Chromatin assembly factor-1 contains a separable domain unrelated to histone deposition, which provides a previously unrecognized ability to maintain nucleolar protein and chromosome associations.
Abstract
Chromatin assembly factor-1 (CAF-1) is a three-subunit protein complex conserved throughout eukaryotes that deposits histones during DNA synthesis. Here we present a novel role for the human p150 subunit in regulating nucleolar macromolecular interactions. Acute depletion of p150 causes redistribution of multiple nucleolar proteins and reduces nucleolar association with several repetitive element–containing loci. Of note, a point mutation in a SUMO-interacting motif (SIM) within p150 abolishes nucleolar associations, whereas PCNA or HP1 interaction sites within p150 are not required for these interactions. In addition, acute depletion of SUMO-2 or the SUMO E2 ligase Ubc9 reduces α-satellite DNA association with nucleoli. The nucleolar functions of p150 are separable from its interactions with the other subunits of the CAF-1 complex because an N-terminal fragment of p150 (p150N) that cannot interact with other CAF-1 subunits is sufficient for maintaining nucleolar chromosome and protein associations. Therefore these data define novel functions for a separable domain of the p150 protein, regulating protein and DNA interactions at the nucleolus.
INTRODUCTION
In eukaryotes, histones are deposited onto DNA by nucleosome assembly proteins, including chromatin assembly factor-1 (CAF-1; reviewed in Ransom et al., 2010). CAF-1 is a three-subunit protein complex conserved throughout eukaryotes. In humans, the three CAF-1 subunits are named based on their gel migration (Smith and Stillman, 1989). The largest subunit, p150, is the platform that binds the other two subunits, the WD-40 repeat proteins p60 and p48; all three subunits are required for in vitro nucleosome assembly activity (Kaufman et al., 1995; Verreault et al., 1996). CAF-1 deposits histones during DNA synthesis and interacts with DNA replication protein proliferating cell nuclear antigen (PCNA) and the replication-linked histone isoform H3.1 (Shibahara and Stillman, 1999; Tagami et al., 2004). Throughout the eukaryotes, CAF-1 is required for normal S-phase progression, heterochromatin formation (Houlard et al., 2006; Takami et al., 2007; Quivy et al., 2008; Klapholz et al., 2009), and chromatin restoration after DNA repair (Gaillard et al., 1996; Green and Almouzni, 2003; Polo et al., 2006). Consistent with its role in chromosome duplication, CAF-1 protein levels correlate with cell proliferation and cancer prognosis (Polo et al., 2004; Staibano et al., 2009; Mascolo et al., 2010).
In all eukaryotes, a dedicated polymerase, RNA polymerase I (Pol I), transcribes the large rRNAs (18S and 28S in humans) as long precursor species (47S in humans) from repeated ribosomal DNA (rDNA) templates. The 47S rRNA–encoding repeats cluster to form the nucleolus, a specialized, non–membrane-bound subnuclear compartment (McStay and Grummt, 2008). Pol I transcription constitutes the majority of RNA synthesis in cells and is regulated by cell growth and energy supply, differentiation, cell cycle progression, tumor suppressors (p53 and Rb), and oncoproteins (c-myc; McStay and Grummt, 2008; Murayama et al., 2008). In addition, the nucleolus is a dynamic hub where many nuclear proteins (Emmott and Hiscox, 2009) and genomic loci (Nemeth et al., 2010; van Koningsbruggen et al., 2010) are tethered (Padeken and Heun, 2014). Of note, nucleolar alterations can be important for cancer diagnoses (Maggi and Weber, 2005). However, our understanding of the structure and function of nucleolar macromolecular networks remains incomplete. Here mass spectrometric analysis of the human CAF-1 p150 subunit led us to discover previously unrecognized roles in regulating macromolecular interactions at the nucleolus.
RESULTS
p150 interacts with nucleolar proteins
To find novel protein interaction partners, we generated a human HeLa S3 cell line expressing an N-terminal tandem affinity–tagged CAF-1 p150 subunit (NTAP-p150). NTAP-p150 localizes normally to PCNA-labeled DNA replication foci during S phase (Campeau et al., 2009), indicating functionality in vivo. Nuclear extracts from asynchronous HeLa S3 cells either containing or lacking the NTAP-tagged p150 gene were subjected to affinity purification under mild (200 mM NaCl) buffer conditions that included 50 μg/ml ethidium bromide to avoid coprecipitation of proteins via bridged nucleic acid interactions (Lai and Herr, 1992). Gel analysis of the purified proteins demonstrated that NTAP-p150 complexed with the other two CAF-1 subunits, p60 and p48 (Figure 1B), and that this tagged complex was recovered in a tag-dependent manner. Proteins from two independent preparations were identified by mass spectrometry (Figure 1A and Supplemental Table S1). In the tagged but not the untagged samples, we detected all three CAF-1 subunits, as well as known CAF-1–binding proteins such as core histones, histone deposition factor Asf1 (Sharp et al., 2001; Tyler et al., 2001; Tang et al., 2006), and heterochromatin protein 1 (HP1; Murzina et al., 1999; Quivy et al., 2004). In addition, the tagged samples contained multiple nucleolar proteins that were not previously known to associate with p150. These included Ki67, a large, proteolytically sensitive protein that is commonly used to mark proliferative cells in pathology studies (Yerushalmi et al., 2010). Of note, Ki67 prominently localizes to nucleoli (Figure 1C; MacCallum and Hall, 2000; Takagi et al., 2001; Espada et al., 2007). This interaction is functionally important because inactivation of Ki67 inhibits rRNA synthesis (Rahmanzadeh et al., 2007). The p150-associated proteins also included nucleophosmin (NPM; also known as B23) and nucleolin (NCL), which are both nucleolar histone chaperones that assist Pol I transcription (Angelov et al., 2006; Rickards et al., 2007; Murano et al., 2008). Our affinity-purified NTAP-p150 preparations additionally included nucleolar protein 58 (Nop5/Nop58), a small nucleolar RNA (snoRNA)–binding protein whose nucleolar localization is regulated by SUMOylation (Westman et al., 2010); DDX17, a DEAD-box protein that contributes to rRNA processing and nucleolar protein localization (Jalal et al., 2007); and RRP1B, a nucleolar targeting protein implicated in metastasis susceptibility (Crawford et al., 2007; Chamousset et al., 2010). In addition, we detected DNAJC9, a DnaJ homologue that was previously isolated as a binding partner of RRP1B, as were the nucleolar proteins NCL, NPM, and Ki67 (Crawford et al., 2009) that we identified here (Supplemental Table S1). Finally, we note that p150 was identified in an independent proteomic analysis of purified nucleoli (Ahmad et al., 2009), supporting our findings.
FIGURE 1:
Nucleolar associations of human p150. (A) Novel nucleolus-related p150-interacting proteins identified by mass spectrometry. Proteins listed were not previously known to interact with p150 and had multiple peptides detected in two different experiments (E1 and E2). Full lists are in Supplemental Table S1. Number of peptides (#Pep) and spectral counts (SpC) are indicated. (B) Purified p150-associated proteins prepared for mass spectrometry. Affinity-purified samples from untagged HeLa S3-Trex (lane 1) and HeLa S3-Trex-NTAP-p150 (lane 2) cells were analyzed on a silver-stained 5–20% SDS–PAGE gel. CAF-1 subunits p150, p60, and p48 are indicated. (C) Confocal microscopy analysis of p150 (red) and Ki67 (green) in HeLa S3 cells detects partial colocalization at nucleoli in the merged image (yellow). Fields of cells were photographed with a 63× objective. Cells had either been blocked in G1 phase by double-thymidine treatment or released into S phase for 2 h. Scale bars, 10 μm. (D) Location of ChIP primer pairs on the human 47S rRNA–encoding rDNA locus. (E) p150 occupancy at rDNA genes in asynchronous HeLa S3-Trex-shRNA cells. Also assayed were the Pol II gene RPS20 and a gene desert from chromosome 16. Cells expressing either a control shRNA (sh-Luc) or the shRNA targeting p150 (sh-p150) were compared. Chromatin was precipitated with either nonimmune rabbit serum (IgG) or anti-p150 serum (α-p150) as indicated. The mean percentages of input chromatin precipitated from three biological replicates are presented, with error bars representing SDs. Asterisks indicate loci at which sh-p150 caused a statistically significant decrease in p150 occupancy (p < 0.05). Dotted line shows p150 signal at the gene desert. (F) p150 occupancy in asynchronous HeLa cells compared with cells blocked with double-thymidine treatment. Asterisks indicate loci at which thymidine arrest caused a statistically significant increase in p150 occupancy (p < 0.05).
p150 interacts with nucleolar DNA
On the basis of the abundance of nucleolar proteins detected, we assessed whether p150 resides in the nucleolus by performing immunolocalization studies. Because CAF-1 localization is regulated by cell cycle progression and is prominently redistributed to DNA replication foci during S phase of the cell cycle (Krude, 1995), we examined synchronized populations, comparing cells blocked at the G1/S-phase border via a double-thymidine block (Whitfield et al., 2002) and those released into S phase. We used Ki67 as a marker for nucleoli, as described previously (Figure 1C; MacCallum and Hall, 2000). In these confocal microscopy images, p150 was detected throughout the nucleoplasm, and in the G1/S-arrested cells a subset of p150 was colocalized with Ki67 at the surface of the nucleoli (Figure 1C). More p150 was observed inside nucleoli in the cells released into mid S phase. We conclude that a subset of cellular p150 colocalized with the nucleolar protein Ki67, even when DNA replication is inhibited.
To assess functional roles of p150, we generated human HeLa S3 cells with a doxycycline-inducible short hairpin RNA (shRNA) targeting p150 (p150shRNA-1). A time course after shRNA induction was analyzed by immunoblotting, confirming p150 depletion (Supplemental Figure S1). On p150 depletion, we observed partial codepletion of CAF-1 p60, as expected from other studies (Hoek and Stillman, 2003; Ye et al., 2003). In contrast, nucleolar protein levels appeared unaltered during this time course, with the exception of a mild increase in UBF1 levels at late time points (Supplemental Figure S1A). Over the first 72 h, there was a mild increase in the doubling time of the p150-depleted cells (from 26 to 33 h) but no dramatic alteration of the cell cycle profile until 96 h, when S/G2-phase cells accumulated (Supplemental Figure S1, B and C). Our acute depletion experiments were therefore limited to the first 72 h after shRNA inductions to minimize the mild growth differences. We also detected no DNA damage checkpoint–mediated phosphorylations (Ciccia and Elledge, 2010) or apoptosis-related PARP cleavage (Duriez and Shah, 1997) during acute depletion of p150 (Supplemental Figure S1D). These data are consistent with previous studies in which depletion of p150 via RNA interference (RNAi) did not activate DNA damage checkpoints (Hoek and Stillman, 2003; Quivy et al., 2008). Similarly, no PARP cleavage was observed in experiments using an alternative depletion reagent, in vitro diced “endoribonuclease-prepared short interfering RNA (esiRNA)” molecules targeting the 3′ untranslated region of the p150 mRNA (Supplemental Figure S1E).
We then tested whether p150 is closely associated with 47S rRNA–encoding chromatin via chromatin immunoprecipitation (ChIP) assays (Figure 1, D–F), comparing cells expressing a control shRNA targeting luciferase with those expressing p150shRNA-1. In control experiments with preimmune sera (immunoglobulin G [IgG]), no specific enrichments were observed in either cell line. In contrast, using anti-p150 polyclonal sera, we detected enrichments at the rDNA enhancer, at multiple sites within the Pol I transcription unit, and within the rDNA intergenic spacer (IGS; Figure 1E). In addition, we examined a gene desert region, which displayed a low level of p150 enrichment. Unlike the other loci tested, the levels of p150 at the gene desert were not significantly reduced upon p150 depletion, suggesting that this represents experimental background. Further, because p150 contributes to genome-wide nucleosome deposition during DNA synthesis, we did not expect rDNA to be the only site of enrichment. Indeed, an RNA polymerase II–transcribed gene, RPS20, also displayed p150 enrichment, as did other Pol II genes (unpublished observations). We conclude that the 47S rRNA–encoding repeats are among the many sites of p150 localization, consistent with the confocal microscopy data (Figure 1C) showing a subset of p150 localized to nucleoli.
The foregoing experiments were performed with populations of asynchronous cells, so that the observed signals could include contributions from CAF-1 complexes associated with loci transiently during genome duplication in S phase of the cell cycle. Our confocal microscopy images suggested that p150’s association with nucleoli would not be limited to S phase (Figure 1C). To explore this via ChIP, we arrested cells at the G1/S phase border via double-thymidine treatment. In the arrested cells, p150 was still associated with 47S rRNA–encoding repeats (Figure 1F), but punctate PCNA foci characteristic of mid and late S phase were absent (Supplemental Figure S1G). Indeed, at each of the locations within the rDNA analyzed, but not at the RPS20 gene, p150 occupancy was significantly increased in the thymidine-arrested cells (Figure 1F). We conclude that p150 is associated with 47S rRNA–encoding repeats and that these associations are not dependent on ongoing DNA replication.
p150 regulates nucleolar protein localization
One of the nucleolar proteins identified in our mass spectrometry data is NPM (also known as B23, encoded by the NPM1 gene; Figure 1A), which is a nucleocytoplasmic shuttling protein important for the localization of multiple proteins to the nucleolus (Korgaonkar et al., 2005; Li and Hann, 2013). We therefore tested whether p150 depletion also affected the localization of nucleolar proteins. We began this analysis with HeLa-derived cells and observed that p150 depletion indeed altered the steady-state localization of multiple nucleolar interaction partners we identified by mass spectrometry (Figure 2, A and B: NPM/B23, Ki67, and nucleolar phosphoprotein Nopp140 (also known as Nopp130; encoded by the NOLC1 gene; Isaac et al., 1998). In addition, the Pol I transcription factor UBF1 (Bell et al., 1988), the Pol I transcription termination/activation protein TTF1 (Langst et al., 1997; Németh et al., 2008), and NCL (also known as C23), which binds rDNA and stimulates Pol I transcription (Rickards et al., 2007), were also redistributed upon p150 depletion. In contrast, the localization of fibrillarin, an rRNA 2′-O-methyltransferase, and histone H2A glutamine methyltransferase, involved in RNA polymerase I transcription and pre-rRNA processing (Reichow et al., 2007; Tessarz et al., 2014), remained unchanged by p150 depletion, indicating that nucleolar structure was not completely disrupted (Figure 2C). Consistent with the unaltered distribution of fibrillarin, the overall appearance of the 47S rRNA–encoding repeats is not altered upon p150 depletion (see later discussion of Figures 6 and 7). In each of the cases we tested (e.g., UBF1, NCL/C23, NPM/B23, Ki67), none of the relocalized proteins displayed reduced total protein levels (Supplemental Figure S1). Protein relocalizations were not limited to HeLa S3 cells; similar results were observed in HT1080 cells (Supplemental Figure S2).
FIGURE 2:
p150 depletion disrupts localization of multiple nucleolar proteins. (A) HeLa S3-Trex-p150-shRNA1 (sh-p150) or luciferase-shRNA cells (sh-Luc) were treated with doxycycline for 72 h to induce shRNA expression and then prepared for indirect immunofluorescence, staining up to two nucleolar proteins simultaneously in each experiment as indicated. Fields of cells photographed with a 63× objective. Scale bar, 20 μm. (B) Representative individual cells from A were enlarged. Scale bar, 10 μm. (C) As in A and B, showing the localization of fibrillarin, which remains unchanged upon p150 depletion. (D) ChIP analysis of TTF-1 in HeLa S3 cells expressing the indicated shRNAs. Chromatin was precipitated with either nonimmune rabbit serum (IgG) or anti–TTF-1 antibodies as indicated and occupancy at the rDNA terminator R1 and the glyceraldehyde-3-phosphate dehydrogenase gene was measured. The mean amounts of precipitated chromatin from three biological replicates are presented as percentages of input. Error bars represent SDs. *p < 0.01 comparing TTF-1 occupancy at R1 in the sh-Luc and sh-p150 samples; p ≥ 0.25 for all other pairs shown.
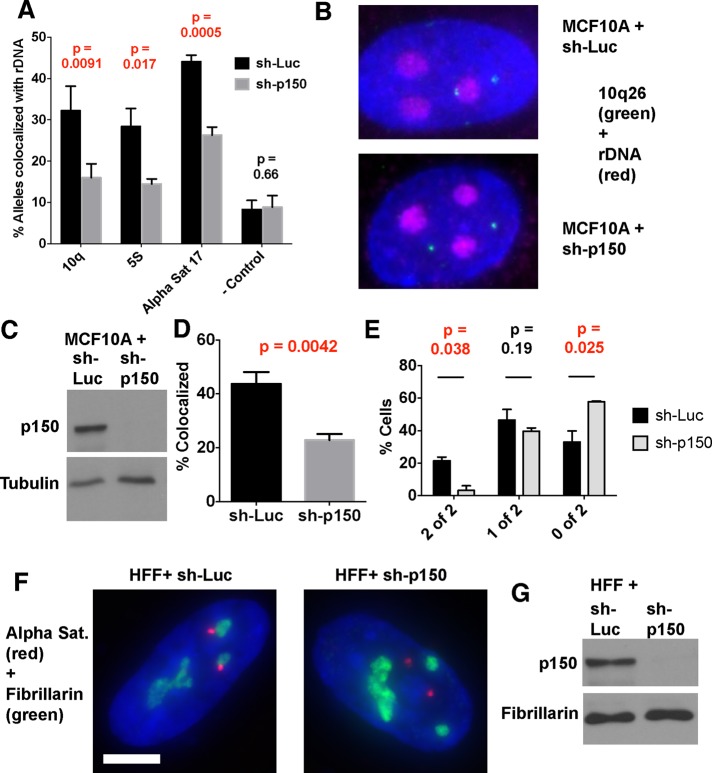
FIGURE 6:
Higher-order interactions of rDNA chromatin are altered upon p150 depletion. (A) DNA FISH analysis of MCF10A-Tet-KRAB cells expressing the indicated shRNAs. Three biological replicate experiments were performed, with mean percentage nucleolar association and SDs graphed. Probes analyzed were a chromosome 10q BAC (10q, total alleles assayed [n] = 332 for sh-Luc, 262 for sh-p150), a 5S rDNA–containing BAC (5S, n = 312 for sh-Luc, 352 for sh-p150), α-satellite DNA from chromosome 17 (αSat 17, n = 288 for sh-Luc, 306 for sh-p150), and negative control BAC (− Control, n = 314 for sh-Luc, 306 for sh-p150) that was previously reported to be unassociated with nucleoli (Nemeth et al., 2010). The p values comparing the sh-Luc and sh-p150 samples for each probe are indicated, with p < 0.05 indicated in red, demonstrating statistical significance. (B) Fluorescence microscopy images of representative cells from an experiment from A. rDNA is colored red, the 10q BAC is green, and DAPI is blue. Scale bar, 5 μm. (C) Immunoblot of representative whole-cell extracts from an experiment in A, with tubulin as loading control. (D) Immuno-FISH analysis of primary human foreskin fibroblasts treated either with luciferase control or p150-targeted sh-RNAs. Cells were treated with anti-fibrillarin antibodies, followed by hybridization with a αSat 17 probe. total alleles assayed = 324 for sh-Luc, 308 for sh-p150. (E) Data from D, plotting the percentage of cells displaying the indicated numbers of αSat 17 alleles associated with fibrillarin. The total cells assayed = 162 for sh-Luc, 154 for sh-p150. (F) Fluorescence microscopy images of representative cells from an experiment from D. Fibrillarin is in green, and the αSat 17 probe is red. Scale bar, 5 μm. (G) Immunoblot of representative whole-cell extracts from an experiment in D, with fibrillarin as loading control.
FIGURE 7:
The p150 N-terminus is necessary and sufficient for nucleolar interchromosomal associations. (A) DNA FISH analysis of the association between αSat 17 and rDNA in HeLa cells expressing the indicated V5-tagged transgenes (luciferase, p150N [aa 1–310], or p150C [aa 245–938]). The percentage of αSat 17 alleles colocalized with the rDNA is indicated, with mean and SDs from three experiments. For V5-luciferase cells, total alleles assayed = 471 for sh-Luc and 480 for sh-p150; for V5-p150N cells, 525 for sh-Luc and 489 for sh-p150; and or V5-p150C cells, 450 for sh-Luc and 468 for sh-p150. p values comparing the sh-Luc and sh-p150 samples are indicated, demonstrating statistically significant rescue by the p150N but not the p150C transgene. (B–D) Data from the experiment in A replotted to display the number of αSat 17 alleles per cell associated with the rDNA. Note that these HeLa-derived lines have three copies of chromosome 17. For V5-luciferase cells, total cells examined = 157 for sh-Luc and 160 for sh-p150; for V5-p150N cells, 175 for sh-Luc and 163 for sh-p150; and for V5-p150C cells, 150 for sh-Luc and 156 for sh-p150. Comparisons for which p ≤ 0.05 are indicated by asterisks, and a number sign indicates p ≤ 0.01. (E) Fluorescence microscopy images of representative cells from an experiment from A. rDNA is colored red, the αSat 17 is green, and DAPI is blue. Scale bar, 5 μm.
We further explored our finding that TTF-1 is among several nucleolar proteins dispersed upon p150 depletion. TTF-1 is a DNA-binding protein that interacts with the enhancer, promoter, and terminator regions in the rDNA repeats and stimulates both rDNA transcription and Pol I transcriptional termination (McStay and Grummt, 2008). Furthermore, TTF-1 promotes loop formation between the promoter and terminator regions (Németh et al., 2008), and this three-dimensional interaction is believed to be of central importance to rDNA gene activation (Denissov et al., 2011). We therefore hypothesized that p150 might contribute to localization of TTF-1 to rDNA. Indeed, ChIP analyses demonstrated that p150 depletion reduced the level of TTF-1 at the rDNA terminator (Figure 2D). We conclude that p150 regulates the nucleolar localization of multiple proteins.
An N-terminal p150 domain incapable of CAF-1 complex formation is sufficient for normal nucleolar protein localization
To determine whether the entire p150 protein is important for nucleolar protein localizations, we generated cloned, stable cell lines that each expresses a different V5 epitope–tagged, shRNA-resistant, p150-derived transgene (Figure 3A). These cell lines were then subjected to acute RNA-mediated protein depletion experiments. These complementation experiments allowed us to define functional domains and determine whether p150 depletion phenotypes could be directly attributed to p150 loss rather than indirect effects. Immunoblot analyses of these cells confirmed expression of the expected protein species and demonstrated that we could achieve efficient depletion of endogenous p150 in these cells (Figure 3B). Many of these cell lines up-regulated transgene-encoded protein levels in response to depletion of endogenous p150, perhaps suggesting a compensatory mechanism.
FIGURE 3:
Domain analysis of human p150. (A) Diagram of transgenes introduced into HeLa cells. Colored boxes indicate the V5 epitope tags (yellow), PEST region (red), low-complexity regions KER (blue) and ED (green), dimerization region (D, orange), the N-terminal PCNA interaction peptide (PIP, cyan), and the HP1-binding site (pink). The I99A point mutation that inactivates the SUMO-interaction motif (SIM, red asterisk) and silent mutations rendering the transgenes resistant to RNAi (shR, black asterisks) are indicated. Regions previously shown to be sufficient for in vitro chromatin assembly and p60 interaction are drawn below the top diagram. (B) Immunoblot analysis of V5-tagged transgene-expressing cell lines in two independent experiments (lanes 1–10 and 11–28). Cells were infected for 72 h with lentiviruses expressing either sh-luciferase (L, odd-numbered lanes) or sh-p150 (P, even-numbered lanes) and whole-cell extracts were separated on 10% SDS–PAGE gels. After transfer, blots were probed with anti-p150 to detect endogenous p150, anti-V5 to detect transgene products, and anti-tubulin (TUBA) or anti-fibrillarin (FBL) antibodies as loading controls. Arrowheads indicate full-length p150 protein. (C) Interaction between p150 transgene products and p60. Extracts from the indicated V5-tagged transgene-expressing cell lines were immunoprecipitated with anti-V5 antibodies. Input extracts (In, odd-numbered lanes) and immunoprecipitates (IP, even-numbered lanes) were resolved on 10% SDS–PAGE gels, transferred to membranes, and probed with anti-p150, V5, and p60 antibodies. Arrowhead indicates full-length p150 protein; asterisks indicate anti-V5 IgG bands in IP samples detected by the anti-mouse secondary antibody. Note that the 1-310 transgene comigrates with the IgG heavy chain. (D) V5-tagged, RNAi-resistant p150 transgenes were tested for the ability to maintain cell proliferation in the presence of either sh-luciferase (sh-Luc) or p150-shRNA1 (sh-p150) in HeLa cells. Cells (4500) were plated in six-well dishes and infected with the indicated lentivirus, and selection for infected cells began 48 h post infection. At 7d postinfection, cell proliferation was assessed by crystal violet staining.
We tested the ability of the transgene-encoded proteins to interact with the CAF-1 p60 subunit via immunoprecipitation. Consistent with published data (Kaufman et al., 1995; Takami et al., 2007), amino acids (aa) 245–938 contained the p60-binding domain (Figure 3C, compare lanes 4 and 10), and deletion of the C-terminal one-third of p150 (aa 642–938) eliminated coprecipitation of p60 (Figure 3C, compare lanes 4 and 6). We also tested transgenes for their ability to maintain cell proliferation over a 7-d period (Figure 3D). Consistent with data from chicken cells (Takami et al., 2007), the C-terminal region (aa 245–938) containing the p60-binding site was required for maintaining cell growth upon loss of endogenous p150. We conclude that these cell lines are useful tools for studying human p150 functional domains.
We then tested the transgene-expressing cells in immunolocalization assays, comparing cells infected with shRNA-encoding lentiviruses to deplete endogenous p150 or luciferase as a control. Immunolocalization of the V5 epitope–tagged proteins confirmed that the majority of cells in these lines expressed detectable transgene products (Figures 4 and 5). We first confirmed that negative control cells expressing a V5-luciferase transgene and an shRNA targeting luciferase displayed normal nucleolar localization of Ki67 and Nopp140 and that depletion of p150 in the same cells delocalized these proteins (Figures 4A and 5A; see also Supplemental Figures S3–S8). Conversely, in positive control cells expressing an shRNA-resistant cDNA encoding full-length p150 (aa 1–938), nucleolar localization of Ki67 and Nopp140 was unaltered upon depletion of endogenous p150, indicating functional complementation. We conclude that p150 depletion and not untargeted alterations caused mislocalization of Ki67 and Nopp140.
FIGURE 4:
The p150 SUMO-interaction motif (SIM) is required to maintain nucleolar Ki67 localization. HeLa cell lines expressing the indicated V5-tagged transgenes (A, luciferase; B, 1-938; C, 1-310; D, 1-310-ΔHP1; E, 1-310-I99A; F, 1-310-ΔPIP1; see Figure 3A) were infected with lentiviruses encoding the indicated shRNAs (sh-luciferase or sh-p150) and prepared for indirect immunofluorescence 72 h later. Scale bar, 20 μm.
FIGURE 5:
The p150 SIM is required to maintain nucleolar Nopp140 localization. As in Figure 4, HeLa cell lines expressing the indicated V5-tagged transgenes were infected with lentiviruses encoding the indicated shRNAs (sh-luciferase or sh-p150) and prepared for indirect immunofluorescence 72 h later. Scale bar, 20 μm.
We also observed that C-terminally deleted p150 transgenes (e.g., 1–641 and 1–310) maintained Ki67 and Nopp140 localization upon depletion of endogenous p150 (Figures 4 and 5 and Supplemental Figures S3–S8). Thus aa 1–310 of p150 sufficed for Ki67 and Nopp140 localization, even though this domain is incapable of p60 binding (Figure 3C, compare lanes 4 and 8), These data indicate that the N-terminal 310 aa of p150 constitute a domain that is functionally separable from the CAF-1 complex, which we will term p150N.
To test for functionally important regions within p150N, we mutated previously described interaction motifs. These included a noncanonical PCNA-interaction peptide (PIP; Moggs et al., 2000; Ben-Shahar et al., 2009), a heterochromatin protein 1 (HP1)–binding site (Murzina et al., 1999), and a SUMO interaction motif (SIM) that binds SUMOylated proteins (Uwada et al., 2010; Sun and Hunter, 2012). Deletion of the PCNA- or HP1-binding site resulted in transgenes that supported less robust growth than the wild-type full-length transgene, whereas a point mutation within the SIM did not appear to reduce growth (Figure 3D).
In protein localization experiments analogous to those described, cells expressing p150 transgenes with mutations in either the PCNA- or HP1-binding motif displayed no defects in localizing Ki67, Nopp140, or TTF-1 (Figures 4 and 5 and Supplemental Figures S3–S8). In contrast, a single I99A amino acid substitution within the SIM reduced the ability of the p150 transgene to maintain Ki67, Nopp140, and TTF-1 localization upon depletion of endogenous p150 (Figures 4 and 5 and Supplemental Figures S3–S8). We conclude that the SIM domain within p150N is a key factor for the normal localization of several nucleolar proteins.
Analysis of additional transgenes showed that Ki67 and Nopp140 display some difference in the regulation of their localization. Nucleolar localization of Ki67 was largely diminished in cells expressing a large C-terminal fragment of p150 (aa 245–938) upon depletion of endogenous p150 (Supplemental Figures S3 and S4). Therefore p150N was necessary and sufficient for Ki67 localization. In contrast, nucleolar localization of Nopp140 was maintained not only by p150N, but by the C-terminus as well (Supplemental Figures S5 and S6). Nevertheless, the Nopp140-localizing function within p150N depended on the SIM motif (Figure 5), and mutation of the SIM motif reduced nucleolar localization of Nopp140 even within the context of the full-length p150 transgene (Supplemental Figure S5). These data indicate that the SIM is an important but not unique domain for localizing Nopp140. In addition, a localization pattern similar to Nopp140 was observed for TTF-1 (Supplemental Figures S7 and S8).
The p150 N-terminus promotes interchromosomal interactions of the 47S rRNA–encoding repeats
Deep sequencing experiments have detected genomic regions preferentially associated with nucleoli (Nemeth et al., 2010; van Koningsbruggen et al., 2010). Among the “nucleolar-associated domains (NADs)” discovered were several types of repetitive DNA, including centromeric satellite DNA, D4Z4 repetitive DNA elements from the telomeric regions of chromosomes 4q and 10q, and the array of 5S rDNA genes from chromosome 1q. Therefore, using two-color DNA fluorescent in situ hybridization (FISH) experiments, we tested whether these long-range interchromosomal associations with 47S rRNA–encoding repeats were affected by p150 depletion. FISH experiments were initially performed in MCF-10A cells because these have a flattened morphology that we found more conducive to rDNA hybridization. In MCF10A cells expressing a control sh-luciferase hairpin RNA, we observed that ∼30–40% of 10q telomere, 5S rDNA array, and α-satellite alleles associated with 47S rRNA–encoding repeats (Figure 6, A–C). In contrast, a negative control BAC probe lacking NAD sequences displayed the expected background of ∼10% association (Nemeth et al., 2010). In cells expressing sh-p150, we observed a statistically significant decrease in nucleolar associations for all three NAD-containing loci tested but no change for the negative control. To extend this finding beyond immortalized cell lines, we then tested whether p150-dependent nucleolar associations could be detected in primary human cells. Using foreskin fibroblasts, we observed that the association of α-satellite DNA with nucleoli was indeed significantly reduced upon depletion of p150 (Figure 6, D–G). In these cells, detection of nucleoli was more robust using an anti-fibrillarin antibody rather than a 47S rRNA–encoding repeat probe. We conclude that p150 is important for nucleolar association of several repetitive DNAs in both primary and transformed human cells.
We then tested whether the p150N protein fragment would suffice for maintaining these chromosomal interactions. In HeLa cells expressing the control V5-luciferase transgene, we again observed frequent associations of α-satellite DNA with the 47S rRNA–encoding repeats, which were reduced upon p150 depletion (Figure 7, A and B). A similar loss of α-satellite–rDNA associations was observed in cells expressing a large C-terminal p150 fragment (aa 245–938; Figure 7, A and C). In contrast, cells expressing p150N (aa 1–310) maintained these associations upon depletion of endogenous p150, measured either as the total number of allele associations (Figure 7A) or number of alleles associated per cell (Figure 7D). Taken together, our data indicate that the p150N domain of p150 is sufficient for promoting efficient nucleolar protein localizations and interchromosomal associations.
We next tested how point mutations within p150N affected α-satellite interactions with the nucleolus. As observed in the protein localization experiments (Figures 4 and 5), the I99A but not the PCNA- or HP1-binding site mutations reduced the nucleolar associations (Figure 8,A–D). To test whether SUMO conjugation itself is important for nucleolar chromosome interactions, we used siRNAs to transiently deplete p150, Ubc9, the E2 enzyme required for all SUMO conjugation events in human cells (Jentsch and Psakhye, 2013), or SUMO2 itself. Reverse-transcriptase-PCR analysis confirmed reduction of UBC9 and SUMO2 mRNA levels (Figure 8G). The p150 depletion confirmed that siRNA-mediated targeting of the p150 mRNA via a different sequence than recognized in shRNA-mediated depletions still significantly reduced α-satellite–nucleolar interactions. In addition, we observed significantly reduced interactions upon either UBC9 or SUMO2 depletion. We conclude that both a SUMO interaction motif within the p150N domain and the SUMO conjugation machinery contribute to efficient nucleolar protein localizations and interchromosomal interactions.
FIGURE 8:
The p150 SIM and sumoylation proteins are required for nucleolar interchromosomal associations. (A) Immuno-FISH analysis of the association between αSat 17 and fibrillarin in HeLa cells expressing the indicated V5-tagged transgene derivatives of full-length p150. The percentage of αSat 17 alleles colocalized with fibrillarin is indicated, with mean and SDs from three experiments. For ΔPIP cells, total alleles assayed = 510 for sh-Luc and 477 for sh-p150; for ΔHP1 cells, 522 for sh-Luc and 513 for sh-p150; and for I99A cells, 486 for sh-Luc and 480 for sh-p150. p values comparing the sh-Luc and sh-p150 samples are indicated, demonstrating statistically significant rescue by the ΔPIP and ΔHP1 but not the I99A transgenes. (B–D) Data from the experiment in A replotted to display the number of αSat 17 alleles per cell associated with the rDNA. Note that these HeLa-derived lines have three copies of chromosome 17. For ΔPIP cells, total cells examined = 170 for sh-Luc and 159 for sh-p150; for ΔHP1 cells, 174 for sh-Luc and 171 for sh-p150; and for I99A cells, 162 for sh-Luc and 160 for sh-p150. Comparisons for which p ≤ 0.05 are indicated by asterisks. (E) Immuno-FISH analysis of MCF10A cells treated with the indicated siRNAs. The percentage of αSat 17 alleles colocalized with fibrillarin is indicated, with mean and SDs from three experiments. The total alleles assayed = 328 for si-luciferase–treated cells, 346 for si-p150–treated cells, 324 for si-UBC9–treated cells, and 342 for si-SUMO2-treated cells. p values comparing the sh-Luc and other samples are indicated, demonstrating statistically significant reductions in association by each of the three test depletions. (F) Data from the experiment in E replotted to display the number of αSat 17 alleles per cell associated with fibrillarin. Note that these MCF10A-derived lines have two copies of chromosome 17. (G) Quantitative PCR analyses of esiRNA treatments demonstrating reduced steady-state mRNA levels in the targeted cells. (H) Immunoblot of representative whole-cell extracts from an experiment in E, with fibrillarin as loading control. (I) Fluorescence microscopy images of representative cells from an experiment from E and F. Fibrillarin is colored green, the αSat 17 is red, and DAPI is blue. Scale bar, 5 μm.
DISCUSSION
A separable functional domain within the CAF-1 p150 N-terminus
We show here that the N-terminal domain of p150 (aa 1–310) is sufficient to maintain nucleolar protein and interchromosomal associations and thereby constitutes a distinct and separable functional domain. Because p150 residues 1–310 cannot bind the CAF-1 p60 subunit and lacks the previously described dimerization motif within p150 (Quivy et al., 2001; Figure 3), these data suggest that the canonical three-subunit CAF-1 complex is not required for these novel functions. We note that previous proteomic analysis of nucleoli detected p150 but not p60 (Ahmad et al., 2009), consistent with the idea of nucleolar functions for p150 separate from CAF-1. We also note that a previous proteomic analysis of p150 detected interacting proteins largely nonoverlapping with those reported here (Hoek et al., 2011), raising the possibility that differences in the epitope tags used or other experimental details can favor retention of distinct subsets of the protein interactions of p150.
In all eukaryotic species, CAF-1 is a complex of three conserved subunits, all of which are required for in vitro nucleosome assembly activity (Kaufman et al., 1995; Verreault et al., 1996). In addition, roles outside of the CAF-1 complex have been described for individual subunits. For example, p60 was recently shown to act as a protamine deposition factor independent of CAF-1 during fruit fly spermatogenesis (Doyen et al., 2013). p48 is a histone H4–binding (Murzina et al., 2008; Song et al., 2008) subunit of many complexes involved in chromosome biology (Martinez-Balbas et al., 1998; Zhang et al., 1999; Kuzmichev et al., 2002). Two functions for p150 independent of CAF-1 have been described. First, p150, but not p60, is required for efficient replication of HP1-rich pericentric heterochromatin in mouse cells, and this activity requires the canonical HP1-binding site on p150 (Quivy et al., 2008). Second, p150 but not p60 stimulates transcription from the major immediate early promoter (MIEP) of human cytomegalovirus (CMV; Lee et al., 2009). In the latter studies, the p150 N-terminal 310 aa (p150N) were shown to be an activation domain for MIEP transcription (Lee et al., 2009). Although subregions within p150N were not tested in that system, those findings are consistent with our conclusion that p150N can act as a separable domain with distinct functions. Here we show that human p150N has important structural roles outside of the context of viral infection, maintaining normal nucleolar protein localization and interchromosomal interactions. Therefore the C-terminal two-thirds of human p150 essential for formation of the nucleosome assembly complex CAF-1 are appended to a separable domain that regulates the structure of the nucleolus and the interconnectivity of the human genome.
The SIM domain links human p150N to nucleolar structures
Our studies show the p150 SIM domain is required for nucleolar macromolecular associations. This p150 SIM domain was previously shown to bind the highly homologous SUMO-2 and SUMO-3 proteins but not SUMO-1 (Uwada et al., 2010). Furthermore, mutational analysis of the p150 SIM was shown to alter association of SUMO-2 and SUMO-3 with DNA replication foci. However, the converse was not true, because acute depletion of SUMO2/3 did not affect p150 association with replication foci. These data suggested that p150 recruits SUMO2/3 to DNA replication foci via the SIM domain. We do not yet know whether those observations are related to those reported here. Our data suggest that the interaction of p150 with the rDNA is not limited to S phase of the cell cycle (Figures 1 and 2), and the p150 SIM domain appears dispensable for p150’s contribution to cell proliferation (Figure 3D). In addition, we observe that strong inhibition of rDNA transcription with actinomycin D does not impair the α-satellite–nucleolar interaction (Supplemental Figure S9), although in other studies different NAD–nucleolar interactions were variably reduced upon similar treatments (Nemeth et al., 2010). Therefore we are investigating how these chromosome interactions are dynamic with regard to transcription and growth control and whether they are cell cycle regulated. We also note that a large number of SUMOylated proteins reside in the nucleolus (Westman et al., 2010; Finkbeiner et al., 2011), and we are exploring whether particular protein interactions depend on the p150 SIM domain.
A previously published protein alignment suggested that Cac1, the budding yeast homologue of human p150, also contains a putative SIM (Uwada et al., 2010). Subsequent analysis in the Hunter laboratory showed that the SIM in the N-terminus of human p150 is a “type B” or “VIDLT” class of SIM that is often found as a dominant SIM in multi-SIM proteins (Sun and Hunter, 2012). The p150 N-terminal SIMs are also of the canonical type B in other animals, including mammals and the sea urchin Strongylocentrotus purpuratus (Supplemental Figure S10). In contrast, this SIM is altered from the type B consensus in frogs, zebrafish and chickens, and insects. The budding yeast SIM sequence lacks the characteristic aspartate at position 3 that is critical for high-affinity binding, and no apparent type B SIM sequences could be identified in fission yeast, worms, or the plants Arabidopsis thaliana and Triticum urartu. Future studies will be required to clarify whether and how nucleolar associations are mediated by p150 homologues in other organisms.
In that regard, it is of particular interest that recent work showed a progressive loss of 45S rDNA copies in A. thaliana mutants in lacking the CAF-1 p150 or p60 subunit (Mozgova et al., 2010). Further, these mutants display a loss of partitioning of silent 45S rDNA copies into the nucleoplasm (Pontvianne et al., 2013), whereas 5S rDNA remains stable in its copy number and localization. We do not observe altered DNA-FISH signals with a 47S-encoding rDNA probe in human cells (Figures 6 and 7 and unpublished data), and so we do not have evidence for a similar mechanism in humans. Further, as mentioned earlier, there is no discernible SIM in A. thaliana p150. However, we cannot rule out less dramatic reorganization of 47S rDNA that would have escaped detection in our FISH experiments, and the full range of contributions of p150 to the structure and function of nucleolar chromatin in human cells remains an open an interesting avenue for exploration.
Higher-order interactions of nucleolar chromatin
Several connections between heterochromatin, centromeric DNA, and the nucleolus have been described. For example, in Drosophila, the nucleolar protein Modulo, an NCL homologue, affects formation of centromeric chromatin (Chen et al., 2012). Conversely, depletion of Drosophila HP1 causes dispersal of the rDNA and nucleolar proteins, including fibrillarin (Peng and Karpen, 2007). We note that vertebrate p150 homologues include an HP1-binding domain (Murzina et al., 1999) that is within the p150N domain, although we do not observe fragmentation of rDNA upon depletion of p150 in human cells (Figure 7). Other links between the nucleolus and centromeric DNA in Drosophila include recent studies showing that NLP, a nucleophosmin-related protein, is required is required for centromere clustering and anchoring of centromeric DNA to nucleoli (Padeken et al., 2013). Further, both Drosophila NLP (Padeken et al., 2013) and human NPM (Foltz et al., 2006) interact with the centromere-specific histone CENP-A. Biochemical interactions between CENP-C and UBF1 (Pluta and Earnshaw, 1996) and colocalization of centromeric proteins with the nucleolus (Ochs and Press, 1992; Wong et al., 2007) have also been described. Together these data, and other data reviewed recently (Padeken and Heun, 2014), suggest conserved links among nucleoli, heterochromatin, and centromeres. However, we do not observe changes in the distribution of CENP-A upon p150 depletion in human cells (Supplemental Figure S10B). Therefore the extent of p150N's contributions to heterochromatin and centromere function remains to be determined. Experiments are underway to determine which partner proteins are involved in the ability of p150N to stimulate the higher-order chromatin interactions of nucleoli and determine whether these interactions are specific to repetitive sequences.
MATERIALS AND METHODS
Preparation of nuclear extracts and affinity chromatography for mass spectroscopy samples
HeLa S3 Trex (untagged) and HeLa S3 Trex-NTAP-p150 cells were maintained as monolayers and then expanded to suspension cultures for large-scale preparations. Suspension cultures were started at a cell density of 3 × 105/ml and maintained between 1.5 × 105 and 6 × 105/ml. NTAP-p150 synthesis was induced by addition of doxycycline (1 μg/ml) for 16–18 h. Untagged samples were also treated with doxycycline in the same manner. Routinely, 6–8 l of suspension cells were grown for each harvest. Suspension cultures were harvested by centrifugation at 1000 × g at 4°C. Pellets were used to generate nuclear extracts by Dounce homogenization. Briefly, suspension cells were collected by centrifugation at 1000 × g for 5 min. Cells were washed with ice-cold phosphate-buffered saline (PBS) and then homogenization buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES]-KOH, pH 8.0, 5 mM KCl, 1.5 mM MgCl2) and then resuspended in 1 ml of homogenization buffer/ml of packed cell volume. Cells were disrupted by 28 strokes of a B pestle (loose) by Dounce homogenization (Wheaton, Millville, NJ), and nuclei were pelleted by centrifugation (5 min at 1000 × g) and washed with nuclei wash buffer (10 mM NaCO3, 150 mM NaCl). Nuclear extracts were made by incubating nuclei with extraction buffer (15 mM Tris-HCl, pH 7.8, 1 mM EDTA, 400 mM NaCl, 10% sucrose, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol [DTT]) for 30 min, rotating at 4°C. Extracts were then clarified by ultracentrifugation at 100,000 × g for 60 min and then frozen in aliquots and stored at −80°C. For samples analyzed by mass spectroscopy, 12.5 mg (experiment 1) or 25 mg (experiment 2) of nuclear extract was used for affinity purification.
Affinity purifications were performed with streptavidin–Sepharose (GE Healthcare). All steps were performed at 4°C. We used 300 μl of resin/25 mg of nuclear extract. Extracts were diluted twofold with 25 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% glycerol, and 0.01% NP40 to reduce the NaCl concentration from 400 to 200 mM and rotated with the resin for 3 h. Beads were washed twice for 20 min with MS200 (100 mM Tris, pH 8.5, 200 mM NaCl) plus 50 μg/ml ethidium bromide (EtBr). Beads were then washed twice more with MS200 without EtBr and twice with MS50 (100 mM Tris, pH 8.5, 50 mM NaCl). Proteins were then eluted from the beads with ME buffer (100 mM Tris, pH 8.5, 8 M urea). Samples were precipitated with 20% trichloracetic acid on ice for 30 min and centrifuged for 10 min at 16,000 × g at 4°C. The supernatants were removed, and the pellets were washed twice with −20°C acetone and air-dried.
Mass spectroscopy
The NTAP-p150 and untagged samples were first denatured in 8 M urea and then reduced and alkylated with 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (Roche Applied Science; Indianapolis, IN) and 55 mM iodoacetamide (Sigma-Aldrich, St. Louis, MO), respectively. The sample was then digested overnight with trypsin (Promega, Madison, WI) according to the manufacturer's specifications.
The protein digest was pressure loaded onto a fused silica capillary (Polymicro Technologies) column of 250-μm inner diameter with a Kasil frit packed with 3 cm of 3-μm C18 resin (Phenomenex, Torrance, CA). After desalting, this column was connected to a fused silica capillary (Polymicro Technologies) analytical column of 100-μm inner diameter with a 5-μm pulled tip, packed with 10 cm of 3-μm C18 resin (Phenomenex).
The column was placed inline with a 1200 quaternary high-performance liquid chromatography (HPLC) pump (Agilent Technologies, Lexington, MA), and the eluted peptides were electrosprayed directly into an LTQ mass spectrometer (Thermo Scientific). The buffer solutions used were 5% acetonitrile/0.1% formic acid (buffer A) and 80% acetonitrile/0.1% formic acid (buffer B). The 180-min elution gradient had the following profile: 10% buffer B beginning at 15 min to 40% buffer B at 105 min, to 70% buffer B at 150 min, to 100% buffer B from 155 to 165 min. A cycle consisted of one full scan mass spectrum (400–1600 m/z), followed by five data-dependent collision-induced dissociation tandem mass spectrometry (MS/MS) spectra. Application of mass spectrometer scan functions and HPLC solvent gradients was controlled by the Xcalibur data system (Thermo Scientific).
MS/MS spectra were extracted using RawXtract (version 1.9.9; McDonald et al., 2004). MS/MS spectra were searched with the Sequest algorithm (Eng et al., 1994) against an European Bioinformatics Institute International Protein Index protein database, version 3.30 (June 2007), concatenated to a decoy database in which the sequence for each entry in the original database was reversed (Peng et al., 2003). The Sequest search was performed using full enzyme specificity, including static modification of cysteine due to carboxyamidomethylation (57.02146). Up to two missed cleavages were considered. Sequest search results were assembled and filtered using the DTASelect (version 2.0) algorithm (Tabb et al., 2002). The peptide identification false-positive rate was kept to <1%.
shRNAs
shRNA expression constructs were made using the system we described previously (Campeau et al., 2009). The shRNA for depletion of p150 (p150shRNA-1) had the targeting sequence AGGGGAAAGCCGATGACAT. Destination vectors for expressing p150shRNA-1 were created by recombining pENTR1A/pTER p150shRNA-1 (435-1) into pLenti RNAi X2 neo (w18-1) to create pLenti RNAi X2 Neo/pTER p150shRNA-1 (w23-2) and pLenti RNAi X2 Puro (w16-1) to create pLenti RNAi X2 Puro/pTer p150shRNA-1 (pPK655). The luciferase shRNA-expressing construct pLenti X2 Neo/pTER shLUC (w181-1) was previously described (Campeau et al., 2009).
p150 derivatives and other transgenes
The p150 amino acid numbers used here are based on the literature (Kaufman et al., 1995; Lee et al., 2009). Various p150 constructs were altered by site-directed mutagenesis to make them resistant to p150shRNA-1. First, we used oligonucleotides ECA552 and ECA553 (Supplemental Table S2) to alter the p150 cDNA in plasmid pPK8 (Kaufman et al., 1995), generating plasmid pPK649, which carries the “shR” version of the p150 cDNA. RNAi-resistant, V5-tagged p150 C-terminal or internal deletion Gateway entry constructs were generated by cloning fragments of pPK649 into pENTR-V5 (w71-3). Lentiviral expression vectors were made by LR recombination of the pENTR-based entry constructs into pLenti CMV Hygro (w117-1). The negative control transgene, V5-tagged luciferase was generated by LR recombination of 468-1 (pENTR vector + V5-luciferase) into w117-1 to generate pPK678 (pLenti CMV V5-luciferase).
esiRNA methods
siRNAs were generated using the Deqor method (Surendranath et al., 2013), with primers designed using the Deqor database (http://deqor.mpi-cbg.de/deqor_new/input.html) to generate T7-tailed dsDNA templates from human cDNA in two rounds of PCR. In the case of the luciferase esiRNA, RNA from a HeLa cell line expressing V5-luciferase was used to generate cDNA; all other esiRNAs derived from cDNA from normal HeLa cells. In the first round of PCR, primers specific to the gene of interest (p150, luciferase, UBC9, or SUMO2) were used in the following program: 95°C for 3 min, 16 cycles of (95°C for 1 min, 65°C − 1°C/cycle for 1 min, and 72°C for 1 min), followed by an additional 15 cycles of (95°C for 1 min, 60°C for 1 min, and 72°C for 1 min) and one final cycle at 72°C for 5 min. In the second round of PCR, a T7 promoter-tailed primer was used to amplify the product of the first round with the following program: 95°C for 2 min, four cycles of (95°C for 30 s, 42°C for 45 s, and 72°C for 1 min), followed by an additional 29 cycles of (95°C for 30 s, 60°C for 45 s, and 72°C for 1 min) and one final cycle at 72°C for 5 min. The product of the second round of PCR is used to transcribe double-stranded esiRNA using T7 RNAP in presence of 25 mM nucleoside triphosphates and 40 mM Tris, pH 7.9, 6 mM MgCl2, 2 mM spermidine, and 10 mM NaCl using the following thermocycler program: 37°C for 5 h, 30 min; 90°C for 3 min; ramp 0.1°C/s to 70°C; 70°C for 3 min; ramp 0.1°C/s to 50°C; 50°C for 3 min; ramp 0.1°C/s to 25°C; 25°C for 3 min. After transcription and annealing, products were digested with 2 U of RQ1 DNase I for 15 min at 37°C. Then esiRNAs were generated by digesting doubled-stranded RNAs with RNase III (Fazzio et al., 2008) and purified as described using Invitrogen RNA Mini Kits.
Cell culture methods
For inducible expression of transgenes, a HeLa S3-Trex cell line expressing the tetracycline repressor was established by infecting HeLa S3 cells with lentiviruses containing pLenti CMV TetR Blast (716-1) and selecting for blasticidin-resistant cells. HeLa S3-Trex cells were maintained either on plates or in suspension in RPMI medium with 5% tetracycline-free fetal bovine serum (FBS). HeLa-S3-Trex cells were infected with lentiviruses carrying CMV/TO promoters driving expression of NTAP-tagged p150 (pPK560) or p150-directed shRNAs. Individual drug-resistant clones were picked for each of these cell lines. The NTAP-p150–expressing clone was chosen because it expressed tagged protein levels similar to endogenous p150 levels and was described previously (Campeau et al., 2009). p150 shRNA clones were selected based on the extent of p150 depletion. Inducible expression of transgenes was achieved by supplementing media with 1 μg/ml (NTAPp150) or 2 μg/ml (shRNAs) doxycycline.
RNAi-resistant, V5-tagged transgene expression constructs were introduced into the HeLa S3 Trex-p150shRNA-1 cell line via lentiviral infection, followed by hygromycin selection. Cloned MCF10A-Tet-KRAB cell lines (Cohet et al., 2010) harboring p150shRNA-1 or sh-luciferase were generated and grown in DMEM/F12 (Lonza) supplemented with 5% horse serum, 20 ng/ml epidermal growth factor, 10 μg/ml insulin, 0.5 μg/ml hydrocortisone, 100 ng/ml cholera toxin, and antibiotic/antimycotic solution (Life Technologies, Grand Island, NY). HT1080 cells (ATCC) were maintained in DMEM with 5% tetracycline-free FBS, and Trex- and sh-RNA expressing cells lines were made as described.
For double-thymidine-block experiments, HeLa S3 cells were blocked in RPMI plus 5% FBS medium supplemented with 2 mM thymidine for 18 h, released into thymidine-free media for 9 h, and blocked again with medium containing 2 mM thymidine for 17 h (Whitfield et al., 2002). For serum starvation studies, MCF10A-Tet-KRAB cells were cultured in complete DMEM/F12 as described or in DMEM/F12 containing only 1 mg/ml bovine serum albumin (BSA) and antibiotic/antimycotic with no additional growth factors for 96 h. The shRNAs were induced by adding 2 μg/ml doxycycline to the medium during the last 48 h.
Human foreskin fibroblasts (HFFs; a kind gift of Jennifer Benanti, University of Massachusetts Medical School; Benanti and Galloway, 2004) were grown in DMEM supplemented with 10% FBS and antibiotic/antimycotic solution (Life Technologies). HFF cells were maintained at >25% confluence at all times and were split 1:4 every 2 d. HFF cells used for the immuno-FISH and DNA-FISH experiments were grown on polylysine-coated coverslips between PD22-30. MCF-10A and HeLa cells were cultured as described on nontreated coverslips for immuno-FISH and DNA-FISH experiments.
Growth analyses
To measure growth rates during p150 depletion, HeLa S3 Trex-p150shRNA-1 and HeLa S3 Trex-shRNA-luciferase cells were plated at 1500 cells/well in 48-well plates. Triplicate wells for each time point (0–120 h) were assayed with CellTiter 96 AQueous One (Promega) per manufacturer's instructions. For growth analysis of cells expressing V5-tagged p150 or luciferase transgenes, 9000 cells/well in six-well dishes were infected with lentiviruses expressing either shRNA-luciferase or p150shRNA-1 at a multiplicity of infection (MOI) of 2. After 48 h, drug selection for infected cells was begun (1 mg/ml G418 or 0.5 μg/ml puromycin, respectively). After 7 d postinfection, cells were stained with crystal violet (Campeau et al., 2009) and photographed.
Antibodies
For p150, mouse monoclonals ss1 and ss48 (Smith and Stillman, 1991) were used for immunoblots and immunofluorescence. In addition, rabbit polyclonal anti-p150 antibodies (sc-10772, Santa Cruz Biotechnology, Santa Cruz, CA; and sera specifically generated for this study) were used for immunoblotting and ChIP. To generate polyclonal antibodies, a codon-optimized version of the first 200 aa of the p150 cDNA was cloned into pGEX6 (GE Healthcare) to generate pPK621. The codon numbering for this construct is based on the full-length p150 open reading frame, which contains an additional 18 aa at the N-terminus not present in the original cDNAs (accession number NM_005483). Synthesis of the glutathione S-transferase (GST)–p150(1-200) fusion protein was induced in BL21(DE3)pLysS Escherichia coli cells grown in Luria broth with 35 μg/ml chloramphenicol and 100 μg/ml ampicillin at 37°C by addition of 0.5 mM isopropyl-β-d-thiogalactoside at an OD600 of 0.4 for 2 h. Pellets were collected, washed in ice-cold PBS, frozen in liquid nitrogen, and stored at −80°C. Glutathione affinity chromatography on Glutathione Sepharose 4 Fast Flow and separation of GST from p150(1-200) by PreScission Protease digestion were performed in PBS at 4°C per manufacturer's instructions (GE Healthcare). Additional purification of p150(1-200) was performed by ion exchange chromatography on a 1-ml Poros 20 HQ column (Life Technologies). Soluble protein collected from the PreScission digestion was loaded onto a HQ column preequilibrated in HQ buffer (25 mM Tris, pH 7.5, 1 mM EDTA, 1 mM DTT) with 100 mM NaCl and eluted using a gradient from 100 mM to 1 M NaCl. p150(1-200) eluted near the beginning of the gradient. This purified antigen was used to immunize rabbits for antisera production (at Pocono Rabbit Farm and Laboratory, under the approval of University of Massachusetts Medical School IACUC Off-Site Animal Protocol A-1692-12). Antibodies to CBP (RCBP-45A; ICL), Ki67 (NB500-170; Novus), CAF1-p60 (ss96; Smith and Stillman, 1991), CENPA (ab13939; Abcam), phospho(S1981)-ATM (200-301-400; Rockland), phospho(S317)-Chk1 (A300-163A; Bethyl), phospho(S139)-γH2AX (05-636; Upstate), HP1α (05-689; Millipore), NPM/B23 (sc-32256; Santa Cruz Biotechnology), NCL/C23 (sc-9893; Santa Cruz Biotechnology), Nopp140 (RS8; a generous gift of U. Thomas Meier, Albert Einstein College of Medicine, New York, NY; Kittur et al., 2007), fibrillarin (FBL, ab5821; Abcam), UBF1 (sc-13125; Santa Cruz Biotechnology), PARP (556362; BD PharMingen), TTF1 (sc-136371; Santa Cruz Biotechnology), tubulin (DM1a; T9026; Sigma-Aldrich), and V5 (R96025; Life Technologies) were used for immunofluorescence and immunoblots. Anti-bromodeoxyuridine (B2850-01G; USBiologicals) was used to detect BrUTP-labeled RNAs. Secondary antibodies were obtained from either Jackson Laboratory (fluorescein isothiocyanate [FITC], Cy5) or Life Technologies (Alexa Fluor 488, 594, and 647).
Immunofluorescence
Cells grown on LabTek cc2 eight-chamber slides were fixed in either methanol (10 min) or 4% paraformaldehyde (5 min) as indicated and then permeabilized with 0.5% Triton X-100 (5 min) at room temperature. Samples were blocked in 0.5% BSA/0.2% fish-skin gelatin for 1 h. Samples were then incubated with primary antibodies (1:500–1:1000) in blocking buffer (1 h), followed by three PBS washes and then secondary antibody incubation (1:800 FITC and Cy5, 1:2000 Alexa Fluor 488 and Alexa Fluor 647). Immunofluorescence images were taken on a Zeiss Axioplan2 microscope using AxioVision, version 4.6, with a 63× objective connected to a Hamamatsu 1394 ORCA-ER HD camera. Confocal images were taken on a Leica SP2 microscope with a 100× objective using Leica imaging software, using 405-, 488-, and 561-nm laser lines. Colocalization images were generated using Imaris x64, version 6.1.3. Densitometry of individual immunostained cells were performed using TIFF files in ImageJ 1.48r (National Institutes of Health) by constructing histograms across nucleoli of shRNA-treated cell lines. Pixel values were normalized to the total pixel density of the histogram and plotted in Prism (GraphPad).
Chromatin immunoprecipitation
HeLa S3-Trex-p150shRNA-1 or sh-luciferase cells were treated with 2 μg/ml doxycycline for 72 h. For each sample, ∼3 × 107 cells were fixed with 1% formaldehyde for 10 min at room temperature and subsequently quenched with 0.125 M glycine. After two washes in PBS, cells were collected by scraping, centrifuged, and then flash frozen in liquid nitrogen. Thawed pellets were resuspended in ChIP lysis buffer (50 mM HEPES-KOH, pH 7.5, 150 mM NaCl, 1 mM EDTA, 0.5% SDS, 0.1% sodium deoxycholate, 1% Triton X-100) and sonicated using a Branson Sonifier 450. After six 20-s pulses (power 3, 30% duty cycle), fractionated chromatin was ultracentrifuged at 100,000 × g for 45 min, and the supernatant was recovered. Bradford assays (Bio-Rad) were used to determine protein content in the soluble chromatin preparations. All samples were precleared with 100 μl of Recombinant Protein G–Sepharose 4B beads (Life Technologies) prelocked with 0.5 mg/ml BSA and 0.2 mg/ml sheared salmon sperm DNA in ChIP lysis buffer overnight at 4°C. For p150 and TTF-I analyses, 500 μg of chromatin was incubated overnight with 5 μg of our own anti-p150 antisera or 2.5 μg of anti–TTF-I (SC-136371; Santa Cruz Biotechnology) antibody. Rabbit preimmune serum or mouse IgG (SC-2025; Santa Cruz Biotechnology) served as negative immunoprecipitation control. After overnight incubation with 100 μl of Protein G beads, beads were washed twice with ChIP lysis buffer, once with high-salt buffer (50 mM HEPES-KOH. pH 7.5, 1 M NaCl, 1 mM EDTA, 0.5% SDS, 0.1% sodium deoxycholate, 1% Triton X-100), three times with RIPA buffer (50 mM Tris-HCl, pH 8.0, 250 mM LiCl, 1 mM EDTA, 0.5% NP40, 0.5% sodium deoxycholate), and once with 10 mM Tris-Cl, pH 8.0, 1 mM EDTA (TE) buffer containing 50 mM NaCl. Protein–DNA complexes were extracted from beads at 65°C in elution buffer (0.1 M NaHCO3, 1% SDS) for 15 min, followed by centrifugation at 16,000 × g for 1 min and collection of supernatants. To reverse cross-linking, eluted chromatin and corresponding input samples were rotated overnight at 65ºC. Samples were then subjected to RNase A (80 μg for 2 h at 37°C) and proteinase K (80 μg for 2 h at 55°C) digestions. Samples were extracted with phenol:chloroform:isoamyl alcohol (25:24:1) and precipitated with 2.5 volumes of ethanol in 0.2 M NaCl plus 30 μg of glycogen. Precipitated DNA was resuspended in 10 mM Tris-HCl, pH 8.0. The Fast SYBR Green reagent (Life Technologies) was used in ChIP-PCR studies to quantify p150 and TTF-1 occupancies. Primer sequences are in Supplemental Table S2. The PCRs were performed using a StepOnePlus machine (Life Technologies) with the following program: hold 95°C for 20 min, followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. To calculate the percentage of bound DNA, unimmunoprecipated input chromatin from each sample was analyzed simultaneously. ∆∆Ct was calculated from the following formula (Life Technologies): Adjusted input = Ct input − 6.644. Percentage input = 100 × 2Adjusted input − Ct(IP). Three biological replicates for each experiment were performed, and comparisons between samples were analyzed by two-tailed, unpaired Student's t tests with equal variance (homoscedastic).
Visualization of 5-ethynyl-uridine–labeled nascent RNA
MCF-10A cells were grown in four-well chamber slides (Fisher) overnight and then in some cases incubated with the RNA polymerase II inhibitor α-amanitin (Sigma-Aldrich), the RNA polymerase I inhibitor actinomycin D (Sigma-Aldrich), or both. α-Amanitin was added at 20 μg/ml for 4 h, and actinomycin D was added at 50 ng/ml for 15 min. Prewarmed MCF-10A medium containing the indicated drug(s) and 500 μM of the UTP analogue 5-ethynyl-uridine (Invitrogen) was added for 15 min, followed by washing with 1× PBS. Cells were then fixed with 3.7% formaldehyde/1× PBS for 15 min at room temperature. After another 1× PBS wash, the cells were permeabilized in 0.5% Triton X-100/1× PBS for 15 min at room temperature. Cells were then washed again in 1× PBS before incubation with the click chemistry cocktail (50 mM carboxyrhodamine 110-Azide [Click Chemistry Tools], 100 mM ascorbic acid [Sigma-Aldrich], and 1 mM CuSO4 [Sigma-Aldrich] in 100 mM Tris-Cl, pH 8.5) for 30 min at room temperature in the dark. The cells were then washed three times in 0.5% Triton X-100/1× PBS for 5 min in the dark and then incubated in 130 ng/ml 4′,6-diamidino-2-phenylindole (DAPI) in 1× PBS for 1 min. The chamber was then manually removed, and a few drops of Vectashield (Vector Labs) were added before a coverslip was placed on top and sealed with nail polish. Images were taken on a Zeiss Axioplan2 microscope with a 63× objective.
Immuno-FISH and DNA-FISH
The bacterial artificial chromosomes RP5-915N17 (1q42.13; “5S rDNA”), RP11-413F20 (10q26.3; “D4Z4 array”), and RP11-123G19 (10q24.1; “Negative Control,” which was previously shown to lack nucleolar localization; Nemeth et al., 2010) were obtained from the BACPAC Resource Center of Children's Hospital Oakland Research Institute (Oakland, CA). The plasmid pB (Wilson, 1982) containing a 5.6-kb EcoRI fragment of human 47S rRNA–encoding repeat was a gift from Sui Huang (Northwestern University School of Medicine, Chicago, IL). The chromosome 17 α-satellite probe was prepared as previously described (Warburton et al., 1991). BAC probes were labeled using the Bioprime Labeling Kit (Invitrogen), and the rDNA and α-satellite probes were labeled using the Nick Translation Mix Kit (Roche).
Expression of either sh-luciferase or sh-p150 in MCF10A-Tet-KRAB cells was induced with 2 μg/ml doxycycline for 72 h, followed by fixation and storage in 1× PBS at 4°C for up to 1 wk. For all transgene experiments involving lentivirus-encoded shRNAs, cells were infected at MOI = 7.5 with 6 μg/ml Polybrene. At 72 h postinfection, cells were fixed and stored for up to 1 wk in 1× PBS at 4°C. For other depletions, siRNAs were constructed as previously described (Fazzio et al., 2008), and 500 ng of siRNAs were transfected in 1 ml Opti-MEM (Life Technologies) with 6 μl of Oligofectamine (Life Technologies). Transfections were performed in six-well dishes containing coverslips, and after 6.5 h, 2.5 ml of appropriate medium was added on top of the transfection cocktail. At 72 h after transfection, cells were fixed and stored for up to 1 wk in 1× PBS at 4°C.
Immuno-FISH and DNA FISH experiments in MCF-10A, HFF, and HeLa cells were performed using previously published methods (Byron et al., 2013) with some modification. Briefly, cells were grown on coverslips, permeabilized in 0.5% Triton X-100 in cytoskeletal (CSK) buffer on ice for 5 min, and fixed in 4% paraformaldehyde in 1× PBS at room temperature for 10 min. For immuno-FISH experiments, cells were then processed using the immunofluorescence protocol described here using an anti-fibrillarin antibody (Abcam). The cells were then fixed again in 4% paraformaldehyde in 1× PBS at room temperature for 10 min before proceeding to DNA denaturation. For DNA-FISH experiments, after initial fixation, RNA was removed by incubating the cells in 0.2 N NaOH in 70% ethanol for 5 min at room temperature before proceeding to genome denaturation. The genome was denatured by placing the coverslips in 70% formamide (Sigma-Aldrich)/2× saline-sodium citrate (SSC) heated to 80°C for 2–3 min. Denaturation was quenched in ice-cold 70% ethanol for 5 min, followed by washing with ice-cold 100% ethanol and air drying. A 50-μg amount of labeled probe was mixed with 1 μg of Cot-1 (Roche), 1 μg of E. coli tRNA (Sigma-Aldrich), and 1 μg of single-stranded salmon sperm DNA (Sigma-Aldrich) in 50% formamide (Sigma-Aldrich) and 50% hybridization buffer. Hybridization buffer comprises 20% dextran sulfate (Sigma-Aldrich) and 1% BSA in 4× SSC. The probe was denatured at 80°C for 10 min and then incubated underneath the coverslips in a 37°C humid chamber overnight. Cells were washed for 20 min in 50% formamide in 2× SSC at 37°C and then for 20 min in 2× SSC at 37°C and 20 min in 1× SSC at room temperature. The probe was detected using either FITC conjugated to an anti-digoxigenin antibody (Roche) or an Alexa Fluor conjugated to streptavidin (Invitrogen) in 1% BSA/4× SSC for 60 min in a 37°C humid chamber. Cells were washed again in 4× SSC three times for 10 min at room temperature in the dark. Coverslips were mounted with Vecta Shield (Vector Labs), and Z-stack images were taken on a Zeiss Axioplan2 microscope with a 63× objective. Z-steps of 200 nm were taken and the maximum intensity projections generated using AxioVision, version 4.6. Association percentages for each of the three biological replicates were transformed into arcsine units, and the unpaired Student's t test was used (with Welch's correction) to generate p values. p < 0.05 was considered statistically significant.
Supplementary Material
Acknowledgments
We thank U. Thomas Meier for anti-Nopp140 sera; Li-Jung Juan for p150 DNA constructs; Anthony Imbalzano, Jeffrey Nickerson, and Nathalie Cohet for the gift of the MCF10A TetR-KRAB cells; Jennifer Benanti for human fibroblasts; Michael Brodsky for generous use of the AxioPlan microscope; and Sui Huang for helpful discussions.
Abbreviations used:
- CAF-1
chromatin assembly factor-1
- HP1
heterochromatin protein 1
- NAD
nucleolus-associated domain
- NTPs
nucleoside triphosphates
- PCNA
proliferating cell nuclear antigen
- PIP
PCNA-interaction peptide
- Pol I
RNA polymerase I
- rDNA
ribosomal DNA
- RT-PCR
reverse transcriptase-PCR
- SIM
SUMO-interaction motif
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E14-05-1029) on July 23, 2014.
*Present address: Zenith Epigenetics Corp., Calgary, AB T3E 6L1, Canada.
REFERENCES
- Ahmad Y, Boisvert FM, Gregor P, Cobley A, Lamond AI. NOPdb: Nucleolar Proteome Database—2008 update. Nucleic Acids Res. 2009;37:D181–184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Bondarenko VA, Almagro S, Menoni H, Mongelard F, Hans F, Mietton F, Studitsky VM, Hamiche A, Dimitrov S, et al. Nucleolin is a histone chaperone with FACT-like activity and assists remodeling of nucleosomes. EMBO J. 2006;25:1669–1679. doi: 10.1038/sj.emboj.7601046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell SP, Learned RM, Jantzen HM, Tjian R. Functional cooperativity between transcription factors UBF1 and SL1 mediates human ribosomal RNA synthesis. Science. 1988;241:1192–1197. doi: 10.1126/science.3413483. [DOI] [PubMed] [Google Scholar]
- Benanti JA, Galloway DA. Normal human fibroblasts are resistant to RAS-induced senescence. Mol Cell Biol. 2004;24:2842–2852. doi: 10.1128/MCB.24.7.2842-2852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Shahar TR, Castillo AG, Osborne MJ, Borden KLB, Kornblatt J, Verreault A. Two fundamentally distinct PCNA interaction peptides contribute to chromatin assembly factor 1 function. Mol Cell Biol. 2009;29:6353–6365. doi: 10.1128/MCB.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byron M, Hall LL, Lawrence JB. A multifaceted FISH approach to study endogenous RNAs and DNAs in native nuclear and cell structures. Curr Protoc Hum Genet. 2013 doi: 10.1002/0471142905.hg0415s76. Chapter 4, Unit 4.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campeau E, Ruhl VE, Rodier F, Smith CL, Rahmberg BL, Fuss JO, Campisi J, Yaswen P, Cooper PK, Kaufman PD. A versatile viral system for expression and depletion of proteins in mammalian cells. PLoS One. 2009;4:e6529. doi: 10.1371/journal.pone.0006529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamousset D, De Wever V, Moorhead GB, Chen Y, Boisvert FM, Lamond AI, Trinkle-Mulcahy L. RRP1B targets PP1 to mammalian cell nucleoli and is associated with Pre-60S ribosomal subunits. Mol Biol Cell. 2010;21:4212–4226. doi: 10.1091/mbc.E10-04-0287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CC, Greene E, Bowers SR, Mellone BG. A role for the CAL1-partner Modulo in centromere integrity and accurate chromosome segregation in Drosophila. PLoS One. 2012;7:e45094. doi: 10.1371/journal.pone.0045094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohet N, Stewart KM, Mudhasani R, Asirvatham AJ, Mallappa C, Imbalzano KM, Weaver VM, Imbalzano AN, Nickerson JA. SWI/SNF chromatin remodeling enzyme ATPases promote cell proliferation in normal mammary epithelial cells. J Cell Physiol. 2010;223:667–678. doi: 10.1002/jcp.22072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NP, Qian X, Ziogas A, Papageorge AG, Boersma BJ, Walker RC, Lukes L, Rowe WL, Zhang J, Ambs S, et al. Rrp1b, a new candidate susceptibility gene for breast cancer progression and metastasis. PLoS Genet. 2007;3:e214. doi: 10.1371/journal.pgen.0030214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NP, Yang H, Mattaini KR, Hunter KW. The metastasis efficiency modifier ribosomal RNA processing 1 homolog B (RRP1B) is a chromatin-associated factor. J Biol Chem. 2009;284:28660–28673. doi: 10.1074/jbc.M109.023457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denissov S, Lessard F, Mayer C, Stefanovsky V, van Driel M, Grummt I, Moss T, Stunnenberg HG. A model for the topology of active ribosomal RNA genes. EMBO Rep. 2011;12:231–237. doi: 10.1038/embor.2011.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyen CM, Moshkin YM, Chalkley GE, Bezstarosti K, Demmers JA, Rathke C, Renkawitz-Pohl R, Verrijzer CP. Subunits of the histone chaperone CAF1 also mediate assembly of protamine-based chromatin. Cell Rep. 2013;4:59–65. doi: 10.1016/j.celrep.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Duriez PJ, Shah GM. Cleavage of poly(ADP-ribose) polymerase: a sensitive parameter to study cell death. Biochem Cell Biol. 1997;75:337–349. [PubMed] [Google Scholar]
- Emmott E, Hiscox JA. Nucleolar targeting: the hub of the matter. EMBO Rep. 2009;10:231–238. doi: 10.1038/embor.2009.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng J, McCormack A, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Espada J, Ballestar E, Santoro R, Fraga MF, Villar-Garea A, Nemeth A, Lopez-Serra L, Ropero S, Aranda A, Orozco H, et al. Epigenetic disruption of ribosomal RNA genes and nucleolar architecture in DNA methyltransferase 1 (Dnmt1) deficient cells. Nucleic Acids Res. 2007;35:2191–2198. doi: 10.1093/nar/gkm118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazzio TG, Huff JT, Panning B. An RNAi screen of chromatin proteins identifies Tip60-p400 as a regulator of embryonic stem cell identity. Cell. 2008;134:162–174. doi: 10.1016/j.cell.2008.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner E, Haindl M, Muller S. The SUMO system controls nucleolar partitioning of a novel mammalian ribosome biogenesis complex. EMBO J. 2011;30:1067–1078. doi: 10.1038/emboj.2011.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foltz DR, Jansen LE, Black BE, Bailey AO, Yates JR, 3rd, Cleveland DW. The human CENP-A centromeric nucleosome-associated complex. Nat Cell Biol. 2006;8:458–469. doi: 10.1038/ncb1397. [DOI] [PubMed] [Google Scholar]
- Gaillard P-HL, Martini EM-D, Kaufman PD, Stillman B, Moustacchi E, Almouzni G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor-I. Cell. 1996;86:887–896. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- Green CM, Almouzni G. Local action of the chromatin assembly factor CAF-1 at sites of nucleotide excision repair in vivo. EMBO J. 2003;22:5163–5174. doi: 10.1093/emboj/cdg478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M, Myers MP, Stillman B. An analysis of CAF-1-interacting proteins reveals dynamic and direct interactions with the KU complex and 14–3-3 proteins. J Biol Chem. 2011;286:10876–10887. doi: 10.1074/jbc.M110.217075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek M, Stillman B. Chromatin assembly factor 1 is essential and couples chromatin assembly to DNA replication in vivo. Proc Natl Acad Sci USA. 2003;100:12183–12188. doi: 10.1073/pnas.1635158100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houlard M, Berlivet S, Probst AV, Quivy JP, Hery P, Almouzni G, Gerard M. CAF-1 is essential for heterochromatin organization in pluripotent embryonic cells. PLoS Genet. 2006;2:e181. doi: 10.1371/journal.pgen.0020181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C, Yang Y, Meier UT. Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J Cell Biol. 1998;142:319–329. doi: 10.1083/jcb.142.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal C, Uhlmann-Schiffler H, Stahl H. Redundant role of DEAD box proteins p68 (Ddx5) and p72/p82 (Ddx17) in ribosome biogenesis and cell proliferation. Nucleic Acids Res. 2007;35:3590–3601. doi: 10.1093/nar/gkm058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jentsch S, Psakhye I. Control of nuclear activities by substrate-selective and protein-group SUMOylation. Annu Rev Genet. 2013;47:167–186. doi: 10.1146/annurev-genet-111212-133453. [DOI] [PubMed] [Google Scholar]
- Kaufman PD, Kobayashi R, Kessler N, Stillman B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–1114. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- Kittur N, Zapantis G, Aubuchon M, Santoro N, Bazett-Jones DP, Meier UT. The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol Biol Cell. 2007;18:2296–2304. doi: 10.1091/mbc.E07-02-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klapholz B, Dietrich BH, Schaffner C, Heredia F, Quivy JP, Almouzni G, Dostatni N. CAF-1 is required for efficient replication of euchromatic DNA in Drosophila larval endocycling cells. Chromosoma. 2009;118:235–248. doi: 10.1007/s00412-008-0192-2. [DOI] [PubMed] [Google Scholar]
- Korgaonkar C, Hagen J, Tompkins V, Frazier AA, Allamargot C, Quelle FW, Quelle DE. Nucleophosmin (B23) targets ARF to nucleoli and inhibits its function. Mol Cell Biol. 2005;25:1258–1271. doi: 10.1128/MCB.25.4.1258-1271.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T. Chromatin assembly factor 1 (CAF-1) colocalizes with replication foci in HeLa cell nuclei. Exp Cell Res. 1995;220:304–311. doi: 10.1006/excr.1995.1320. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D. Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev. 2002;16:2893–2905. doi: 10.1101/gad.1035902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci USA. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langst G, Blank TA, Becker PB, Grummt I. RNA polymerase I transcription on nucleosomal templates: the transcription termination factor TTF-I induces chromatin remodeling and relieves transcriptional repression. EMBO J. 1997;16:760–768. doi: 10.1093/emboj/16.4.760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Ou DS, Lee CF, Juan LJ. Gene-specific transcriptional activation mediated by the p150 subunit of the chromatin assembly factor 1. J Biol Chem. 2009;284:14040–14049. doi: 10.1074/jbc.M901833200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Hann SR. Nucleophosmin is essential for c-Myc nucleolar localization and c-Myc-mediated rDNA transcription. Oncogene. 2013;32:1988–1994. doi: 10.1038/onc.2012.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacCallum DE, Hall PA. The location of pKi67 in the outer dense fibrillary compartment of the nucleolus points to a role in ribosome biogenesis during the cell division cycle. J Pathol. 2000;190:537–544. doi: 10.1002/(SICI)1096-9896(200004)190:5<537::AID-PATH577>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Maggi LB, Jr, Weber JD. Nucleolar adaptation in human cancer. Cancer Invest. 2005;23:599–608. doi: 10.1080/07357900500283085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Balbas MA, Tsukiyama T, Gdula D, Wu C. Drosophila NURF-55, a WD repeat protein involved in histone metabolism. Proc Natl Acad Sci USA. 1998;95:132–137. doi: 10.1073/pnas.95.1.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascolo M, Vecchione ML, Ilardi G, Scalvenzi M, Molea G, Di Benedetto M, Nugnes L, Siano M, De Rosa G, Staibano S. Overexpression of chromatin assembly factor-1/p60 helps to predict the prognosis of melanoma patients. BMC Cancer. 2010;10:63. doi: 10.1186/1471-2407-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald WH, Tabb DL, Sadygov RG, MacCoss MJ, Venable J, Graumann J, Johnson JR, Cociorva D, Yates JR., 3rd MS1, MS2, and SQT-three unified, compact, and easily parsed file formats for the storage of shotgun proteomic spectra and identifications. Rapid Commun Mass Spectrom. 2004;18:2162–2168. doi: 10.1002/rcm.1603. [DOI] [PubMed] [Google Scholar]
- McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- Moggs JG, Grandi P, Quivy JP, Jonsson ZO, Hubscher U, Becker PB, Almouzni G. A CAF-1-PCNA-mediated chromatin assembly pathway triggered by sensing DNA damage. Mol Cell Biol. 2000;20:1206–1218. doi: 10.1128/mcb.20.4.1206-1218.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mozgova I, Mokros P, Fajkus J. Dysfunction of chromatin assembly factor 1 induces shortening of telomeres and loss of 45S rDNA in Arabidopsis thaliana. Plant Cell. 2010;22:2768–2780. doi: 10.1105/tpc.110.076182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murano K, Okuwaki M, Hisaoka M, Nagata K. Transcription regulation of the rRNA gene by a multifunctional nucleolar protein, B23/nucleophosmin, through its histone chaperone activity. Mol Cell Biol. 2008;28:3114–3126. doi: 10.1128/MCB.02078-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama A, Ohmori K, Fujimura A, Minami H, Yasuzawa-Tanaka K, Kuroda T, Oie S, Daitoku H, Okuwaki M, Nagata K, et al. Epigenetic control of rDNA loci in response to intracellular energy status. Cell. 2008;133:627–639. doi: 10.1016/j.cell.2008.03.030. [DOI] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–540. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- Nemeth A, Conesa A, Santoyo-Lopez J, Medina I, Montaner D, Peterfia B, Solovei I, Cremer T, Dopazo J, Langst G. Initial genomics of the human nucleolus. PLoS Genet. 2010;6:e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Németh A, Guibert S, Tiwari VK, Ohlsson R, Längst G. Epigenetic regulation of TTF-I-mediated promoter–terminator interactions of rRNA genes. EMBO J. 2008;27:1255–1265. doi: 10.1038/emboj.2008.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs RL, Press RI. Centromere autoantigens are associated with the nucleolus. Exp Cell Res. 1992;200:339–350. doi: 10.1016/0014-4827(92)90181-7. [DOI] [PubMed] [Google Scholar]
- Padeken J, Heun P. Nucleolus and nuclear periphery: Velcro for heterochromatin. Curr Opin Cell Biol. 2014;28C:54–60. doi: 10.1016/j.ceb.2014.03.001. [DOI] [PubMed] [Google Scholar]
- Padeken J, Mendiburo MJ, Chlamydas S, Schwarz HJ, Kremmer E, Heun P. The Nucleoplasmin homolog NLP mediates centromere clustering and anchoring to the nucleolus. Mol Cell. 2013;50:236–249. doi: 10.1016/j.molcel.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pluta AF, Earnshaw WC. Specific interaction between human kinetochore protein CENP-C and a nucleolar transcriptional regulator. J Biol Chem. 1996;271:18767–18774. doi: 10.1074/jbc.271.31.18767. [DOI] [PubMed] [Google Scholar]
- Polo SE, Roche D, Almouzni G. New histone incorporation marks sites of UV repair in human cells. Cell. 2006;127:481–493. doi: 10.1016/j.cell.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Polo SE, Theocharis SE, Klijanienko J, Savignoni A, Asselain B, Vielh P, Almouzni G. Chromatin assembly factor-1, a marker of clinical value to distinguish quiescent from proliferating cells. Cancer Res. 2004;64:2371–2381. doi: 10.1158/0008-5472.can-03-2893. [DOI] [PubMed] [Google Scholar]
- Pontvianne F, Blevins T, Chandrasekhara C, Mozgova I, Hassel C, Pontes OM, Tucker S, Mokros P, Muchova V, Fajkus J, et al. Subnuclear partitioning of rRNA genes between the nucleolus and nucleoplasm reflects alternative epiallelic states. Genes Dev. 2013;27:1545–1550. doi: 10.1101/gad.221648.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Gerard A, Cook AJ, Roche D, Almouzni G. The HP1-p150/CAF-1 interaction is required for pericentric heterochromatin replication and S-phase progression in mouse cells. Nat Struct Mol Biol. 2008;15:972–979. doi: 10.1038/nsmb.1470. [DOI] [PubMed] [Google Scholar]
- Quivy JP, Grandi P, Almouzni G. Dimerization of the largest subunit of chromatin assembly factor 1: importance in vitro and during Xenopus early development. EMBO J. 2001;20:2015–2027. doi: 10.1093/emboj/20.8.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. EMBO J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmanzadeh R, Huttmann G, Gerdes J, Scholzen T. Chromophore-assisted light inactivation of pKi-67 leads to inhibition of ribosomal RNA synthesis. Cell Prolif. 2007;40:422–430. doi: 10.1111/j.1365-2184.2007.00433.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ransom M, Dennehey BK, Tyler JK. Chaperoning histones during DNA replication and repair. Cell. 2010;140:183–195. doi: 10.1016/j.cell.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichow SL, Hamma T, Ferre-D'Amare AR, Varani G. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 2007;35:1452–1464. doi: 10.1093/nar/gkl1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickards B, Flint SJ, Cole MD, LeRoy G. Nucleolin is required for RNA polymerase I transcription in vivo. Mol Cell Biol. 2007;27:937–948. doi: 10.1128/MCB.01584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell. 1999;96:575–585. doi: 10.1016/s0092-8674(00)80661-3. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- Smith S, Stillman B. Immunological characterization of chromatin assembly factor I, a human cell factor required for chromatin assembly during DNA replication in vitro. J Biol Chem. 1991;266:12041–12047. [PubMed] [Google Scholar]
- Song JJ, Garlick JD, Kingston RE. Structural basis of histone H4 recognition by p55. Genes Dev. 2008;22:1313–1318. doi: 10.1101/gad.1653308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staibano S, Mascolo M, Mancini FP, Kisslinger A, Salvatore G, Di Benedetto M, Chieffi P, Altieri V, Prezioso D, Ilardi G, et al. Overexpression of chromatin assembly factor-1 (CAF-1) p60 is predictive of adverse behaviour of prostatic cancer. Histopathology. 2009;54:580–589. doi: 10.1111/j.1365-2559.2009.03266.x. [DOI] [PubMed] [Google Scholar]
- Sun H, Hunter T. Poly-small ubiquitin-like modifier (PolySUMO)-binding proteins identified through a string search. J Biol Chem. 2012;287:42071–42083. doi: 10.1074/jbc.M112.410985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surendranath V, Theis M, Habermann BH, Buchholz F. Designing efficient and specific endoribonuclease-prepared siRNAs. Methods Mol Biol. 2013;942:193–204. doi: 10.1007/978-1-62703-119-6_11. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagami H, Ray-Gallet D, Almouzni G, Nakatani Y. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell. 2004;116:51–61. doi: 10.1016/s0092-8674(03)01064-x. [DOI] [PubMed] [Google Scholar]
- Takagi M, Sueishi M, Saiwaki T, Kametaka A, Yoneda Y. A novel nucleolar protein, NIFK, interacts with the forkhead associated domain of Ki-67 antigen in mitosis. J Biol Chem. 2001;276:25386–25391. doi: 10.1074/jbc.M102227200. [DOI] [PubMed] [Google Scholar]
- Takami Y, Ono T, Fukagawa T, Shibahara K, Nakayama T. Essential role of chromatin assembly factor-1-mediated rapid nucleosome assembly for DNA replication and cell division in vertebrate cells. Mol Biol Cell. 2007;18:129–141. doi: 10.1091/mbc.E06-05-0426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Poustovoitov MV, Zhao K, Garfinkel M, Canutescu A, Dunbrack R, Adams PD, Marmorstein R. Structure of a human ASF1a-HIRA complex and insights into specificity of histone chaperone complex assembly. Nat Struct Mol Biol. 2006;13:921–929. doi: 10.1038/nsmb1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessarz P, Santos-Rosa H, Robson SC, Sylvestersen KB, Nelson CJ, Nielsen ML, Kouzarides T. Glutamine methylation in histone H2A is an RNA-polymerase-I-dedicated modification. Nature. 2014;505:564–568. doi: 10.1038/nature12819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasad-Sinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT. Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol. 2001;21:6574–6584. doi: 10.1128/MCB.21.19.6574-6584.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwada J, Tanaka N, Yamaguchi Y, Uchimura Y, Shibahara K-I, Nakao M, Saitoh H. The p150 subunit of CAF-1 causes association of SUMO2/3 with the DNA replication foci. Biochem Biophys Res Commun. 2010;391:407–413. doi: 10.1016/j.bbrc.2009.11.071. [DOI] [PubMed] [Google Scholar]
- van Koningsbruggen S, Gierlinski M, Schofield P, Martin D, Barton GJ, Ariyurek Y, den Dunnen JT, Lamond AI. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21:3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Warburton PE, Greig GM, Haaf T, Willard HF. PCR amplification of chromosome-specific alpha satellite DNA: definition of centromeric STS markers and polymorphic analysis. Genomics. 1991;11:324–333. doi: 10.1016/0888-7543(91)90139-6. [DOI] [PubMed] [Google Scholar]
- Westman BJ, Verheggen C, Hutten S, Lam YW, Bertrand E, Lamond AI. A proteomic screen for nucleolar SUMO targets shows SUMOylation modulates the function of Nop5/Nop58. Mol Cell. 2010;39:618–631. doi: 10.1016/j.molcel.2010.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Sherlock G, Saldanha AJ, Murray JI, Ball CA, Alexander KE, Matese JC, Perou CM, Hurt MM, Brown PO, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GN. The structure and organization of human ribosomal genes. In: Busch ed. H, Rothblum and L., editors. In: The Cell Nucleus. Vol. 10A. New York: Academic Press; 1982. pp. 289–318. [Google Scholar]
- Wong LH, Brettingham-Moore KH, Chan L, Quach JM, Anderson MA, Northrop EL, Hannan R, Saffery R, Shaw ML, Williams E, et al. Centromere RNA is a key component for the assembly of nucleoproteins at the nucleolus and centromere. Genome Res. 2007;17:1146–1160. doi: 10.1101/gr.6022807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- Yerushalmi R, Woods R, Ravdin PM, Hayes MM. Gelmon KA (2010). Ki67 in breast cancer: prognostic and predictive potential. Lancet Oncol. 11:174–183. doi: 10.1016/S1470-2045(09)70262-1. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Ng HH, Erdjument-Bromage H, Tempst P, Bird A, Reinberg D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999;13:1924–1935. doi: 10.1101/gad.13.15.1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.