Abstract
Objective: To establish a human hepatoma HepG2 cell line with stable expression of Prolyl hydroxylase domain 3 (PHD3) gene and study its effect of growth and proliferation in nude mice xenograft tumor. Methods: Eukaryotic expression vectors of pcDNA 3.1-PHD3 was constructed. HepG2 cells were transfected with recombinant plasmid pcDNA 3.1-PHD3 and empty vector plasmid pcDNA 3.1 by lipofectamine 2000 as transfected group, control group respectively, while the HepG2 cell without any operation was considered as parental group. Steady expression cells were gotten by G418 selecting. RT-PCR and agarose gel electrophoresis were used to confirm the expression of PHD3 in HepG2 cells and transfection successfully. The growth of these cells in vivo were also observed by injecting three groups of cell into nude mice, and volume were measured and compared. Results: The recombinant plasmid pcDNA 3.1-PHD3 and empty vector plasmid pcDNA 3.1 were successfully transfected into human hepatoma HepG2 cell line and showed stable expression in this cell line. Tumors were observed in nude mice when the transfectant cells were xenografted successfully, The average tumor size of PCDNA (3.1)-PHD3 groups are significant different compared with other two groups (P < 0.001). Conclusion: The PHD3 gene may have negative influence of growth and proliferation on HepG2 cells in vitro. The PHD3 may be a potentially tumor suppressor.
Keywords: Prolyl hydroxylase domain 3 (PHD3), hypoxia inducible factor (HIF), hepatocellular cancer (HCC), transfection
Introduction
The genesis of most solid tumors are closely associated with local tissue hypoxia [1]. In previous study, hypoxia has been shown to correlate with tumor therapeutic resistance and metastasis, increased invasion and poor prognosis [2,3]. Hypoxia inducible factor (HIF) plays a central role in the transcriptional response to altered oxygen level [4]. Prolyl hydroxylase domains (PHDs, HIF-PHDs or Egl-9 homologues) are the key regulators of oxygen-dependent degradation of HIFα regulator which include three family members: PHD1, PHD2 and PHD3 [4-6]. PHDs are Fe2+- and 2-oxoglutarate-dependent dioxygenases belonging to oxygenase superfamily that require oxygen in order to function [7]. Hypoxia-induced PHD3 expression primarily mediates degradation of HIF-2a and PHD3 appears through a negative feedback mechanism [8,9]. Recently, numerous studies have indicated that PHD3 expression has been downregulated in a number of cancers [9-12]. PHD3 also has been reported to be a tumor suppressor as its HIF-independent roles in regulating cell proliferation and apoptosis [13]. However, the study on PHD3’s molecular mechanism as tumor suppressor is still lacking, especially for hepatocellular cancer (HCC). In our previous study, Liang successfully constructed a eukaryotic expression vector containing the PHD3 gene and detected its expression of transient transfection in a human hepatoma derived cell line, HepG2 and the function of apoptosis induction, and the data suggested that PHD3 may be a tumor suppressor in HepG2 cells [14]. For further research, we constructed a HepG2 cell line with stable expression of PHD3 gene and study its biological behaviors of the cells in nude mice xenograft tumor.
Materials and methods
Materials
The recombinant plasmid pcDNA 3.1-PHD3 was constructed by inserting the cDNA sequence of PHD3 into suitable site of multiple cloning sites *MCS of the eukaryotic vector plasmid pcDNA 3.1 as described previously. Human hepatoma derived cell line, HepG2 was obtained from the Cell Bank of Institute of Life Science, Chinese Academy of Science (CAS), Shanghai, China; RNAiso Plus, PrimeScript® RT reagent Kit With gDNA Eraser (Perfect Real Time), SYBR® Premix Ex TaqTM II, DL10,000 DNA Marker, Hind III, Xho I were all purchased from Takara (Japan); G418 was purchased from Merck (Germany); Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were purchased from Hyclone Company (USA); Lipofectamine™ 2000 was purchased from Invitrogen Biotechnology (USA); DMSO was purchased from Sigma (USA); MTT reagent was purchased from Sangon Biotech (Shanghai) Co., Ltd (China). TIANprep Midi Plasmid Kit was purchased from TIANGEN Biotech (Beijing) CO., Ltd (China). Balb/c mice were purchased from SLAC Laboratory Animal LLC, Shanghai.
Methods
Construction and expression of an eukaryotic expression vector containing the PHD3 gene
This construction of the recombinant PHD3 expressor vector was done as described [14]. A pair of specific primers was designed according the sequence of PHD3 cDNA obtained from GenBank (Accession Number NM_022073) were amplified by RT-PCR with indicated primers. The obtained pcDNA 3.1(+)-PHD3 plasmid clones were veried by restriction enzyme digestion and DNA sequencing. T were reported in Liang et al. [14].
Stable transfection
Plasmid extraction and cell culture
The recombinant plasmid pcDNA 3.1(+)-PHD3 and the empty vector plasmid pcDNA 3.1 were transformed to competent E.coli cells, and the bacterial strains were amplified in 1xLB supplemented with ampicillin, and then plasmids were extracted by TIANprep Midi Plasmid Kit. The recombinant plasmid pcDNA 3.1(+)-PHD3 were digested by Hind III and Xho I, and then the digested products were analyzed by agarose gel electrophoresis to confirm the presence of the insert of PHD3 cDNA. HepG2 cells were cultured in DMEM medium containing 10% fetal bovine serum at 37°C in a humidified atmosphere of 5% CO2. G418 dose used for selection of positive clones was determined by challenging HepG2 cells with gradient concentration of G418. The optimal dose was determined by titration to be 500 ug/ml.
Transfection and selection of positive clones
Plasmid pcDNA 3.1-PHD3 and plasmid pcDNA 3.1 were transfected into the HepG2 cells by mixing with Lipofectamine™ 2000 reagent according to the instructions of manufacturer Forty-eight hours after transfection, cells were passaged and were cultured in DMEM medium containing 10% fetal bovine serum and 500 ug/ml G418. Ten days later the concentration of G418 changed to 200 ug/ml in order to maintain screening pressure. G418-resistant clones was visible approximately 2 weeks after the selection. Then the G418-resistant clones were picked up and cultured, after 4 weeks of cloning finally we got 2 cell lines with stable expression of pcDNA 3.1(+)-PHD3 and pcDNA 3.1(+) respectively. Cells were divided into three groups parental group (HepG2 cell), pcDNA 3.1(+) group and pcDNA 3.1(+)-PHD3 group.
RNA isolation and real-time PCR
Total RNA was extracted from cells of three groups using RNAiso Plus according to the manufacturer’s protocol. Real-time reverse transcriptase (RT) PCR was performed as follows: cDNA synthesis was performed with an PrimeScript® RT reagent Kit With gDNA Eraser (Perfect Real Time) using 1 μg total RNA for each reaction and the following condion: 37°C 15 minutes, 85°C 5 seconds. The primers were used to synthesize the cDNA. Primer sequences used for PCR are: the primers PHD3 forward 5’-CATCAGCTTCCTCCTGTC-3’, reverse 5’-CCACCATTGCCTTAGACC-3’ and β-actin forward 5’-CTGTGCCCATCTACGAGG-3’, reverse 5’-ATGTCACGCACGATTTCC-3’. PHD3 and β-actin were amplified with SYBR® Premix Ex TaqTM II for 45 cycles in LightCycler 480 PCR Machine using the following conditions: stage 1: 95°C 30 s 1 Cycle, stage 2: 95°C 5 s, 60°C 20 s, 45 cycles. The parental group was used as an internal control. SYBR Green was employed to detect the accumulation of amplification products in the reactions. For each sample, the difference in threshold cycles for each PHD gene was calculated (ΔCT, ΔCT = Avg. PHD3 CT -Avg. β-actin CT), and a calibrated ΔCT value (ΔΔCT, ΔΔCT = Avg. PHD3 ΔCT -Avg. β-actin ΔCT) was analyzed. The relative expression of each group PHD3 mRNA in HepG2 cell was calculated using the 2-ΔΔCT method. And then the PHD3 mRNA was detected by agarose gel electrophoresis.
Construction and evaluation of xenograft tumor in nude mice
Sixty Balb/c mice in a SPF grade aged 6 weeks (body weight 18-20 g,) half male and half female, were randomly divided into three groups (parental group, pcDNA (3.1) group and pcDNA (3.1)-PHD3 group), each group then divided into two subgroups (subcutaneous tumor, tail vein injection), 10 mice in each subgroups. Subconfluent monolayer cells were used, they were detached from the bottom of the flask with trypsin and suspended in sterile PBS with density of (1-2) ×108 /ml.The mice in three groups received injection of HepG2 cell, and HepG2 cell stably expressing pcDNA (3.1), and pcDNA (3.1)-PHD3 stable expressing HepG2 cells respectively. After injection, the mice were marked with picric acid on different location of the body, and then continued to be fed at SPF level. General conditions of nude mice were recorded every day.
Subcutaneous tumor
0.2 mL of each suspension was subcutaneously injected into the right back of the nude mice. The subcutaneous tumor formation, dynamic observation tumor size and weight of nude mice were observed. Long and perpendicular dimension (a, b) of the tumor were measured by using a caliper, the volume of the tumor was calculated according to the formula: v = a×b2/2. After 5 weeks, mice were killed by cervical dislocation.
Tail vein injection
0.5 ml of each cell suspension was quickly injected through the tail vein. After 5 weeks, mice were killed by cervical dislocation.
Pathology examination
The subcutaneous and visceral metastases of the tumor were observed. The thin sections were prepared with HE staining and photographs were taken under optical microscope.
Statistical analysis
All statistical analyses were conducted using the SPSS 17.0 software package. If the variance was aligned, SNK q test was used for comparison between three groups. If variances was not aligned, using rank sum test. P < 0.05 was considered statistically significant.
Results
Identification of the recombinant plasmid pcDNA 3.1(+)-PHD3
Two fragments of 721 bp (insert) and 5400 bp (linear DNA strand of the vector) was generated when the recombinant plasmid pcDNA 3.1(+)-PHD3 was digested by Hind III and Xho I, as revealed by agarose Gel electrophoresis (Figure 1A), suggesting that PHD3 fragments was inserted into the pcDNA 3.1(+) vector and the PHD3 stably expressing clones could be produced by transfection with this construct.
Figure 1.
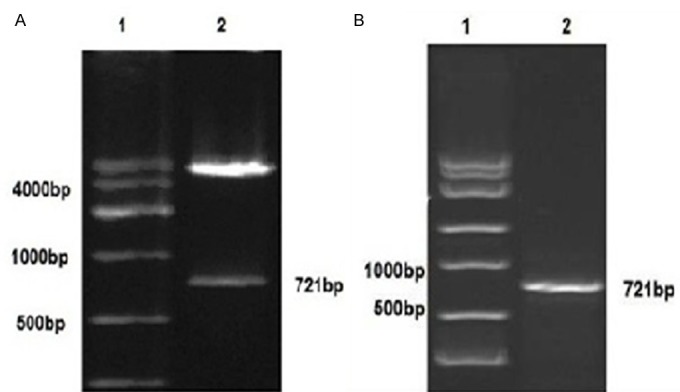
Verification of the PHD3 cDNA insert with restriction enzyme digestion. A: Plasmid pcDNA 3.1(+)-PHD3 was digested by Hind III and Xho I, 1: DNA Marker DL 10,000; 2: Two fragments of 721 bp 5400 bp. The small band corresponds with the size of the insert. B: The PHD3 mRNA was detected by agarose gel electrophoresis. 1: DNA Marker DL 10,000; 2: PHD3 mRNA 721 bp.
The expression of PHD3mRNA in three groups HepG2 cells
To study the expression pattern of the PHD3 gene in HepG2 cells, the levels of the PHD3 mRNAs were quantified by real-time RT-PCR in three groups. Expression levels of mRNA extracted from each sample were assessed in the three groups with calibration of β-actin amounts. The parental group (HepG2 cell), pcDNA 3.1(+) group and pcDNA 3.1(+)-PHD3 group PHD3 mRNA expression relative to the expression of the parental group were 1.000 ± 0.000, 0.987 ± 0.045, and 38.156 ± 1.795 respectively. A significant difference was observed between pcDNA 3.1(+)-PHD3 group and the parental group (HepG2 cell), pcDNA 3.1(+) group were 0.000, 0.000, respectively (P = 0.013). But there was no Statistical difference between the parental group (HepG2 cell), pcDNA 3.1(+) group (P = 0.991) (Table 1; Figure 2). The RT-PCR products were loaded on 1.5% agarose gels, the band for full-length PHD3 was located at 721 bp (Figure 1B), suggesting that the PHD3 was high expressed in PHD3 steady transfection cell line in HepG2 cells and the transfection was constructed successfully.
Table 1.
The volume change of subcutaneous tumor after subcutaneous inoculation
| Group N | The volume of subcutaneous tumor: X̅ ± s (mm3) | ||||
|---|---|---|---|---|---|
|
| |||||
| 14 d | 21 d | 28 d | 35 d | ||
| pcDNA 3.1(+)-PHD3 group | 10 | 17.56 ± 1.92 | 61.07 ± 5.04 | 159.29 ± 7.24 | 294.79 ± 8.34 |
| pcDNA 3.1(+) group | 10 | 28.37 ± 3.50 | 97.89 ± 6.63 | 225.92 ± 2.9 | 457.88 ± 3.43 |
| blank group | 10 | 32.05 ± 2.75 | 104.81 ± 5.97 | 228.44 ± 5.81 | 462.22 ± 3.32 |
Figure 2.
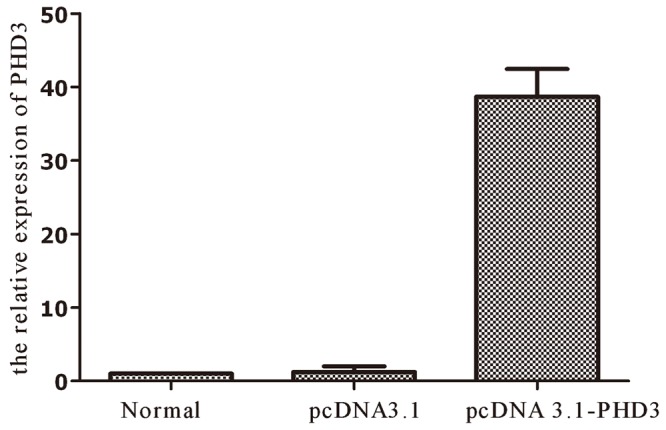
Comparison of the expression levels of PHD3 in three groups of parental, mock or empty vector transfectant and PHD3 transfectant HepG2 cells. The expression of PHD3 mRNA was much higher in pcDNA 3.1(+)-PHD3 group than in the parental and pcDNA 3.1(+) groups expressing endogenous PHD3 (P = 0.000). Normal: The parental group (HepG2 cell), PC3.1: pcDNA 3.1(+) group, PHD3: pcDNA 3.1(+)-PHD3 groups.
General condition of nude mouse
The life state of nude mouse in the pcDNA 3.1(+)-PHD3 group carrying xenograft was normal all the time, voracious and active. The pcDNA 3.1(+) group and parental group carrying xenograft lost appetite gradually, listlessness and oscitancy.
The volume change of subcutaneous tumor
Subcutaneous tumors were palpable on 6th day after subcutaneous inoculation in parental group and pcDNA 3.1(+) group, But in pcDNA 3.1(+)-PHD3 group it was palpable in 9th days after inoculation.
We chose the volume of 14th day, 21th day, 28th day and 35th days for comparison (Figure 3). As the variance of each set of data was aligned, SNK q test was used for multiple comparison at different time point. On the 14th day, the average volume of tumor was 17.56 mm3 in pcDNA 3.1(+)-PHD3 group and in pcDNA 3.1(+) group it was 28.37 mm3, the difference was statistically significant between pcDNA 3.1(+)-PHD3 group and pcDNA 3.1(+) group (p < 0.0001). The average volume of tumor was 32.05 mm3 in parental group, the difference was statistically significant between pcDNA 3.1(+)-PHD3 group and parental group (p < 0.0001). But the difference was not statistically significant between pcDNA 3.1(+) group and parental group (p > 0.05). On the 21th day, 28th day and 35th day, the values of p for multiple comparison were familiar with the 14th day.
Figure 3.
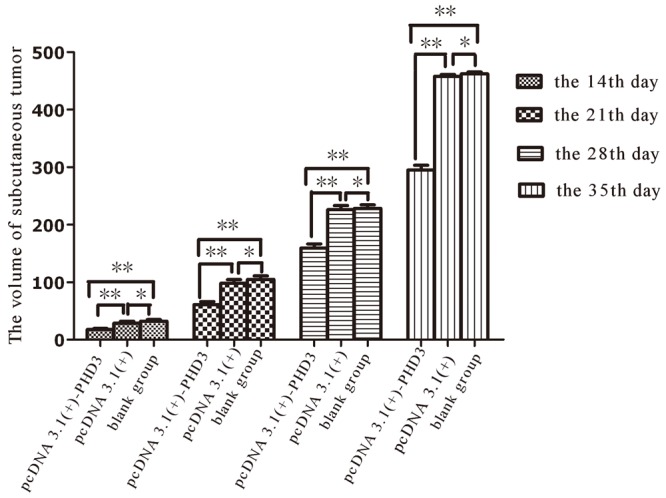
Subcutaneous tumors were palpable on 6th day after subcutaneous inoculation in parental group and pcDNA 3.1(+) group, But in pcDNA 3.1(+)-PHD3 group it was palpable in 9th days after inoculation. SNK q test was used to compared the differences between pcDNA 3.1(+)-PHD3 group and pcDNA 3.1(+) group, blank group at the 14th day, 21th day, 28th day and 35th day. *indicated P > 0.05, **indicate p < 0.0001.
Finally when the nude mices were killed, the average volume of tumor was 462.22 mm3, 457.88 mm3 and 294.79 mm3 in parental group, pcDNA 3.1(+) group and pcDNA 3.1(+)-PHD3 group respectively.
Discussion
Hepatocellular carcinoma (HCC) contributes to a global health problem with high malignant degree, rapid progression and poor prognosis [15]. At molecular level the development of HCC the implication of activation of oncogenes or inactivation of tumor suppressor genes has been suggested, therefore study on new tumor suppressor gene has a great significance for clinical treatment and predicting the prognosis of HCC. Hypoxia may enhance the expansion and metastasis of tumor cells by diminishing apoptotic potential, HIF as the key molecular regulator in this process of adaptation to low oxygen levels [16]. PHD3 was considered as an HIF-1α regulator by targeting the prolyl hydroxylated of HIFα. A number of recent studies have shown that PHD3 played an important role in the progression and prognosis of cancer, independent of its hydroxylase activity.
The expression level of PHD3 was inconsistent or varied in tumor tissues and tumor cell lines of different origins, including gastric, pancreatic and colorectal colorectal cancer and other gastrointestinal cancers [12,16]. Xue [11] found that PHD3 was lowly expressed in colorectal cancer tissues, relating to differentiation and metastasis. Su [12] detected that PHD3 was highly expressed in gastric cancer, but was not expressed in adjacent normal tissue. Similarly, in non-small cell lung cancer PHD3 was also highly expressed [17,18]. In pancreatic cancer cells Su et al. [16] found that PHD3 overexpression promoted caspase-3 activation and increased apoptosis rate; in contrast, inhibition of PHD3 expression with siRNA reduces the caspase-3 activation. Variant expression level of PHD3 in different cancer types remains to be elucidated mechanistically.
The relationship between PHD3 and HCC is still unclear. To clarify the possible roleof PHD3 on the genesis of HCC, we transfected a human hepatoma HepG2 cell line to obtain stable expression of PHD3 gene and study the biological behaviors of the cells in vitro and in vivo. In preliminary research of our study [14], we analyzed the Caspase-3 activity in transiently transfected HepG2 cells, and found that cleaved 17 kD fragment of active Caspase-3 was significantly higher expressed in PHD3 group than the other control group. Similarly, cells with PHD3 stable expression were prone to apoptosis, the gapping among these cell masses was scattered and the growing state were bad, as Figure 2 shows. The observations suggested of the apoptotic induction potential of PHD3 in HepG2 cells. In the present study, the effect on the growth of these cells in vivo was also investigated by injecting three groups of cell into nude mice. The data suggested that PHD3 remarkably reduced the uptake of the tumor. In conclusion, we have successfully constructed a human hepatoma HepG2 cell line with stable expression of PHD3 gene and have shown that PHD3 could inhibit tumor growth and proliferation in nude mice xenograft tumor. To a certain extent, it suggested PHD3 expression can inhibit growth, invasion and metastasis of liver cancer cells in vitro and in vivo. Our study has formed the basis for further research of the mechanism of PHD3-induced apoptosis and anticancer gene therapy.
Acknowledgements
This work was supported by a grant from The National Nature Science Foundation of China (No. 81273185), Science and Technology Innovation Fund of Guangdong Medical College, China (No. STIF201126) and Excellent Master’s Thesis Fostering Fund of Affiliated Hospital of Guangdong Medical College, China (No. YS1108).
Disclosure of conflict of interest
None declared.
References
- 1.Vaupel P, Mayer A. Hypoxia in cancer: significance and impact on clinical outcome. Cancer Metastasis Rev. 2007;26:225–239. doi: 10.1007/s10555-007-9055-1. [DOI] [PubMed] [Google Scholar]
- 2.Mayer A, Vaupel P. Hypoxia, lactate accumulation, and acidosis: siblings or accomplices driving tumor progression and resistance to therapy? Adv Exp Med Biol. 2013;789:203–209. doi: 10.1007/978-1-4614-7411-1_28. [DOI] [PubMed] [Google Scholar]
- 3.Ghattass K, Assah R, El-Sabban M, Gali-Muhtasib H. Targeting hypoxia for sensitization of tumors to radio- and chemo-therapy. Curr Cancer Drug Targets. 2013;13:670–685. doi: 10.2174/15680096113139990004. [DOI] [PubMed] [Google Scholar]
- 4.Kaelin WG Jr, Ratcliffe PJ. Oxygen sensing by metazoans: the central role of the HIF hydroxylase pathway. Mol Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 6.Epstein AC, Gleadle JM, McNeill LA, Hewitson KS, O’Rourke J, Mole DR, Mukherji M, Metzen E, Wilson MI, Dhanda A, Tian YM, Masson N, Hamilton DL, Jaakkola P, Barstead R, Hodgkin J, Maxwell PH, Pugh CW, Schofield CJ, Ratcliffe PJ. C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell. 2001;107:43–54. doi: 10.1016/s0092-8674(01)00507-4. [DOI] [PubMed] [Google Scholar]
- 7.Hirsilä M, Koivunen P, Günzler V, Kivirikko KI, Myllyharju J. Characterization of the human prolyl 4-hydroxylases that modify the hypoxia-inducible factor. J Biol Chem. 2003;278:30772–30780. doi: 10.1074/jbc.M304982200. [DOI] [PubMed] [Google Scholar]
- 8.Marxsen JH, Stengel P, Doege K, Heikkinen P, Jokilehto T, Wagner T, Jelkmann W, Jaakkola P, Metzen E. Hypoxia-inducible factor-1 (HIF-1) promotes its degradation by induction of HIF-alpha-prolyl-4-hydroxylases. Biochem J. 2004;381:761–767. doi: 10.1042/BJ20040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henze AT, Riedel J, Diem T, Wenner J, Flamme I, Pouyseqqur J, Plate KH, Acker T. Prolyl hydroxylases 2 and 3 act in gliomas as protective negative feedback regulators of hypoxiainducible factors. Cancer Res. 2010;70:357–366. doi: 10.1158/0008-5472.CAN-09-1876. [DOI] [PubMed] [Google Scholar]
- 10.Gossage L, Zaitoun A, Fareed KR, Turley H, Aloysius M, Lobo DN, Harris AL, Madhusudan S. Expression of key hypoxia sensing prolyl-hydroxylase PHD1, -2 and -3 in pancreaticobiliary cancer. Histopathology. 2010;56:908–920. doi: 10.1111/j.1365-2559.2010.03566.x. [DOI] [PubMed] [Google Scholar]
- 11.Xue J, Li X, Jiao S, Wei Y, Wu G, Fang J. Prolyl hydroxylase-3 is downregulated in colorectal cancer cells and inhibits IKKbeta independent of hydroxylase activity. Gastroenterology. 2010;138:606–615. doi: 10.1053/j.gastro.2009.09.049. [DOI] [PubMed] [Google Scholar]
- 12.Su C, Huang K, Sun L, Yang D, Zheng H, Gao C, Tong J, Zhang Q. Overexpression of the HIF hydroxylase PHD3 is a favorable prognosticator for gastric cancer. Med Oncol. 2012;29:2710–5. doi: 10.1007/s12032-012-0171-6. [DOI] [PubMed] [Google Scholar]
- 13.Nakayama K, Frew IJ, Hagensen M, Skals M, Habelhah H, Bhoumik A, Kadoya T, Erdjument-Bromaqe H, Temost P, Frappell PB, Bowtell DD, Ronai Z. Siah2 regulates stability of prolyl-hydroxylases, controls HIF1alpha abundance, and modulates physiological responses to hypoxia. Cell. 2004;117:941–952. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 14.Liang QL, Li ZY, Zhou Y, Liu QL, Ou WT, Huang ZG. Construction of a recombinant eukaryotic expression vector containing PHD3 gene and its expression in HepG2 cells. J Exp Clin Cancer Res. 2012;31:64. doi: 10.1186/1756-9966-31-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi: 10.1038/nrgastro.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su Y, Loos M, Giese N, Hines OJ, Diebold I, Görlach A, Metzen E, Pastorekova S, Friess H, Büchler P. PHD3 regulates differentiation, tumour growth and angiogenesis in pancreatic cancer. Br J Cancer. 2010;103:1571–1579. doi: 10.1038/sj.bjc.6605936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen S, Zhang J, Li X, Luo X, Fang J, Chen H. The expression of prolyl hydroxylase domain enzymes are up-regulated and negatively correlated with Bcl-2 in non-small cell lung cancer. Mol Cell Biochem. 2011;358:257–263. doi: 10.1007/s11010-011-0976-1. [DOI] [PubMed] [Google Scholar]
- 18.Andersen S, Donnem T, Stenvold H, Al-Saad S, Al-Shibli K, Busund LT, Bremnes RM. Overexpression of the HIF hydroxylases PHD1, PHD2, PHD3 and FIH are individually and collectively unfavorable prognosticators for NSCLC survival. PLoS One. 2011;6:e23847. doi: 10.1371/journal.pone.0023847. [DOI] [PMC free article] [PubMed] [Google Scholar]


