Abstract
Methicillin resistant Staphylococcus aureus (MRSA) strains lead to severe infections in immunosupressive patients, geriatric population and premature infants. 27 MRSA strains isolated in the Neonatal Intensive Care Unit was considered as an outbreak and it was aimed to investigate the genetic and epidemiologic relation of the MRSA outbreak. MecA gene was investigated in the S. aureus strains and pulsed field gel electrophoresis (PFGE) was used to investigate the genetic relation between outbreak strains. MecA gene was showed in all isolates. PFGE revealed that there were two different strains and most of the isolates (25/27) were owing to same clone. One of the samples were found closely related with the common strain and the other sample was found genetically unrelated. To terminate the outbreak; liquid baby food was gained to the baby food kitchen, no more new patient was imported to the neonatal unit and none of the patients were exported from neonatal unit to other clinics during outbreak, education about infection control precautions was given to all the staff and nursing bottle dishwasher was obtained. To manage and terminate the outbreak, besides the infection control precautions, tests to determine the genetic relation between outbreak strains which are done in the microbiology laboratory are needed. Molecular analysis of outbreak strains will contribute to prove the epidemiologic and evolution of outbreaks.
Keywords: Staphylococcus aureus, neonatal, outbreak, PFGE
Introduction
Staphylococci are one of the major causative agents of nosocomial infections. Increase in the number of methicillin resistant Staphylococcus aureus (MRSA) strains lead to difficulties in treatment and increase morbidity and mortality rates [1]. MRSA, may lead to wide variety of clinical manifestations like skin and soft tissue infections, bacteremia, pneumonia, endocarditis, toxic shock syndrome [2]. MRSA leads to severe infections in immun-compromised hosts, especially in geriatric and premature patients. Broad spectrum antibiotic therapies, invasive procedures and surgical interventions are the major reasons of MRSA outbreaks [3,4]. The route of nosocomial infections are, endogenous infections caused by the microorganisms inhabiting the patient’s pharynx, nasal cavity and skin and exogenous infections from other patients, medical devices and medical personnel. Nosocomial infections are considered to be caused by direct contact with the hands of medical personnel [4].
In the management of outbreaks, rapid and reliable determination of genetic relation between outbreak strains is a very important tool to organize the infection control precautions [5]. Pulsed field gel electrophoresis is the gold standard method to investigate the genetic relation [1,6,7]. In this study we aimed to analyze the clonal relation of the MRSA outbreak seen at the Neonate Intensive Care Unit (NICU) of Uludag University Hospital.
Materials and methods
Culture samples and Staphylococcus aureus strains
In this study we investigated 26 gastric aspiration specimen (GAS) and one tracheal aspiration specimen (TAS) from 10 patients at the NICU. Bacterial identification and antibiotic susceptibility tests were done in BD Phoenix 100 (Becton Dickinson, USA) system. Methicillin resistance tests were repeated according to Clinical and Laboratory Standards Institute (CLSI) recommendations by cefoxitin disc in the Kirby Bauer disc diffusion method [8]. Baby food samples and nasal samples of medical personnel were cultured as a possibble source of MRSA.
MecA gene detection by polymerase chain reaction (PCR)
Bacterial colonies grown on solid media were transferred to tubes containing 500 μl distilled water and after 24 hours of incubation at -80°C and then brought to room temperature. Suspensions were transferred to heat blocks and incubated at 100°C for 10 minutes and then centrifuged at 12.000 g at 4°C for three minutes. Supernatant was used for PCR. MecA gene was researched according to the method done by Murakami et al. [9]. PCR products were visualized after electrophoresis on 2% agarose.
Pulsed field gel electrophoresis (PFGE)
Colonies grown on solid media were transferred to cell suspension buffer including 10 mM Tris-HCl, 50 mM EDTA and 20 mM NaCl and then centrifuged at 13.000 g at 4°C for two minutes. Pellet was added into 2% low melt agarose and after adding 2 μl lysostaphine (10 mg/ml), it was incubated at 4°C until solidification. Cells in the agarose were lysed by cell lysis solution 1 including 10 mM Tris-HCl, 50 mM NaCl, 50 mM EDTA-0.2% sodyum deoxicholate-0.5% sarkosyl. After lysis agarose was incubated on ice for solidification. Cells in the agarose were lysed by cell lysis solution 2 including 30 μl proteinase K (10 mg/ml) and 5970 μl ES solution (250 mM EDTA, 1% sarkosyl). Agarose molds were washed thrice at 50°C for 30 minutes by Tris-HCl-EDTA buffer (10 mM Tris-HCl, 0.1 mM EDTA). Plug molds were divided into two by lancet of which, one was used for PFGE and the other was kept as a spare. Plug molds were incubated at 30°C by SmaI in water bath. Plug molds were loaded on electrophoresis gel. All gels were electrophoresed in 0.5x TBE buffer at 6 V/cm for 20 hours 14°C with a pulse duration of 5.3 to 34.9 seconds ramped linearly in CHEF-DR II system (Bio-Rad Laboratories, Belgium) [10,11]. Gels were stained with etidium bromide, destained in distilled water and photographed with Biometra Gel Documentation Module (Biometra GmBH, Germany). Band profiles were inspected by Bio Doc Analyze Software (Biometra GmBH, Almanya). These data were used to produce dendograms showing unwighted pair group method with mathematical averaging (UPGMA) cluster analysis of Dice similarity coefficients produced from pair-wise comparisons of the coded profiles which was taken as 1%. Results were interpreted accoding to Tenover criteria [12].
Results
S. aureus grown in all patient samples were identified as MRSA according to CLSI [8]. All of the strains were resistant against methicillin, penicillin, rifampicin and tetracycline and susceptible to vancomycin, teicoplanin, trimethoprim/sulfamethoxazole and erythromycin. Nine of 10 MRSA isolates were evaluated as infectious agents and treated by glycopeptides. One of the MRSA isolates, which was isolated from patient five was evaluated as colonisation and glycopeptid was not administered to the patient who was being empirically treated by amikacin and cefotaxime. All of the patients survived after the MRSA outbreak. MecA gene was found in all of the MRSA isolates. PFGE band patterns are shown in Figure 1 and dendograms are shown in Figure 2. Dendogram profiles revealed two different strains, of which the majority of them (25/27) had the same clonal origin. One of the strains (patient 9 and strain 26) were found to be closely related with the common strain. One of the strains (patient 1 and strain 6) were found unrelated to other strains and the results are summarized in Table 1 and Figure 2. S. aureus was not grown in nasal specimens from the staff and baby food samples.
Figure 1.
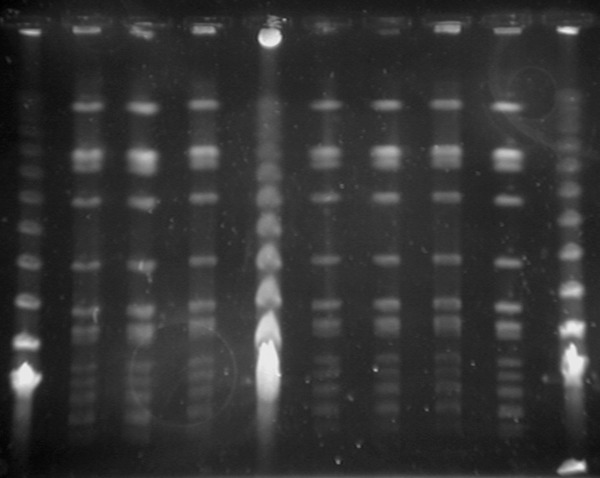
Strain relation according to PFGE (Lane 1, 5 and 10: Molecular marker) (PCR 20 bp Low Ladder, Sigma-Aldrich, USA).
Figure 2.
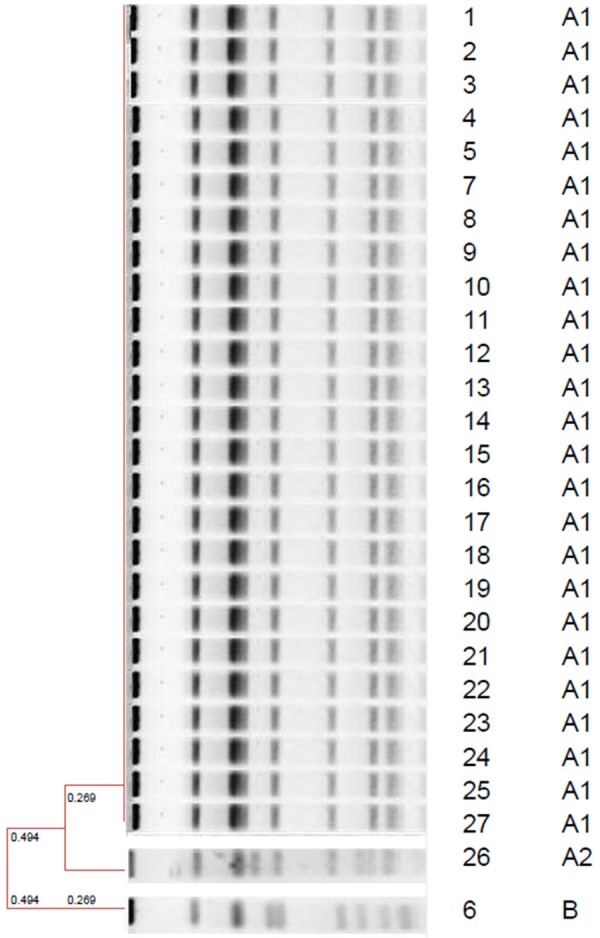
UPGMA patterns of strains; strain numbers and pulsotypes.
Table 1.
Specimen type, number and pulsotype according to patients
| Patient No | Strain No | Sample | Pulsotype |
|---|---|---|---|
| 1 | 1 | GAS | A1 |
| 1 | 2 | GAS | A1 |
| 1 | 3 | GAS | A1 |
| 1 | 4 | GAS | A1 |
| 1 | 5 | GAS | A1 |
| 1 | 6 | GAS | B |
| 2 | 7 | GAS | A1 |
| 2 | 8 | GAS | A1 |
| 2 | 9 | GAS | A1 |
| 2 | 10 | GAS | A1 |
| 2 | 11 | GAS | A1 |
| 3 | 12 | GAS | A1 |
| 4 | 13 | GAS | A1 |
| 4 | 14 | GAS | A1 |
| 4 | 15 | GAS | A1 |
| 5 | 16 | GAS | A1 |
| 5 | 17 | GAS | A1 |
| 5 | 18 | GAS | A1 |
| 6 | 19 | GAS | A1 |
| 6 | 20 | GAS | A1 |
| 6 | 21 | GAS | A1 |
| 7 | 22 | GAS | A1 |
| 7 | 23 | GAS | A1 |
| 8 | 24 | GAS | A1 |
| 9 | 25 | GAS | A1 |
| 9 | 26 | TAS | A2 |
| 10 | 27 | GAS | A1 |
GAS: gastric aspiration specimen, TAS: tracheal aspiration specimen.
Discussion
MRSA outbreaks are usually nosocomial infections and especially seen in intensive care units, and burn units. These infections are usually seen as pneumoniae and sepsis which increase morbidity and mortality [13-15]. S. aureus can survive on environmental places for a long time which is an important reservoir. MRSA infected and colonized patients are the other important reservoir for nosocomial infections and insufficient practice of infection control precautions lead to spread of these strains [16-18]. MRSA incidence was found related with the workload of nurses in a study performed in an adult intensive care unit [19]. Haley et al. [20] found that increase in the number of patients in the intensive care unit and decrease in the number of medical staff prevents medical staff to wash hands which results in spread of infections. Hand hygiene of the medical staff is the most important precaution for nosocomial infections. Other precautions in the control of MRSA outbreaks are intranasal mupirocin ointment and vancomycin treatment of patients [17,21,22]. Andersen et al. [23] had provided contact isolation, treatment of infected and colonised patients and medical staff, disinfected all the patient rooms and educated the staff about infection control precautions to stop the MRSA outbreak which was seen in a NICU. In or hospital, patients were treated with glycopeptide antibiotics, contact isolation was carried out for the patients, To terminate the outbreak, liquid baby food was gained to the baby food kitchen, no more new patient was imported to the neonatal unit and none of the patients were exported from neonatal unit to other clinics during outbreak, education about infection control precautions was given to all workers and nursing bottle dishwasher was obtained. After the treatment of the last outbreak patient and no more MRSA growth in culture plates, it was thought that the outbreak was terminated.
Typing of bacterial strains is important in the suspicion of an outbreak and proving the nosocomial spread of infections. Phenotypic and genotypic methods can be used for strain typing, but now molecular methods are being used generally [12,24,25]. PFGE, rep-PCR and RFLP methods are the common methods for molecular typing. In this study we used PFGE to fingerprint DNA of MRSA. DNA was digested by SmaI restriction enzyme and divided into 10 to 20 pieces differing from 10 to 800 kilobase pairs which was electrophoresed by iso-electric focusing [24-26]. PFGE results revealed that 25 (93%) of 27 strains were genetically identical. One of the samples were found closely related with the common clone and it was thought that this strain had changed its genetic structure because of antibiotic therapies [12]. The other sample was genetically unrelated among the common clone. The source of the outbreak was thought as the first isolate of patient number one. Infection control precautions could not be implemented until the time passed during isolation, identification and reporting of this strain, which resulted in spread of MRSA to other patients and led to an outbreak. Isolate six, which was genetically unrelated was thought to be due to a new infection after the treatment of the outbreak strain. Though all the strains were phenotypically the same, PFGE revealed genetical discrepancy between the strains. It was thought that, phenotypical methods are incapable of determining genetical discrepancies.
Implementation of molecular methods have an important role in the management and analysis of outbreaks. PFGE, which is used in outbreaks has a high correlation with epidemiological data and used as the gold standard molecular method. PFGE usage is restricted because of the facts that; PFGE is labor intensive and it needs an expensive device.
As a result; to manage and terminate the outbreak, besides the infection control precautions, tests to determine the genetic relation between outbreak strains which are done in the microbiology laboratory are needed. PFGE was showed as a reliable method in the microbiologic analysis of outbreaks in this study. Molecular microbiologic analysis of outbreak strains will contribute to prove the epidemiologic and evolution of outbreaks.
Disclosure of conflict of interest
None declared.
References
- 1.Tekerekoglu MS, Ay S, Otlu B, Cicek A, Kayabas U, Durmaz R. Molecular epidemiology of methicillin-resistant Staphylococcus aureus isolates from clinical specimens of patients with nosocomial infection: are there unnoticed silent outbreaks? New Microbiol. 2007;30:131–137. [PubMed] [Google Scholar]
- 2.Chen H, Liu Y, Jiang X, Chen M, Wang H. Rapid change of methicillin-resistant Staphylococcus aureus isolates in a Chinese tertiary care hospital over a 15-year period. Antimicrob Agents Chemother. 2010;54:1842–1847. doi: 10.1128/AAC.01563-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang JT, Chang SC, Ko WJ, Chang YY, Chen ML, Pan HJ, Luh KT. A hospital-acquired outbreak of methicillin-resistant Staphylococcus aureus infection initiated by a surgeon carrier. J Hosp Infect. 2001;47:104–109. doi: 10.1053/jhin.2000.0878. [DOI] [PubMed] [Google Scholar]
- 4.Takei Y, Yokoyama K, Katano H, Tsukiji M, Ezaki T. Molecular epidemiological analysis of methicillin-resistant Staphylococci in a neonatal intensive care unit. Biocontrol Sci. 2010;15:129–138. doi: 10.4265/bio.15.129. [DOI] [PubMed] [Google Scholar]
- 5.Grisold AJ, Zarfel G, Strenger V, Feierl G, Leitner E, Masoud L, Hoenigl M, Raggam RB, Dosch V, Marth E. Use of automated repetitive-sequence-based PCR for rapid laboratory confirmation of nosocomial outbreaks. J Infect. 2010;60:44–51. doi: 10.1016/j.jinf.2009.10.045. [DOI] [PubMed] [Google Scholar]
- 6.Raimundo O, Heussler H, Bruhn JB, Suntrarachun S, Kelly N, Deighton MA, Garland SM. Molecular epidemiology of coagulase-negative staphyloccal bacteraemia in a newborn intensive care unit. J Hosp Infect. 2002;51:33–42. doi: 10.1053/jhin.2002.1203. [DOI] [PubMed] [Google Scholar]
- 7.Hidaka H, Miura M, Mansunaga K, Qin L, Uemura Y, Sakai Y, Hashimoto K, Kawano S, Yamashita N, Sakamoto T, Watanabe H. Infection control for a methicillin-resistant Staphylococcus aureus outbreak in an advanced emergency medical service center, as monitored by molecular analysis. J Infect Chemother. 2013;19:884–90. doi: 10.1007/s10156-013-0587-8. [DOI] [PubMed] [Google Scholar]
- 8.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Testing. 22nd Informational Supplement, M100-S22. Pennsylvania, USA: Wayne; 2012. [Google Scholar]
- 9.Murakami A, Minamide W, Wada K, Nakamura E, Teraoka H, Watanabe S. Identification of methicillin-resistant strains of staphylococci by polymerase chain reaction. J Clin Microbiol. 1991;29:2240–2244. doi: 10.1128/jcm.29.10.2240-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tekerekoglu MS, Durmaz R, Ay S, Cicek A, Kutlu O. Epidemiologic and clinical features of a sepsis caused by methicillin-resistant Staphylococcus epidermidis (MRSE) in a pediatric intensive care unit. Am J Infect Control. 2004;32:362–364. doi: 10.1016/j.ajic.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Duman Y, Tekerekoglu MS, Otlu B. Investigation of the presence of Panton-Valentine leukocidin and clonal relationship of community- and hospital-acquired clinical isolates of Staphylococcus aureus. Mikrobiyol Bul. 2013;47:389–400. doi: 10.5578/mb.5760. [DOI] [PubMed] [Google Scholar]
- 12.Tenover FC, Arbeit RD, Goering RV, Mickelsen PA, Murray BE, Persing DH, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vincent JL, Bihari DJ, Suter PM, Bruining HA, White J, Nicolas-Chanoin MH, Wolff M, Spencer RC, Hemmer M. The prevalance of nosocomial infection in intensive care units in Europe. Results of the European Prevalance of Infection in Intensive Care (EPIC) Study. EPIC International Advisory Comitee. JAMA. 1995;274:639–44. [PubMed] [Google Scholar]
- 14.Song X, Cheung S, Klontz K, Short B, Campos J, Singh N. A step wise approach to control an outbreak and ongoing transmission of methicillin-resistant Staphylococcus aureus in a neonatal intensive care unit. Am J Infect Control. 2010;38:607–611. doi: 10.1016/j.ajic.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 15.Lee YT, Lin DB, Wang WY, Tsao SM, Yu SF, Wei MJ, Yang SF, Lu MC, Chiou HL, Chen SC, Lee MC. First identification of methicillin-resistant Staphylococcus aureus MLST types ST5 and ST45 and SCCmec types IV and Vt by multiplex PCR during an outbreak in a respiratory care ward in central Taiwan. Diagn Microbiol Infect Dis. 2011;70:175–182. doi: 10.1016/j.diagmicrobio.2010.12.021. [DOI] [PubMed] [Google Scholar]
- 16.Teare L, Shelley OP, Millership S, Kearns A. Outbreak of Panton-Valentine leucocidine-positive methicillin-resistant Staphylococcus aureus outbreak in a regional burns unit. J Hosp Infect. 2010;76:220–224. doi: 10.1016/j.jhin.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 17.Ho ML, Seto WH, Wong LC, Wong TY. Effectiveness of multi faceted hand hygiene interventions in long-term care facilities in Hong Kong: a cluster-randomized controlled trial. Infect Control Hosp Epidemiol. 2012;33:761–7. doi: 10.1086/666740. [DOI] [PubMed] [Google Scholar]
- 18.Lin YC, Lauderdale TL, Lin HM, Chen PC, Cheng MF, Hsieh KS, Liu YC. An outbreak of methicillin-resistant Staphylococcus aureus infection in patients of a pediatric intensive care unit and high carriage rate among health care workers. J Microbiol Immunol Infect. 2007;40:325–334. [PubMed] [Google Scholar]
- 19.Vicca AF. Nursing staff workload as a determinant of methicillin-resistant Staphylococcus aureus. J Hosp Infect. 1999;43:109–113. doi: 10.1053/jhin.1999.0246. [DOI] [PubMed] [Google Scholar]
- 20.Haley R, Bergman D. The role of understanding and overcrowding in recurrent outbreaks of staphylococcal infections in a neonatal special-care unit. J Infect Dis. 1982;145:875–885. doi: 10.1093/infdis/145.6.875. [DOI] [PubMed] [Google Scholar]
- 21.Mori N, Hitomi S, Nakajima J, Okuzumi K, Murakami A, Kimura S. Unselective use of intranasal mupirocin ointment for controlling propagation of methicillin-resistant Staphylococcus aureus in a thoracic surgery ward. J Infect Chemother. 2005;11:231–233. doi: 10.1007/s10156-005-0396-9. [DOI] [PubMed] [Google Scholar]
- 22.Silvestri L, Milanese M, Oblach L, Fontana F, Gregori D, Guerra R, van Saene HK. Enteral vancomycin to control endemicity of methicillin-resistant Staphylococcus aureus outbreak in mechanically ventilated patients. Am J Infect Control. 2002;30:391–399. doi: 10.1067/mic.2002.122255. [DOI] [PubMed] [Google Scholar]
- 23.Andersen BM, Lindemann R, Bergh K, Nesheim BI, Syversen G, Solheim N, Laugerud F. Spread of methicillin-resistant Staphylococcus aureus in a neonatal intensive unit associated with understaffing, overcrowding and mixing of patients. J Hosp Infect. 2002;50:18–24. doi: 10.1053/jhin.2001.1128. [DOI] [PubMed] [Google Scholar]
- 24.Cantor CR, Smith CL, Mathew M. Pulsed-field gel electrophoresis of very large DNA molecules. Annu Rev Biophys Biophys Chem. 1988;17:287–304. doi: 10.1146/annurev.bb.17.060188.001443. [DOI] [PubMed] [Google Scholar]
- 25.Ostojic M. Epidemiologic genotyping of methicillin-resistant Staphylococcus aureus (MRSA) by pulsed field gel electrophoresis (PFGE) Bosn J Basic Med Sci. 2008;8:259–265. doi: 10.17305/bjbms.2008.2930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grisold AJ, Zarfel G, Strenger V, Feierl G, Leitner E, Masoud L, Hoenigl M, Raggam RB, Dosch V, Marth E. Use of automated repetitive-sequence-based PCR for rapid laboratory confirmation of nosocomial outbreaks. J Infect. 2010;60:44–51. doi: 10.1016/j.jinf.2009.10.045. [DOI] [PubMed] [Google Scholar]


