccharomyces cerevisiae: in situ hybridization analysis. Mol Biol Cell. 1998;9:3041–3055. doi: 10.1091/mbc.9.11.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlenstedt G, Saavedra C, Loeb JD, Cole CN, Silver PA. The GTP-bound form of the yeast Ran/TC4 homologue blocks nuclear protein import and appearance of poly(A)+ RNA in the cytoplasm. Proc Natl Acad Sci USA. 1995;92:225–229. doi: 10.1073/pnas.92.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwoebel ED, Ho TH, Moore MS. The mechanism of inhibition of Ran-dependent nuclear transport by cellular ATP depletion. J Cell Biol. 2002;157:963–974. doi: 10.1083/jcb.200111077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger B, Simos G, Bischoff FR, Podtelejnikov A, Mann M, Hurt E. Mtr10p functions as a nuclear import receptor for the mRNA-binding protein Npl3p. EMBO J. 1998;17:2196–2207. doi: 10.1093/emboj/17.8.2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaheen HH, Hopper AK. Retrograde movement of tRNAs from the cytoplasm to the nucleus in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 2005;102:11290–11295. doi: 10.1073/pnas.0503836102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata S, Sasaki M, Miki T, Shimamoto A, Furuichi Y, Katahira J, Yoneda Y. Exportin-5 orthologues are functionally divergent among species. Nucleic Acids Res. 2006;34:4711–4721. doi: 10.1093/nar/gkl663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanford DR, Whitney ML, Hurto RL, Eisaman DM, Shen WC, Hopper AK. Division of labor among the yeast Sol proteins implicated in tRNA nuclear export and carbohydrate metabolism. Genetics. 2004;168:117–127. doi: 10.1534/genetics.104.030452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stochaj U, Rassadi R, Chiu J. Stress-mediated inhibition of the classical nuclear protein import pathway and nuclear accumulation of the small GTPase Gsp1p. FASEB J. 2000;14:2130–2132. doi: 10.1096/fj.99-0751fje. [DOI] [PubMed] [Google Scholar]
- Strom AC, Weis K. Importin-beta-like nuclear transport receptors. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-6-reviews3008. REVIEWS3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano A, Endo T, Yoshihisa T. tRNA actively shuttles between the nucleus and cytosol in yeast. Science. 2005;309:140–142. doi: 10.1126/science.1113346. [DOI] [PubMed] [Google Scholar]
- Tkach JM, Yimit A, Lee AY, Riffle M, Costanzo M, Jaschob D, Hendry JA, Ou J, Moffat J, Boone C, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14:966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolerico LH, Benko AL, Aris JP, Stanford DR, Martin NC, Hopper AK. Saccharomyces cerevisiae Mod5p-II contains sequences antagonistic for nuclear and cytosolic locations. Genetics. 1999;151:57–75. doi: 10.1093/genetics/151.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya R, Lee J, Willis IM. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol Cell. 2002;10:1489–1494. doi: 10.1016/s1097-2765(02)00787-6. [DOI] [PubMed] [Google Scholar]
- Verheggen C, Bertrand E. CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes. Nucleus. 2012;3:132–137. doi: 10.4161/nucl.19266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldron C, Lacroute F. Effect of growth rate on the amounts of ribosomal and transfer ribonucleic acids in yeast. J Bacteriol. 1975;122:855–865. doi: 10.1128/jb.122.3.855-865.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney ML, Hurto RL, Shaheen HH, Hopper AK. Rapid and reversible nuclear accumulation of cytoplasmic tRNA in response to nutrient availability. Mol Biol Cell. 2007;18:2678–2686. doi: 10.1091/mbc.E07-01-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson WA, Hawley SA, Hardie DG. Glucose repression/derepression in budding yeast: SNF1 protein kinase is activated by phosphorylation under derepressing conditions, and this correlates with a high AMP:ATP ratio. Curr Biol. 1996;6:1426–1434. doi: 10.1016/s0960-9822(96)00747-6. [DOI] [PubMed] [Google Scholar]
- Wu J, Huang HY, Hopper AK. A rapid and sensitive non-radioactive method applicable for genome-wide analysis of Saccharomyces cerevisiae genes involved in small RNA biology. Yeast. 2013;30:119–128. doi: 10.1002/yea.2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- Wu JQ, Sirotkin V, Kovar DR, Lord M, Beltzner CC, Kuhn JR, Pollard TD. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J Cell Biol. 2006;174:391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihisa T, Ohshima C, Yunoki-Esaki K, Endo T. Cytoplasmic splicing of tRNA in Saccharomyces cerevisiae. Genes Cells. 2007;12:285–297. doi: 10.1111/j.1365-2443.2007.01056.x. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Yunoki-Esaki K, Ohshima C, Tanaka N, Endo T. Possibility of cytoplasmic pre-tRNA splicing: the yeast tRNA splicing endonuclease mainly localizes on the mitochondria. Mol Biol Cell. 2003;14:3266–3279. doi: 10.1091/mbc.E02-11-0757. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.