Abstract
Aberrant expression of miR-148b has been found in several types of cancer, but its expression and potential biologic role in non-small cell lung cancer (NSCLC) are still largely unknown. Here, we found that miR-148b was commonly under-expressed in human non-small celllung cancer (NSCLC) specimens and cell lines. The overexpression of miR-148b dramatically suppressed NSCLC cell proliferation and migration. Furthermore, miR-148b could regulate carcinoembryonic antigen (CEA) expression by luciferase reporter assay. On the other hand, CEA was widely up-regulated in NSCLC specimens, and its mRNA levels were inversely correlated with miR-148b expression. These suggest that CEA expression may be regulated by miR-148b. Collectively, our findings indicate miR-148b is low expression in NSCLC cells, which results in CEA overexpression and disease progression in NSCLC patients.
Keywords: NSCLC, CEA, miR-148b, migration, tumor suppressors
Introduction
Lung cancer has a higher death rate among all cancers and continues to be the leading cause of cancer-related death in both men and women throughout the world [1]. Lung tumors are usually classified into non-small cell lung cancer (NSCLC) and small-cell lung cancer (SCLC) depending on their pathological and histological characteristics [2]. Non-small cell lung cancer (NSCLC) accounts for the majority (85%) of lung cancers and its five-year survival is only 15% [3]. More than half of the patients (56%) with lung cancer at the time of diagnosis have advanced or metastatic disease [4] and even with chemotherapy have a median survival of one year or less [5]. Therefore, a good understanding of the molecular mechanisms underlying NSCLC development and progression is urgently needed.
miRNAs are small non-coding RNA molecules that suppress gene expression by interacting with the 3’-untranslated regions (3’UTRs) of target mRNAs. These interactions may result in either inhibition of translation of the targeted mRNAs or their degradation [6]. Dysregulation of miRNAs may lead to alterations in cellular differentiation, proliferation, apoptosis and metastasis processes that are important in the development of cancer [7]. In lung cancer, multiple miRNAs, such as let-7 family, miR-200, miR-486, miR-124 and miR-146a have been identified as tumor suppressors [8-12]; on the other hand, miR-31, miR-212 and miR-196a were found to promote NSCLC carcinogenesis [13-15].
miR-148b is located at chromosome 12q13 and recent studies have found it is down-regulated in oral, pancreatic, colon and gastric cancer tissues [16-21]. Furthermore, an increasing number of reports indicate that miR-148b acts as a tumor suppressor by targeting specific oncogenes. However, the function and mechanism of miR-148b in NSCLC development remain unknown. In this study, we identified that miR-148b was down-regulated in NSCLC cell lines and tissues. Functionally, miR-148b suppresses the proliferation and migration of NSCLC cells by targeting CEA pro-oncogene. Our finding will help to better understand the biological activities of miR-148b in NSCLC.
Materials and methods
Cell lines and culture
The NSCLC cell lines PC14/B and A549 were maintained in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, USA) supplemented with 10% fetal bovine serum (HyClone, Logan, USA). The cells were grown in a humidified incubator at 37°C with 5% CO2 and split every two days.
Oligonucleotide transfection
The miR-148b mimics and a nonspecific miRNA control were synthesized by GenePharma (Shanghai, China). miRNAs were transfected at a working concentration of 50 nmol/L using RNAiMAX reagent (Invitrogen, USA) according to the manufacturer’s instructions. The transfected cells were incubated at 37°C for 24 hrs in complete medium and harvested at the indicated time points.
RNA extraction, reverse transcription, and quantitative real-time PCR
Total RNA was extracted using TRIzol (Invitrogen, USA) according to the manufacturer’s instructions. cDNA was synthesized with the MLV transcriptase Kit (Invitrogen, USA). The quantitative analysis of miR-148b expression was assayed using a Bulge-LoopTM miRNA qRT-PCR primer (RIBOBIO, Guangzhou, China) and Platinum® SYBR® Green qPCR SuperMix-UDG with ROX (Invitrogen, USA) on an ABI 7900HT instrument (Applied Biosystems, Foster, CA) according to the manufacturer’s instructions, and U6 small nuclear RNA (U6-snRNA) purchased from Ribobio was used as an internal control. The fold changes were calculated through relative quantification with 2-ΔΔCt [22]. All of the reactions were performed in a 20 μL reaction volume in triplicate.
Western blot analysis
This procedure was detailed previously [23]. Briefly, protein lysates were resolved through 10% SDS-PAGE and electrophoretically transferred to a PVDF (polyvinylidene difluoride) membrane (Millipore, USA). Then, the membrane was probed with an antibody against human CEA (1:1,000 dilution, Cell Signaling Technology) or β-actin (Bioworld, USA) as a protein loading control, followed by peroxidase-conjugated goat anti-mouse IgG (H+L) (DingGuo, China) as the secondary antibody. The intensity of the protein fragments was visualized with an X-ray image film processing machine (Kodak, Japan).
MTT and colony formation assay
For cell proliferation assays, cells transfected with miRNAs or co-transfected with miRNAs and plasmids for 24 hrs were reseeded in 96-well plates at 1.5 × 103 cells/well in a final volume of 150 μL and incubated overnight. The effect of miR-148b/CEA on cell growth and proliferation was determined with an MTT assay as described previously [24].
For colony-forming assays, after 24 hrs of transfection, the cells were reseeded in 6-well plates at 1,000 cells per well, and the medium was replaced every 3 days. After incubation at 37°C for 2 weeks, the cells were washed twice with PBS, fixed and stained with 0.5% crystal violet. The number of colonies was counted under a microscope.
Transwell assay
Transwell assays were carried out in modified Boyden chambers with 8 μm pore filter inserts for 24-well plates as described previously [12]. Briefly, 105 cells suspended in serum free DMEM were added to the up chamber (BD Biosciences, USA) present in the insert of a 24-well culture plate. FBS was added to the lower chamber at the final concentration of 10% as a chemoattractant. After 8 hrs, the non-filtered cells were gently removed with a cotton swab. Filtered cells located on the lower side of the chamber were stained with crystal violet, air dried and photographed.
Luciferase reporter assay
The 3’UTR sequence of CEA containing the putative miR-148b-binding site was amplified with PCR and cloned into the pGL3 vector (Promega) downstream the firefly luciferase gene, which was designated as wild-type 3’UTR (wt 3’UTR). Mutagenesis was performed using a Quik-Change Site-Directed Mutagenesis Kit (Stratagene, USA) according to the manufacturer’s instructions, resulting in mutated 3’UTR (mut 3’UTR). The primers used for the construction of luciferase reporters are CEA wt (forward, GCTCTAGACAGCCCTGGTGTAGTTTCTTC; reverse, GCTCTAGACGACATTCTAGTCAAAGTCTTGG) and CEA mut (forward, AGTTTCTGATACCACAGGAGTCTCTGAGAATTTCCAA; reverse, AACCTTTAAGAGTCTCTGAGGACACCATAGTCTTTGA). Wt or mut 3’UTR vector and the control vector pRL-TK (Promega, USA) coding for Renilla luciferase were cotransfected with miR-148b mimics or miR-control into PC14/B cells using Lipofectamine 2000 (Invitrogen). The luciferase activity was measured 48 hrs later using the Dual-Luciferase Reporter Assay System (Promega). The firefly luciferase values were normalized to Renilla, and the ratio of firefly/Renilla values is presented. The experiments were performed independently in triplicate.
Rescue experiments
The open reading frame (ORF) encoding CEA protein was amplified and cloned into pCDNA6B in frame with the Myc-tag coding frame. The primers used for amplified CEA were forward, TAGGTACCATGGAGTCTCCCTCGGCCC and reverse, GTCTCGAGTATCAGAGCAACCCCAACCA. NSCLC cells cotransfected with miR-148b mimics and CEA expressing vector or empty plasmid for 24 hrs were trypsinized and subjected to MTT, colony formation or transwell assay.
Statistical analysis
Statistical analyses were performed using SPSS 16.0 software (SPSS Inc., Chicago, IL). The values were presented as the mean ± standard deviation (SD) of three independent experiments. Differences between two groups were evaluated by a two-tailed Student’s t test. The relationship between CEA and mir-148b expression was assessed with 2-tailed Pearson’s correlation. Differences were considered to be statistically significant at P<0.05, *P<0.05 and **P<0.01.
Results
miR-148b was down-regulated in human NSCLC cell lines and clinical samples
To test whether miR-148b was down-regulated in NSCLC, the level of miR-148b was evaluated with qRT-PCR in 13 NSCLC specimens and 12 normal lung tissues. As shown in Figure 1A, miR-148b was obviously down-regulated in NSCLC compared with normal lung tissues. We then examined miR-148b level in NSCLC cells. Consistent with the observation that miR-148b level reduced in the clinical samples, miR-148b expression was lower in NSCLC cells than in normal cells (Figure 1B). These data indicate that miR-148b was down-regulated in NSCLC and may be functional as a tumor suppressor in NSCLC.
Figure 1.
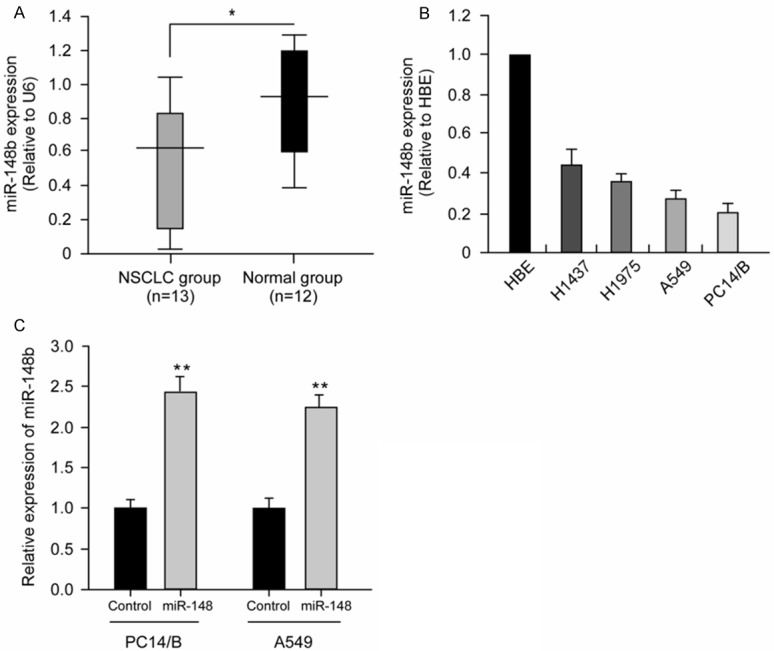
miR-148b was down-regulated in NSCLC cell lines and clinical specimens. A. miR-148b expression was examined in human NSCLC specimens (n = 13) and unmatched normal lung tissues (n = 12) using real-time PCR (*P<0.05). B. The expression levels of miR-148b was lower in 4 NSCLC cell lines (H1437, H1975, A549 and PC14/B) than in normal lung cells (HBE) (*P<0.05). Each experiment was performed in triplicate. C. Altered miR-148b expression after transfection with miR-148b mimic in PC14/B and A549 cells. Total RNA was extracted from PC14/B and A549 cells transfected with miRNAs as indicated and analyzed by qRT-PCR. Each value represents the mean ± SD for three independent experiments and the data were normalized with U6 content. **P<0.01 compared with control.
miR-148b regulated cell proliferation and migration of NSCLC cells
To test the functional contribution of miR-148b on NSCLC cells, we transiently transfected miR-148b mimics or miR-control into PC14/B and A549 cells and the overexpression of miR-148b in PC14/B and A549 cells was evaluated with qRT-PCR (Figure 1C). The proliferation of NSCLC cells was tested by MTT assay. As shown in Figure 2A, the overexpression of mir-148b led to an obvious growth arrest in PC14/B and A549 cells (P<0.05). Along with the observation that miR-148b induced growth inhibition of NSCLC cells, the colony formation assay demonstrated that miR-148b overexpression in PC14/B and A549 cells showed obvious colony inhibition compared with control transfectants (Figure 2B; P<0.01). We then examined the effect of miR-148b on the migratory ability of NSCLC cells. As shown in Figure 2C, ectopic expression of miR-148b in PC14/B and A549 cells notably reduced the filtered cells compared with control cells. Taken together, these results indicate that miR-148b may function as a tumor suppressor in NSCLC and inhibit the cell proliferation and migration of NSCLC cells.
Figure 2.
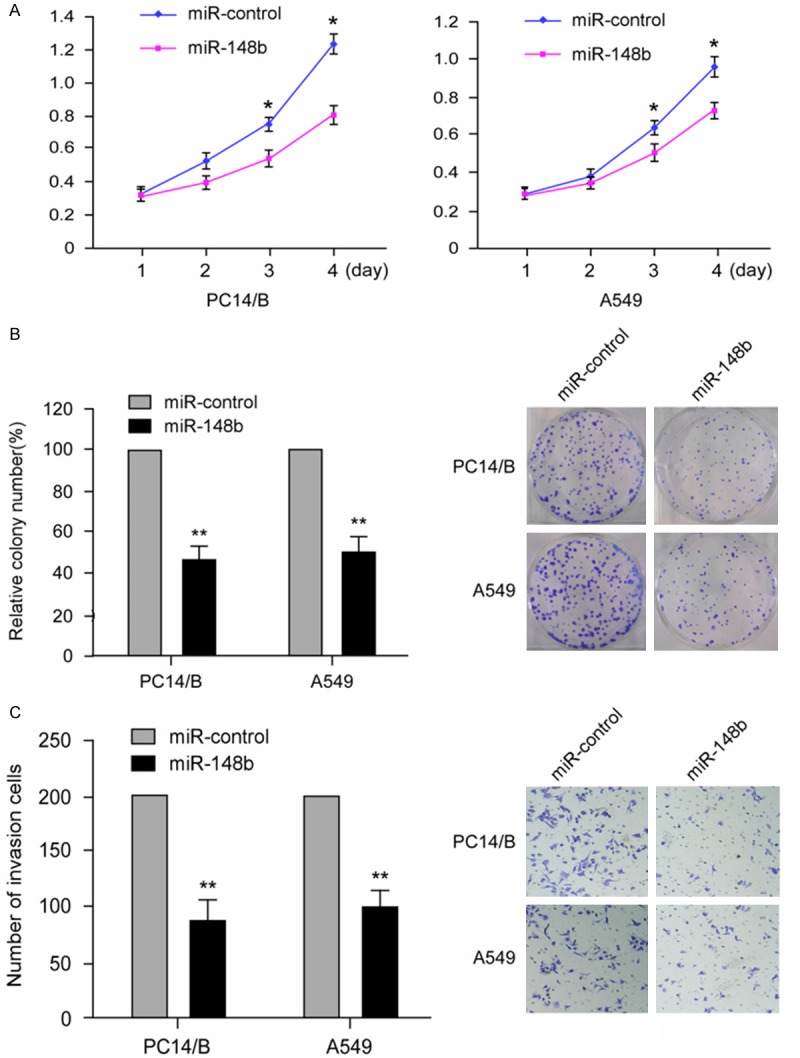
miR-148b induced growth inhibition and inhibited migration of NSCLC cells. A. The MTT assay showed that miR-148b overexpression inhibited the growth and proliferation of PC14/B and A549. The data are expressed as absorbance values (*P<0.05). The data is a represent of at least three independent experiments. B. The colony formation assay for mir-148b-transfected PC14/B and A549 cells compared with control transfectants. The results are expressed as the means ± SD; n = 3, **P<0.01 compared with the control. C. PC14/B and A549 cells transfected with miR-148b and miR-control for 48 hrs were subjected to transwell assay. Quantification was conducted by counting the stained cells invading the lower chamber under a light microscopy. The results are expressed as the means ± SD; n = 3, **P<0.01 compared with the control.
miR-148b directly regulated CEA expression in NSCLC cells
To determine the mechanism by which miR-148b suppresses the growth and migration of NSCLC cells, we used the bioinformatic prediction algorithm Target Scan to analyze the direct mRNA targets of miR-148b. Of all of the hypothetical targets of miR-148b, CEA, an important oncogene in NSCLC, provoked our interest. To further characterize whether CEA responds to miR-148b through direct 3’UTR interaction in NSCLC cells, we cloned the wild-type 3’UTR of the putative miR-148b target (wt 3’UTR) or a mutated sequence (mut 3’UTR) into a reporter plasmid downstream of the luciferase gene (Figure 3A). These plasmids, together with miR-148b mimics or miR-control and the Renilla luciferase vector (pRL-TK) (internal control), were transiently transfected into PC14/B cells. A dual-luciferase reporter assay system was used to detect luciferase expression 48 hrs after transfection. The results showed that miR-148b mimics led to the attenuation of luciferase activity of the wt 3’UTR by more than 2 fold compared with the negative control (Figure 3B, lanes 2 and 3; P<0.05), whereas mut 3’UTR showed no response to miR-148b (Figure 3B, lanes 5 and 6). To further confirm the potential role of miR-148b in the regulation of CEA, we evaluated CEA mRNA and protein expression levels in PC14/B cell when introduction of miR-148b mimics. As shown in Figure 3C, the overexpression of miR-148b with miR-148b mimics led to a notable decrease in CEA mRNA and protein levels (P<0.05). These results indicated that CEA is a direct target of mir-148b in NSCLC cells.
Figure 3.
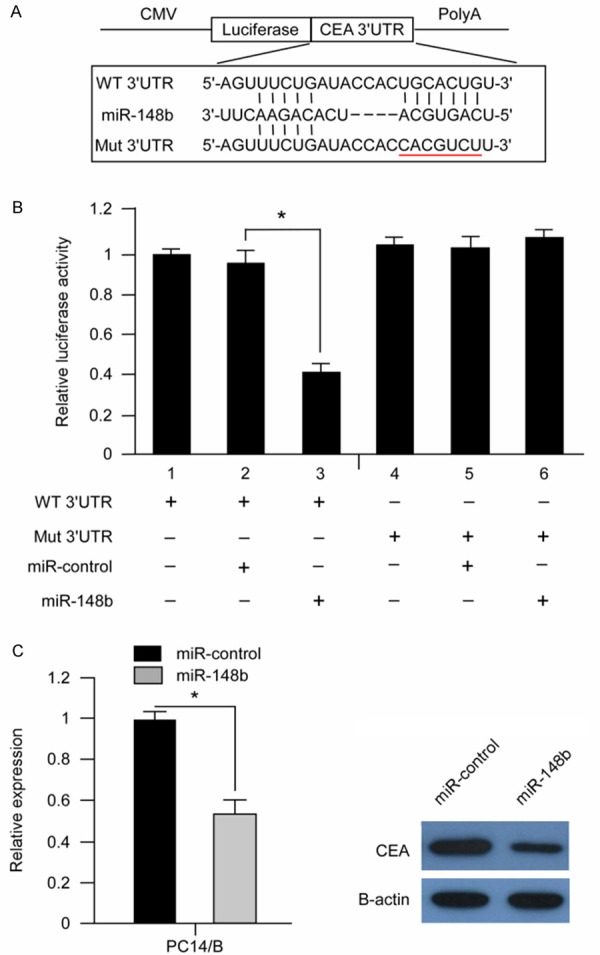
CEA was a direct target of mir-148b in NSCLC cells. A. Schematic diagram of the reporter constructs containing the predicted mir-148b-binding site in the 3’UTR of CEA. B. The overexpression of mir-148b with the miRNA mimics significantly attenuated the luciferase activitiy of the CEA 3’UTR in PC14/B cells. The co-transfection of wt or mut 3’UTR and miRNAs is indicated. *P<0.05 compared with the control. C. The overexpression of mir-148b reduced endogenous CEA protein and mRNA levels. PC14/B cell was transfected with miRNAs for 48 hrs. The CEA protein level was measured with Western blotting, and the target mRNA levels were evaluated with real-time quantitative PCR.
miR-148b regulates the proliferating and migratory ability of NSCLC through CEA signal pathway
We then examined whether CEA could counteract the tumor suppressing function of miR-148b on NSCLC cells. Recombinant vectors delivering CEA or empty vector were co-transfected with miR-148b mimics into PC14/B or A549 cells and the overexpression of CEA were evaluated by western blot (Figure 4A). MTT assay showed that overexpression of CEA abrogated the miR-148b mediated proliferation inhibition of PC14/B and A549 cells (Figure 4B). Besides, ectopic expression CEA reverses the colony formation and migration suppressing effect on PC14/B and A549 cells exerted by miR-148b (Figure 4C and 4D). Taken together, these results indicate that CEA mediated the tumor suppressing function of miR-148b on NSCLC cells.
Figure 4.
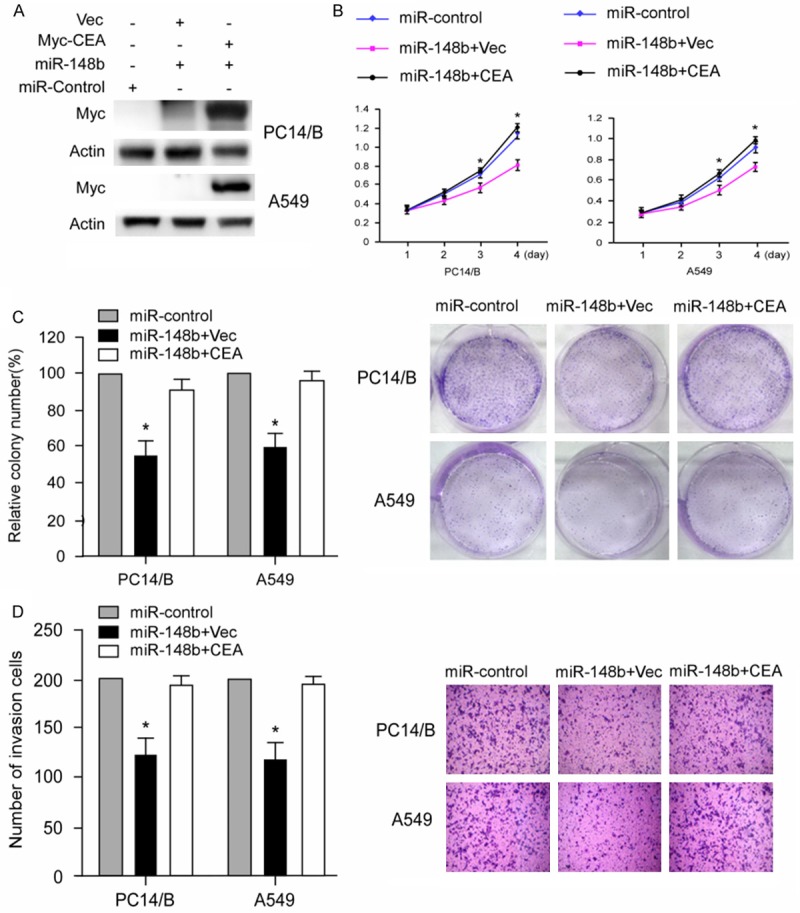
Overexpression of CEA counteracted the miR-148b mediated tumor-suppressing effect on NSCLC cells. (A) Overexpression of CEA in PC14/B and A549 cells was evaluated by western blot. (B-D) Overexpression of CEA reversed inhibiting effect of proliferation (B), colony formation (C) and migration (D) on PC14/B and A549 cells mediated by miR-148b. The results are expressed as the means ± SD; n = 3, *P<0.05 compared with the control.
CEA was up-regulated and inversely correlated with miR-148b levels in NSCLC
To better address the important role of miR-148b/CEA signal pathway in NSCLC, we assessed the significance of the mir-148b/CEA correlation in NSCLC. The qRT-PCR results indicated that the mean expression level of CEA was markedly higher in human NSCLC tissues than that in normal lung tissues (Figure 5A; P<0.05). Next, we determined the correlation between CEA and mir-148b expression in the same NSCLC specimens. As shown in Figure 5B, when the CEA mRNA levels were plotted against mir-148b expression, a significant inverse correlation was obtained (2-tailed Pearson’s correlation, Figure 5B; r = -0.803; P<0.01).
Figure 5.
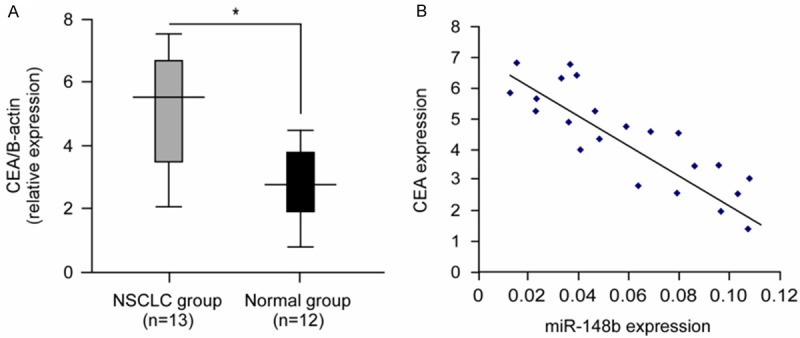
The correlation between mir-148b and CEA expression in human NSCLC. A. The average expression level of CEA (analyzed with q-PCR) in FFPETs of human NSCLC and unmatched normal lung tissue. The CEA level was normalized to β-actin (*P<0.05). B. mir-148b expression levels were inversely correlated with CEA mRNA levels in NSCLC FFPETs (2-tailed Pearson’s correlation analysis, r = -0.803; **P<0.01).
Discussion
The down-regulation of miR-148b is a frequent event in various cancers, suggesting that miR-148b may play an important role in tumorigenesis and tumor progression. Indeed, several key oncogenic functions have been attributed to miR-148b in the context of tumorigenesis. Zhao et al reported that overexpression of miR-148b induced the apoptosis and cell-cycle arrest of pancreatic cancer cells by targeting AMPKa1 [20]. Furthermore, miR-148b was found to play an important role in regulating the metastasis of hepatocellular carcinoma cells through targeting CCK2R [25]. In breast cancer, ectopical expression of miR-148b inhibits the invasion, survival to anoikis, extravasation, and lung metastasis formation of breast cancer cells [26]. In non-Hodgkin’s Lymphoma (NHL), miR-148b was reported to increase the radiosensitivity of NHL cells [27]. However, the potential function of this microRNA in human NSCLC remains unknown. In the present study, we are interested in the potential role of miR-148b in suppressing the progression of NSCLC cells.
We first analyzed the expression level of miR-148b in NSCLC clinical samples and cells and found that miR-148b was notably down-regulated in NSCLC. These data indicate that miR-148b may function as a tumor suppressor. We then investigated the function of miR-148b in NSCLC. The results showed that the re-introduction of miR-148b dramatically repressed the cell proliferation and migration of NSCLC cells in vitro. Therefore, our data suggested that the decreased expression of miR-148b may contribute to the progression of NSCLC cells. We further characterized CEA as a direct target of miR-148b by luciferase reporter assays, RT-PCR and Western blot analysis, respectively. CEA expression was first characterized in gastrointestinal cancers [28] and was considered to involve cell-cell recognition and modulation of cellular processes [29]. This biomarker has been extensively studied in a variety of neoplasms, such as colorectal [30], gastric [31], esophageal [32], pancreatic [33], and breast carcinoma [34], in regard to its potential role as a prognostic factor. For NSCLC in particular, CEA levels have been widely reported to be correlated with advanced disease, early relapse, pathological upstaging, poor therapeutic response and survival [35-38]. In the present study, we found that CEA mRNA was highly expressed and reversely correlated with miR-148b level in NSCLC tissues. Notably, overexpression of CEA could reverse the proliferating and migratory inhibition effect on NSCLC cells mediated by miR-148b, indicating that miR-148b may exert functions in proliferation and migration of NSCLC cells through modulating CEA level. Therefore, our data provide the evidence that miR-148b regulates CEA and that low miR-148b level contributes to the overexpression of CEA in NSCLC. These findings suggest another mechanism for the upregulation of CEA in NSCLC. In summary, we investigated the role of miR-148b in NSCLC development. Our finding suggests that miR-148b was down-regulated and acted as a tumor suppressor in human NSCLC. mir-148b blocks the proliferation and migration of NSCLC cells through targeting CEA. Our data provide new insight into the mechanism responsible for the development of human NSCLC. Additionally, miR-148b may serve as a potential therapeutic candidate in the treatment of NSCLC.
In summary, we investigated the role of miR-148b in NSCLC development. Our finding suggests that miR-148b was down-regulated and acted as a tumor suppressor in human NSCLC. miR-148b blocks the proliferation and migration of NSCLC cells through targeting CEA. Our data provide new insight into the mechanism responsible for the development of human NSCLC. Additionally, miR-148b may serve as a potential therapeutic candidate in the treatment of NSCLC.
Acknowledgements
This work was supportedby the Project of Science and Information Technology of Guangzhou (7411788560328), Guangzhou Medical College and Project of Guangdong Key Laboratory of Clinical Molecular Medicine and Diagnostics.
Disclosure of conflict of interest
None.
References
- 1.Chen Y, Peng W, Lu Y, Chen J, Zhu YY, Xi T. MiR-200a enhances the migrations of A549 and SK-MES-1 cells by regulating the expression of TSPAN1. J Biosci. 2013;38:523–532. doi: 10.1007/s12038-013-9351-6. [DOI] [PubMed] [Google Scholar]
- 2.Calbo J, Meuwissen R, van Montfort E, van Tellingen O, Berns A. Genotype-phenotype relationships in a mouse model for human small-cell lung cancer. Cold Spring Harb Symp Quant Biol. 2005;70:225–232. doi: 10.1101/sqb.2005.70.026. [DOI] [PubMed] [Google Scholar]
- 3.Verdecchia A, Francisci S, Brenner H, Gatta G, Micheli A, Mangone L, Kunkler I. Recent cancer survival in Europe: a 2000-02 period analysis of EUROCARE-4 data. Lancet oncol. 2007;8:784–796. doi: 10.1016/S1470-2045(07)70246-2. [DOI] [PubMed] [Google Scholar]
- 4.Jafri SH, Shi R, Mills G. Advance lung cancer inflammation index (ALI) at diagnosis is a prognostic marker in patients with metastatic non-small cell lung cancer (NSCLC): a retrospective review. BMC Cancer. 2013;13:158. doi: 10.1186/1471-2407-13-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sandler A, Gray R, Perry MC, Brahmer J, Schiller JH, Dowlati A, Lilenbaum R, Johnson DH. Paclitaxel-carboplatin alone or with bevacizumab for non-small-cell lung cancer. N Engl J Med. 2006;355:2542–2550. doi: 10.1056/NEJMoa061884. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Chitwood DH, Timmermans MC. Small RNAs are on the move. Nature. 2010;467:415–419. doi: 10.1038/nature09351. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, Zheng Q, Ma Y, Zhang J, Wu N, Yang Y. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51. doi: 10.1016/j.canlet.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 9.Pacurari M, Addison JB, Bondalapati N, Wan YW, Luo D, Qian Y, Castranova V, Ivanov AV, Guo NL. The microRNA-200 family targets multiple non-small cell lung cancer prognostic markers in H1299 cells and BEAS-2B cells. Int J Oncol. 2013;43:548–560. doi: 10.3892/ijo.2013.1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang J, Tian X, Han R, Zhang X, Wang X, Shen H, Xue L, Liu Y, Yan X, Shen J, Mannoor K, Deepak J, Donahue JM, Stass SA, Xing L, Jiang F. Downregulation of miR-486-5p contributes to tumor progression and metastasis by targeting protumorigenic ARHGAP5 in lung cancer. Oncogene. 2014;33:1181–1189. doi: 10.1038/onc.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen G, Umelo IA, Lv S, Teugels E, Fostier K, Kronenberger P, Dewaele A, Sadones J, Geers C, De Greve J. miR-146a inhibits cell growth, cell migration and induces apoptosis in non-small cell lung cancer cells. PLoS One. 2013;8:e60317. doi: 10.1371/journal.pone.0060317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv XB, Jiao Y, Qing Y, Hu H, Cui X, Lin T, Song E, Yu F. miR-124 suppresses multiple steps of breast cancer metastasis by targeting a cohort of pro-metastatic genes in vitro. Chin J Cancer. 2011;30:821–830. doi: 10.5732/cjc.011.10289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhong Z, Dong Z, Yang L, Chen X, Gong Z. MicroRNA-31-5p modulates cell cycle by targeting human mutL homolog 1 in human cancer cells. tumour biology the journal of the International Society for Oncodevelopmental Biology and Medicine. Tumour Biol. 2013;34:1959–1965. doi: 10.1007/s13277-013-0741-z. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zhang D, Chen C, Ruan Z, Huang Y. MicroRNA-212 displays tumor-promoting properties in non-small cell lung cancer cells and targets the hedgehog pathway receptor PTCH1. Mol Biol Cell. 2012;23:1423–1434. doi: 10.1091/mbc.E11-09-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu XH, Lu KH, Wang KM, Sun M, Zhang EB, Yang JS, Yin DD, Liu ZL, Zhou J, Liu ZJ, De W, Wang ZX. MicroRNA-196a promotes non-small cell lung cancer cell proliferation and invasion through targeting HOXA5. BMC Cancer. 2012;12:348. doi: 10.1186/1471-2407-12-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu T, Wang XY, Gong RG, Li A, Yang S, Cao YT, Wen YM, Wang CM, Yi XZ. The expression profile of microRNAs in a model of 7,12-dimethyl-benz[a] anthrance-induced oral carcinogenesis in Syrian hamster. J Exp Clin Cancer Res. 2009;28:64. doi: 10.1186/1756-9966-28-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, Liu CG, Bhatt D, Taccioli C, Croce CM. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. doi: 10.1001/jama.297.17.1901. [DOI] [PubMed] [Google Scholar]
- 18.Schetter AJ, Leung SY, Sohn JJ, Zanetti KA, Bowman ED, Yanaihara N, Yuen ST, Chan TL, Kwong DL, Au GK, Liu CG, Calin GA, Croce CM, Harris CC. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299:425–436. doi: 10.1001/jama.299.4.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, Alder H, Liu CG, Oue N, Yasui W, Yoshida K, Sasaki H, Nomura S, Seto Y, Kaminishi M, Calin GA, Croce CM. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010;11:136–146. doi: 10.1016/S1470-2045(09)70343-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao G, Zhang JG, Liu Y, Qin Q, Wang B, Tian K, Liu L, Li X, Niu Y, Deng SC, Wang CY. miR-148b functions as a tumor suppressor in pancreatic cancer by targeting AMPKa1. Mol Cancer Ther. 2013;12:83–93. doi: 10.1158/1535-7163.MCT-12-0534-T. [DOI] [PubMed] [Google Scholar]
- 21.Song YX, Yue ZY, Wang ZN, Xu YY, Luo Y, Xu HM, Zhang X, Jiang L, Xing CZ, Zhang Y. MicroRNA-148b is frequently down-regulated in gastric cancer and acts as a tumor suppressor by inhibiting cell proliferation. Mol Cancer. 2011;10:1. doi: 10.1186/1476-4598-10-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Lv XB, Xie F, Hu K, Wu Y, Cao LL, Han X, Sang Y, Zeng YX, Kang T. Damaged DNA-binding protein 1 (DDB1) interacts with Cdh1 and modulates the function of APC/CCdh1. J Biol Chem. 2010;285:18234–18240. doi: 10.1074/jbc.M109.094144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X, Lv XB, Wang XP, Sang Y, Xu S, Hu K, Wu M, Liang Y, Liu P, Tang J, Lu WH, Feng QS, Chen LZ, Qian CN, Bei JX, Kang T, Zeng YX. MiR-138 suppressed nasopharyngeal carcinoma growth and tumorigenesis by targeting the CCND1 oncogene. Cell Cycle. 2012;11:2495–2506. doi: 10.4161/cc.20898. [DOI] [PubMed] [Google Scholar]
- 25.Song Y, Xu Y, Wang Z, Chen Y, Yue Z, Gao P, Xing C, Xu H. MicroRNA-148b suppresses cell growth by targeting cholecystokinin-2 receptor in colorectal cancer. Int J Cancer. 2012;131:1042–1051. doi: 10.1002/ijc.26485. [DOI] [PubMed] [Google Scholar]
- 26.Cimino D, De Pitta C, Orso F, Zampini M, Casara S, Penna E, Quaglino E, Forni M, Damasco C, Pinatel E, Ponzone R, Romualdi C, Brisken C, De Bortoli M, Biglia N, Provero P, Lanfranchi G, Taverna D. miR148b is a major coordinator of breast cancer progression in a relapse-associated microRNA signature by targeting ITGA5, ROCK1, PIK3CA, NRAS, and CSF1. FASEB J. 2013;27:1223–1235. doi: 10.1096/fj.12-214692. [DOI] [PubMed] [Google Scholar]
- 27.Wu Y, Liu GL, Liu SH, Wang CX, Xu YL, Ying Y, Mao P. MicroRNA-148b enhances the radiosensitivity of non-Hodgkin’s Lymphoma cells by promoting radiation-induced apoptosis. J Radiat Res. 2012;53:516–525. doi: 10.1093/jrr/rrs002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sikorska H, Shuster J, Gold P. Clinical applications of carcinoembryonic antigen. Cancer Detect Prev. 1988;12:321–355. [PubMed] [Google Scholar]
- 29.Kuespert K, Pils S, Hauck CR. CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol. 2006;18:565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li M, Li JY, Zhao AL, He JS, Zhou LX, Li YA, Gu J. Comparison of carcinoembryonic antigen prognostic value in serum and tumour tissue of patients with colorectal cancer. Colorectal Dis. 2009;11:276–281. doi: 10.1111/j.1463-1318.2008.01591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han SU, Kwak TH, Her KH, Cho YH, Choi C, Lee HJ, Hong S, Park YS, Kim YS, Kim TA, Kim SJ. CEACAM5 and CEACAM6 are major target genes for Smad3-mediated TGF-beta signaling. Oncogene. 2008;27:675–683. doi: 10.1038/sj.onc.1210686. [DOI] [PubMed] [Google Scholar]
- 32.Mroczko B, Kozlowski M, Groblewska M, Lukaszewicz M, Niklinski J, Jelski W, Laudanski J, Chyczewski L, Szmitkowski M. The diagnostic value of the measurement of matrix metalloproteinase 9 (MMP-9), squamous cell cancer antigen (SCC) and carcinoembryonic antigen (CEA) in the sera of esophageal cancer patients. Clin Chim Acta. 2008;389:61–66. doi: 10.1016/j.cca.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 33.Kelly KJ, Wong J, Gladdy R, Moore-Dalal K, Woo Y, Gonen M, Brennan M, Allen P, Fong Y, Coit D. Prognostic impact of RT-PCR-based detection of peritoneal micrometastases in patients with pancreatic cancer undergoing curative resection. Ann Surg Oncol. 2009;16:3333–3339. doi: 10.1245/s10434-009-0683-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charalabopoulos K, Kotsalos A, Batistatou A, Charalabopoulos A, Vezyraki P, Peschos D, Kalfakakou V, Evangelou A. Selenium in serum and neoplastic tissue in breast cancer: correlation with CEA. Br J Cancer. 2006;95:674–676. doi: 10.1038/sj.bjc.6603292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsuoka K, Sumitomo S, Nakashima N, Nakajima D, Misaki N. Prognostic value of carcinoembryonic antigen and CYFRA21-1 in patients with pathological stage I non-small cell lung cancer. Eur J Cardiothorac Surg. 2007;32:435–439. doi: 10.1016/j.ejcts.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 36.Tomita M, Shimizu T, Hara M, Ayabe T, Onitsuka T. Serum carcinoembryonic antigen level in non-small-cell lung cancer patients with preoperative normal serum level. Gen Thorac Cardiovasc Surg. 2009;57:303–306. doi: 10.1007/s11748-008-0397-6. [DOI] [PubMed] [Google Scholar]
- 37.Molina R, Filella X, Auge JM, Fuentes R, Bover I, Rifa J, Moreno V, Canals E, Vinolas N, Marquez A, Barreiro E, Borras J, Viladiu P. Tumor markers (CEA, CA 125, CYFRA 21-1, SCC and NSE) in patients with non-small cell lung cancer as an aid in histological diagnosis and prognosis. Comparison with the main clinical and pathological prognostic factors. Tumour Biol. 2003;24:209–218. doi: 10.1159/000074432. [DOI] [PubMed] [Google Scholar]
- 38.Lee S, Lee CY, Kim DJ, Hong DJ, Lee JG, Chung KY. Pathologic correlation of serum carcinoembryonic antigen and cytokeratin 19 fragment in resected nonsmall cell lung cancer. Korean J Thorac Cardiovasc Surg. 2013;46:192–196. doi: 10.5090/kjtcs.2013.46.3.192. [DOI] [PMC free article] [PubMed] [Google Scholar]


