The nucleoplasmic domain of the Caenorhabditis elegans SUN protein UNC-84 interacts with lamin. If this interaction is disrupted, a partial failure in nuclear migration occurs.
Abstract
Nuclear migration is a critical component of many cellular and developmental processes. The nuclear envelope forms a barrier between the cytoplasm, where mechanical forces are generated, and the nucleoskeleton. The LINC complex consists of KASH proteins in the outer nuclear membrane and SUN proteins in the inner nuclear membrane that bridge the nuclear envelope. How forces are transferred from the LINC complex to the nucleoskeleton is poorly understood. The Caenorhabditis elegans lamin, LMN-1, is required for nuclear migration and interacts with the nucleoplasmic domain of the SUN protein UNC-84. This interaction is weakened by the unc-84(P91S) missense mutation. These mutant nuclei have an intermediate nuclear migration defect—live imaging of nuclei or LMN-1::GFP shows that many nuclei migrate normally, others initiate migration before subsequently failing, and others fail to begin migration. At least one other component of the nucleoskeleton, the NET5/Samp1/Ima1 homologue SAMP-1, plays a role in nuclear migration. We propose a nut-and-bolt model to explain how forces are dissipated across the nuclear envelope during nuclear migration. In this model, SUN/KASH bridges serve as bolts through the nuclear envelope, and nucleoskeleton components LMN-1 and SAMP-1 act as both nuts and washers on the inside of the nucleus.
INTRODUCTION
Nuclear migration is essential for a wide variety of cellular processes conserved across eukaryotes, including cell migration, cell division, and polarity establishment (Morris, 2000; Burke and Roux, 2009; Starr and Fridolfsson, 2010; Gundersen and Worman, 2013). Nuclear migrations are also central to many developmental processes, including fertilization (Reinsch and Gonczy, 1998), neurogenesis (Tsai et al., 2007; Zhang et al., 2009), and muscle development (Bruusgaard et al., 2006). In addition, nuclear migration is an important process in metastasizing cancer cells (Chow et al., 2012; Fu et al., 2012).
Nuclear migration requires mechanical forces generated by the cytoskeleton to be transferred to the nucleoskeleton—the structural network of the nucleus consisting of lamins, actin, and inner nuclear membrane components (Simon and Wilson, 2011). The nuclear envelope is a specialized extension of the endoplasmic reticulum consisting of an outer nuclear membrane, an inner nuclear membrane, and the lumen between the membranes, called the perinuclear space (Franke et al., 1981). The inner nuclear membrane is tightly associated with the underlying nucleoskeleton. The unique architecture of the nuclear envelope presents a special challenge to force transfer from the cytoskeleton to the nucleoskeleton.
A nuclear membrane complex of Sad1 and UNC-84 (SUN) and Klarsicht, ANC-1, and Syne homology (KASH) proteins, termed the linker of nucleoskeleton and cytoskeleton (LINC) complex, traverses the barrier created by the nuclear envelope and allows for forces generated in the cytoplasm to be transduced into the nucleus (Starr and Fridolfsson, 2010; Tapley and Starr, 2013). SUN proteins are single-pass transmembrane proteins specifically localized to the inner nuclear membrane. They consist of an N-terminal nucleoplasmic domain and a C-terminal domain in the perinuclear space containing the conserved SUN domain (Turgay et al., 2010; Tapley et al., 2011; Tapley and Starr, 2013). The SUN domain functions to recruit KASH proteins to the outer nuclear membrane through a direct interaction between conserved SUN and KASH domains in the perinuclear space (Crisp et al., 2006; McGee et al., 2006; Sosa et al., 2012; Tapley and Starr, 2013). KASH proteins are the only known integral membrane proteins that are specifically localized to the cytoplasmic surface of the nucleus. They are classified by a small conserved KASH peptide at the C-terminus of the protein (Starr and Han, 2002; Starr and Fridolfsson, 2010). The large cytoplasmic domains of KASH proteins interact with a variety of cytoskeletal elements, including microtubule motors, actin, and intermediate filaments (Luxton and Starr, 2014). Thus KASH proteins interact with the cytoskeleton and then partner with SUN proteins to form a bridge across both membranes of the nuclear envelope, allowing the transfer of force to position nuclei.
Interactions between the cytoskeleton and KASH proteins and between SUN and KASH proteins are relatively well understood (Tapley and Starr, 2013; Luxton and Starr, 2014). However, it is much less clear how SUN proteins interact with the nucleoskeleton. The major component of the nucleoskeleton is the intermediate filament lamin, which provides structure and strength to the nuclear envelope. Vertebrates have two types of lamin proteins; B-type lamins are broadly expressed, and A/C-type lamins are expressed in differentiated tissues (Gruenbaum et al., 2005; Dittmer and Misteli, 2011; Simon and Wilson, 2011). A large class of diseases, called laminopathies, has been linked to mutations primarily in lamin A/C (Worman, 2012). Because lamin A/C is involved in disease, most studies on interactions between lamins and SUN proteins have focused on lamin A/C rather than the more broadly expressed lamin B. Therefore how SUN proteins interact with the nuclear lamina and especially lamin B remains an open question. Here we test the hypothesis that SUN proteins interact with lamin B during nuclear migration.
Reports of interactions between SUN proteins and lamin A/C are limited to in vitro glutathione S-transferase (GST) pull-down assays and fluorescence recovery after photobleaching and fluorescence resonance energy transfer assays in transfected tissue culture cells. These data show that SUNs interact with lamin A/C, but conflict as to whether mammalian SUN1 or SUN2 binds more tightly (Crisp et al., 2006; Ostlund et al., 2009). Other studies show that some lamin A disease mutations disrupt the ability of lamin A to bind SUN proteins, whereas other mutations increase the interaction between lamin A and SUN1 (Haque et al., 2010). Nonetheless, SUN proteins properly localize to the nuclear envelope in lamin A mutant cells (Crisp et al., 2006; Haque et al., 2010; Chen et al., 2012). Lamin A is also required for nuclear migrations in polarizing fibroblasts (Folker et al., 2011). Depletion of SUN1 in lamin A–knockout mice increases longevity and the health of the mice, suggesting that the interaction between lamin A and SUN1 is important in the progression of laminopathies (Chen et al., 2012; Chen et al., 2014). One possible explanation for this finding is that by removing SUN1, the forces transferred to the weakened nucleoskeleton are reduced, which may lead to less mechanical damage to already fragile nuclei (Starr, 2012). Such a model fits well with our hypothesis that SUN proteins interact with lamins to move nuclei.
SUN protein interactions with lamin B are less well understood at a biochemical level than their lamin A/C counterparts. In Caenorhabditis elegans and Drosophila, lamin B is required for the localization of SUN proteins (Lee et al., 2002; Kracklauer et al., 2007). However, the extent to which SUN localization to the nuclear envelope requires direct interaction with lamin B is not clear. There is conflicting evidence from in vitro pull-down assays as to whether lamin B interacts with mammalian SUN1 or SUN2 (Crisp et al., 2006; Haque et al., 2006). However, two important developmental genetic experiments suggest that lamin B functions in some of the same nuclear migration events as SUN and KASH proteins. Mice with knockout mutations in lamin B2 have nuclear migration defects in the CNS similar to SUN- and KASH-knockout defects (Zhang et al., 2009; Coffinier et al., 2010a, b, 2011). Similarly, null mutations in the Drosophila lamin B gene Lam Dmo cause a nuclear migration defect in the developing eye disk very similar to that in SUN and KASH mutants (Patterson et al., 2004; Kracklauer et al., 2007). Taken together, these data are consistent with a model in which SUN proteins interact with lamin B to mediate nuclear migration.
Here we used nuclear migration in C. elegans embryonic hypodermal cells (Starr and Han, 2005; Zhou and Hanna-Rose, 2010) as a model for studying the interaction between SUN proteins and lamins. C. elegans has a single lamin gene, as compared with three to four lamins in vertebrate systems. Invertebrate lamins are widely considered as B-type lamins, but unrooted phylogenetic trees place invertebrate lamins in their own clade nearly equal distant from vertebrate lamin As and Bs (Liu et al., 2000; Dittmer and Misteli, 2011). Having a single lamin gene is both an advantage and a disadvantage for this study. It makes the study feasible but complicates the significance of the study when thinking about vertebrate cells. The C. elegans lamin protein LMN-1, also known as Ce-lamin and CeLam-1, is broadly expressed and required for early embryonic cell divisions; lmn-1(RNAi) embryos die at around the 100-cell stage with multiple mitotic defects (Liu et al., 2000). Furthermore, only one SUN protein, UNC-84, is present in the cell at the time of hyp7 nuclear migration (Fridkin et al., 2004; Minn et al., 2009; Wang et al., 2009). Finally, C. elegans hyp7 nuclear migration is amenable to the use of many genetic and live-imaging tools (Starr et al., 2001; Fridolfsson et al., 2010; Fridolfsson and Starr, 2010). Here we combine C. elegans genetics and yeast two-hybrid assays to test our hypothesis that the SUN protein UNC-84 binds to the lamin B protein LMN-1. Furthermore, we use live imaging to carefully describe the nuclear migration phenotypes of unc-84 mutants that disrupt the interaction with lamin B. Our data strongly support that SUN proteins bind directly to lamin B to transfer forces generated in the cytoskeleton to the nucleoskeleton, thus facilitating nuclear migration.
RESULTS
Mutations in the nucleoplasmic domain of UNC-84 lead to an intermediate nuclear migration defect
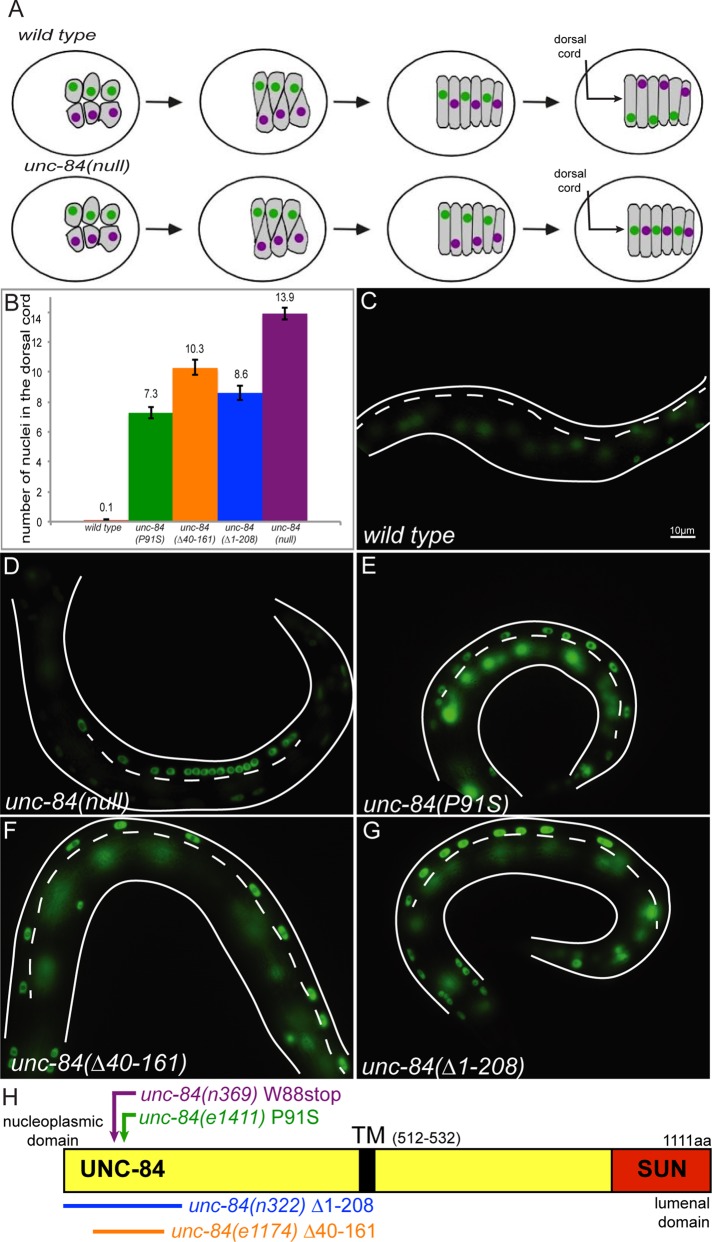
Null mutations in unc-84 cause a complete block of nuclear migration in hyp7 precursors, resulting in ∼14 nuclei residing abnormally in the dorsal cord of larvae (Figure 1; Malone et al., 1999; Fridolfsson and Starr, 2010). However, three alleles—unc-84(e1411, e1174, and n322)—were originally reported to cause an intermediate hyp7 precursor nuclear migration defect (Malone et al., 1999). Normally, nuclei migrate across the length of the dorsal hyp7 precursors during C. elegans embryogenesis. After the bulk of embryonic cell division and just before the initiation of morphogenesis, 15 dorsal epithelial cells intercalate, and their nuclei migrate across the dorsal midline to the contralateral side of the embryo (Figure 1A; Sulston et al., 1983). Nuclei are pulled along polarized microtubules by kinesin-1 and dynein, which are recruited to the surface of the nuclear envelope by the KASH protein UNC-83 (Meyerzon et al., 2009a; Fridolfsson et al., 2010; Fridolfsson and Starr, 2010). These cells subsequently fuse to form the embryonic dorsal hyp7 syncytium (Sulston et al., 1983; Altun, 2009). Mutations that block nuclear migration in hyp7 precursors result in nuclei abnormally residing in the dorsal cord of newly hatched L1 larvae, having been pushed there by underlying body wall muscles (Figure 1A; Sulston and Horvitz, 1981; Malone et al., 1999). About 90% of the nuclei that fail to migrate end up in the dorsal cord (Fridolfsson and Starr, 2010).
FIGURE 1:
Mutations in the nucleoplasmic domain of UNC-84 lead to an intermediate nuclear migration defect. (A) Cartoon describing hyp7 precursor nuclear migration on the dorsal surface of the pre–comma-stage embryo. In wild-type embryos (top), two rows of hyp7 precursors (gray) intercalate to form a row of column-shaped cells. Nuclei then migrate from right to left (green) or left to right (purple). In unc-84(null) mutant embryos, intercalation occurs normally, but the nuclei fail to migrate. Instead, underlying body wall muscle migrations push unc-84 nuclei to the dorsal cord (arrow). The dorsal surface is shown; anterior is left. (B) Average number of nuclei present in the dorsal cord of L1 larvae, which approximates the number of failed nuclear migrations. Error bars, 95% CI. (C–G) The number of nuclei in the larval dorsal cord was counted following hypodermal nuclei that express a nucleoplasmic GFP from ycIs10[pcol-10nls::gfp::lacZ]. Lateral views of L1 larvae. Dorsal is up, and anterior is left; the dorsal cord (arrow in A) is demarcated by the white dotted line. Scale bar, 10 μm. Representative images of (C) wild type, (D) unc-84(null), (E) unc-84(P91S), (F) unc-84(Δ40-161), and (G) unc-84(Δ1-208). (H) Schematic of the domain structure of UNC-84. The conserved SUN domain is red, and the transmembrane span is black. The mutants discussed in the text are indicated.
The unc-84 alleles e1411, e1174, and n322 resulting in an intermediate hyp7 precursor nuclear migration defect all disrupt the N-terminal nucleoplasmic domain of UNC-84. unc-84(e1411) is a P91S missense mutation, unc-84(e1174) is a deletion removing residues 40–161 of UNC-84, and unc-84(n322) is a small deletion of the ATG and is predicted to use the ATG at residue 209 (Figure 1H; Malone et al., 1999). We henceforth refer to these alleles as unc-84(P91S), unc-84(Δ40-161), and unc-84(Δ1-208).
To quantify partial nuclear migration defects, we created a transgenic line expressing nuclear GFP specifically in hypodermal nuclei (ycIs10[pcol-10nls::gfp::lacz]). The unc-84 alleles P91S, Δ40-161, and Δ1-208 caused nuclear migration defects in which 7.3 ± 0.4, 10.3 ± 0.5, and 8.6 ± 0.5 (mean ± 95% confidence interval [CI]; Figure 1, B and E–G) nuclei fail to migrate, respectively. This intermediate phenotype is significantly different from either the null allele unc-84(n369), for which 13.9 ± 0.4 nuclei failed to migrate, or wild-type animals, for which only 0.1 ± 0.1 nucleus was mispositioned to the dorsal cord (Figure 1). The intermediate nuclear migration defect of at least unc-84(P91S) is unlikely due to a reduction in the levels of the mutant UNC-84 protein as compared with wild type. Quantification of immunofluorescence intensity showed that approximately equal levels of UNC-83 protein were found at the nuclear envelope in both the unc-84(P91S) mutant and wild-type embryos (Supplemental Figure S1). Because unc-84(n369)-null mutations disrupt both migration and anchorage (Malone et al., 1999), we next asked about the extent to which these three mutant lines caused any anchorage defects. The nuclei that failed to migrate and are abnormally found in the dorsal cord of the hyp7 syncytium are usually clumped together in unc-84(n369) mutant larvae (Figure 1D). We classified a nuclear anchorage defect (Anc−) if an L1 larva had a row of at least three nuclei touching each other. In the null unc-84(n369) allele, 43% (n = 14) of larvae were Anc−. In contrast, 0% of unc-84(P91S), 6% of unc-84(Δ40-161), and 0% of unc-84(Δ1-208) L1 larvae were Anc− (n ≥ 30). Our data therefore suggest that disruption of the nucleoplasmic domain of UNC-84 results in partial nuclear migration, but not nuclear anchorage, defects.
The nucleoplasmic domain of UNC-84 binds to lamin
We hypothesized that the P91S mutation in the nucleoplasmic domain of UNC-84 disrupted an interaction between UNC-84 and some unknown component of the nucleoskeleton. A yeast two-hybrid screen of a C. elegans mixed-stage cDNA library was conducted to identify proteins interacting with the nucleoplasmic domain of UNC-84. As bait we used the first 385 amino acids of UNC-84 fused to the GAL4 DNA–binding domain. This construct incorporates the majority of the nucleoplasmic domain of UNC-84 upstream of the transmembrane domain located at residues 512–532 (Figure 1H; Tapley et al., 2011). Approximately 4 × 106 yeast clones were screened, and the prey inserts of 106 positive colonies were sequenced. Sixteen different proteins were identified as potential interacting partners of UNC-84. LMN-1, the sole C. elegans lamin protein (Liu et al., 2000), was found in 16 independent clones. No other known component of the nucleoskeleton was identified.
We used the yeast two-hybrid assay to further map the LMN-1 interaction domain of UNC-84 (Figure 2A). The assay was repeated five times with UNC-84(1-385) and the empty vector to verify the interaction. The other constructs containing smaller regions of UNC-84 were examined at least twice. The original bait used for the screen, UNC-84(1-385), strongly interacted with the LMN-1 prey. A smaller bait, UNC-84(1-100), also interacted with LMN-1. However, UNC-84(1-59), UNC-84(59-385), and UNC-84(385-510) did not interact with LMN-1. These data suggest that the minimal interaction domain is somewhere in the first 100 amino acids of UNC-84. Of interest, all three unc-84 alleles with the intermediate hyp7 nuclear migration phenotype disrupt this portion of UNC-84 (Figure 1H). We therefore tested the hypothesis that the unc-84(P91S) mutation disrupted the two-hybrid interaction with LMN-1. We used quantitative β-galactosidase liquid assays to measure the yeast two-hybrid interaction between LMN-1 and wild-type or P91S mutant UNC-84. The P91S mutation significantly reduced the strength of the interaction between LMN-1 and UNC-84, as determined by Student's t tests (Figure 2B).
FIGURE 2:
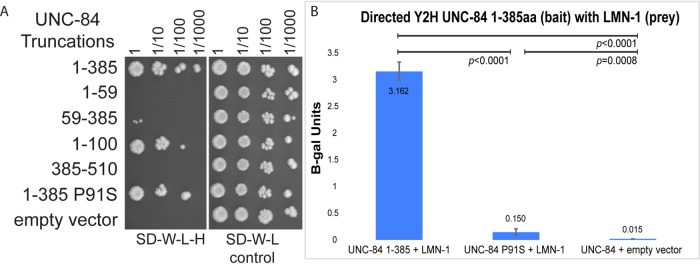
UNC-84 and LMN-1 interact in a yeast two-hybrid assay. (A) Yeast growing in a directed yeast two-hybrid assay. All yeast express the LMN-1::Gal4AD prey construct and the UNC-84::Gal4BD bait construct indicated on the left. Yeast were grown to the same concentration, serially diluted (as indicated at the top), and plated on SD−Trp−Leu−His medium, which requires an interaction to grow (left), or SD−Trp−Leu medium as control (right). (B) Activity of the lacZ gene as activated by a liquid o-nitrophenyl-β-galactoside assay that represents a two-hybrid interaction. Average β-galactosidase units (ΔOD420/min/ml of cells) from three different experiments, each done in triplicate, and the associated 95% CI error bars. Significant statistical differences as determined by Student's t test are noted at the top.
lmn-1(RNAi) leads to a nuclear migration defect
The yeast two-hybrid data are consistent with a hypothesis that the unc-84(P91S) intermediate nuclear migration defect is due to a reduced interaction between UNC-84 and LMN-1. One prediction of this model is that disruptions of lmn-1 should lead to similar nuclear migration defects. lmn-1 is an essential gene required for the earliest embryonic cell divisions. Adults fed double-stranded RNA (dsRNA) against lmn-1 for >24 h produce embryos that have small pronuclei and chromosomal segregation defects, leading to embryonic lethality before the 100-cell stage (Liu et al., 2000; Meyerzon et al., 2009b). To study the effect of lmn-1(RNAi) later in embryogenesis, at the time of nuclear migration in hyp7 precursors, we fed young adults dsRNA against lmn-1 over shorter windows, which allowed for the survival of 10–40 larvae per mother. These larvae demonstrated a nuclear migration defect in which screening a total of 121 larvae from four different experiments resulted in an average of 2.4 ± 0.5 (mean ± 95% CI) hyp7 nuclei in the dorsal cord (Figure 3). An example of an animal with ∼50% hyp7 nuclear migration failure is depicted in Figure 3B. The lmn-1(RNAi) hyp7 nuclear migration failure is statistically more severe than in wild type (p < 0.0001 when using an unpaired t test with Welch's correction). The number of nuclei in the dorsal cord per animal ranges from 0 to 10. The range is large because individuals with no nuclei in the dorsal cord were likely subjected to little or no dsRNA, leading to incomplete knockdown of lmn-1. Finally, lmn-1(RNAi) treatment of the three UNC-84 N-terminal mutant lines resulted in minor enhancement. Given the hypomorphic nature of both the N-terminal mutations and lmn-1(RNAi), this is consistent with our model that UNC-84 and LMN-1 function in the same pathway. This enhancement was not significant for either of the two deletion alleles: unc-84(∆40-161) increased by an average of 0.35 nuclei, and unc-84(∆1-208) increased by an average of 0.45 nuclei. The average enhancement in unc-84(P91S) of 1.9 nuclei was significant (p < 0.0001 with Student's t test) but did not increase to anywhere near the levels observed in unc-84(null) animals.
FIGURE 3:
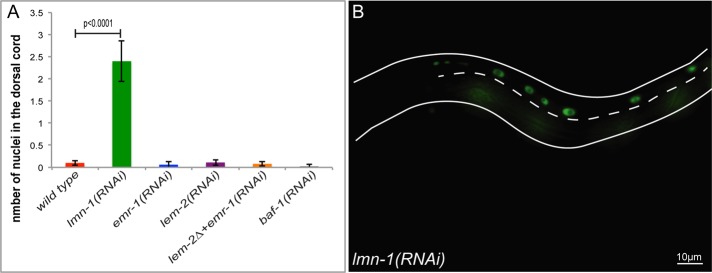
lmn-1(RNAi) animals have a nuclear migration defect. (A) Mean number of nuclei present in the dorsal cord of wild type, lmn-1(RNAi), emr-1(RNAi), lem-2(RNAi), lem-2(tm1582); emr-1(RNAi), and baf-1(RNAi). Error bars, 95% CI. (B) A representative lmn-1(RNAi) L1 larva depicting nuclei in the dorsal cord that have failed to migrate. The dashed line marks the dorsal cord, anterior is to the left, and dorsal is up. Scale bar, 10 μm.
LMN-1 is part of a network of C. elegans proteins at the nucleoskeleton, including the Baf homologue BAF-1 and the LEM proteins emr-1 and lem-2, that are believed to function in a mutually dependent network. Mutants with baf-1 or emr-1; lem-2 double mutations have identical early embryonic phenotypes as lmn-1(RNAi) (Liu et al., 2003; Margalit et al., 2005; Meyerzon et al., 2009b). We therefore examined the extent to which baf-1, emr-1, and lem-2 functioned similarly to lmn-1 during nuclear migration in hyp7 precursors. We observed the previously reported early embryonic lethality in baf-1(RNAi) and lem-2(tm1582); emr-1(RNAi) embryos (unpublished data). However, when fed dsRNA for shorter periods of time as in our lmn-1(RNAi) experiment, we did not observe any nuclear migration defects in baf-1(RNAi) or lem-2(tm1582); emr-1(RNAi) larvae (Figure 3A). We therefore conclude that LMN-1 functions independently of BAF-1 and the LEM proteins during embryonic hyp7 nuclear migration.
Live imaging of the partial nuclear migration defect in unc-84(P91S) embryos
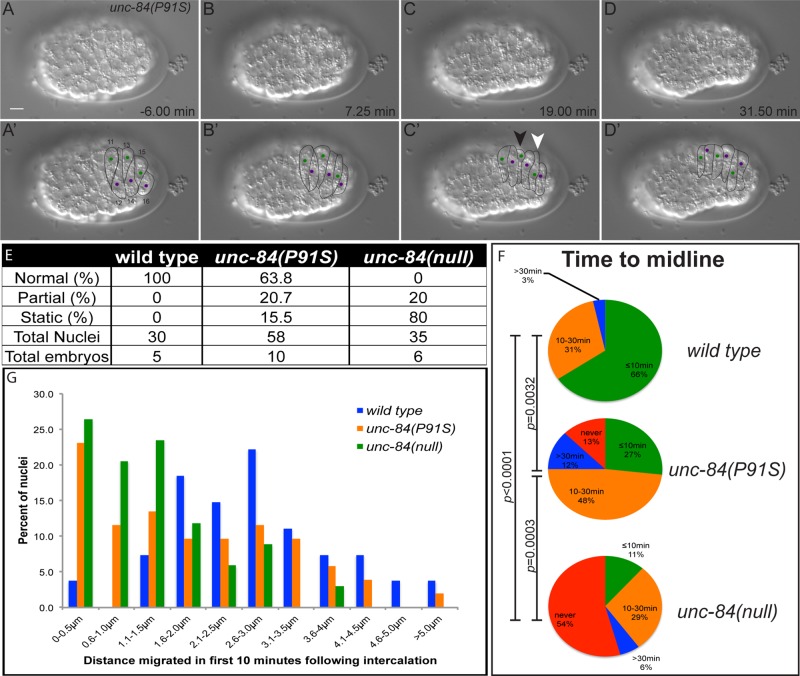
We envisioned three possible ways that the unc-84(P91S) mutation could partially disrupt nuclear migration. First, half of the nuclei could migrate normally, whereas the other half failed to even initiate nuclear migration. Alternatively, unc-84(P91S) mutant nuclei could migrate at a slower velocity than wild-type nuclei, such that only half finish migration in the available period of time. Finally, unc-84(P91S) mutant nuclei might initiate nuclear migration at the normal time and velocity and then fail at some point later in nuclear migration. To distinguish among these possibilities, we observed unc-84(P91S) mutant nuclei by time-lapse imaging during hyp7 precursor development. Time-lapse imaging of hyp7 nuclear migration in wild-type, unc-84(null), or microtubule motor mutant embryos previously led to great insights into the mechanisms of nuclear migration (Fridolfsson and Starr, 2010; Zhang and Han, 2010; Starr, 2011). For example, see our previous studies for details on wild-type and unc-84(null) nuclear migrations (Fridolfsson and Starr, 2010). Here we used both differential interference contrast (DIC) microscopy and fluorescence imaging of LMN-1::green fluorescent protein (GFP) to follow nuclear migration in a subset of hyp7 precursor cells on the dorsal surface of the embryo (Figures 1A and 4, A–D; hyp7 precursors 11–16, following designations of Sulston et al., 1983). Nuclear migration defects in wild-type, unc-84(null), and unc-84(P91S) embryos were quantified by four different assays. For the first three assays, DIC time-lapse microscopy was used to characterize the complete nuclear migration in embryonic hyp7 precursors (Figure 4, A–D). Images were taken at 15-s intervals over 35–50 min. Representative time-lapse movies of wild-type, unc-84(null), and unc-84(P91S) embryos at the time of nuclear migration are shown in Supplemental Movies S1–S3.
FIGURE 4:
Time-lapse imaging of unc-84 mutant nuclear migration events. (A–D) A time-lapse series showing nuclear migration in an unc-84(P91S) mutant embryo. Dorsal views with anterior on the left. Top row, raw DIC images; bottom row, marked cell–cell boundaries and nuclei. The green nuclei in cells 11, 13, and 15 migrate from right to left, and the purple nuclei in cells 12, 14, and 16 migrate left to right. The nucleus of cell 13 (white arrowhead) fails to migrate, whereas the nucleus in cell 15 (black arrowhead) migrated halfway across and then stopped. Scale bar, 5 µm. This is the same embryo as in Supplemental Movie S3. (E) Quantification of percentage of nuclei that migrated normally, initiated nuclear migration but failed to complete it (partial), or failed to move at all (static). (F) Quantification of the time it took nuclei to reach the dorsal midline of the embryo. Nuclei were categorized into those that reached the midline within 10 min of the completion of intercalation (green), at 10–30 min (orange), at >30 min (blue), or never (red). Significant statistical differences as determined by χ2 contingency tests are noted on the left. (G) The distance a nucleus traveled in the first 10 min after completion of intercalation plotted in a histogram. Each individual nucleus was binned into 0.5-μm increments.
Nuclear migration events were scored as normal if full contralateral movement was completed within 30 min, partial if migration initiated but failed to complete, or static if initiation did not occur (Figure 4E). See Supplemental Movie S3 for an example of a static nucleus (white arrowhead) and a nucleus that partially migrated (black arrowhead) in an unc-84(P91S) embryo. One hundred percent of wild-type nuclei completed their migrations normally. Eighty percent of the unc-84(null) nuclei were static, whereas 20% had partial migrations. The unc-84(P91S) nuclei had an intermediate phenotype for which 63.8% of nuclei migrated normally, 20.7% initiated but failed to complete migration, and 15.5% were static.
The time it took a nucleus to reach the midline was also analyzed (Figure 4F). We previously showed that wild-type nuclei reached the midline ∼11 min after intercalation, whereas most unc-84(null) nuclei failed to reach the midline, and those that did took an average of 37 min (Fridolfsson and Starr, 2010). Here, the time a migrating nucleus took to reach the midline was binned into <10, 10–30, or >30 min or not within the length of the film (films were 35–50 min in length). Late during filming, embryos began to rotate in the egg shell. This often resulted in the midline moving to or past a subset of nuclei, thus making the unc-84(null) phenotype described in Figure 4F appear less severe than the phenotype presented in Figure 4E. Consistent with our previous results, the unc-84(P91S) mutant nuclei had an intermediate defect; 27% migrated to the midline in <10 min, but 25% took >30 min or never reached the midline. This intermediate phenotype was significantly different from wild-type or unc-84(null) nuclei as determined by χ2 contingency tests (Figure 4F).
As a third assay, the distance nuclei moved in the first 10 min of migration was binned into half-micrometer increments and plotted (Figure 4G). Wild-type nuclei moved an average of 2.7 ± 0.4 μm and had the highest frequency of nuclei that migrated 2.6–3.0 μm in the first 10 min of migration (blue in Figure 4G). unc-84(null) nuclei migrated an average of 1.2 ± 0.3 μm and were shifted greatly toward the three lowest categories that migrated <1.5 μm in 10 min (green in Figure 4G). unc-84(P91S) nuclei migrated an average of 1.9 ± 0.4 μm, which is statistically less than wild-type movements (p = 0.005 in a t test) and more than unc-84(null) nuclei move (p = 0.016).
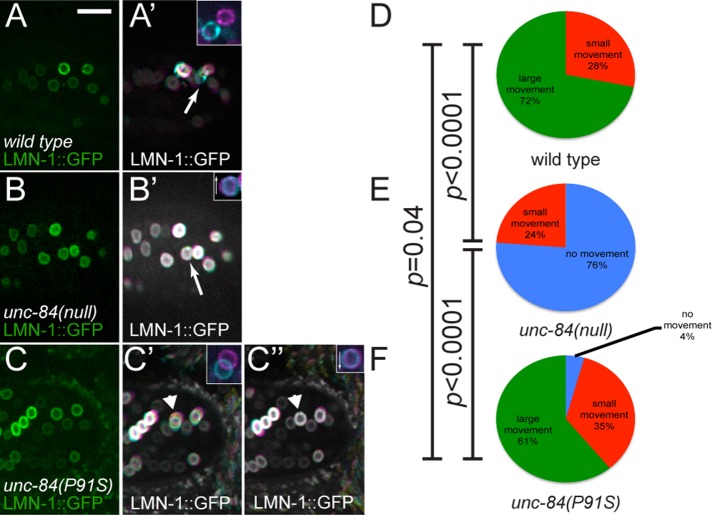
To visualize changes in nuclear envelope morphology over shorter time frames, we performed time-lapse fluorescence imaging to follow LMN-1::GFP in migrating nuclei. A transgenic line expressing lmn-1::gfp under the control of a hypodermal-specific promoter from an extrachromosomal array (Fridolfsson and Starr, 2010) was crossed to unc-84(P91S) and unc-84(null) animals. Embryos at the stage at which hyp7 nuclear migration would normally occur were identified using DIC microscopy. LMN-1::GFP was then imaged in these embryos at 1-s intervals for 3–9 min to follow changes in nuclear envelope morphology during nuclear migration. Movies of LMN-1::GFP in wild-type, unc-84(null), and unc-84(P91S) embryos were visually different (Supplemental Movies S4–S6). Nuclei in wild-type embryos underwent large movements—greater than half the width of a nucleus—and were almost constantly moving (Figure 5A and Supplemental Movie S4). In contrast, unc-84(null) nuclei tended to remain in place over several minutes of filming (Figure 5B and Supplemental Movie S5); most movements were due to the drift of the entire embryo within its eggshell. Of interest, in unc-84(P91S) nuclei, both phenotypes were visualized. Some nuclei were observed undergoing large directional movements of up to 1 μm/min, whereas other nuclei did not move at all.
FIGURE 5:
LMN-1::GFP shows dynamic nuclear morphology during nuclear migration. (A–C) Images of embryos expressing LMN-1::GFP specifically in hypodermal cells at the start of time-lapse imaging. Dorsal views; anterior is left. Insets show the identified nucleus at the beginning (magenta) and end (cyan) of the 8 min, 20 s film. Arrows in insets show the direction the nucleus is supposed to be moving. (A) Wild-type, (B) unc-84(null), and (C) unc-84(P91S) embryos. (A′–C′) Time projections of 500 frames taken at 1-s intervals. In these projections, frames 1–166 are colored magenta, 167–333 are yellow, and 334–500 are cyan to show the direction of movement (A′–C′). A second time-lapse projection of the same embryo for unc-84(P91S) (C′′). The arrowheads in C′ and C′′ mark a unc-84(P91S) nucleus that was migrating normally in time-lapse 1 (C′) but then failed to continue migration in time lapse 2 (C′′). Scale bar, 10 μm. (D–F) Nuclei were classified into three categories: no movement, small movement, and large movement. The percentage in each category is depicted. Significant statistical differences as determined by χ2 contingency tests are noted on the left. The arrow in A′ is an example of a large movement, and the arrow in B′ demonstrates no movement.
To categorize the movements of LMN-1::GFP during nuclear migration, we made projections combining each frame of an 8 min, 20 s time-lapse series (Figure 5, A′–C′). The projections were split into three colors to show the direction of movement. Magenta signifies the first third of the series, yellow the second, and cyan the final third. Using the time-lapse projections of LMN-1::GFP, we binned nuclei into three categories based on the size of an individual nuclear projection (Figure 5, D–F). To better visualize movement, insets show the nuclei identified in the projections in the first frame (magenta) and the last frame (cyan) of the film. Many nuclei had large directional movements over the course of imaging, as visualized by lack of overlap between the initial and final positions of the nucleus of at least half the width of the nucleus (arrow and inset in Figure 5A′; green in Figure 5, D–F). Other nuclei that moved small amounts but the projections of which remained mostly circular were classified as small movements. Finally, nuclei that did not move in up to 9 min of imaging were scored as static when the time-lapse projection remained circular, and when the projection was split into thirds, the colors were merged to white (arrow in Figure 5B′). The same identified nucleus is shown in the inset, which demonstrates slight embryo drift, as the first and last images are not directly superimposed (inset in Figure 5B′). In summary of these data, 72% of wild-type nuclei moved large distances, whereas 28% had small movements (Figure 5D). Seventy-six percent of unc-84(null) nuclei did not move, whereas the remaining 24% had only small movements (Figure 5E). In unc-84(P91S) animals, large movements were seen 61% of the time, and small movements were seen in 35% of nuclei; the remaining 4% of nuclei did not move (Figure 5F). Our LMN-1::GFP movement assay demonstrated statistically significant differences when comparing unc-84(null) nuclear migrations to both wild-type and unc-84(P91S) embryos (p < 0.0001 using a χ2 contingency test). However, wild type and unc-84(P91S) were not significantly different (p = 0.4). The lack of statistical significance may result from the relatively short duration of the time-lapse series, such that only a snapshot of nuclear migration was visualized as compared with the longer analyses in Figure 4. Nonetheless, the unc-84(P91S) phenotype followed the trend of intermediate nuclear migration phenotypes. Multiple time-lapse series were taken of some embryos. Occasionally unc-84(P91S) nuclei were observed to move in one series but then failed to migrate in the subsequent series (arrowhead and insets in Figure 4, C′ and C′′). In another unc-84(P91S) time-lapse movie, a nucleus was observed in which a large and rapid invagination appeared to push the nucleus just before the time of nuclear migration initiation (Supplemental Movie S7). This rapid change may have resulted from abrupt microtubule motor activity acting against a weakened UNC-84–LMN-1 interaction. Together these data are consistent with our hypothesis that a weakened connection between UNC-84 and LMN-1 could lead to a nucleus that initiates migration normally but then fails to complete its migration.
The inner nuclear membrane component SAMP-1 functions during nuclear migration
In our working model, forces generated in the cytoplasm are transmitted across the nuclear envelope by SUN/KASH bridges and then dissipated across the nucleoskeleton by lamin. The nucleoskeleton consists of lamins, scores of inner nuclear membrane proteins, and other proteins that mediate interactions between the nuclear envelope and chromatin (Simon and Wilson, 2011). We therefore hypothesized that other components of the nucleoskeleton play roles in connecting the nucleus to the nuclear envelope to allow for force dissipation during nuclear migration. An attractive candidate to play such a role is the Samp1/NET5/Ima1 C. elegans homologue T24F1.2, which we named SAMP-1. The mammalian putative orthologue was originally found in a proteomic screen for integral components of the inner nuclear membrane and named NET5 (Schirmer et al., 2003). NET5 was subsequently named Samp1 and shown to play a role in positioning nuclei in polarizing NIH 3T3 cells. Nuclear migration in polarizing mouse NIH3T3 cells relies on SUN-KASH bridges to couple moving actin arrays in the cytoplasm to the nucleoskeleton (Luxton et al., 2010; Folker et al., 2011). This nuclear migration also requires Samp1, which partially colocalizes and coimmunoprecipitates with SUN proteins in transmembrane actin-associated nuclear (TAN) lines (Borrego-Pinto et al., 2012). The homologous protein in Schizosaccharomyces pombe, Ima1, interacts in yeast two-hybrid assays with the SUN protein Sad1 and has been implicated in the maintenance of nuclear morphology (Hiraoka et al., 2011). Previously a broad bioinformatics study predicted that C. elegans SAMP-1 would be a component of the nuclear envelope and confirmed this localization in the early embryo using a transgenic SAMP-1::GFP fusion protein (Gunsalus et al., 2005). However, nothing else is known about the function of C. elegans SAMP-1. We therefore set out to examine the role of C. elegans SAMP-1 in nuclear migration.
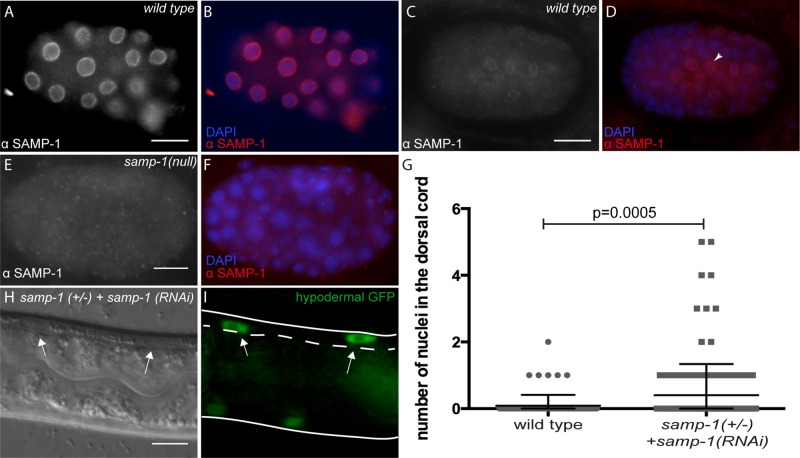
We first characterized the intracellular localization pattern of endogenous SAMP-1 to see whether it was plausible that SAMP-1 functions at the nuclear envelope during nuclear migration in embryonic hyp7 precursor. Antibodies were raised against the C-terminus of SAMP-1. Anti–SAMP-1 antibodies recognized a band of the predicted size on a Western blot. The band intensity was greatly reduced in samp-1(RNAi) extracts (Supplemental Figure S2). SAMP-1 antibodies localized strongly to a ring around 4′,6-diamidino-2-phenylindole (DAPI)–stained nuclei, consistent with nuclear envelope staining, in all cells of wild-type early embryos but not in samp-1(tm2710) likely null embryos (Figure 6 and Supplemental Figure S2). Therefore the antibody is specific for SAMP-1, with a localization pattern expected for a nuclear membrane protein. Although we did not test the specific localization within the nuclear envelope, we hypothesize that SAMP-1 is an inner nuclear membrane protein based on the published localization of the mouse orthologue, Samp1 (Buch et al., 2009). In later embryos at the time of hyp7 nuclear migration, SAMP-1 localization at the nuclear envelope was less strong and limited to a subset of cells that included hyp7 precursors (Figure 6, C and D).
FIGURE 6:
samp-1(RNAi) animals have a weak nuclear migration defect. (A–F) Embryos were stained for SAMP-1 localization. Lateral views, with anterior left and dorsal up. Scale bars, 10 μm. For each pair of images, SAMP-1 immunostaining is shown in white on the left and in red on the right when it is merged with DAPI staining of nuclei in blue. (A, B) An early wild-type embryo. (C, D) A later, pre–comma-stage embryo. Arrowhead points to a hyp7 precursor nucleus. (E, F) A samp-1(tm2710)-null embryo is shown to demonstrate specificity of the antibody. (G) Numbers of nuclei in the dorsal cords of wild-type or samp-1(tm2710)/(+); samp-1(RNAi) L1 larvae. Each gray dot represents an individual animal. The mean and 95% CI error bars are shown. (H, I) DIC and GFP images showing two hyp7 nuclei abnormally in the dorsal cord (arrows) of a samp-1(tm2710)/(+); samp-1(RNAi) L1 larva. The dorsal cord is up and is demarcated by the dotted line. Scale bar, 10 μm.
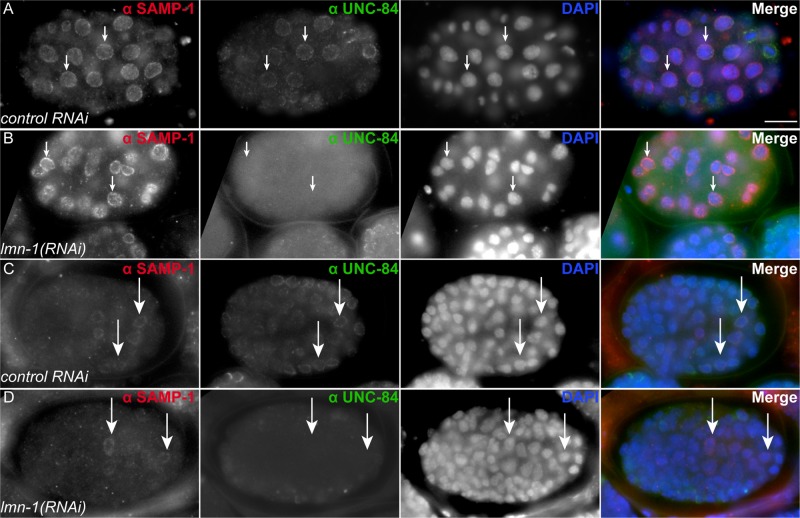
In mammalian tissue culture, Samp1 requires lamin A/C for localization to the nuclear envelope (Borrego-Pinto et al., 2012). It has also been demonstrated that C. elegans UNC-84 requires LMN-1 for nuclear envelope localization (Lee et al., 2002). Surprisingly, SAMP-1 localized to the nuclear envelope in lmn-1(RNAi) embryos (Figure 7). In both early embryos (Figure 7, A and B) and embryos around the time of migration (Figure 7, C and D), SAMP-1 was able to localize in lamin-knockdown animals, whereas UNC-84 was not. LMN-1 staining was used as a control to confirm that the lmn-1(RNAi) knockdown was efficient.
FIGURE 7:
SAMP-1 localizes independently of LMN-1. (A–D) Embryos were stained for SAMP-1 and UNC-84 localization. Lateral views, with anterior left and dorsal up. For each row, SAMP-1 immunostaining is shown in white in the left column and in red on the right when all channels are merged. UNC-84 is shown in white in the second column from the left and in green when merged. DAPI staining of nuclei is shown in white in the third column and in blue when merged. (A) An early embryo fed bacteria containing the empty L4440 vector as control. (B) An early embryo fed lmn-1(RNAi). (C) A later, pre–comma-stage embryo fed bacteria containing the empty L4440 vector as control. (D) A later, pre–comma-stage embryo fed lmn-1(RNAi). Arrows highlight specific nuclei to provide reference points in all four columns. Scale bar, 10 μm.
After showing that SAMP-1 localizes to the nuclear envelope in migrating nuclei, we tested the extent to which SAMP-1 functions to move nuclei. Homozygous samp-1(tm2710) was embryonic lethal. We therefore fed samp-1(tm2710)/+ adults dsRNA against samp-1 for 48–72 h and examined their offspring. Nuclei abnormally located in the dorsal cord were counted in 266 samp-1(tm2710)/+; samp-1(RNAi) L1 larvae. On average, 0.4 ± 0.1 nuclei (mean ± 95% CI) were observed in the dorsal cord (Figure 6, G–I), which is statistically significantly when compared with wild type (p = 0.005 by unpaired t test with Welch's correction). Occasionally, samp-1(RNAi) L1 larvae had up to five nuclei/worm that failed to migrate (Figure 6G). We therefore concluded that samp-1 plays a small but significant role in nuclear migration.
DISCUSSION
The results presented here combine genetic analyses, time-lapse imaging of nuclear migration, and a yeast two-hybrid screen. Together the data provide mechanistic insights into both the molecular interaction between the SUN protein UNC-84 and lamin and the functional implications of disruption of this interaction during nuclear migration. We showed that alleles disrupting the N-terminal nucleoplasmic domain of UNC-84 led to an intermediate nuclear migration defect. We then performed a yeast two-hybrid screen to find candidate interacting partners of the nucleoplasmic domain of UNC-84. Of interest, we identified an interaction between UNC-84 and the C. elegans lamin protein LMN-1. Furthermore, the point mutation UNC-84(P91S) that led to an intermediate nuclear migration phenotype also disrupted the interaction between UNC-84 and LMN-1. As predicted from these data, lmn-1(RNAi) led to a similar nuclear migration defect. Knockdown of another member of the nucleoskeleton, samp-1, led to a weak nuclear migration phenotype. Nuclear migrations in unc-84(P91S) embryos were carefully analyzed by time-lapse imaging to provide insight into the intermediate nuclear migration defect. Some nuclei in the unc-84(P91S) mutant background migrated normally, whereas others failed. Of the failed nuclei, many initiated migration normally before stopping part way through, whereas others failed to move at all. Of interest, we did not see slow-moving nuclei in unc-84(P91S) embryos; the nuclei that did move did so similarly to wild-type nuclei.
Our proposed model is shown in Figure 8. In this model, KASH/SUN bridges serve as molecular bolts through the nuclear envelope to transfer forces generated in the cytoplasm across the nuclear envelope. UNC-83 on the cytoplasmic surface of the nuclear envelope interacts with microtubule motors kinesin-1 and dynein (Meyerzon et al., 2009a; Fridolfsson et al., 2010). The forces generated by the motors are transmitted across the outer nuclear membrane by UNC-83 and then to the SUN protein UNC-84 through an interaction between KASH and SUN domains (Sosa et al., 2012; Tapley and Starr, 2013). Finally, UNC-84 spans the inner nuclear membrane (Tapley et al., 2011) and interacts with lamin (Figure 2) to complete the connection between the cytoskeleton to the nucleoskeleton (Figure 8). However, the amount of force that can be transferred across the nuclear envelope by the KASH/SUN molecular bolt without stabilization by an interaction with the nucleoskeleton is limited. It is useful to think of this complex as analogous to a nut and bolt in drywall, where the interaction of UNC-84 with nucleoskeletal components dissipates forces across a larger area along the inside of the wall, reducing the likelihood of failures under tension. In our model, LMN-1 functions as a key component of the molecular nut and washer to dissipate the forces transmitted across the KASH/SUN bolt to structural elements inside the nucleus. The LMN-1 nut and washer are only as strong as the interaction with the UNC-84 nucleoplasmic domain of the bolt. If the UNC-84– LMN-1 interaction fails, as in the UNC-84(P91S) mutant, the nucleus could continue to migrate for some time without lamin serving as a nut and washer. However, at some critical point the forces are too strong, and the KASH/SUN bolt is catastrophically detached, leading to a failure in nuclear migration. The stability of the bolt depends on many factors. It is therefore difficult to predict when the KASH/SUN bridge will fail, explaining why some UNC-84(P91S) nuclei fail at the initiation of migration, whereas others migrate part way before stalling and many successfully complete migration. Although the LMN-1 nut is required to ensure normal nuclear migration, it does not appear necessary for nuclear anchorage, because unc-84(P91S) nuclei are normally anchored (Figure 1; Malone et al., 1999).
FIGURE 8:
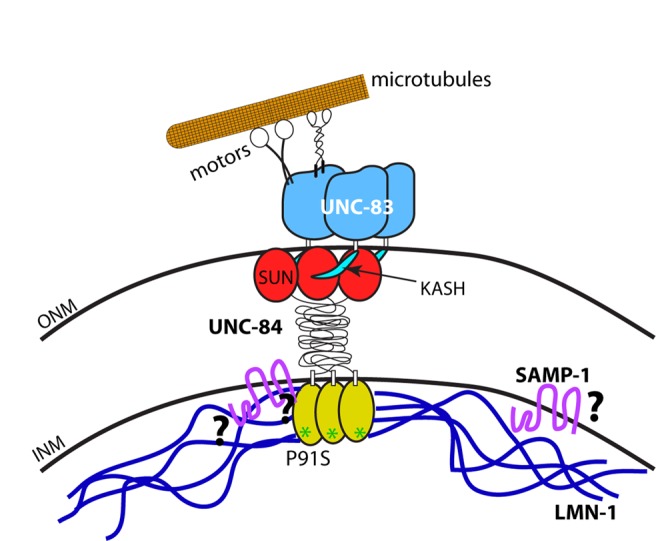
Nut-and-bolt model for nuclear migration. Cartoon of the KASH/SUN nuclear envelope bridge during nuclear migration. UNC-83 is shown in blue, with the KASH peptide in teal. UNC-84 is shown with the SUN domain in red, the domain spanning the perinuclear space in black, and the nucleoplasmic domain in yellow. The green asterisks indicate the P91S mutation in UNC-84. LMN-1 is shown in dark blue and SAMP-1 in fuchsia. Microtubule motors are shown in black and white interacting with a single microtubule in orange. Question marks symbolize open questions regarding protein interactions.
It is difficult to translate our findings from the C. elegans system, in which there is a single lamin, to vertebrate cells, in which there are three to four lamins. However, mutations in lamins in other developmental systems lead to nuclear migration phenotypes consistent with our model. Mice carrying knockout mutations in lamin B2 have nuclear migration defects in the developing CNS similar to defects in SUN or KASH mutant mice (Coffinier et al., 2010b). Similarly, mutations in Drosophila Lam Dmo have nuclear migration defects in the developing eye disk similar to SUN and KASH mutants (Patterson et al., 2004). Furthermore, heterozygous, dominant loss-of-function mutations in Drosophila Lam Dmo were identified as enhancers of phenotypes caused by overexpression of the KASH protein in the developing eye disk, suggesting that the function of SUN-KASH bridges is sensitive to the levels of lamin (Patterson et al. 2004). These nuclear migration defects are consistent with the model that lamin serves an evolutionarily conserved role as a molecular nut and washer during many nuclear migration events in developing tissues. Defects in human lamin B have been associated with disease, although nowhere near to the extent of lamin A mutations (Coffinier et al., 2010b; Worman, 2012). A duplication of LMNB1 is associated with a leukodystrophy, in which myelin is progressively lost (Padiath et al., 2006). Mutations in the LMNB2 gene are linked to a lipodystrophy (Hegele et al., 2006). Given the mouse knockout phenotypes, it is likely that additional mutations in LMNB1 and LMNB2 will be found to be associated with neuronal diseases (Coffinier et al., 2010b).
The nucleoskeleton is a complex network consisting of lamins, inner nuclear membrane proteins, and small proteins that link the nuclear envelope to chromatin (Simon and Wilson, 2011). It is therefore unlikely that lamin B is the only component of the molecular nut and washer that dissipates forces throughout the nucleoskeleton during nuclear migration. Obvious candidates to participate with lamins during nuclear migration include the LEM proteins, Baf, and Ima1/NET5/Samp1. In C. elegans, mutations in baf-1 or double mutations in emr-1 and lem-2 phenocopy all previously described lmn-1 phenotypes (Liu et al., 2003; Margalit et al., 2005; Meyerzon et al., 2009b). We were therefore surprised that disrupting baf-1 or emr-1 and lem-2 did not lead to a nuclear migration phenotype. The inner nuclear membrane protein Ima1/NET5/Samp1 was previously shown to play a role in nuclear migration in mammalian tissue culture cells (Borrego-Pinto et al., 2012). It was also shown to interact with a SUN protein in both yeast and mammalian tissue culture (Hiraoka et al., 2011; Borrego-Pinto et al., 2012). Here we showed that the C. elegans homologue SAMP-1 plays a minor role in nuclear migration. Of interest, we found despite the requirement of lamin A/C in tissue culture (Borrego-Pinto et al., 2012), C. elegans SAMP-1 localized independently of LMN-1. The SAMP-1 binding partners in the C. elegans nuclear envelope remain to be determined (question marks in Figure 8). We suspect that baf-1, emr-1, lem-2, samp-1, and other components of the nucleoskeleton play partially redundant roles with lamin during nuclear migration. Therefore characterization of how the network of proteins forming the nucleoskeleton functions as a unit during nuclear migration requires further investigation.
MATERIALS AND METHODS
C. elegans strains and RNA interference
C. elegans were cultured using standard conditions, and N2 was used as wild type (Brenner, 1974). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis, MN), which is funded by the National Institutes of Health National Center for Research Resources. The unc-84(n322), unc-84(e1174), unc-84(e1411), and unc-84(n369) alleles, from strains MT322, CB1174, CB1411, and MT369, respectively, were previously described (Malone et al., 1999). The construct pSL589 was created by cloning the XmaI/SphI fragment of the col-10 promoter from pOS12 (Spencer et al., 2001) into pPD96.04 to specifically drive expression of NLS::GFP::LacZ in embryonic hypodermal cells. This construct, pSL589, was injected into N2 worms and then integrated (Kage-Nakadai et al., 2012) to create strain UD469 (ycIs10[pcol-10nls::gfp::lacZ]). This outcrossed line was then crossed to unc-84(n322), unc-84(e1174), unc-84(e1411), and unc-84(n369) strains to create UD414, UD412, UD396, and UD399, respectively. UD87, expressing the full-length UNC-84 rescue construct, was used as the starting strain for SAMP-1 localization studies in lmn-1(RNAi) (McGee et al., 2006; Chang et al., 2013. The lem-2(tm1582) deletion strain FX1582 and the samp-1(tm2710) deletion strain FX2710 were kindly supplied by Shohei Mitani (National Bioresource Project at the Tokyo Women's Medical University, Tokyo, Japan). samp-1(tm2710) was balanced with mIn1 [dpy-10(e128) mIs14] II from strain BS3493 to make strain UD470. The plbp-1 lmn-1::gfp strain UD324 was previously described (Fridolfsson and Starr, 2010). UD324 was crossed to the unc-84(n369) and unc-84(e1411) strains to create UD437 and UD436, respectively.
For the lmn-1(RNAi) experiments, ycIs10 animals in an otherwise wild-type background were staged for 24 h post L4 at 20°C and fed bacteria expressing dsRNA against lmn-1 from the Ahringer library (Fraser et al., 2000). Worms were transferred after 24 h, and the plate from the first 24 h was counted at 48 h. For emr-1(RNAi); lem-2(1582) treatment the first 24 h was discarded and the second day was counted at 72 h (Meyerzon et al., 2009b). Empty vector L4440 was used as control RNA interference (RNAi) for all feeding experiments. For baf-1(RNAi), dsRNA was transcribed in vitro from the EST yk333d11 and subsequently injected into young adults (Meyerzon et al., 2009b). Progeny laid from 12 to 24 h postinjection were screened. RNAi efficiency was assessed by progeny inviability.
Yeast two-hybrid assay
Constructs expressing GAL-4 DNA–binding domain::UNC-84 fusion proteins for yeast two-hybrid baits were created by amplifying inserts with PCR from the unc-84 cDNA, yk402g1 (Kohara, 1996; McGee et al., 2006), and cloning the inserts into pDEST32 using Gateway Technology (Invitrogen, Grand Island, NY). pSL242 expresses residues 1–385 of UNC-84, pSL244 has 59–385, pSL593 has 1–100, pSL592 has 1–59, and pSL595 has 385–510. The P91S mutation was introduced into pSL242 using PCR SOEing to create the mutant bait construct pSL596. The ProQuest C. elegans mixed-stage cDNA library (Invitrogen) was screened using the UNC-84(1-385) as a bait as previously described (Fridolfsson et al., 2010). Positives with candidate interacting partners were selected on SD−Trp−Leu−His. To map the LMN-1 interaction domain of UNC-84, full-length LMN-1 prey, pSL719, obtained from the screen, was transformed into yeast strain Y187 (Clontech Laboratories, Mountain View, CA). The various UNC-84 baits were transformed into yeast strain Y2HGold (Clontech Laboratories). The bait strains were then mated to the prey-containing Y187 strains. Spot assays were conducted by spotting 2 μl of yeast serial dilutions; growth was then imaged with an AlphaImager 3400 (Alpha Innotech Corporation, San Leandro, CA). Liquid β-galactosidase assays were conducted following Clontech protocol PT1020-1 (Schneider et al., 1996).
Nuclear migration assays
To count abnormally positioned hyp7 nuclei in the dorsal cord, L1 animals carrying ycIs10[nls::gfp::lacZ] were examined on a 2% agar pad in M9 solution with 1 mM tetramisole (Starr et al., 2001). DIC time-lapse imaging of migrating embryonic hypodermal nuclei was performed as previously reported (Fridolfsson and Starr, 2010). Images were taken every 15 s; time 0 was defined as the time that cell 12 hit the opposite seam cell boundary (Fridolfsson and Starr, 2010). Nuclear position was determined by measuring the leading edge, lagging edge, and midpoint of migrating nuclei using the manual tracking plug-in for ImageJ (National Institutes of Health, Bethesda, MD). The distance a nucleus migrated in the first 10 min determined in a cell-specific manner such that time 0 was when the respective cell's leading edge reached the opposite seam cell boundary. Images were obtained using DIC optics and a 63× Plan Apo 1.40 numerical aperture (NA) objective on a Leica DM6000 compound microscope equipped with a Leica DC350 camera operated by Leica LAS AF software (Leica Microsystems, Wetzlar, Germany). Images were processed using ImageJ.
LMN-1::GFP time-lapse images were captured at a frame rate of 1/s using the 488-nm laser on a spinning disk confocal microscope (Marianas Real Time Confocal SDC Workstation; Intelligent Imaging Innovations, Denver, CO), using a scan head (CSU-X1: Yokogawa, Tokyo, Japan), an autofocusing laser, Definite Focus (Zeiss, Oberkochen, Germany), and a modified electron multiplying charge-coupled device camera (Cascade QuantEM 512SC; Photometrics). A 63×/NA 1.4 oil immersion objective was used (Zeiss). Images were acquired using SlideBook software (version 5.5; Intelligent Imaging Innovations).
Antibodies and immunofluorescence
Polyclonal antibodies (HJA2) against the C. elegans SAMP-1 were raised in rabbits injected with a histidine-NusA–tagged C-terminal fragment of SAMP-1 comprising residues 338–555. SAMP-1 antibodies were affinity purified using the same C-terminal SAMP-1 fragment tagged with GST. For immunofluorescence, embryos were extruded from hermaphrodites, permeabilized by the freeze-crack method, fixed for 10 min in −20˚C methanol, blocked in PBST (phosphate-buffered saline [PBS] and 0.1% Triton X-100) with 5% milk, and stained as previously described (Miller and Shakes, 1995). The rabbit antibody against SAMP-1 was diluted 1:200 in PBS. UNC-83 monoclonal 1209D7D5 was used undiluted (Starr et al. 2001). UNC-84 IgM monoclonal L 72 6 was diluted 1:100 in PBS (Cain et al., 2014). Alexa Fluor antibodies 594 donkey anti-rabbit immunoglobulin G (IgG), goat 488 anti-mouse IgM, and 488 goat anti-mouse IgG diluted 1:500 (Life Technologies, Carlsbad, CA) were used as secondary antibodies. DNA was visualized by a 5-min stain in 1 μg/ml DAPI in PBS.
Supplementary Material
Acknowledgments
We thank Kevin Hart for generating the integrated hypodermal GFP::NLS transgenic strain and Ben Lorton, Allen Wang, and Joe Nguyen for assistance in the two-hybrid screen. We thank Heidi Fridolfsson, Michael Paddy, and the Molecular and Cellular Biology Imaging Facility at the University of California, Davis, for advice and assistance in microscopy. We thank David Fay and Dan Levy (University of Wyoming, Laramie, WY) for helpful discussions and hosting D.A.S. on sabbatical. We thank members of the Starr lab and our colleagues in the Molecular and Cellular Biology Department at the University of California, Davis, for helpful discussions. C.R.B. was supported by National Institutes of Health Training Grant T32 GM007377. This study was supported by Grant R01 GM073874 from the National Institutes of Health/National Institute of General Medical Sciences.
Abbreviations used:
- KASH
Klarsicht, ANC-1, and Syne homology
- LINC
linker of nucleoskeleton and the cytoskeleton
- SUN
Sad1 and UNC-84
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-05-0971) on July 23, 2014.
*Present address: Therapeutic Research Group Oncology, Bayer HealthCare Pharmaceuticals, Bayer Pharma AG, 13353 Berlin, Germany.
REFERENCES
- Altun ZF, Hall DH. Epithelial system, hypodermis. 2009 In: WormAtlas, DOI:10.3908/wormatlas.1.13. [Google Scholar]
- Borrego-Pinto J, Jegou T, Osorio DS, Aurade F, Gorjanacz M, Koch B, Mattaj IW, Gomes ER. Samp1 is a component of TAN lines and is required for nuclear movement. J Cell Sci. 2012;125:1099–1105. doi: 10.1242/jcs.087049. [DOI] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruusgaard JC, Liestol K, Gundersen K. Distribution of myonuclei and microtubules in live muscle fibers of young, middle-aged, and old mice. J Appl Physiol. 2006;100:2024–2030. doi: 10.1152/japplphysiol.00913.2005. [DOI] [PubMed] [Google Scholar]
- Buch C, Lindberg R, Figueroa R, Gudise S, Onischenko E, Hallberg E. An integral protein of the inner nuclear membrane localizes to the mitotic spindle in mammalian cells. J Cell Sci. 2009;122:2100–2107. doi: 10.1242/jcs.047373. [DOI] [PubMed] [Google Scholar]
- Burke B, Roux KJ. Nuclei take a position: managing nuclear location. Dev Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Cain NE, Tapley EC, McDonald KL, BM C, Starr DA. The SUN protein UNC-84 is required only in force-bearing cells to maintain nuclear envelope architecture. J Cell Biol. 2014:206, 163–172. doi: 10.1083/jcb.201405081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YT, Dranow D, Kuhn J, Meyerzon M, Ngo M, Ratner D, Warltier K, Starr DA. toca-1 is in a novel pathway that functions in parallel with a SUN-KASH nuclear envelope bridge to move nuclei in Caenorhabditis elegans. Genetics. 2013;193:187–200. doi: 10.1534/genetics.112.146589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Chi YH, Mutalif RA, Starost MF, Myers TG, Anderson SA, Stewart CL, Jeang KT. Accumulation of the inner nuclear envelope protein SUN is pathogenic in progeric and dystrophic laminopathies. Cell. 2012;149:565–577. doi: 10.1016/j.cell.2012.01.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Wang WP, Chen YC, Wang JY, Lin WH, Tai LA, Liou GG, Yang CS, Chi YH. Dysregulated interactions between lamin A and SUN1 induce abnormalities in the nuclear envelope and endoplasmic reticulum in progeric laminopathies. J Cell Sci. 2014;127:1792–1804. doi: 10.1242/jcs.139683. [DOI] [PubMed] [Google Scholar]
- Chow KH, Factor RE, Ullman KS. The nuclear envelope environment and its cancer connections. Nat Rev Cancer. 2012;12:196–209. doi: 10.1038/nrc3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Chang SY, Nobumori C, Tu Y, Farber EA, Toth JI, Fong LG, Young SG. Abnormal development of the cerebral cortex and cerebellum in the setting of lamin B2 deficiency. Proc Natl Acad Sci USA. 2010a;107:5076–5081. doi: 10.1073/pnas.0908790107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Fong LG, Young SG. LINCing lamin B2 to neuronal migration: growing evidence for cell-specific roles of B-type lamins. Nucleus. 2010b;1:407–411. doi: 10.4161/nucl.1.5.12830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffinier C, Jung HJ, Nobumori C, Chang S, Tu Y, Barnes RH, 2nd, Yoshinaga Y, Jong PJ de, Vergnes L, Reue K, et al. Deficiencies in lamin B1 and lamin B2 cause neurodevelopmental defects and distinct nuclear shape abnormalities in neurons. Mol Biol Cell. 2011;22:4683–4693. doi: 10.1091/mbc.E11-06-0504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp M, Liu Q, Roux K, Rattner JB, Shanahan C, Burke B, Stahl PD, Hodzic D. Coupling of the nucleus and cytoplasm: role of the LINC complex. J Cell Biol. 2006;172:41–53. doi: 10.1083/jcb.200509124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer TA, Misteli T. The lamin protein family. Genome Biol. 2011;12:222. doi: 10.1186/gb-2011-12-5-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folker ES, Ostlund C, Luxton GW, Worman HJ, Gundersen GG. Lamin A variants that cause striated muscle disease are defective in anchoring transmembrane actin-associated nuclear lines for nuclear movement. Proc Natl Acad Sci USA. 2011;108:131–136. doi: 10.1073/pnas.1000824108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke WW, Scheer U, Krohne G, Jarasch ED. The nuclear envelope and the architecture of the nuclear periphery. J Cell Biol. 1981;91:39s–50s. doi: 10.1083/jcb.91.3.39s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fridkin A, Mills E, Margalit A, Neufeld E, Lee KK, Feinstein N, Cohen M, Wilson KL, Gruenbaum Y. Matefin, a Caenorhabditis elegans germ line-specific SUN-domain nuclear membrane protein, is essential for early embryonic and germ cell development. Proc Natl Acad Sci USA. 2004;101:6987–6992. doi: 10.1073/pnas.0307880101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Ly N, Meyerzon M, Starr DA. UNC-83 coordinates kinesin-1 and dynein activities at the nuclear envelope during nuclear migration. Dev Biol. 2010;338:237–250. doi: 10.1016/j.ydbio.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fridolfsson HN, Starr DA. Kinesin-1 and dynein at the nuclear envelope mediate the bidirectional migrations of nuclei. J Cell Biol. 2010;191:115–128. doi: 10.1083/jcb.201004118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Chin LK, Bourouina T, Liu AQ, VanDongen AM. Nuclear deformation during breast cancer cell transmigration. Lab Chip. 2012;12:3774–3778. doi: 10.1039/c2lc40477j. [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y, Margalit A, Goldman RD, Shumaker DK, Wilson KL. The nuclear lamina comes of age. Nat Rev Mol Cell Biol. 2005;6:21–31. doi: 10.1038/nrm1550. [DOI] [PubMed] [Google Scholar]
- Gundersen GG, Worman HJ. Nuclear positioning. Cell. 2013;152:1376–1389. doi: 10.1016/j.cell.2013.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus KC, Ge H, Schetter AJ, Goldberg DS, Han JD, Hao T, Berriz GF, Bertin N, Huang J, Chuang LS, et al. Predictive models of molecular machines involved in Caenorhabditis elegans early embryogenesis. Nature. 2005;436:861–865. doi: 10.1038/nature03876. [DOI] [PubMed] [Google Scholar]
- Haque F, Lloyd DJ, Smallwood DT, Dent CL, Shanahan CM, Fry AM, Trembath RC, Shackleton S. SUN1 interacts with nuclear lamin A and cytoplasmic nesprins to provide a physical connection between the nuclear lamina and the cytoskeleton. Mol Cell Biol. 2006;26:3738–3751. doi: 10.1128/MCB.26.10.3738-3751.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haque F, Mazzeo D, Patel JT, Smallwood DT, Ellis JA, Shanahan CM, Shackleton S. Mammalian SUN protein interaction networks at the inner nuclear membrane and their role in laminopathy disease processes. J Biol Chem. 2010;285:3487–3498. doi: 10.1074/jbc.M109.071910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegele RA, Cao H, Liu DM, Costain GA, Charlton-Menys V, Rodger NW, Durrington PN. Sequencing of the reannotated LMNB2 gene reveals novel mutations in patients with acquired partial lipodystrophy. Am J Hum Genet. 2006;79:383–389. doi: 10.1086/505885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Maekawa H, Asakawa H, Chikashige Y, Kojidani T, Osakada H, Matsuda A, Haraguchi T. Inner nuclear membrane protein Ima1 is dispensable for intranuclear positioning of centromeres. Genes Cells. 2011;16:1000–1011. doi: 10.1111/j.1365-2443.2011.01544.x. [DOI] [PubMed] [Google Scholar]
- Kage-Nakadai E, Kobuna H, Funatsu O, Otori M, Gengyo-Ando K, Yoshina S, Hori S, Mitani S. Single/low-copy integration of transgenes in Caenorhabditis elegans using an ultraviolet trimethylpsoralen method. BMC Biotechnol. 2012;12:1. doi: 10.1186/1472-6750-12-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y. Large scale analysis of C. elegans cDNA [in Japanese] Tanpakushitsu Kakusan Koso. 1996;41:715–720. [PubMed] [Google Scholar]
- Kracklauer MP, Banks SM, Xie X, Wu Y, Fischer JA. Drosophila klaroid encodes a SUN domain protein required for Klarsicht localization to the nuclear envelope and nuclear migration in the eye. Fly. 2007;1:75–85. doi: 10.4161/fly.4254. [DOI] [PubMed] [Google Scholar]
- Lee KK, Starr D, Cohen M, Liu J, Han M, Wilson KL, Gruenbaum Y. Lamin-dependent localization of UNC-84, a protein required for nuclear migration in Caenorhabditis elegans. Mol Biol Cell. 2002;13:892–901. doi: 10.1091/mbc.01-06-0294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Ben-Shahar T Rolef, Riemer D, Treinin M, Spann P, Weber K, Fire A, Gruenbaum Y. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol Biol Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lee KK, Segura-Totten M, Neufeld E, Wilson KL, Gruenbaum Y. MAN1 and emerin have overlapping function(s) essential for chromosome segregation and cell division in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2003;100:4598–4603. doi: 10.1073/pnas.0730821100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GG, Starr DA. KASHing up with the nucleus: novel functional roles of KASH proteins at the cytoplasmic surface of the nucleus. Curr Opin Cell Biol. 2014;28C:69–75. doi: 10.1016/j.ceb.2014.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luxton GW, Gomes ER, Folker ES, Vintinner E, Gundersen GG. Linear arrays of nuclear envelope proteins harness retrograde actin flow for nuclear movement. Science. 2010;329:956–959. doi: 10.1126/science.1189072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malone CJ, Fixsen WD, Horvitz HR, Han M. UNC-84 localizes to the nuclear envelope and is required for nuclear migration and anchoring during C. elegans development. Development. 1999;126:3171–3181. doi: 10.1242/dev.126.14.3171. [DOI] [PubMed] [Google Scholar]
- Margalit A, Segura-Totten M, Gruenbaum Y, Wilson KL. Barrier-to-autointegration factor is required to segregate and enclose chromosomes within the nuclear envelope and assemble the nuclear lamina. Proc Natl Acad Sci USA. 2005;102:3290–3295. doi: 10.1073/pnas.0408364102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee MD, Rillo R, Anderson AS, Starr DA. UNC-83 IS a KASH protein required for nuclear migration and is recruited to the outer nuclear membrane by a physical interaction with the SUN protein UNC-84. Mol Biol Cell. 2006;17:1790–1801. doi: 10.1091/mbc.E05-09-0894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Fridolfsson HN, Ly N, McNally FJ, Starr DA. UNC-83 is a nuclear-specific cargo adaptor for kinesin-1-mediated nuclear migration. Development. 2009a;136:2725–2733. doi: 10.1242/dev.038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerzon M, Gao Z, Liu J, Wu JC, Malone CJ, Starr DA. Centrosome attachment to the C. elegans male pronucleus is dependent on the surface area of the nuclear envelope. Dev Biol. 2009b;327:433–446. doi: 10.1016/j.ydbio.2008.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller DM, Shakes DC. Immunofluorescence microscopy. Methods Cell Biol. 1995;48:365–394. [PubMed] [Google Scholar]
- Minn IL, Rolls MM, Hanna-Rose W, Malone CJ. SUN-1 and ZYG-12, mediators of centrosome-nucleus attachment, are a functional SUN/KASH pair in Caenorhabditis elegans. Mol Biol Cell. 2009;20:4586–4595. doi: 10.1091/mbc.E08-10-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris NR. Nuclear migration. From fungi to the mammalian brain. J Cell Biol. 2000;148:1097–1101. doi: 10.1083/jcb.148.6.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostlund C, Folker ES, Choi JC, Gomes ER, Gundersen GG, Worman HJ. Dynamics and molecular interactions of linker of nucleoskeleton and cytoskeleton (LINC) complex proteins. J Cell Sci. 2009;122:4099–4108. doi: 10.1242/jcs.057075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padiath QS, Saigoh K, Schiffmann R, Asahara H, Yamada T, Koeppen A, Hogan K, Ptacek LJ, Fu YH. Lamin B1 duplications cause autosomal dominant leukodystrophy. Nat Genet. 2006;38:1114–1123. doi: 10.1038/ng1872. [DOI] [PubMed] [Google Scholar]
- Patterson K, Molofsky AB, Robinson C, Acosta S, Cater C, Fischer JA. The functions of Klarsicht and nuclear lamin in developmentally regulated nuclear migrations of photoreceptor cells in the Drosophila eye. Mol Biol Cell. 2004;15:600–610. doi: 10.1091/mbc.E03-06-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinsch S, Gonczy P. Mechanisms of nuclear positioning. J Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Schirmer EC, Florens L, Guan T, Yates JR, 3rd, Gerace L. Nuclear membrane proteins with potential disease links found by subtractive proteomics. Science. 2003;301:1380–1382. doi: 10.1126/science.1088176. [DOI] [PubMed] [Google Scholar]
- Schneider S, Buchert M, Hovens CM. An in vitro assay of beta-galactosidase from yeast. BioTechniques. 1996;20:960–962. doi: 10.2144/96206bm03. [DOI] [PubMed] [Google Scholar]
- Simon DN, Wilson KL. The nucleoskeleton as a genome-associated dynamic ‘network of networks’. Nat Rev Mol Cell Biol. 2011;12:695–708. doi: 10.1038/nrm3207. [DOI] [PubMed] [Google Scholar]
- Sosa BA, Rothballer A, Kutay U, Schwartz TU. LINC complexes form by binding of three KASH peptides to domain interfaces of trimeric SUN proteins. Cell. 2012;149:1035–1047. doi: 10.1016/j.cell.2012.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer AG, Orita S, Malone CJ, Han M. A RHO GTPase-mediated pathway is required during P cell migration in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2001;98:13132–13137. doi: 10.1073/pnas.241504098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA. Watching nuclei move: insights into how kinesin-1 and dynein function together. Bioarchitecture. 2011;1:9–13. doi: 10.4161/bioa.1.1.14629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA. Laminopathies: too much SUN is a bad thing. Curr Biol. 2012;22:R678–R680. doi: 10.1016/j.cub.2012.06.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Fridolfsson HN. Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu Rev Cell Dev Biol. 2010;26:421–444. doi: 10.1146/annurev-cellbio-100109-104037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr DA, Han M. Role of ANC-1 in tethering nuclei to the actin cytoskeleton. Science. 2002;298:406–409. doi: 10.1126/science.1075119. [DOI] [PubMed] [Google Scholar]
- Starr DA, Han M. A genetic approach to study the role of nuclear envelope components in nuclear positioning. Novartis Found Symp. 2005;264:208–219. discussion, 219–230. [PubMed] [Google Scholar]
- Starr DA, Hermann GJ, Malone CJ, Fixsen W, Priess JR, Horvitz HR, Han M. unc-83 encodes a novel component of the nuclear envelope and is essential for proper nuclear migration. Development. 2001;128:5039–5050. doi: 10.1242/dev.128.24.5039. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Horvitz HR. Abnormal cell lineages in mutants of the nematode Caenorhabditis elegans. Dev Biol. 1981;82:41–55. doi: 10.1016/0012-1606(81)90427-9. [DOI] [PubMed] [Google Scholar]
- Sulston JE, Schierenberg E, White JG, Thomson JN. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev Biol. 1983;100:64–119. doi: 10.1016/0012-1606(83)90201-4. [DOI] [PubMed] [Google Scholar]
- Tapley EC, Ly N, Starr DA. Multiple mechanisms actively target the SUN protein UNC-84 to the inner nuclear membrane. Mol Biol Cell. 2011;22:1739–1752. doi: 10.1091/mbc.E10-08-0733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapley EC, Starr DA. Connecting the nucleus to the cytoskeleton by SUN-KASH bridges across the nuclear envelope. Curr Opin Cell Biol. 2013;25:57–62. doi: 10.1016/j.ceb.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai JW, Bremner KH, Vallee RB. Dual subcellular roles for LIS1 and dynein in radial neuronal migration in live brain tissue. Nat Neurosci. 2007;10:970–979. doi: 10.1038/nn1934. [DOI] [PubMed] [Google Scholar]
- Turgay Y, Ungricht R, Rothballer A, Kiss A, Csucs G, Horvath P, Kutay U. A classical NLS and the SUN domain contribute to the targeting of SUN2 to the inner nuclear membrane. EMBO J. 2010;29:2262–2275. doi: 10.1038/emboj.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Zhao Y, Wong K, Ehlers P, Kohara Y, Jones SJ, Marra MA, Holt RA, Moerman DG, Hansen D. Identification of genes expressed in the hermaphrodite germ line of C. elegans using SAGE. BMC Genomics. 2009;10:213. doi: 10.1186/1471-2164-10-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worman HJ. Nuclear lamins and laminopathies. J Pathol. 2012;226:316–325. doi: 10.1002/path.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Han M. Nuclear migration: rock and roll facilitated by dynein and kinesin. Curr Biol. 2010;20:R1027–1029. doi: 10.1016/j.cub.2010.10.047. [DOI] [PubMed] [Google Scholar]
- Zhang X, Lei K, Yuan X, Wu X, Zhuang Y, Xu T, Xu R, Han M. SUN1/2 and Syne/Nesprin-1/2 complexes connect centrosome to the nucleus during neurogenesis and neuronal migration in mice. Neuron. 2009;64:173–187. doi: 10.1016/j.neuron.2009.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou K, Hanna-Rose W. Movers and shakers or anchored: Caenorhabditis elegans nuclei achieve it with KASH/SUN. Dev Dyn. 2010;239:1352–1364. doi: 10.1002/dvdy.22226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.