1. Introduction
The family of sialic acids comprises more than 50 members, different in particular on the hydroxyl substituents of the common precursor N-acetylneuraminic acid (Neu5Ac). The biosynthesis of sialic acids is therefore a complex process, which can generally be divided into two parts. The first steps comprise the de novo synthesis of Neu5Ac and its activated nucleotide sugar CMP-Neu5Ac. The second part covers the introduction of the several substituents to Neu5Ac, including formation of N-glycolylneuraminic acid (Neu5Gc) at the stage of the nucleotide sugar (see Varki chapter) and modifications of the hydroxyl groups of glycan-bound sialic acids (see Vlasak chapter). In the following review we will focus on the initial biosynthesis of CMP-Neu5Ac from UDP-N-acetylglucosamine (UDP-GlcNAc), and in particular on the key enzyme of this pathway in all vertebrates, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (UDP-GlcNAc 2-epimerase/ManNAc kinase; GNE).
First, we will give a brief overview of the sialic acid biosynthesis (Section 2). Then we will summarize the research on GNE in a historical outline (Section 3). The following three sections will describe structure, biochemistry and genetics of GNE in detail. We will further place emphasis on the very complex regulation of GNE (Section 7), which will highlight the enzyme as a master regulator of sialic acid synthesis. The last section will focus on diseases related to GNE, the so called GNE-opathies, a fast growing field of research with fascinating new insights into the role of GNE and of sialic acids.
2. Sialic acid biosynthesis pathway
The starting compound for the biosynthesis of sialic acids is UDP-GlcNAc. This nucleotide sugar is the key compound of the whole amino sugar metabolism, and it is formed by the hexosamine pathway, which splits off from glycolysis at the stage of fructose 6-phosphate. In a particular context the hexosamine pathway may also be included to the sialic acid biosynthetic pathway, but UDP-GlcNAc is a substrate for several reactions in glycan metabolism and is not exclusive for sialic acid formation.
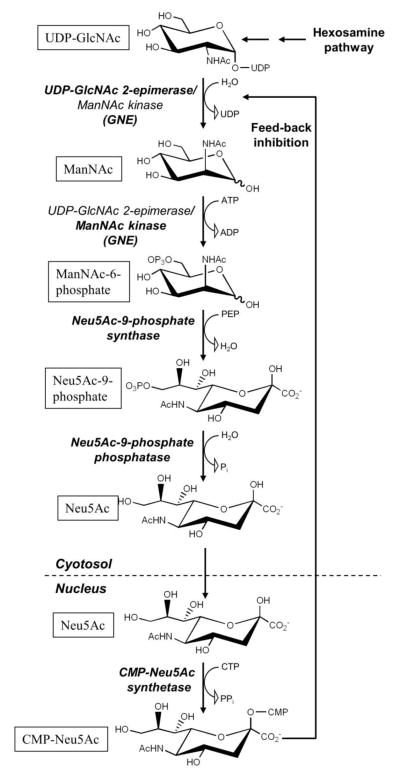
The synthesis of CMP-Neu5Ac from UDP-GlcNAc requires five enzymatic steps (Fig. 1). Four of them are necessary to generate Neu5Ac, and they are localized in the cytosol. The final formation of CMP-Neu5Ac takes place in the nucleus (for details about this particular exception see Muenster/Weinhold chapter). Every step is combined with cleavage of one energy-rich bond, which directs the de novo pathway straightforward to product formation.
Figure 1. Sialic acid biosynthesis in vertebrates.
GNE catalyzes the first two steps of this pathway. The respective enzymatic reactions are indicated in bold. Note, that UDP-GlcNAc is supplied from fructose-6-phosphate by the hexosamine pathway, and that CMP-Neu5Ac is formed in the nucleus. Feed-back inhibition of the UDP-GlcNAc 2-epimerase activity of GNE by CMP-Neu5Ac is further indicated.
The biosynthesis of Neu5Ac begins with the formation of ManNAc from UDP-GlcNAc by simultaneous release of UDP. This initial step is catalyzed by UDP-GlcNAc 2-epimerase. ManNAc is then phosphorylated at C-6 by the specific ManNAc kinase. Other potential sources of ManNAc or ManNAc-6-phosphate would be GlcNAc and GlcNAc-6-phosphate, respectively. However, a GlcNAc-6-phosphate epimerase only exists in prokaryotes [1], and the formation of ManNAc from GlcNAc by GlcNAc 2-epimerase does not occur under physiological circumstances [2]. The only salvage pathway known is the introduction of ManNAc by direct phosphorylation via a kinase [3], e.g. from nutritional sources (which, however, contain only trace amounts of ManNAc) or from the degradation of Neu5Ac [4].
Neu5Ac is a condensation product of ManNAc and pyruvate. The activated form of pyruvate, phosphoenolpyruvate (PEP), is derived from glycolysis. The Neu5Ac-9-phosphate synthase uses ManNAc-6-phosphate instead of ManNAc, and catalyzes an aldol addition with PEP for the formation of Neu5Ac-9-phosphate [5]. The reaction is thereby driven by the release of the phosphate group from PEP. This enzymatic reaction is followed by the release of the 9-phosphate group from Neu5Ac-9-phosphate by a specific phosphatase [6]. The final step of the de novo pathway is the formation of CMP-Neu5Ac in the nucleus of vertebrate cells by the CMP-Neu5Ac synthetase [7]. The nucleotide sugar is then transported to the Golgi apparatus by the CMP-sialic acid transporter and is used there as a substrate for the different sialyltransferases.
A catabolic pathway of Neu5Ac metabolism also exists. As there is no direct involvement of GNE in this pathway, we only decribed it briefly. Neu5Ac can be transported as a monosaccharide from the lysosome to the cytosol [8]. There it is cleaved by the Neu5Ac aldolase, which forms ManNAc and pyruvate [4]. Whereas pyruvate can reach the energy metabolism or gluconeogenesis, ManNAc is further converted to GlcNAc and introduced into the hexosamine pathway [2]. For CMP-Neu5Ac a degradation mechanism is not known, as no enzymatic activity for cleavage of the nucleotide sugar has been identified [9]. The only way for degradation is the transfer of the sugar moiety to glycans in the Golgi apparatus.
3. History of GNE research
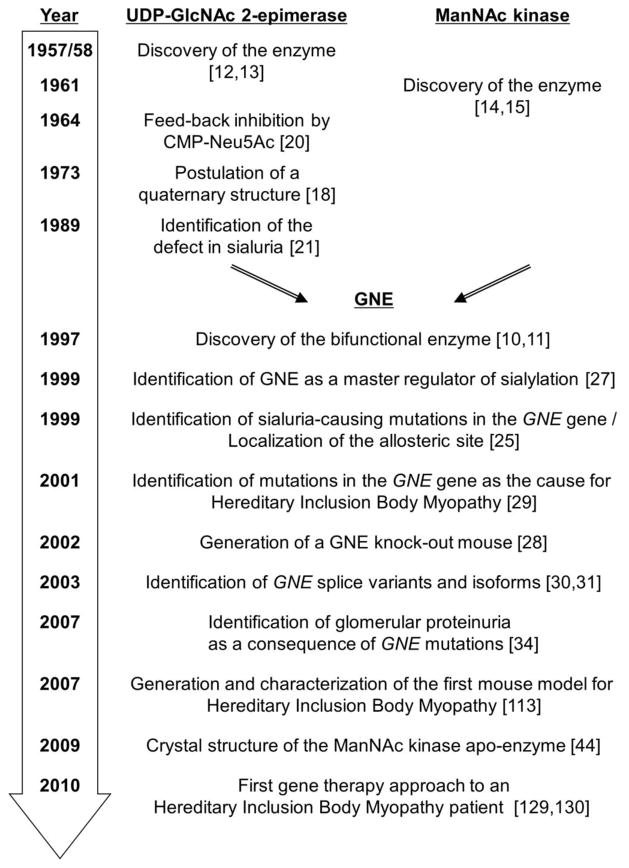
A historical view on GNE (Fig. 2) can be divided into two different eras. The first one covers the description of the two functional parts of GNE, the UDP-GlcNAc 2-epimerase and the ManNAc kinase, as separate entities. The second one started in 1997, when Reutter and coworkers discovered the bifunctionality of GNE [10, 11]. From then on all following studies either directly investigated the bifunctional enzyme or at least, kept this structure in mind.
Figure 2. Milestones in GNE research.
UDP-GlcNAc 2-epimerase (E.C. 5.1.3.14) was first observed by Cardini and Leloir [12]. In 1958 Comb and Roseman [13] discovered that the products of the enzyme are ManNAc and UDP, which was not completely clarified in the previous report. In 1961, discovery of ManNAc kinase (E.C. 2.7.1.60) was reported simultaneously by Gosh and Roseman [14] and Warren and Felsenfeld [15]. Initial biochemical characterization of the enzymes were performed in the 60-ies and early 70-ies for the epimerase [16–18] and the kinase [19]. The ManNAc kinase was not an object of outstanding interest in the following years, due to its biochemical features comparable to several other sugar kinases. On the other hand, the UDP-GlcNAc 2-epimerase was already discovered as a regulator in sialic acid biosynthesis due to its feed-back inhibition by CMP-Neu5Ac in 1964 by Kornfeld et al. [20]. A complex quaternary structure involved in this regulation mechanism was postulated in 1973 by Kikuchi and Tsuiki [18], underlining the key role of UDP-GlcNAc 2-epimerase. At that time, first speculations came out, that a recently discovered disease, sialuria, might be due to misfunctioning of UDP-GlcNAc 2-epimerase. However, it needed more than ten years, before Ashwell and co-workers proofed a defect in the feed-back inhibition mechanism of this enzyme as disease-causing [21].
In 1997 GNE was purified to homogeneity by independent isolation of the same polypeptide using procedures either focusing on UDP-GlcNAc 2-epimerase or on ManNAc kinase, accompanied by detection of both enzyme activities in all fractions [10]. In a parallel study the GNE cDNA was cloned from rat and functionally expressed in a heterologous system [11]. This experiment unequivocally showed the bifunctional character of GNE. From this time research on GNE started to speed up. The same gene was identified and studied in a variety of mammalian species [22–24], and later also in other vertebrate species by genome sequencing, leading to the rapid identification of the genetic defect in sialuria [25] and the determination of the allosteric site in the epimerase domain [26]. In the following years, biochemical characterization revealed an array of novel structural and enzymatic GNE features (see sections 5 and 6). The role of GNE as a master regulator for sialylation was proofed by Keppler et al. [27] in a number of different cell types, and later by the lethal effect of targeted mutagensis of the Gne gene in mice [28]. The latter study overlapped with the completely unexpected identification of a novel congenital disease caused by mutations in the GNE gene, hereditary inclusion body myopathy (HIBM) [29]. GNE now became the focus of researchers who were no glycobiologists before. They contributed with novel findings and approaches, including the discovery of GNE splice variants [30] and, as a consequence, additional protein isoforms [31]; GNE-dependent genome [32] and proteome analysis [33]; the discovery of other GNE-opathies [34]; the establishment of a set of mouse models, and, importantly, the development of metabolic and genetic therapies for GNE-related diseases (see section 8).
4. The GNE gene
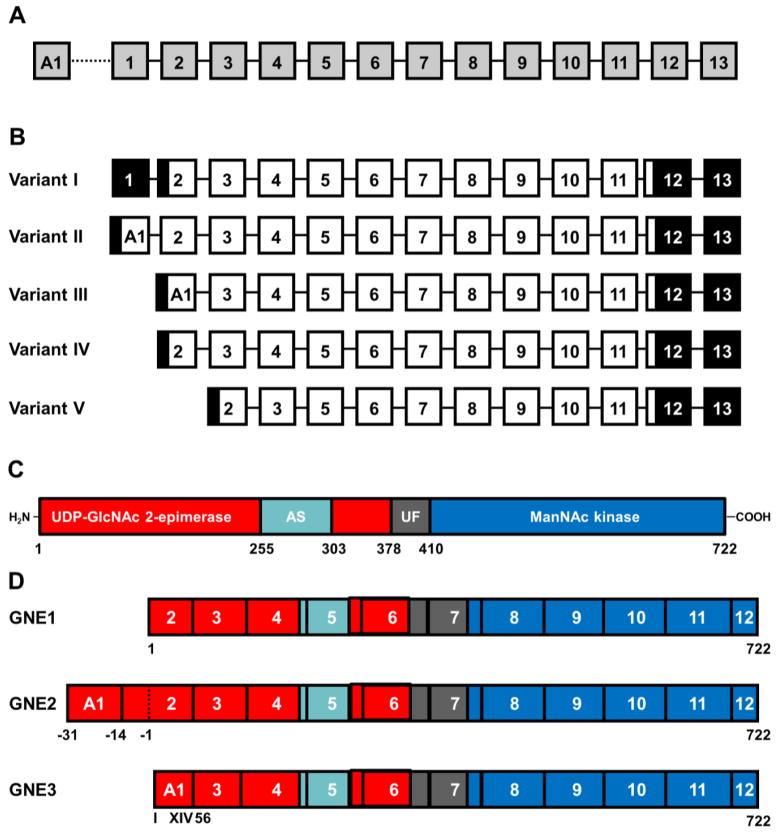
The human GNE gene is localized on chromosome 9p12–13. It consists of fourteen exons (Fig. 3A). Thirteen of these exons are assembled together with twelve introns in a gene stretching across a length of about 45,000 bp. In 2003 Watts et al. [30] discovered an additional exon, named A1, which is localized 20,000 bp upstream of exon 1 and extended the actual GNE gene to more than 65.000 bp [30]. GNE gene transcription and splicing results in mRNA of about 5,000 bp. Alternative splicing of exons A1, 1, and 2 leads to four different mRNA variants, described by Watts et al. [30] and later an exon 4-skipping splice variant was found in humans by Tomimitsu et al. [35], now named splice variant V (Fig. 3B). A similar formation of GNE splice variants can be postutated for other mammalian species, since they have a comparable exon structure and distribution as the human GNE gene. Overall GNE transcription can be regulated by CpG methylation [36] (see also section 7). Interestingly, the CpG islets responsible for this mechanism are localized upstream of exon A1 and not of exon 1 [37]. Although CpG methylation of the GNE gene was unequivocally shown only in lymphocytes, it is very likely, that the same mechanismoccurs in other tissues and contributes to tissue-specific adjustment of GNE expression. The non-coding part of human GNE exon 12 together with exon 13 form a non-coding 3′-end with an extraordinary lenght of more than 2000 bp in human transcripts [23]. However, GNE transcripts of other mammalian species do contain much shorter 3′-ends, as shown by Northern blot analysis [11, 22]. It is therefore likely, that there is a recently unknown regulatory function of the 3′-end of GNE mRNA specific for humans.
Figure 3. Structure of the human GNE gene, mRNA and protein.
(A) Schematic exon structure of the human GNE gene.
(B) Predicted GNE mRNAs by alternative splicing.
(C) Protein domain structure of GNE.
(D) Schematic structure of the GNE isoforms with proven enzymatic activity (for details see text).
5. The GNE protein
5.1. Primary structure
GNE is a very highly conserved protein. In all mammalian species with a known genome GNE (actually GNE1; see below), consists of 722 amino acids (or in the case of giant panda 721 amino acids) with a sequence identity of >98%. Generally, homology of other proteins between several vertebrate species is lower. Moreover, even the amino acid sequence identities between human and chicken or frog GNE are still high, 92% and 86%, respectively. A sequence comparison of human GNE and that of different fish species also revealed a sequence identity of about 80%. In addition, all vertebrates possess the bifunctional GNE protein, and no evidences for separately expressed enzymes have been found within the available sequence information. The following detailed description of GNE features could therefore be applied to the enzyme from all vertebrate species.
GNE consists of two functional domains, an N-terminal part responsible for the UDP-GlcNAc 2-epimerase activity and a C-terminal part covering the ManNAc kinase activity (Fig. 3C). The two parts are about equal in size, but the exact domain boundaries cannot be defined, because one of the last challenges in GNE research, the resolution of the three-dimensional structure of the entire enzyme, has not been accomplished so far. However, due to sequence comparisons with other enzymes of homologous functions [38] and the construction of deletion mutants [39] the epimerase domain can be specified to the amino acids 1 to 378, and the kinase domain covers at least the amino acids 410 to 684. A further hinge domain has been postulated between amino acids 378 and 410, but this remains speculative without reliable structural data. The binding site of the feed-back inhibitor CMP-Neu5Ac has been mapped first by sialuria patients’ mutations at amino acids 263 and 266 [25]. Yarema et al. [26] identified further sialuria-like mutants in cultivated cells in between amino acids 255 and 275. Whether this area forms a separate regulatory subdomain within the epimerase domain, or “only” defines the allosteric binding site for CMP-Neu5Ac remains to be clarified.
The five different splice variants of the GNE mRNA altogether encode (at least theoretically) for four different protein isoforms (Fig. 3D) [31]. The originally described GNE protein is therefore also called GNE1. It covers 722 amino acids with a calculated molecular mass of about 79 kDa. GNE2 is an isoform with 31 additional amino acids at the N-terminus, which are encoded by the exon A1. These amino acids extend the epimerase domain for about 3 kDa. The third isoform, GNE3, lacks the first 55 amino acids of GNE1 due to exon 2 elimination. On the other hand, 14 new amino acids are encoded by another reading frame of exon A1, resulting in a total GNE3 size of about 75 kDa. Interestingly, GNE3 seems to be restricted to primates, whereas GNE1 and GNE2 appear ubiquitous among vertebrates. However, the partial deletion of the epimerase domain in GNE3 leads to the total loss of the respective enzymatic activity, whereas the kinase domain is still intact [40]. Together with the fact that the GNE3 protein has been analyzed as a recombinant protein only, the question arises whether GNE3 exists as a protein in vivo at all. In this case a function in fine-tuning of the sialic acid pathway might be postulated. On the other hand, also a role of the GNE3 mRNA itself, likely in translation regulation, is possible, and the GNE3 protein may be an artifact of recombinant technologies. The fourth GNE isoform, which is encoded by the exon 4-skipping splice variant [35], is completely inactive when expressed as a recombinant protein (S. Hinderlich, unpublished data). Due to an unclear biological function, we hesitate to call this isoform GNE4.
5.2. Quaternary structure
One of the extraordinary features of GNE is its ability to form different oligomeric structures. Already in early GNE research an oligomeric structure was postulated by Kikuchi and Tsuiki [18], who proposed a hexameric structure for rat UDP-GlcNAc 2-epimerase, derived from kinetic data of the allosteric enzyme inhibition by CMP-Neu5Ac. Hinderlich et al. [10] confirmed these data by gel filtration analysis of purified rat liver GNE, which revealed a native molecular mass for the fully active enzyme of 450 kDa, corresponding to a hexamer of 75 kDa subunits. However, determination of the oligomerization of purified rat liver GNE by the identical kinetic calculating a Hill plot from the CMP-Neu5Ac inhibition curve as performed by Kikuchi and Tsuiki, suggested a tetrameric protein [10]. Besides the fully active hexameric/tetrameric GNE, also a second, dimeric state was observed. This GNE dimer possesses only ManNAc kinase activity, but can be reassembled to the higher oligomeric state by incubation with the epimerase-substrate UDP-GlcNAc, revealing a potential regulation mechanism (see also section 7). A more detailed analysis of GNE oligomerization using recombinant rat GNE and modern biophysical methods confirmed the existence of the two different oligomeric states whereby the higher one was unequivocally assigned to a tetramer [41]. Moreover, these experiments revealed a dynamic interplay between dimers and tetramers, also influenced by different GNE substrates. Interestingly, also higher, but less defined oligomers were observed, which, at least in part, tend to precipitation. The latter observation might be due to the unphysiologically high concentration of GNE by its recombinant expression. But it may also explain the observation of hexamers of the native rat liver enzyme, which might reflect a mixed population of tetramers and higher oligomers, which appear as a hexamer in size exclusion chromatography. Another potential explanation for GNE hexamers in the physiological environment of the cell might be different posttranslational modifications (see also section 7), which promote hexamer formation, but did not occur in the heterologous baculovirus expression system used for the study of Ghaderi et al. [41].
5.3. Three-dimensional structure
One of the unresolved questions of GNE research is the three-dimensional structure of the bifunctional enzyme. Due to the lack of a crystal structure two different alternative approaches were made. First, models of the structure of the two different functional domains were generated basing on similarities to related enzymes, mostly of prokaryote origin, and their known crystal structures [42, 43]. These studies were motivated in particular by the huge set of mutations causing HIBM and its unclear influence on GNE function (see below). Second, the functional domains were expressed as separate recombinant proteins and exerted in crystallization experiments. Whereas the epimerase domain so far refused to form diffractable crystals, the structure of human ManNAc kinase in its ligand-free state has been solved recently [44]. We will therefore describe the ManNAc kinase structure mainly based on data of this study, whereas our overview of the epimerase domain uses the model structures. However, the assembly of the two domains within the bifunctional enzyme or even the quaternary structure remains speculative and will not be a topic here.
The GNE kinase domain belongs to the ROK family (repressor/open reading frame/kinase family, PF00480). This protein family in particular consists of prokaryotic members, and the ManNAc kinase is its only human member, clearly differing from other sugar kinases with related functions. The crystal structure of the human ManNAc kinase domain revealed the typical bi-lobal structure with the postulated ATP-binding side in a deep cleft between the two lobes typical for all ROK kinases [44]. Both lobes consist of / folds with central -sheets surrounded by -helices. Furthermore, the ManNAc kinase contains a ROK-characteristic complex-bound zinc ion, integrated into a HC3-type zinc finger. As zinc fingers are often found as DNA-binding structures of transcription factors, speculations emerged that potential functions of GNE outside the sialic acid metabolism might be due to transcription regulation. However, the zinc ion seems to be more likely essential for substrate binding of ManNAc kinase ([44]; S. Hinderlich, unpublished observation). Recombinant ManNAc kinase assembles as a dimer, which was also observed in the crystal structure [44]. The dimerization site was localized within the C-terminal lobe of the protein. Interestingly, some hexameric structures were observed by gel-filtration analysis of the purified ManNAc kinase, and a crystallographic hexamer could also be postulated [44]. However, the authors could not completely rule out artifacts of recombinant protein expression or crystallization, so that the discussion about the oligomeric structure of GNE (see above) remains open.
The potential structure of the UDP-GlcNAc 2-epimerase domain of GNE was evaluated by models based on similarities to prokaryotic UDP-GlcNAc 2-epimerases. The prokaryotic enzymes cover non-hydrolyzing epimerases which produce UDP-ManNAc and hydrolyzing epimerases resulting in UDP and ManNAc as products. However, the overall three-dimensional structures are similar and can be superimposed to the GNE epimerase domain due to a common secondary structure. Based on these data sets, an initial model was generated by Penner et al. [42], and a strongly improved calculation was published by Kurochkina et al. [43], who also integrated the structures of phosphoglycosyltransferases. The GNE epimerase domain consists of two domains, which form a cleft between the domains harboring the active site; a structure, which was found similarly also for the ManNAc kinase domain. The N-terminal domain consists of seven -sheets surrounded by seven -helices, and the C-terminal domain contains further six -sheets and six -helices. Both domains form structures similar to Rossman dinucleotide binding folds. Although some of the three-dimensional structures of the prokaryotic epimerases display dimers, conclusions about the contributions of the epimerase domain to GNE oligomerization were not possible from the models.
6. GNE enzymatic function
The two enzymatic functions of GNE could be considered independently. This is comparable to the protein structure, which can be described as two independent functional domains, and the physical separation of the two GNE domains results in enzymes with remaining activity [39]. Therefore, detailed descriptions about the two enzymatic mechanisms and active site architecture will be discussed separately in this section. The functional consequences of the two-domain structure are also discussed here, although some points can only be clarified by a still pending three-dimensional GNE structure. One of the basic features of bifunctional enzymes is the physical connection of two active sites for substrate channeling, when two successive steps of one pathway are catalyzed [45]. This mechanism avoids release of intermediates from the enzymes, which may either be instable or should be prevented from further conversion by competing enzymes of other pathways. The latter point might be true for GNE, as ManNAc is also a substrate of GlcNAc 2-epimerase which would lead the sugar to amino sugar catabolism [2]. Furthermore, substrate channeling can accelerate the overall enzyme reaction, which is reasonable for GNE as the rate-limiting enzyme of sialic acid biosynthesis. Another important point of bifunctionality is the concerted regulation of two enzymatic steps. Although this may be the case mainly for bifunctional enzymes catalyzing opposed reactions, in the case of GNE, a tight regulation mechanisms between the epimerase and the kinase function might be desirable (see also section 7).
6.1. Enzymatic properties of UDP-GlcNAc 2-epimerase
UDP-GlcNAc is the physiological substrate of UDP-GlcNAc 2-epimerase. In an early study Salo and Fletcher [46] identified UDP-ManNAc as an additional in vitro substrate of the enzyme, resulting in hydrolysis of the nucleotide sugar. However, UDP-ManNAc occurs only in prokaryotes as a substrate in cell wall construction and is not a physiological substrate of GNE. Interestingly, Blume et al. [47] also identified UDP-GalNAc as a GNE binding partner in STD-NMR analysis, but the binding mode of the substrate did not favor an enzymatic reaction. In the same study UDP was found as a substrate with an even higher affinity than UDP-GlcNAc to the epimerase site. UDP is therefore a competitive inhibitor of UDP-GlcNAc 2-epimerase. This is less important under physiological conditions, as the intracellular UDP-GlcNAc concentration exceeds the UDP concentration up to two orders of magnitude. However, in in vitro assays UDP is often used as a GNE stabilizing agent, as it maintains the fully active oligomeric state of the enzyme [10, 41] and its influence on activity assays has to be taken into account. Furthermore, a mechanism can be postulated where UDP remains in the active site after the enzymatic reaction and the release of ManNAc, and a new UDP-GlcNAc molecule enters the active site by a UDP/UDP-GlcNAc exchange.
The epimerization mechanism of GNE starts with the release of the non-acidic proton at C-2 [48]. A set of histidine residues come into consideration as the acting base during this step, whereby the favorite one is His-220, which is homologous to the histidine residue essential for the activity of E.coli UDP-GlcNAc 2-epimerase [49]. The second step covers the elimination of the UDP moiety and the formation of 2-acetamidoglucal. Whereas the nucleotide most likely remains in the active site (see above), the sugar derivative is further converted to ManNAc by stereospecific addition of a water molecule to C-1 and C-2 [48]. Interestingly, 2-acetamidoglucal was already observed by Sommar and Ellis [50] as a byproduct of the epimerase reaction, and later as a minor, but significant unusual component of the urine of sialuria patients [51](see also Section 8).
The exact architecture of the epimerase active site remains to be clarified by a three-dimensional structure. However, several relevant amino acids could be postulated by the combination of mutation analyses and structural models ([38, 42, 43]; S. Hinderlich, unpublished data). Besides His-220, which was already described as a possible general catalyst, His-45 also seems to be essential for the epimerase activity. From the amino acids addressed by site-directed mutagenesis, the following ones showed reduced enzyme activities and are located at least in vicinity of the active site, suggesting a further role in the catalytic mechanism or in substrate binding: Lys-24, Asp-112, His-132, Glu-134, Asp-143, C-303. Several other amino acids affect the epimerase activity by mutation, in particular in HIBM patients, but an indirect effect via structural changes also seems to be likely and remains to be clarified. The epimerase domain models suggest additional amino acids as Arg-19, Asp-225, Ser-301, Arg-306 and Glu-307 involved in substrate binding and are therefore worth to be investigated in the future.
The epimerase domain is able to bind the strong feed-back inhibitor CMP-Neu5Ac (see also Section 7.2). The respective allosteric site at least covers the amino acids 255 to 275, as identified by a mutation screen for cells with high levels of sialic acid [26]. This area also includes the two recently identified mutated amino acids, Arg-263 and Arg-266, in the disease sialuria (see Section 8.1). In vitro site-directed mutagenesis studies identified C303 as another amino acid which mutation resulted in reduced feedback inhibition by CMP-Neu5Ac [42]. It is still unclear if the allosteric site forms a discrete subdomain or the binding area for CMP-Neu5Ac or is integrated within the structure of the epimerase domain. The three-dimensional models of the epimerase domain were unable to address these questions, as the bacterial counterparts, serving as a calculation basis, do not bind allosteric regulators. Nevertheless, we define the area of the amino acids 255 to 303 as the provisional allosteric binding site (see Fig. 3).
6.2. Enzymatic properties of ManNAc kinase
The enzymatic phosphorylation of ManNAc requires ATP as its phosphate donor. Kundig et al. [19] already excluded other nucleotidetriposphates as further substrates. Nevertheless, ADP is also able to bind to the active site of ManNAc kinase with an affinity in about the same range as ATP [52]. In in vitro assays ADP therefore may act as a competitive inhibitor. However, under physiological conditions the cytosolic concentration of ADP is very low due to the action of the ubiquitous adenylate kinase. In addition to ManNAc N-glycolylmannosamine (ManNGc) is a further sugar substrate of ManNAc kinase [19]. ManNGc derives from the degradation of N-glycolylneuraminic acid and is salvaged by the sialic acid biosynthesis pathway. The metabolization of ManNGc suggests a promiscuity of ManNAc kinase and the following enzymes in this pathway for structural modifications of the N-Acyl side chain. This feature was used for the introduction of unphysiological functional groups into sialic acids by metabolic oligosaccharide engineering [53]. Benie et al. [52] showed that two of the ManNAc derivatives often used for this method, N-propanoylmannosamine and N-butanoylmannosamine, are additional substrates of ManNAc kinase. In the same study GlcNAc was also suggested to be phosphorylated by ManNAc kinase. However, affinity of GlcNAc to the enzyme and the presence of a specific GlcNAc kinase [3] most likely exclude a physiological function of ManNAc kinase in GlcNAc phosphorylation.
The enzymatic mechanism of ManNAc kinase has not been investigated directly. However, it can be assumed, that it is closely related to the mechamism of other 6-kinases such as hexokinase. This enzyme transfers the -phosphate from ATP to the 6-hydroxyl group of the sugar via a trigonal-bipyramidal intermediate. The reaction finally results in inversion of the phosphate configuration [54, 55]. The binding, and most likely also the transfer, of the phosphate group is supported by arginine and aspartate residues in 6-kinases. The mutation of the two highly conserved residues Asp-413 and Arg-420 of ManNAc kinase result in complete loss of enzymatic activity [38]. Besides these two amino acids, Asp-517 can be suggested as an additional essential amino acid, as it may act as a catalytic base in abstraction of the proton from the 6-hydroxyl group of ManNAc [43]. The sugar binding site of ManNAc kinase consists of the amino acids Asn-516, Asp-517, Glu-566, His-569 and Glu-588, whereby His-569, together with the three cysteine residues at position 579, 581 and 586, is also involved in binding of the zinc ion of the active side.
7. Regulation of GNE
There are several possible mechanisms to interfere and regulate an enzyme-catalyzed reaction. These include the expression level of the enzyme (quantity and/or isoform expression = enzyme induction), the control of the catalytic reaction by substrates or products (e.g. feed-back control), by posttranslational modification of specific amino acids (such as phosphorylation on serine, threonine or tyrosine residues = enzyme interconversion) or by alteration of chemical or physiological conditions. All these options are explored to control GNE enzymatic activities and will be discussed in detail in this section. The regulation of GNE seems to be of the same complexity as its structure and, interestingly, almost all described GNE regulation mechanisms specifically affect the UDP-GlcNAc 2-epimerase activity and not the ManNAc kinase activity. This might reflect the epimerization of UDP-GlcNAc to ManNAc as the key step of sialic acid biosynthesis.
A total knock-out of the GNE gene (and subsequenctly its enzymatic acitivities) results in complete loss of endogenous sialic acid production in cells or animals. Cultivated cells are able to tolerate a GNE gene knock-out because sialic acids seem to be not essential for survival and only particular sialic acid-dependent functions of these cultured cells appeared to be affected [27]. In a multicellular organism a GNE gene knock-out is lethal. A targeted mutagenesis of the Gne gene in mice results in early embryonal lethality at developmental day E8.5 [28]. Interestingly, heterozygous Gne knock-out mice displayed slightly reduced UDP-GlcNAc 2-epimerase activity and up to 25% reduced sialylation of glycans compared to wild-type mice, but no significant phenotype was observed [56]. This suggests a range of reduced sialylation that appears to be tolerated by an animal, and consequently a threshold of up- or down-regulation of GNE expression and sialylation status, which has to be exceeded by the different regulation mechanisms.
7.1. Regulation by mRNA and protein expression
Sialylation occurs at all stages of mammalian development and even embryonic stem cells already express sialylated glycoconjugates [57–59]. The expression of GNE mRNA, encoding the key enzyme of the sialic acid biosynthesis, strongly correlates with the occurrence of sialic acid. GNE mRNA is ubiquitously expressed in all organs and during all stages of development, as shown in Northern blot and in situ-hybridization experiments. However the major expression of GNE mRNA is found in liver [11, 22, 23], likely because liver is the organ with the highest biosynthesis rate of secreted sialylated glycoproteins (e.g. serum glycoproteins).
The detailed mechanisms of GNE mRNA transcription and GNE protein translation regulation have not been clarified so far. However, transcription of the GNE gene is unequivocally influenced by an epigenetic event. Methylation of CpG islets in the GNE promoter region leads to down regulation of GNE transcription in several cancer cell lines [36] and in HIV-infected lymphocytes [37]. GNE protein expression and enzyme activity are reduced below detectable levels in these cell lines. The same mechanism seems to be involved in down-regulation of GNE gene expression in a Morris hepatoma-derived cell line [36]. Morris hepatoma displayed less than 10% of the GNE activity of rat liver [60]. This is obviously due to reduced production of secretory sialoglycoproteins and a lower need for intracellular sialic acid production. A similar correlation was also found in regenerating rat liver after partial hepatectomy [61, 62], but without assigning this observation to CpG methylation.
Recently it became obvious that GNE exists in several isoforms, which are generated by alternative splicing (for details see sections 4 and 5). However, the existence of each isoform in vivo has not been demonstrated yet on the protein level. Recombinant over-expression of the human isoforms revealed that GNE1, which is the original described 722 amino acids containing isoform, has the highest epimerase activity, whereas the two other isoforms with 31 additional N-terminal amino acids (GNE2) or a different N-terminus (GNE3) displayed strongly reduced epimerase activity [40]. Since the C-terminus of GNE is known to harbor the kinase domain, it was not surprising that ManNAc kinase activity was similar in all isoforms. It was therefore suggested, that the ubiquitous isoform GNE1 serves as a general catalyst in sialic acid production, whereas GNE2 and GNE3 are isoforms involved in fine-tuning the sialic acid biosynthesis. This is supported by a tissue-specific expression of GNE2 and GNE3 mRNA [40]. However the regulation of isoform expression in vivo remains unknown.
As described earlier, the biosynthesis of sialic acids takes place in the cytosol. Therefore GNE is mainly located within the cytosol and standard purification of GNE starts from cytosolic preparations [10]. In one report, GNE was also found in the nucleus and further associated with the Golgi membrane [63]. Although the detection of endogenous GNE is difficult, since most available antibodies do not detect endogenous GNE due to its very low expression level, this observation suggests an additional regulation mechanism via differential subcellular localization. Direct detection of GNE in cellular compartments would also be possible by overexpression of tagged protein (e.g. via fluorescence), and is worth pursuing to clarify this point.
7.2. Regulation by substrates and multimerization
The main regulation mechanism of UDP-GlcNAc 2-epimerase activity is the feed-back inhibition by CMP-Neu5Ac [20]. This allosteric inhibitor affects the epimerase activity in a concentration dependent manner, which connects the production of sialic acids to glycan synthesis. A complete inhibition of the epimerase activity is reached at a CMP-Neu5Ac concentration of about 60 μM [21], which fits well with the intracellular concentration of this nucleotide sugar in cells under normal circumstances. This suggests a sophisticated mechanism of feed-back inhibition, which responds even to minor variations of CMP-Neu5Ac concentration, due to varying rates of biosynthesis of sialylated glycans and subsequent CMP-sialic acid use by the Golgi.
The feed-back inhibition mechanism can be circumvented by extracellular application of ManNAc. ManNAc treatment of, for example, Madin Darby canine kidney cells results in an intracellular CMP-Neu5Ac concentration up to 10-fold over the normal level [64]. These data indicate not only that the ManNAc kinase activity of GNE is not affected by CMP-Neu5Ac inhibition, but also that no additional metabolic regulation mechanisms exists within the sialic acid biosynthesis pathway.
A second regulation mechanism using GNE substrates includes the epimerase educt UDP-GlcNAc and its product UDP. As mentioned above (Section 6.1.) UDP binds with a high affinity to the active site of the epimerase enzyme and might act, at least in vitro, as a competitive inhibitor. Moreover, physiological regulation via UDP and UDP-GlcNAc is strongly connected to GNE oligomerization. Whereas the GNE dimer is only able to catalyze the phosphorylation of ManNAc, the epimerase activity requires the highest possible oligomeric state (tetramer or hexamer). This structure is maintained by UDP, which usually remains in the active site after one catalytic cycle, and can be stabilized in vitro by addition of 100 μM UDP to buffers. More interestingly, once GNE is decayed to a dimer, it can be reassociated into tetrameric states by addition of UDP-GlcNAc (but not UDP) to the system [10], suggesting a direct regulation of the ability of GNE to catalyze its first enzymatic step by the respective substrate. Intracellular UDP-GlcNAc concentrations are usually sufficient to maintain the tetrameric state. The interchange between the oligomeric states may therefore regulate the UDP-GlcNAc 2-epimerase activity only under certain conditions with low UDP-GlcNAc concentrations. This might be in pathological situations or in events which require a high amount of UDP-GlcNAc for other purposes, such as the synthesis of non-sialylated glycans or O-GlcNAcylation of proteins.
7.3. Regulation by posttranslational modification
Posttranslational modifications (PTM) are chemical modifications of proteins after its biosynthesis (translation). PTMs include glycosylation, phosphorylation, acetylation, and addition of lipids, ubiquitinylation, SUMOylation or sulfatation. PTMs often occur at specific residues of the primary sequence. Typically amino acids with additional amino- or hydroxyl groups such as Lysine, Asparagine, Threonine, Tyrosine or Serine bear PTMs. As mentioned earlier, GNE consists of 722 amino acids and 144 of these are Lysine (41), Asparagine (29), Threonine (33) or Serine (41) residues. When analyzing the sequences around these amino acids in more detail it becomes obvious that GNE contains 42 possible phosphorylation sites (thereof 13 PKC-sites with the highest possible score using the tool NetPhosK). These analyses are supported by earlier studies where it was demonstrated that GNE is phosphorylated in vivo and interacts with several isoforms of protein kinase C in mouse liver [65]. Consequently, protein kinase C is one candidate enzyme acting on GNE and its phosphorylation of GNE results in an activation of the UDP-GlcNAc 2-epimerase activity. In this light it is remarkable that about 20 of the known mutations responsible for Hereditary Inclusion Body Myopathy (HIBM) (see Section 8.2.) are mutations where a novel Serine or Threonine is introduced (Table 1), making GNE accessible for further posphorylation and likely misregulation involved in the pathogenesis of these HIBM-related GNE mutations.
Table 1.
HIBM-related GNE mutations, which introduce novel Serine or Threonine residues and their phosphoryation probability.
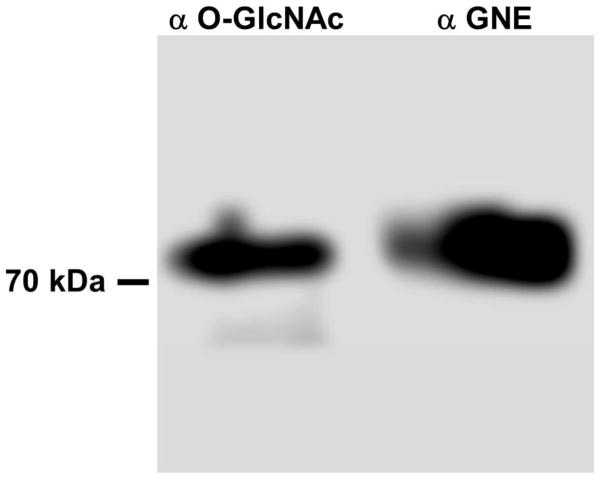
A unique PTM is the modification of Serine or Threonine with O-GlcNAc [66]. There are five potential O-GlcNAc sites in GNE (S122, T223, S505, S689, T718; calculated by YinOYang_1.2). These residues are always very close to phosphorylation sites, but never represent the same residue. Therefore we tested GNE for its content of O-GlcNAc by Western blot analysis after immunoprecipitation from rat liver and demonstrated that GNE is modified by O-GlcNAc (Fig. 4). Together with a phosphorylation likely close to the O-GlcNAcylation site this suggests another level of fine-tuning of the enzyme function in response to cellular changes, as observed for other proteins simultaneously modified by phosphorylation and O-GlcNAcylation [66]. Further analysis revealed that GNE contains three potential sites for SUMOylation (K33, K427, K618; calculated by SUMOsp_2.0), but no consensus sequence for ubiquitinylation (www.UbPred.org). A regulation via SUMOylation is therefore likely. Finally, GNE possesses three potential N-glycosylation sites (N300, N395, N661; calculated by NetNGlyc_1.0). However, N-glycosylation was already ruled out for the purified rat liver enzyme [11], and is in agreement with the usual absence of this PTM for intracellular proteins.
Figure 4. GNE is modified by O-GlcNAcylation.
GNE was immunoprecipitated from rat liver by a GNE-specific antibody and Western-blotted. Immunodetection was performed with O-GlcNAc- and GNE-specific antibodies, indicating O-GlcNAcylation of the GNE protein.
7.4. Regulation potential by interacting with other proteins
Protein-protein interactions are well-documented events to regulate the function of a protein or enzyme. During the last five years several GNE-interacting proteins have been identified. Using 5600 cDNAs from a human brain library in a yeast two-hybrid system four proteins were identified as potential interacting partners: promyelocytic leukemia zinc finger protein (PLZF), collapsin response mediator protein 1 (CRMP1), receptor interacting factor 1 (RIF1) and KIAA 1549, a protein of unknown function [67]. The interaction of CRMP1 and GNE could be verified by co-immunoprecipitation from PC12 cell lysates, and the interaction of PLZF and GNE was confirmed by a pull-down assay. A fragment based approach for identification of the GNE domain involved in these protein-protein interaction revealed that only the full length enzyme as well as the separately expressed ManNAc kinase domain showed positive results, indicating the importance of the ManNAc kinase domain and likely also the oligomeric state of the full length enzyme for these interactions [67].
CRMP1 is a member of the TUC (Toad-64/Ulip/CRMP) protein family and is involved in growth cone collapse [68]. As a modulator of the Rho-A dependent signaling pathway it regulates the F-actin depolymerization [69]. CRMP1 analogs in humans are called dihydropyrimidinase-related proteins (DHPase) and their different isoforms are tissue specific expressed [70]. A connection between sialic acids and members of the TUC family was shown by Büttner et al. [71]. They used unnatural sialic acid ManNProp to induce neurite outgrowth in PC12 cells. During this process a protein was downregulated, which was identified as the TUC protein family member Ulip by MALDI-TOF MS analysis. In a recently unpublished study it was shown that CRMP1 interacts with Huntingtin and modulates its aggregation and neuronal toxicity (Y. Bounab, personal communication). This is of special interest because in affected muscles of HIBM patients, insoluble inclusion bodies are found, which are worthwile investigating for presence of Huntingtin accumulation.
PLZF was identifiedas fusion partner of retinoic acid receptor α, causing acute promyelocytic leukemia (APL). PLZF is a transcription factor and regulates Cyclin A2 and Hox gene expression [72, 73]. Krause et al. [63] showed co-localization of GNE with nuclear markers and support the interaction of GNE and PLZF as a potential transcription regulator. The N-terminal BTB/POZ domain of PLZF interacts also with E3 ligase Cullin 3 mediating ubiquitinylation [74]. This interaction may link GNE with protein degradation via the proteasome pathway. A deregulation of protein degradation could also play a role in formation of inclusion bodies in HIBM. Furthermore, PLZF is found in the cytosol where it is associated with the angiotensin 2 receptor (AT2R) [75] and with the prorenin/renin-receptor [76]. AT2R and AT1R play also roles in mitochondrial biogenesis in skeletal muscle, and Eisenberg et al. [32] could show a potential connection by identifying impaired mitochondrial processes in HIBM.
Very recently, Amsili et al. [77] found a direct interaction between GNE and -actinin. They used a Surface Plasmon Resonance (SPR)-based biosensor assay to search for potent GNE interacting proteins and verified the interaction by co-immunoprecipitation and immunohistochemistry. But Amsili et al. [77] could not identify a difference in the binding kinetics of wild-type and mutant GNE to α-actinin using the SPR biosensor analysis. It was therefore suggested that GNE has a specific function in muscle physiology, but its role in HIBM pathology remains unclear.
Although it is convincingly demonstrated that GNE interacts with these binding proteins, there is no evidence that these interactions alter GNE enzymatic activity or subcellular distribution. Further work is needed to elucidate the function of these protein-protein interactions and their impact on sialic acid biosynthesis.
8. GNE-opathies
Mutations or aberrant expression of the GNE gene can underlie distinct diseases, collectively called GNE-opathies. Since GNE has a central role in sialic acid metabolism, most GNE-opathies are characterized by access or paucity of sialic acid synthesis, resulting in hyper- or hypo-sialylation of glycoproteins or glycolipids. These processes may occur tissue-specific, glycan-specific, or globally (most glycans and/or most tissues affected). This section describes the intriguing group of human GNE-opathies, their molecular and cellular features, disease mechanisms, established cellular or animal models for these disorders as well as proposed therapeutic options.
8.1. Sialuria
8.1.1. Clinical features
Sialuria (also called ‘French type’ sialuria; OMIM 269921) is a rare, autosomal dominant inherited inborn error of metabolism which was first described in 1968 by Montrieul et al. [78] and Fontaine et al. [79]. Sialuria patients clinically manifest with mild coarse facies and slight motor delay, with additional sporadic features of hepatosplenomegaly, delayed skeletal development, microcytic anemia, and mild intellectual impairment. They are diagnosed by the detection of gram quantities (> 1 gram/day) of free sialic acid in urine and significantly increased concentrations of free sialic acid in the cytoplasm of cultured fibroblasts [25, 80–82]
So far, only 7 sialuria patients have been reported worldwide, summarized in Table 2. The prevalence of sialuria is probably underestimated due to the mildness of the disorder. Moreover, assaying urinary free sialic acid is not a routine laboratory procedure, but should be considered as part of the metabolic screening of young children with mild developmental delay and in patients with a phenotype suggestive of mucopolysaccharidosis or oligosaccharidosis [83]. Such free sialic acid screening would not only diagnose patients with sialuria, but also with the lysosomal storage disorders Salla disease (MIM 604369) and Infantile Free Sialic Acid Storage Disorder (ISSD, MIM 269920), both caused by recessive mutations in the lysosomal sialic acid transporter SLC17A5 [84].
Table 2.
Summary of ethnicity, molecular and cellular aspects of sialuria patients reported to date.
| Patient No. | GNE mutation | Ethnicity | Urinary sialic acid content1 (μmol/mmol creatinine) | Sialic acid content in fibroblasts2 (nmol/mg protein) | Reference |
|---|---|---|---|---|---|
| 1 | R263L | Caucasian (French) | 13-15 g/24h 3 | NR | [78, 79] |
| 2 | R266W | Caucasian (Australian/British) | 8,500-8,900 | NR | [25, 91] |
| 3 | R266Q | Caucasian | 5,436 | 65.2 | [21, 25, 83] |
| 4 | R263L | Caucasian | 10,453 | 95.9 ± 54.3 | [25, 144] |
| 5 | R266Q | Caucasian (Portuguese) | 1,693 | 17.4 ± 7.4 | [81] |
| 6 | R266Q | Caucasian (Belgium) | 8,950-14,680 | NR | [82] |
| 7 | R266Q | Caucasian (Belgium) | 4,278 | NR | [82] |
NR, reported.
Urinary free sialic acid normal range: < 74 μmol/mmol creatinine
Fibroblast free sialic acid normal range: 0.2-3.2 nmol/mg protein
Not reported per mmol creatinine
8.1.2. Molecular and cellular features
The biochemical defect of sialuria involves failure to regulate sialic acid synthesis, due to impaired allosteric feed-back inhibition of the epimerase domain of GNE by CMP-sialic acid as outlined in section 5.1. Sialuria has an autosomal dominant mode of inheritance; a heterozygous missense mutation in the allosteric site of GNE leads to loss of feedback inhibition of GNE activity by CMP-sialic acid, resulting in cytoplasmic accumulation and urinary excretion of large quantities of free sialic acid [25, 80, 82]. All known patients are heterozygous for a missense mutation in one of two amino acids, arginine at position 263 (R263L) or arginine at position 266 (R266Q; R266W). The clustering of these mutations in the region of codons 263 to 266 marks this region as the allosteric site for CMP-sialic acid binding. However, the full extent of the allosteric site has not been formally defined (see sections 5.1 and 6.1).
To our knowledge, there are no known animal models of sialuria (including rat, mouse, zebrafish). However, sialuria cell lines have been employed for a variety of in vitro studies, including Jurkat cells [26], Chinese Hamster Ovary (CHO) cells [85, 86], Spodoptera frugiperda (Sf9) insect cells [42], and human epithelial kidney (HEK) cells [87]. These mutant cell lines either arose through mutagenesis screens, or were created by transfection of a GNE plasmid containing a sialuria mutation into normal cells. The latter ones can potentially be used to produce highly sialylated proteins for biotechnological purposes [88].
8.1.3. Disease mechanism and therapy
Increased cytoplasmic free sialic acid in sialuria patients’ cells may contribute to the clinical symptoms due to increased acidic and negatively charged properties. Moreover, it is likely that specific glycans or groups of glycoconjugates in sialuria cells are hypersialylated which may also explain some of the clinical features of sialuria. Iso-electric focusing studies of serum from three sialuria patients demonstrated significant hypersialylation of apoC-III (a marker for O-glycan sialylation) and slight hypersialylation of serum transferrin (a marker for N-glycan sialylation) [89]. Levels of lysosomal enzymes, plasma amino acids, and urine organic acids are normal in sialuria patients, and there is no microscopic evidence of stored material in fibroblasts, as seen in lysosomal storage disorders.
Dominant human disorders due to failed allosteric inhibition as sialuria are extremely rare [90]. Therapies for these disorders are lacking. Currently, only symptomatic therapy is available also for sialuria patients. The lowering of sialic acid levels in fibroblasts by cytidine feeding has been suggested [80, 91]. However, recent in vitro studies have shown that allele-specific silencing of disease genes might provide treatment for dominant disorders [92]. Allele-specific silencing studies with siRNAs targeting a specific sialuria mutation in patient’s fibroblasts (c.797G>A; p.R266Q) demonstrated successful down regulation of the mutant allele, a significant decrease of free sialic acid (to the normal range), and recovery of feed-back inhibition of UDP-GlcNAc 2-epimerase activity by CMP-sialic acid [93]. Even though these findings indicate that allele-specific silencing of the mutated allele in sialuria is a viable therapeutic strategy, further investigations are required for the use of siRNA-based therapies in humans [92, 94].
8.2. Hereditary Inclusion Body Myopathy (HIBM)
8.1.1. Clinical features
HIBM (OMIM 600737) is a rare autosomal recessive adult onset progressive neuromuscular disorder. The disease was originally described in Japanese patients as Nonaka Distal Myopathy [95] now commonly referred to Distal Myopathy with Rimmed Vacuoles (DMRV) (OMIM 605820). Later, a similar disorder was described as vacuolar myopathy sparing the quadriceps in Iranian-Jewish patients [96] now commonly referred to as Inclusion Body Myopathy 2 (IBM2) or Hereditary Inclusion Body Myopathy (HIBM). Only after the HIBM causing gene, GNE, was mapped and identified, it was recognized that HIBM was allelic to DMRV [97]. We henceforth refer to the disorder as HIBM. Apart from the Persian-Jewish and Japanese isolates, HIBM patients have now been described in a wide variety of ethnicities, including Caucasian, Indian, Thai, Mexican and African [98]. The diagnosis of HIBM is based upon clinical features, muscle pathology, and, ultimately, the presence of GNE gene mutations.
HIBM is clinically characterized by progressive proximal and distal muscle weakness and atrophy of the upper and lower limbs, in most patients beginning after age 20. After onset, progression of muscle weakness is relentless and continues over the next decades, most patients become wheelchair bound within 10–20 years after onset. A unique feature of HIBM is sparing of the quadriceps muscle, partially or completely, even in advanced stages of the disease [96, 99, 100]. Eye, throat and respiratory muscles seem not to be affected either. HIBM patients have normal cognition, sensation and coordination. Serum creatine kinase levels are normal to slightly elevated and myopathic and neuropathic patterns in electromyograms are diverse among patients. Magnetic resonance imaging (MRI) T1 weighted images of the tight of HIBM patients are generally showing fatty or fibrous replacement of the hamstring muscles with sparing of the quadriceps muscle [99, 101]. HIBM muscle tissue generally does not show inflammation, although a few patients are described with inflammatory symptoms [102]. Histologically, muscle fibers degenerate with fiber size changes and formation of central nuclei. Affected muscle fibers develop characteristic cytoplasmic or nuclear filamentous inclusions as well as cytoplasmic rimmed vacuoles which are immunoreactive to various proteins, and contain clusters of autophagic vacuoles [96, 99, 103, 104]. HIBM degenerating muscle fibers contain abnormal accumulations of -amyloid protein and other Congo-red-positive pathologic markers also found in brain specimens from neurodegenerative disorders, suggesting a common pathogenetic mechanism [96, 100, 105]. However, HIBM patients do not develop central nervous system disease.
8.2.2. Molecular and cellular features
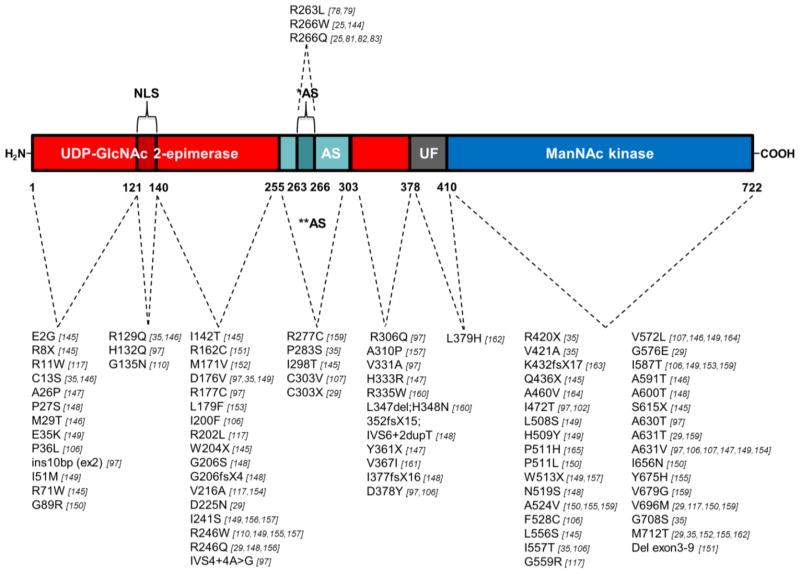
Homozygosity mapping in several HIBM families of Persian-Jewish and Kurdish-Iranian-Jewish origin aided identification of GNE as the gene responsible for HIBM [29]. All patients of Middle Eastern origin have since been found to harbor a p.M712T GNE founder mutation [29, 106]. Two GNE founder mutations are recognized within the Japanese population, p.V572L and p.D176V [97, 107]. At present, more than 80 other GNE mutations have been reported in HIBM patients worldwide, summarized in Figure 5.
Figure 5. GNE mutations associated with HIBM and Sialuria.
Locations and characteristics of all reported human GNE mutations within the functional domains of the GNE protein (as of April 2011). Mutations associated with sialuria are printed above the protein cartoon, and HIBM-associated mutations below, including their references. Red bar, UDP-GlcNAc 2-epimerase domain (GNE, Ep); dark red bar, the putative GNE nuclear export signal (Ep (N)); green bar, experimental allosteric region based on in vitro studies (**AS); dark green bar, allosteric site based on human sialuria mutations (*AS); gray bar, region of unknown function (UF); blue bar, ManNAc kinase domain (MNK, Kin). Note, that references [145–165] were only given in this figure.
Out of the 84 reported human HIBM-related GNE mutations, 68 are missense variants, 8 are nonsense mutations, 5 frame-shift mutations leading to a premature stop codon, 2 splice site variants, and 1 in-frame 3-bp deletion. GNE null mutations have never been identified on both alleles in a patient; this would most likely be lethal; a Gne ‘knock-out’ mouse model did not survive past the embryonic stage [28]. HIBM-related GNE mutations are scattered throughout the UDP-GlcNAc 2-epimerase and ManNAc kinase coding domains. Interestingly, there are 3 missense variants located in the putative nuclear export signal (Ep(N) in Fig. 5), which may play a role in nuclear localization of GNE [63]. There are no HIBM mutations in the allosteric site defined by human sialuria mutations (amino acid 263–266), however, 5 HIBM mutations are located in the ‘experimental’ allosteric region (amino acid 255–303, AS in Fig. 5) and may need further research regarding their effect on allosteric feedback inhibition of CMP-sialic acid. For a large number of mutations, secondary structure predictions are described [42, 43].
The effects of selected GNE mutations on the enzymatic properties of both UDP-GlcNAc 2-epimerase and ManNAc kinase were assessed, which were reduced, but not absent, in cultured HIBM fibroblasts, lymphoblasts, and myoblasts [108–111]. In addition, in vitro studies assessing enzymatic activities by expressing specific human GNE mutations in COS-7 cells [108], Sf9 insect cells [42], or in a cell-free in vitro transcription-translation system [110], revealed that the reductions in UDP-GlcNAc 2-epimerase and ManNAc kinase enzymatic activities are mutation-dependent. Moreover, mutations in one enzymatic domain affect not only that domain’s enzyme activity but also the activity of the other domain. Compared with enzyme activities in a cell-free system, fibroblasts exhibited higher residual activities of both UDP-GlcNAc 2-epimerase and ManNAc kinase, suggesting the presence in fibroblasts of additional sugar epimerases and kinases with overlapping substrate specificity [110].
8.2.3. Disease models
Mouse and cellular models for HIBM have been established. A complete knock-out mouse of the Gne gene displayed early embryonic lethality [28]. Heterozygous Gne-deficient mice were found to be vital and did not show a significant phenotype, although their overall sialylation was reduced by 25% [56]. Even though the Gne knock-out model could not be utilized to study HIBM pathology in mice, the Gne deficient embryonic stem (ES) cells of this model have successfully been employed for additional studies. For example, polysialylation of the neural cell adhesion molecule (NCAM) was affected in Gne deficient ES-cells, which could be restored with supplementation of ManNAc to the growth medium [28]. And recent studies of Gne deficient ES-cell showed that Gne, apart from its role in sialic acid synthesis, may also play a role in cell proliferation, gene expression, and cell differentiation [59].
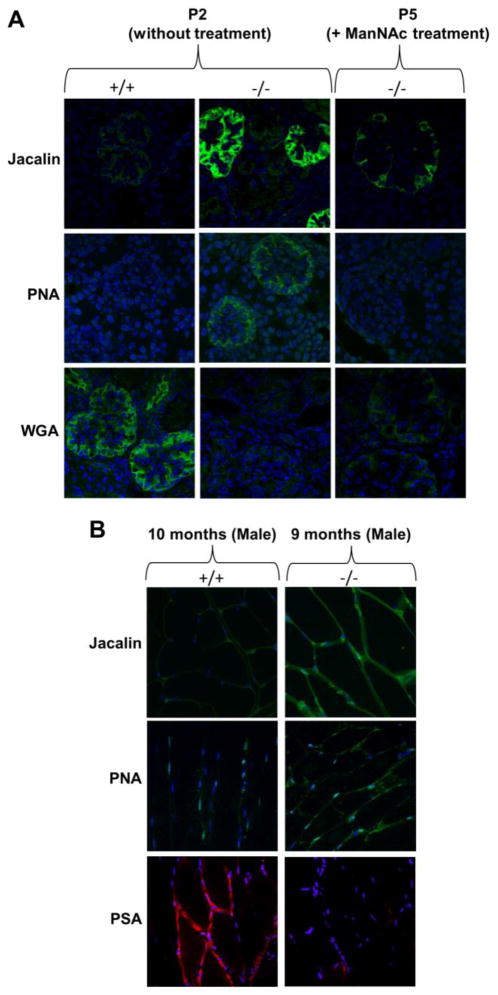
The first HIBM Gne knock-in mouse model was created by Galeano et al. [34], by homologous recombination, introducing the Persian-Jews founder mutation p.M712T into the endogenous mouse Gne gene. Unexpectedly, mutant mice died within 3 days after birth of severe glomerular disease including proteinuria, hematuria, effacement of the podocyte foot processes and segmental splitting of the glomerular basement membrane. Biochemical analysis of mutant mice kidneys revealed decreased UDP-GlcNAc 2-epimerase activity, deficient overall glomerular sialylation (Fig. 6A) and poor sialylation of the major podocyte sialoprotein, podocalyxin, suggesting that decreased renal sialic acid production led to lethality in these mice. Oral supplementation of ManNAc to pregnant and nursing mothers resulted in survival of 43% of mutant pups beyond 3 days of life. Mutant survivors displayed improved kidney histology, increased overall sialylation (Fig. 6A) as well as podocalyxin sialylation, increased GNE protein expression and UDP-GlcNAc 2-epimerase activities. HIBM patients, however, have no indications of renal abnormalities. The importance of sialic acid to the kidney may differ between humans and mice, and protein glycosylation patterns also vary. It is known that O- and N-linked glycosylation patterns of podoxalyxin differ among species, including different types of sialic acids [112].
Figure 6. Fluorescence lectin and antibody imaging demonstrating hyposialylation of kidney glomeruli and quadriceps muscle in mutant Gne M712T knock-in HIBM mice.
Representative images of kidney glomeruli and quadriceps muscle cells in paraffin embedded mouse slides stained with FITC-labeled lectins (green) Jacalin (jackfruit agglutinin) and PNA (peanut agglutinin), which both bind predominantly to terminal galactose residues residues, and WGA (wheat germ agglutinin), which predominantly recognizes terminal Neu5Ac and GlcNAc. Muscle samples were also treated with antibodies to polysialic acid (PSA) (red). All slides were counterstained with the nuclear dye DAPI (blue). Confocal imaging intensity settings were the same across all ages and genotypes for each lectin or antibody.
(A) Glomeruli from Gne M712T knock-in HIBM pups at postnatal day 2 (P2) showed hyposialylation in mutant (−/ −) compared to wild type (+/+) kidneys, as demonstrated by increased Jacalin and PNA signals and decreased WGA signal in −/ − glomeruli compared to +/+. After ManNAc treatment at P5, −/ − glomeruli show decreased Jacalin and PNA signals a more intense WGA signal compared to −/ − glomeruli at P2 without treatment, suggestive of increased sialylation after ManNAc treatment.
(B) Quadriceps muscles from adult male mice (+/+, 10 months; −/ −, 9 months old) showed increased Jacalin and PNA signals and decreased PSA staining in −/ − compared to +/+ tissues, indicating hyposialylation of muscle glycans in adult HIBM mice mimicking the human disorder.
Mutant p.M712T knock-in pups did not live long enough to develop a muscle phenotype. However, in mutant pups rescued from neonatal lethality by ManNAc administration, and not further receiving ManNAc after weaning (onward of age 3 weeks) hyposialylation of muscle tissue can be detected by lectin staining at age 9 months (Fig. 6B). Further studies are pending regarding development of muscle pathology. The findings in this knock-in mouse model not only established these mice as a genetic model for hyposialylation-related podocyte injury, but also supported evaluation of ManNAc as a therapy for HIBM as well as for renal disorders involving proteinuria and hematuria due to podocytopathy and/or segmental splitting of the glomerular basement membrane [34].
A second HIBM Gne mouse model was created in 2007 by Malicdan et al. [113]. This model was a transgenic mouse which expressed the human GNE cDNA with the p.D176V epimerase domain mutation, common among Japanese patients, on a mouse background with a disrupted mouse Gne gene; Gne(−/ −)hGNED176V-Tg. Mutant offspring appeared normal at birth, with no apparent renal issues, but had decreased sialic acid levels in serum and different organs. These mice recapitulated the clinical adult onset features of human HIBM over time. They developed poor motor performance and signs of muscle weakness/atrophy at ~21 weeks of age, and by ~40 weeks the mice showed significant changes in muscle pathology with intracytoplasmic rimmed vacuoles which were immunoreactive to lysosomal markers, amyloid and phosphorylated tau and neurofilaments. Ultrastructural and immunohistochemical studies confirmed the presence of autophagosomes in affected mouse muscle [114]. Importantly, oral prophylactic treatment (starting at week 10–20) of ManNAc, sialic acid or the sialic acid conjugate sialyllactose (containing ~45% of sialic acid) rescued the muscle phenotype in these mice. Compared to untreated mutant mice, mutant mice in all treatment groups at 54–57 weeks of age showed higher survival rates, increased body weight and muscle mass, increased sialic acid levels in serum, muscle and other organs, decreased serum creatine kinase, better overall motor performance (treadmill and hanging wire test) and a marked improvement in pathology (decreased number of rimmed vacuoles and congophilic, amyloid-positive and tau-positive inclusions) [115]. These results strongly support consideration of ManNAc, sialic acid or sialic acid conjugates as a treatment for HIBM. Further research is required to elucidate phenotypic differences between the transgenic Gne(−/ −)hGNED176V-Tg and the knock-in p.M712T Gne models.
To our knowledge, no other multiple cell organisms are reported as models for HIBM. A few cell lines with GNE mutations or decreased GNE expression have been used for in vitro studies. Two Lec3 Chinese Hamster Ovary (CHO) cell lines with either GNE nonsense mutation E35X or missense mutation G135E were shown to have loss of UDP-GlcNAc 2-epimerase enzymatic and dramatically reduced/absent sialic acid on glycans, including polysialylation of NCAM. The hyposialylation was rescued by exogenously added ManNAc or mannosamine [24]. Similar results were found in studies of human HIBM/DMRV cultured myotubes [108] or human lymphoid (B lymphoma cell line BJA-B) or hematopoietic (HL-60 myeloid leukemia) cell lines with no detectable UDP-GlcNAc 2-epimerase activity [27]. Although ManNAc kinase activity was also severely decreased or absent in these mutant cells, it was demonstrated that ancillary kinases (e.g., GlcNAc kinase) exist that can convert ManNAc to ManNAc-6-P and aid sialic acid synthesis [3].
8.2.4. Disease mechanism and therapy
The exact pathophysiology of HIBM remains unknown, but the GNE mutations associated with HIBM suggest involvement of impaired sialylation. Since HIBM is an adult onset disease, and patients have residual UDP-GlcNAc 2-epimerase and ManNAc kinase enzymatic activities, the effects of sialic acid deficiency may appear gradually. Some glycoconjugates, (such as N-linked glycans), might be more readily sialylated than others, for example O-linked or polysiaylated glycans. Furthermore, a preference for the glycosidic linkage ( 2,3, 2,6 and 2,8) is likely. It was suggested that when a shortage of sialic acid occurs, specific proteins may be inadequately glycosylated, contributing to the pathology of HIBM.
Although overall sialylation of HIBM cells and tissues appears normal [109, 111, 116], specific glycoproteins and glycolipids were found to be hyposialylated in HIBM muscle, including alpha-dystroglycan [117], polysialic acid on neural crest adhesion molecule (PSA-NCAM) [118], neprilysin and other O-glycans [119, 120], and the GM3 ganglioside [121]. The contributions of these findings to the pathophysiology of HIBM remain under investigation. More evidence that hyposialylation a key factor in the pathomechanism of HIBM came from the transgenic Gne(−/ −)hGNED176V-Tg mouse model, in which muscle atrophy and weakness could be prevented by treatment with sialic acid metabolites [115].
Apart from hyposialylation, other hypotheses exist for a role of mutated GNE in the pathology of HIBM. These include the unusual sub-cellular distribution (nuclear versus cytoplasmic) of the GNE protein in cells [63, 122], existence of different GNE isoforms with tissue-specific expression [31], involvement of mutated GNE in apoptotic pathways [123] and mitochondrial processes [32]. Other intriguing findings that may contribute to disease pathology are that GNE may control sialyltransferase expression, ganglioside production and modulation of proliferation and apoptosis, independent of sialic acid production [87], and interactions of GNE with alpha-actinin 1, an actin binding and crosslinking protein [77], with the transcription factor PLZF [67], the collapsin response mediator protein CRMP1 which is involved in growth cone collapse and F-actin depolymerization [67], with receptor interacting factor 1 [67], and with KIAA 1549, a protein of unknown function [67].
No therapies are currently available for HIBM. Some dietary modifications were proposed, including administration or avoidance of some nutrients or trace elements which may influence GNE activity [124]. Significant progress toward HIBM treatment was made during the last 5 years, including development of clinical treatment protocols. Increasing sialic acid levels through exogenous means was tested in a pilot study on four affected HIBM patients by administration of intravenous immunoglobulin G (IVIG), which has a large sialic acid content on IgG (~ 8 μmol of sialic acid/g) that could potentially be utilized to sialylate other glycoproteins (http://clinicaltrials.gov: Identifier NCT00195637). This study showed selective improvement of muscle strength, but no improvement of hyposialylation could biochemically be demonstrated [125]. Since treatment studies of HIBM mouse models convincingly showed that oral administration of sialic acid itself, the sialic acid conjugate sialyllactose, or the sialic acid precursor ManNAc could rescue hyposialylation [34, 115], these compounds are good candidate drugs to pursue in patients. However, so far none of these compounds are approved for human use and therefore need to undergo costly and time consuming preclinical studies before treatment trials can be designed. In this regard, the development of sialic acid as a treatment is progressing, a phase I trial in patients with DMRV, the Japanese disorder allelelic to HIBM, is planned, focusing on the pharmacokinetics of sialic acid in humans (http://clinicaltrials.gov: Identifier NCT01236898). The development of ManNAc for treatment of HIBM patients is in the preclinical phase and has yet to be approved by regulatory authorities.
Apart from manipulating products and/or substrates in the sialic acid pathway, other HIBM therapies could lay in the delivery of a healthy GNE gene, gene therapy [126], or a healthy GNE enzyme via (stem) cell transplantation to patients’ cells and tissues, in particular to the muscle [127, 128]. A single patient with severe HIBM was treated by gene therapy, by intramuscular injections or by intravenous delivery of a healthy GNE cDNA embedded in liposomes (GNE-Lipoplex) [129, 130]. The results were encouraging but inconclusive and more patients with different severities of HIBM muscle symptoms may need to be assessed next. With HIBM research and treatment trials quickly evolving, it is likely that elucidating the pathophysiology and therapeutic interventions of this devastating disorder will rapidly progress within the next few years.
8.3. Other GNE-opathies
Although not directly related to GNE mutations, some disorders and cell lines are reported with aberrant GNE expression, likely influenced by epigenetic [36] or other factors. Altered GNE expression can lead to altered sialylation patterns. Altered sialylation (whether or not GNE related) can cause a variety of disease phenotypes including cancers, infectious diseases, neurodegenerative and renal disorders [131]. We will address some disorders and cell lines for which a direct link to aberrant GNE expression (not due to GNE mutations) was demonstrated.
Some cancer cell lines were found with significant reduced GNE mRNA and protein expression and UDP-GlcNAc 2-epimerase enzyme activities below detectable levels, resulting in hyposialylation of cell surface glycans. Such lines include a human B lymphoma cell line and a human myeloid leukemia cell line. Both these lines showed abnormal hypermethylation of the CpG-rich GNE promoter, leading to silencing of GNE expression [27, 36]. In addition, Morris hepatoma cell lines were reported with decreased GNE expression [60], as did a chicken hepatoma cell line [132], although no direct links to GNE promoter methylation status were reported for these lines. Human Burkitt's lymphoma cell lines demonstrated sensitivity to ceramide-induced cell death regulated by GNE expression levels [133]. Hyposialylation due to GNE down-regulation was demonstrated in at least two non-cancerous cell lines. In HIV-1 infected T lymphocytes, hypomethylation of the GNE promoter was demonstrated to underlie decreased UDP-GlcNAc 2-epimerase enzyme activity and hyposialylation. Interestingly, the changes in this cell line could be entirely corrected by complementation with ManNAc in the growth medium [37]. Regenerating rat hepatocytes were shown to be hyposialylated due to decreased UDP-GlcNAc 2-epimerase enzyme activity. It was postulated that an intracellular surge of calmodulin in these cells inhibited UDP-GlcNAc 2-epimerase activity [134].
Despite the above-mentioned hyposialylated lines, in most cancers increased sialic acid levels correlate with increased malignancy, tumorigenicity and metastatic status [131]. This fits the hypothesis that genome-wide hypomethylation, found in most in cancer cells, likely yields a hypomethylated GNE promoter, resulting in increased GNE mRNA transcription, increased GNE protein translation, and increased UDP-GlcNAc 2-epimerase activities, leading to the hypersialylated metastatic phenotype of these cells. However, this hypothesis remains to be verified. Regardless of the exact mechanism of hypersialylation of metastatic cells, it is apparent that a decrease of sialylation in these cells is promising for therapy. Enzymatic removal of sialic acid by neuraminidase or immunization with neuraminidase-treated tumor cells was tested with limited success [135, 136]. Therefore, inhibition of sialic acid synthesis might be more advantageous, which can be achieved through inhibition of GNE mRNA transcription, protein translation, or enzyme activity. Identification of sialic acid synthesis inhibitors has been pursuit for several decades. It was shown that 2-deoxy-2-propionamido-D-glucose (GlcNProp) and, to a lesser extent, 2-deoxy-2-propionamido-D-mannose (ManNProp) were inhibitors of sialic acid synthesis in a cell-free system [137]. Periodate-oxidized UDP-N-acetylglucosamine, 2′,3′-Dialdehydo-UDP-N-acetylglucosamine, was described as an irreversible inhibitor of UDP-GlcNAc-2-epimerase [138], and 3-O-methyl-GlcNAc was found to inhibit ManNAc kinase activity, resulting in decreased sialic acid synthesis [139]. After identification the binding epitopes of the substrates UDP-GlcNAc and ManNAc to the GNE protein by NMR spectroscopy [47, 52], several substrate analogs were synthesized as inhibitors of GNE enzymatic activities, including exo-glycal- and thioglycoside-based UDP-sugar derivatives [140, 141], and deoxyiminosugars [142]. Further research is pending on applicability of most of these inhibitors, including testing on cellular and animal systems.
So far, no other disorders with aberrant sialylation have been reported to be associated with abnormal GNE expression. For disorders where the origin of sialylation defects remain unknown, investigations into GNE expression may be worthwhile. Especially since therapeutic options are emerging, such as sialic acid (or ManNAc or sialylated glycan) therapy for disorders involving hyposialylation, and GNE inhibitors for disorders involving hypersialylation.
Acknowledgments
This work was performed in partial fulfillment of the requirements for a PhD degree of TY, Sackler Faculty of Medicine, Tel Aviv University, Israel. This study was supported by the Intramural Research Program of the National Human Genome Research Institute, National Institutes of Health, Bethesda, MD, USA, to T.Y. and M.H. S.H. acknowledges financial support of the Bundesministerium für Bildung und Forschung and the German-Israeli Foundation for Scientific Research and Development, and R.H. acknowledges support of the Deutsche Forschungsgemeinschaft.
References
- 1.Vimr ER, Kalivoda KA, Deszo EL, Steenbergen SM. Diversity of microbial sialic acid metabolism. Microbiol Mol Biol Rev. 2004;68:132–153. doi: 10.1128/MMBR.68.1.132-153.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luchansky SJ, Yarema KJ, Takahashi S, Bertozzi CR. GlcNAc 2-epimerase can serve a catabolic role in sialic acid metabolism. J Biol Chem. 2003;278:8035–8042. doi: 10.1074/jbc.M212127200. [DOI] [PubMed] [Google Scholar]
- 3.Hinderlich S, Berger M, Keppler OT, Pawlita M, Reutter W. Biosynthesis of N-acetylneuraminic acid in cells lacking UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biol Chem. 2001;382:291–297. doi: 10.1515/BC.2001.036. [DOI] [PubMed] [Google Scholar]
- 4.Schauer R, Wember M. Isolation and characterization of sialate lyase from pig kidney. Biol Chem Hoppe Seyler. 1996;377:293–299. doi: 10.1515/bchm3.1996.377.5.293. [DOI] [PubMed] [Google Scholar]
- 5.Roseman S, Jourdian GW, Watson D, Rood R. Enzymatic synthesis of sialic acid 9-phosphates. Proc Natl Acad Sci U S A. 1961;47:958–961. doi: 10.1073/pnas.47.7.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maliekal P, Vertommen D, Delpierre G, Van Schaftingen E. Identification of the sequence encoding N-acetylneuraminate-9-phosphate phosphatase. Glycobiology. 2006;16:165–172. doi: 10.1093/glycob/cwj050. [DOI] [PubMed] [Google Scholar]
- 7.Münster-Kühnel AK, Tiralongo J, Krapp S, Weinhold B, Ritz-Sedlacek V, Jacob U, Gerardy-Schahn R. Structure and function of vertebrate CMP-sialic acid synthetases. Glycobiology. 2004;14:43R–51R. doi: 10.1093/glycob/cwh113. [DOI] [PubMed] [Google Scholar]
- 8.Sagne C, Gasnier B. Molecular physiology and pathophysiology of lysosomal membrane transporters. J Inherit Metab Dis. 2008;15:15. doi: 10.1007/s10545-008-0879-9. [DOI] [PubMed] [Google Scholar]
- 9.Kean EL. Sialic acid activation. Glycobiology. 1991;1:441–447. doi: 10.1093/glycob/1.5.441. [DOI] [PubMed] [Google Scholar]
- 10.Hinderlich S, Stasche R, Zeitler R, Reutter W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Purification and characterization of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24313–24318. doi: 10.1074/jbc.272.39.24313. [DOI] [PubMed] [Google Scholar]
- 11.Stäsche R, Hinderlich S, Weise C, Effertz K, Lucka L, Moormann P, Reutter W. A bifunctional enzyme catalyzes the first two steps in N-acetylneuraminic acid biosynthesis of rat liver. Molecular cloning and functional expression of UDP-N-acetyl-glucosamine 2-epimerase/N-acetylmannosamine kinase. J Biol Chem. 1997;272:24319–24324. doi: 10.1074/jbc.272.39.24319. [DOI] [PubMed] [Google Scholar]
- 12.Cardini CE, Leloir LF. Enzymatic formation of acetylgalactosamine. J Biol Chem. 1957;225:317–324. [PubMed] [Google Scholar]
- 13.Comb DG, Roseman S. Enzymatic synthesis of N-acetyl-D-mannosamine. Biochim Biophys Acta. 1958;29:653–654. doi: 10.1016/0006-3002(58)90031-3. [DOI] [PubMed] [Google Scholar]
- 14.Gosh S, Roseman S. Enzymatic phosphorylation of N-acetyl-D-mannosamine. Proc Natl Acad Sci U S A. 1961;47:955–958. doi: 10.1073/pnas.47.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Warren L, Felsenfeld H. N-Acetylmannosamine-6-phosphate and N-acetylneuraminic acid-9-phosphate as intermediates in sialic acid biosynthesis. Biochem Biophys Res Commun. 1961;5:185–190. doi: 10.1016/0006-291x(61)90107-3. [DOI] [PubMed] [Google Scholar]
- 16.Spivak CT, Roseman S. UDP-N-acetyl-D-glucosamine 2′-epimerase. Methods Enzymol. 1966;9:612–615. [Google Scholar]
- 17.Sommar KM, Ellis DB. Uridine diphosphate N-acetyl-D-glucosamine 2-epimerase from rat liver. I. Catalytic and regulatory properties. Biochim Biophys Acta. 1972;268:581–589. doi: 10.1016/0005-2744(72)90355-5. [DOI] [PubMed] [Google Scholar]
- 18.Kikuchi K, Tsuiki S. Purification and properties of UDP-N-acetylglucosamine 2'-epimerase from rat liver. Biochim Biophys Acta. 1973;327:193–206. doi: 10.1016/0005-2744(73)90117-4. [DOI] [PubMed] [Google Scholar]
- 19.Kundig W, Gosh S, Roseman S. The sialic acids. VII. N-Acyl-D-mannosamine kinase from rat liver. J Biol Chem. 1966;241:5619–5626. [PubMed] [Google Scholar]
- 20.Kornfeld S, Kornfeld R, Neufeld E, O’ Brien PJ. The feedback control of sugar nucleotide biosynthesis in liver. Proc Natl Acad Sci U S A. 1964;52:371–379. doi: 10.1073/pnas.52.2.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weiss P, Tietze F, Gahl WA, Seppala R, Ashwell G. Identification of the metabolic defect in sialuria. J Biol Chem. 1989;264:17635–17636. [PubMed] [Google Scholar]
- 22.Horstkorte R, Nohring S, Wiechens N, Schwarzkopf M, Danker K, Reutter W, Lucka L. Tissue expression and amino acid sequence of murine UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase. Eur J Biochem. 1999;260:923–927. doi: 10.1046/j.1432-1327.1999.00253.x. [DOI] [PubMed] [Google Scholar]
- 23.Lucka L, Krause M, Danker K, Reutter W, Horstkorte R. Primary structure and expression analysis of human UDP-N-acetyl-glucosamine-2-epimerase/N-acetylmannosamine kinase, the bifunctional enzyme in neuraminic acid biosynthesis. FEBS Lett. 1999;454:341–344. doi: 10.1016/s0014-5793(99)00837-6. [DOI] [PubMed] [Google Scholar]
- 24.Hong Y, Stanley P. Lec3 Chinese hamster ovary mutants lack UDP-N-acetylglucosamine 2-epimerase activity because of mutations in the epimerase domain of the Gne gene. J Biol Chem. 2003;278:53045–53054. doi: 10.1074/jbc.M309967200. [DOI] [PubMed] [Google Scholar]
- 25.Seppala R, Lehto VP, Gahl WA. Mutations in the human UDP-N-acetylglucosamine 2-epimerase gene define the disease sialuria and the allosteric site of the enzyme. Am J Hum Genet. 1999;64:1563–1569. doi: 10.1086/302411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarema KJ, Goon S, Bertozzi CR. Metabolic selection of glycosylation defects in human cells. Nat Biotechnol. 2001;19:553–558. doi: 10.1038/89305. [DOI] [PubMed] [Google Scholar]
- 27.Keppler OT, Hinderlich S, Langner J, Schwartz-Albiez R, Reutter W, Pawlita M. UDP-GlcNAc 2-epimerase: a regulator of cell surface sialylation. Science. 1999;284:1372–1376. doi: 10.1126/science.284.5418.1372. [DOI] [PubMed] [Google Scholar]
- 28.Schwarzkopf M, Knobeloch KP, Rohde E, Hinderlich S, Wiechens N, Lucka L, Horak I, Reutter W, Horstkorte R. Sialylation is essential for early development in mice. Proc Natl Acad Sci U S A. 2002;99:5267–5270. doi: 10.1073/pnas.072066199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eisenberg I, Avidan N, Potikha T, Hochner H, Chen M, Olender T, Barash M, Shemesh M, Sadeh M, Grabov-Nardini G, Shmilevich I, Friedmann A, Karpati G, Bradley WG, Baumbach L, Lancet D, Asher EB, Beckmann JS, Argov Z, Mitrani-Rosenbaum S. The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet. 2001;29:83–87. doi: 10.1038/ng718. [DOI] [PubMed] [Google Scholar]
- 30.Watts GD, Thorne M, Kovach MJ, Pestronk A, Kimonis VE. Clinical and genetic heterogeneity in chromosome 9p associated hereditary inclusion body myopathy: exclusion of GNE and three other candidate genes. Neuromuscul Disord. 2003;13:559–567. doi: 10.1016/s0960-8966(03)00070-1. [DOI] [PubMed] [Google Scholar]
- 31.Reinke SO, Hinderlich S. Prediction of three different isoforms of the human UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. FEBS Lett. 2007;581:3327–3331. doi: 10.1016/j.febslet.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 32.Eisenberg I, Novershtern N, Itzhaki Z, Becker-Cohen M, Sadeh M, Willems PH, Friedman N, Koopman WJ, Mitrani-Rosenbaum S. Mitochondrial processes are impaired in hereditary inclusion body myopathy. Hum Mol Genet. 2008;17:3663–3674. doi: 10.1093/hmg/ddn261. [DOI] [PubMed] [Google Scholar]
- 33.Sela I, Milman Krentsis I, Shlomai Z, Sadeh M, Dabby R, Argov Z, Ben-Bassat H, Mitrani-Rosenbaum S. The proteomic profile of hereditary inclusion body myopathy. PLoS One. 2011;6:e16334. doi: 10.1371/journal.pone.0016334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Galeano B, Klootwijk R, Manoli I, Sun M, Ciccone C, Darvish D, Starost MF, Zerfas PM, Hoffmann VJ, Hoogstraten-Miller S, Krasnewich DM, Gahl WA, Huizing M. Mutation in the key enzyme of sialic acid biosynthesis causes severe glomerular proteinuria and is rescued by N-acetylmannosamine. J Clin Invest. 2007;117:1585–1594. doi: 10.1172/JCI30954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tomimitsu H, Shimizu J, Ishikawa K, Ohkoshi N, Kanazawa I, Mizusawa H. Distal myopathy with rimmed vacuoles (DMRV): new GNE mutations and splice variant. Neurology. 2004;62:1607–1610. doi: 10.1212/01.wnl.0000123115.23652.6c. [DOI] [PubMed] [Google Scholar]
- 36.Oetke C, Hinderlich S, Reutter W, Pawlita M. Epigenetically mediated loss of UDP-GlcNAc 2-epimerase/ManNAc kinase expression in hyposialylated cell lines. Biochem Biophys Res Commun. 2003;308:892–898. doi: 10.1016/s0006-291x(03)01471-2. [DOI] [PubMed] [Google Scholar]
- 37.Giordanengo V, Ollier L, Lanteri M, Lesimple J, March D, Thyss S, Lefebvre JC. Epigenetic reprogramming of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) in HIV-1-infected CEM T cells. FASEB J. 2004;18:1961–1963. doi: 10.1096/fj.04-2467fje. [DOI] [PubMed] [Google Scholar]
- 38.Effertz K, Hinderlich S, Reutter W. Selective loss of either the epimerase or kinase activity of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase due to site-directed mutagenesis based on sequence alignments. J Biol Chem. 1999;274:28771–28778. doi: 10.1074/jbc.274.40.28771. [DOI] [PubMed] [Google Scholar]
- 39.Blume A, Weidemann W, Stelzl U, Wanker EE, Lucka L, Donner P, Reutter W, Horstkorte R, Hinderlich S. Domain-specific characteristics of the bifunctional key enzyme of sialic acid biosynthesis, UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase. Biochem J. 2004;384:599–607. doi: 10.1042/BJ20040917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reinke SO, Eidenschink C, Jay CM, Hinderlich S. Biochemical characterization of human and murine isoforms of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) Glycoconj J. 2009;26:415–422. doi: 10.1007/s10719-008-9189-6. [DOI] [PubMed] [Google Scholar]
- 41.Ghaderi D, Strauss HM, Reinke S, Cirak S, Reutter W, Lucka L, Hinderlich S. Evidence for dynamic interplay of different oligomeric states of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase by biophysical methods. J Mol Biol. 2007;369:746–758. doi: 10.1016/j.jmb.2007.03.037. [DOI] [PubMed] [Google Scholar]
- 42.Penner J, Mantey LR, Elgavish S, Ghaderi D, Cirak S, Berger M, Krause S, Lucka L, Voit T, Mitrani-Rosenbaum S, Hinderlich S. Influence of UDP-GlcNAc 2-epimerase/ManNAc kinase mutant proteins on hereditary inclusion body myopathy. Biochemistry. 2006;45:2968–2977. doi: 10.1021/bi0522504. [DOI] [PubMed] [Google Scholar]
- 43.Kurochkina N, Yardeni T, Huizing M. Molecular modeling of the bifunctional enzyme UDP-GlcNAc 2-epimerase/ ManNAc kinase and predictions of structural effects of mutations associated with HIBM and sialuria. Glycobiology. 2009;16:16. doi: 10.1093/glycob/cwp176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tong Y, Tempel W, Nedyalkova L, Mackenzie F, Park HW. Crystal structure of the N-acetylmannosamine kinase domain of GNE. PLoS One. 2009;4:e7165. doi: 10.1371/journal.pone.0007165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagradova N. Interdomain communications in bifunctional enzymes: how are different activities coordinated? IUBMB Life. 2003;55:459–466. doi: 10.1080/15216540310001602805. [DOI] [PubMed] [Google Scholar]
- 46.Salo WL, Fletcher HG. Studies on the mechanism of action of uridine diphosphate N-acetylglucosamine 2-epimerase. Biochemistry. 1970;9:878–881. doi: 10.1021/bi00806a024. [DOI] [PubMed] [Google Scholar]
- 47.Blume A, Benie AJ, Stolz F, Schmidt RR, Reutter W, Hinderlich S, Peters T. Characterization of ligand binding to the bifunctional key enzyme in the sialic acid biosynthesis by NMR: I. Investigation of the UDP-GlcNAc 2-epimerase functionality. J Biol Chem. 2004;279:55715–55721. doi: 10.1074/jbc.M410238200. [DOI] [PubMed] [Google Scholar]
- 48.Chou WK, Hinderlich S, Reutter W, Tanner ME. Sialic acid biosynthesis: stereochemistry and mechanism of the reaction catalyzed by the mammalian UDP-N-acetylglucosamine 2-epimerase. J Am Chem Soc. 2003;125:2455–2461. doi: 10.1021/ja021309g. [DOI] [PubMed] [Google Scholar]
- 49.Campbell RE, Mosimann SC, Tanner ME, Strynadka NC. The structure of UDP-N-acetylglucosamine 2-epimerase reveals homology to phosphoglycosyl transferases. Biochemistry. 2000;39:14993–15001. doi: 10.1021/bi001627x. [DOI] [PubMed] [Google Scholar]
- 50.Sommar KM, Ellis DG. Uridine diphosphate N-acetyl-D-glucosamine 2-epimerase from rat liver. II. Studies on the mechanism of action. Biochim Biophys Acta. 1972;268:590–595. doi: 10.1016/0005-2744(72)90356-7. [DOI] [PubMed] [Google Scholar]
- 51.Kamerling JP, Strecker G, Farriaux JP, Dorland L, Haverkamp J, Vliegenthart JF. 2-Acetamidoglucal, a new metabolite isolated from the urine of a patient with sialuria. Biochim Biophys Acta. 1979;583:403–408. doi: 10.1016/0304-4165(79)90465-3. [DOI] [PubMed] [Google Scholar]
- 52.Benie AJ, Blume A, Schmidt RR, Reutter W, Hinderlich S, Peters T. Characterization of ligand binding to the bifunctional key enzyme in the sialic acid biosynthesis by NMR: II. Investigation of the ManNAc kinase functionality. J Biol Chem. 2004;279:55722–55727. doi: 10.1074/jbc.M410239200. [DOI] [PubMed] [Google Scholar]
- 53.Du J, Meledeo MA, Wang Z, Khanna HS, Paruchuri VD, Yarema KJ. Metabolic glycoengineering: sialic acid and beyond. Glycobiology. 2009;19:1382–1401. doi: 10.1093/glycob/cwp115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lowe G, Potter BV. The stereochemical course of yeast hexokinase-catalysed phosphoryl transfer by using adenosine 5'[gamma(S)-16O,17O,18O]triphosphate as substrate. Biochem J. 1981;199:227–233. doi: 10.1042/bj1990227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pollard-Knight D, Potter BV, Cullis PM, Lowe G, Cornish-Bowden A. The stereochemical course of phosphoryl transfer catalysed by glucokinase. Biochem J. 1982;201:421–423. doi: 10.1042/bj2010421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gagiannis D, Orthmann A, Danssmann I, Schwarzkopf M, Weidemann W, Horstkorte R. Reduced sialylation status in UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (GNE)-deficient mice. Glycoconj J. 2007;24:125–130. doi: 10.1007/s10719-006-9019-7. [DOI] [PubMed] [Google Scholar]
- 57.Ivatt RJ. Transient expression of sialylated glycans during glycoprotein processing by embryonal carcinomas. Biochem Biophys Res Commun. 1987;142:489–495. doi: 10.1016/0006-291x(87)90301-9. [DOI] [PubMed] [Google Scholar]
- 58.Lackie PM, Zuber C, Roth J. Polysialic acid of the neural cell adhesion molecule (N-CAM) is widely expressed during organogenesis in mesodermal and endodermal derivatives. Differentiation. 1994;57:119–131. doi: 10.1046/j.1432-0436.1994.5720119.x. [DOI] [PubMed] [Google Scholar]
- 59.Weidemann W, Klukas C, Klein A, Simm A, Schreiber F, Horstkorte R. Lessons from GNE-deficient embryonic stem cells: Sialic acid biosynthesis is involved in proliferation and gene expression. Glycobiology. 2010;30:107–117. doi: 10.1093/glycob/cwp153. [DOI] [PubMed] [Google Scholar]
- 60.Harms E, Kreisel W, Morris HP, Reutter W. Biosynthesis of N-acetylneuraminic acid in Morris hepatomas. Eur J Biochem. 1973;32:254–262. doi: 10.1111/j.1432-1033.1973.tb02605.x. [DOI] [PubMed] [Google Scholar]
- 61.Okamoto Y, Akamatsu N. Metabolism of sialic acid in regenerating rat liver. Biochem J. 1980;188:905–911. doi: 10.1042/bj1880905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okubo H, Shibata K, Ishibashi H, Yanase T. Regulation of N-acetylneuraminic acid synthesis following injury and partial hepatectomy. Proc Soc Exp Biol Med. 1977;155:152–156. doi: 10.3181/00379727-155-39763. [DOI] [PubMed] [Google Scholar]
- 63.Krause S, Hinderlich S, Amsili S, Horstkorte R, Wiendl H, Argov Z, Mitrani-Rosenbaum S, Lochmuller H. Localization of UDP-GlcNAc 2-epimerase/ManAc kinase (GNE) in the Golgi complex and the nucleus of mammalian cells. Exp Cell Res. 2005;304:365–379. doi: 10.1016/j.yexcr.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 64.Fritsch M, Geilen CC, Reutter W. Determination of cytidine 5'-monophospho-N-acetylneuraminic acid pool size in cell culture scale using high-performance anion-exchange chromatography with pulsed amperometric detection. J Chromatogr A. 1996;727:223–230. doi: 10.1016/0021-9673(95)01157-9. [DOI] [PubMed] [Google Scholar]
- 65.Horstkorte R, Nohring S, Danker K, Effertz K, Reutter W, Lucka L. Protein kinase C phosphorylates and regulates UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase. FEBS Lett. 2000;470:315–318. doi: 10.1016/s0014-5793(00)01331-4. [DOI] [PubMed] [Google Scholar]
- 66.Hart GW, Slawson C, Ramirez-Correa G, Lagerlof O. Cross Talk Between O-GlcNAcylation and Phosphorylation: Roles in Signaling, Transcription, and Chronic Disease. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-060608-102511. Epub Jun 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weidemann W, Stelzl U, Lisewski U, Bork K, Wanker EE, Hinderlich S, Horstkorte R. The collapsin response mediator protein 1 (CRMP-1) and the promyelocytic leukemia zinc finger protein (PLZF) bind to UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE), the key enzyme of sialic acid biosynthesis. FEBS Lett. 2006;580:6649–6654. doi: 10.1016/j.febslet.2006.11.015. [DOI] [PubMed] [Google Scholar]
- 68.Goshima Y, Nakamura F, Strittmatter P, Strittmatter SM. Collapsin-induced growth cone collapse mediated by an intracellular protein related to UNC-33. Nature. 1995;376:509–514. doi: 10.1038/376509a0. [DOI] [PubMed] [Google Scholar]
- 69.Shih JY, Lee YC, Yang SC, Hong TM, Huang CY, Yang PC. Collapsin response mediator protein-1: a novel invasion-suppressor gene. Clin Exp Metastasis. 2003;20:69–76. doi: 10.1023/a:1022598604565. [DOI] [PubMed] [Google Scholar]
- 70.Hamajima N, Matsuda K, Sakata S, Tamaki N, Sasaki M, Nonaka M. A novel gene family defined by human dihydropyrimidinase and three related proteins with differential tissue distribution. Gene. 1996;180:157–163. doi: 10.1016/s0378-1119(96)00445-3. [DOI] [PubMed] [Google Scholar]
- 71.Büttner B, Kannicht C, Löster K, Reutter W, Lee H-Y, Nöhring S, Horstkorte R. Biochemical engineering of cell surface sialic acids stimulates axonal outgrowth. J Neurosci. 2002;22:8869–8875. doi: 10.1523/JNEUROSCI.22-20-08869.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yeyati PL, Shaknovich R, Boterashvili S, Li J, Ball HJ, Waxman S, Nason-Burchenal K, Dmitrovsky E, Zelent A, Licht JD. Leukemia translocation protein PLZF inhibits cell growth and expression of cyclin A. Oncogene. 1999;18:925–934. doi: 10.1038/sj.onc.1202375. [DOI] [PubMed] [Google Scholar]
- 73.Barna M, Merghoub T, Costoya JA, Ruggero D, Branford M, Bergia A, Samori B, Pandolfi PP. Plzf mediates transcriptional repression of HoxD gene expression through chromatin remodeling. Dev Cell. 2002;3:499–510. doi: 10.1016/s1534-5807(02)00289-7. [DOI] [PubMed] [Google Scholar]
- 74.Pintard L, Willems A, Peter M. Cullin-based ubiquitin ligases: Cul3-BTB complexes join the family. EMBO J. 2004;23:1681–1687. doi: 10.1038/sj.emboj.7600186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Senbonmatsu T, Saito T, Landon EJ, Watanabe O, Price E, Jr, Roberts RL, Imboden H, Fitzgerald TG, Gaffney FA, Inagami T. A novel angiotensin II type 2 receptor signaling pathway: possible role in cardiac hypertrophy. EMBO J. 2003;22:6471–6482. doi: 10.1093/emboj/cdg637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seidel K, Kirsch S, Lucht K, Zaade D, Reinemund J, Schmitz J, Klare S, Li Y, Schefe JH, Schmerbach K, Goldin-Lang P, Zollmann FS, Thone-Reineke C, Unger T, Funke-Kaiser H. The promyelocytic leukemia zinc finger (PLZF) protein exerts neuroprotective effects in neuronal cells and is dysregulated in experimental stroke. Brain Pathol. 2011;21:31–43. doi: 10.1111/j.1750-3639.2010.00427.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Amsili S, Zer H, Hinderlich S, Krause S, Becker-Cohen M, MacArthur DG, North KN, Mitrani-Rosenbaum S. UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE) binds to alpha-actinin 1: novel pathways in skeletal muscle? PLoS One. 2008;3:e2477. doi: 10.1371/journal.pone.0002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Montreuil J, Biserte G, Strecker G, Spik G, Fontaine G, Farriaux JP. Description of a new type of melituria, called sialuria. Clin Chim Acta. 1968;21:61–69. doi: 10.1016/0009-8981(68)90011-9. [DOI] [PubMed] [Google Scholar]
- 79.Fontaine G, Biserte G, Montreuil J, Dupont A, Farriaux JP. Sialuria: an original metabolic disorder. Helv Paediatr Acta. 1968;17:1–32. [PubMed] [Google Scholar]
- 80.Seppala R, Tietze F, Krasnewich D, Weiss P, Ashwell G, Barsh G, Thomas GH, Packman S, Gahl WA. Sialic acid metabolism in sialuria fibroblasts. J Biol Chem. 1991;266:7456–7461. [PubMed] [Google Scholar]
- 81.Ferreira H, Seppala R, Pinto R, Huizing M, Martins E, Braga AC, Gomes L, Krasnewich DM, Sa Miranda MC, Gahl WA. Sialuria in a Portuguese girl: clinical, biochemical, and molecular characteristics. Mol Genet Metab. 1999;67:131–137. doi: 10.1006/mgme.1999.2852. [DOI] [PubMed] [Google Scholar]
- 82.Leroy JG, Seppala R, Huizing M, Dacremont G, De Simpel H, Van Coster RN, Orvisky E, Krasnewich DM, Gahl WA. Dominant inheritance of sialuria, an inborn error of feedback inhibition. Am J Hum Genet. 2001;68:1419–1427. doi: 10.1086/320598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Enns GM, Seppala R, Musci TJ, Weisiger K, Ferrell LD, Wenger DA, Gahl WA, Packman S. Clinical course and biochemistry of sialuria. J Inherit Metab Dis. 2001;24:328–336. doi: 10.1023/a:1010588115479. [DOI] [PubMed] [Google Scholar]
- 84.Strehle EM. Sialic acid storage disease and related disorders. Genet Test. 2003;7:113–121. doi: 10.1089/109065703322146795. [DOI] [PubMed] [Google Scholar]
- 85.Bork K, Reutter W, Gerardy-Schahn R, Horstkorte R. The intracellular concentration of sialic acid regulates the polysialylation of the neural cell adhesion molecule. FEBS Lett. 2005;579:5079–5083. doi: 10.1016/j.febslet.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 86.Son YD, Jeong YT, Park SY, Kim JH. Enhanced sialylation of recombinant human EPO in Chinese hamster ovary cells by combinatorial engineering of selected genes. Glycobiology. 2011 doi: 10.1093/glycob/cwr034. Epub Mar 24. [DOI] [PubMed] [Google Scholar]
- 87.Wang Z, Sun Z, Li AV, Yarema KJ. Roles for UDP-GlcNAc 2-epimerase/ManNAc 6-kinase outside of sialic acid biosynthesis: modulation of sialyltransferase and BiP expression, GM3 and GD3 biosynthesis, proliferation, and apoptosis, and ERK1/2 phosphorylation. J Biol Chem. 2006;281:27016–27028. doi: 10.1074/jbc.M604903200. [DOI] [PubMed] [Google Scholar]
- 88.Bork K, Horstkorte R, Weidemann W. Increasing the sialylation of therapeutic glycoproteins: the potential of the sialic acid biosynthetic pathway. J Pharm Sci. 2009;98:3499–3508. doi: 10.1002/jps.21684. [DOI] [PubMed] [Google Scholar]
- 89.Wopereis S, Abd Hamid UM, Critchley A, Royle L, Dwek RA, Morava E, Leroy JG, Wilcken B, Lagerwerf AJ, Huijben KM, Lefeber DJ, Rudd PM, Wevers RA. Abnormal glycosylation with hypersialylated O-glycans in patients with Sialuria. Biochim Biophys Acta. 2006;1762:598–607. doi: 10.1016/j.bbadis.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 90.Stanley CA, Lieu YK, Hsu BY, Burlina AB, Greenberg CR, Hopwood NJ, Perlman K, Rich BH, Zammarchi E, Poncz M. Hyperinsulinism and hyperammonemia in infants with regulatory mutations of the glutamate dehydrogenase gene. N Engl J Med. 1998;338:1352–1357. doi: 10.1056/NEJM199805073381904. [DOI] [PubMed] [Google Scholar]
- 91.Krasnewich DM, Tietze F, Krause W, Pretzlaff R, Wenger DA, Diwadkar V, Gahl WA. Clinical and biochemical studies in an American child with sialuria. Biochem Med Metab Biol. 1993;49:90–96. doi: 10.1006/bmmb.1993.1010. [DOI] [PubMed] [Google Scholar]
- 92.Miller VM, Xia H, Marrs GL, Gouvion CM, Lee G, Davidson BL, Paulson HL. Allele-specific silencing of dominant disease genes. Proc Natl Acad Sci U S A. 2003;100:7195–7200. doi: 10.1073/pnas.1231012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Klootwijk RD, Savelkoul PJ, Ciccone C, Manoli I, Caplen NJ, Krasnewich DM, Gahl WA, Huizing M. Allele-specific silencing of the dominant disease allele in sialuria by RNA interference. FASEB J. 2008;22:3846–3852. doi: 10.1096/fj.08-110890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Grimm D, Kay MA. Therapeutic application of RNAi: is mRNA targeting finally ready for prime time? J Clin Invest. 2007;117:3633–3641. doi: 10.1172/JCI34129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nonaka I, Sunohara N, Ishiura S, Satoyoshi E. Familial distal myopathy with rimmed vacuole and lamellar (myeloid) body formation. J Neurol Sci. 1981;51:141–155. doi: 10.1016/0022-510x(81)90067-8. [DOI] [PubMed] [Google Scholar]
- 96.Argov Z, Yarom R. “Rimmed vacuole myopathy” sparing the quadriceps. A unique disorder in Iranian Jews. J Neurol Sci. 1984;64:33–43. doi: 10.1016/0022-510x(84)90053-4. [DOI] [PubMed] [Google Scholar]
- 97.Nishino I, Noguchi S, Murayama K, Driss A, Sugie K, Oya Y, Nagata T, Chida K, Takahashi T, Takusa Y, Ohi T, Nishimiya J, Sunohara N, Ciafaloni E, Kawai M, Aoki M, Nonaka I. Distal myopathy with rimmed vacuoles is allelic to hereditary inclusion body myopathy. Neurology. 2002;59:1689–1693. doi: 10.1212/01.wnl.0000041631.28557.c6. [DOI] [PubMed] [Google Scholar]
- 98.Huizing M, Krasnewich DM. Hereditary inclusion body myopathy: a decade of progress. Biochim Biophys Acta. 2009;1792:881–887. doi: 10.1016/j.bbadis.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sivakumar K, Dalakas MC. The spectrum of familial inclusion body myopathies in 13 families and a description of a quadriceps-sparing phenotype in non-Iranian Jews. Neurology. 1996;47:977–984. doi: 10.1212/wnl.47.4.977. [DOI] [PubMed] [Google Scholar]
- 100.Askanas V, Engel WK. New advances in the understanding of sporadic inclusion-body myositis and hereditary inclusion-body myopathies. Curr Opin Rheumatol. 1995;7:486–496. doi: 10.1097/00002281-199511000-00005. [DOI] [PubMed] [Google Scholar]
- 101.Sadeh M, Gadoth N, Hadar H, Ben-David E. Vacuolar myopathy sparing the quadriceps. Brain. 1993;116:217–232. doi: 10.1093/brain/116.1.217. [DOI] [PubMed] [Google Scholar]
- 102.Yabe I, Higashi T, Kikuchi S, Sasaki H, Fukazawa T, Yoshida K, Tashiro K. GNE mutations causing distal myopathy with rimmed vacuoles with inflammation. Neurology. 2003;61:384–386. doi: 10.1212/01.wnl.0000061520.63546.8f. [DOI] [PubMed] [Google Scholar]
- 103.Griggs RC, Askanas V, DiMauro S, Engel A, Karpati G, Mendell JR, Rowland LP. Inclusion body myositis and myopathies. Ann Neurol. 1995;38:705–713. doi: 10.1002/ana.410380504. [DOI] [PubMed] [Google Scholar]
- 104.Malicdan MC, Noguchi S, Nishino I. Autophagy in a mouse model of distal myopathy with rimmed vacuoles or hereditary inclusion body myopathy. Autophagy. 2007;3:396–398. doi: 10.4161/auto.4270. [DOI] [PubMed] [Google Scholar]
- 105.Malicdan MC, Noguchi S, Nishino I. Recent advances in distal myopathy with rimmed vacuoles (DMRV) or hIBM: treatment perspectives. Curr Opin Neurol. 2008;21:596–600. doi: 10.1097/WCO.0b013e32830dd595. [DOI] [PubMed] [Google Scholar]
- 106.Eisenberg I, Grabov-Nardini G, Hochner H, Korner M, Sadeh M, Bertorini T, Bushby K, Castellan C, Felice K, Mendell J, Merlini L, Shilling C, Wirguin I, Argov Z, Mitrani-Rosenbaum S. Mutations Spectrum of the GNE gene in Hereditary Inclusion Body Myopathy sparing the quadriceps. Hum Mutat. 2003;21:99. doi: 10.1002/humu.9100. [DOI] [PubMed] [Google Scholar]
- 107.Tomimitsu H, Ishikawa K, Shimizu J, Ohkoshi N, Kanazawa I, Mizusawa H. Distal myopathy with rimmed vacuoles: novel mutations in the GNE gene. Neurology. 2002;59:451–454. doi: 10.1212/wnl.59.3.451. [DOI] [PubMed] [Google Scholar]
- 108.Noguchi S, Keira Y, Murayama K, Ogawa M, Fujita M, Kawahara G, Oya Y, Imazawa M, Goto Y, Hayashi YK, Nonaka I, Nishino I. Reduction of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase activity and sialylation in distal myopathy with rimmed vacuoles. J Biol Chem. 2004;279:11402–11407. doi: 10.1074/jbc.M313171200. [DOI] [PubMed] [Google Scholar]
- 109.Hinderlich S, Salama I, Eisenberg I, Potikha T, Mantey LR, Yarema KJ, Horstkorte R, Argov Z, Sadeh M, Reutter W, Mitrani-Rosenbaum S. The homozygous M712T mutation of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase results in reduced enzyme activities but not in altered overall cellular sialylation in hereditary inclusion body myopathy. FEBS Lett. 2004;566:105–109. doi: 10.1016/j.febslet.2004.04.013. [DOI] [PubMed] [Google Scholar]
- 110.Sparks SE, Ciccone C, Lalor M, Orvisky E, Klootwijk R, Savelkoul PJ, Dalakas MC, Krasnewich DM, Gahl WA, Huizing M. Use of a cell-free system to determine UDP-N-acetylglucosamine 2-epimerase and N-acetylmannosamine kinase activities in human hereditary inclusion body myopathy. Glycobiology. 2005;15:1102–1110. doi: 10.1093/glycob/cwi100. [DOI] [PubMed] [Google Scholar]
- 111.Salama I, Hinderlich S, Shlomai Z, Eisenberg I, Krause S, Yarema K, Argov Z, Lochmuller H, Reutter W, Dabby R, Sadeh M, Ben-Bassat H, Mitrani-Rosenbaum S. No overall hyposialylation in hereditary inclusion body myopathy myoblasts carrying the homozygous M712T GNE mutation. Biochem Biophys Res Commun. 2005;328:221–226. doi: 10.1016/j.bbrc.2004.12.157. [DOI] [PubMed] [Google Scholar]
- 112.Kershaw DB, Beck SG, Wharram BL, Wiggins JE, Goyal M, Thomas PE, Wiggins RC. Molecular cloning and characterization of human podocalyxin-like protein. Orthologous relationship to rabbit PCLP1 and rat podocalyxin. J Biol Chem. 1997;272:15708–15714. doi: 10.1074/jbc.272.25.15708. [DOI] [PubMed] [Google Scholar]
- 113.Malicdan MC, Noguchi S, Nonaka I, Hayashi YK, Nishino I. A Gne knockout mouse expressing human GNE D176V mutation develops features similar to distal myopathy with rimmed vacuoles or hereditary inclusion body myopathy. Hum Mol Genet. 2007;16:2669–2682. doi: 10.1093/hmg/ddm220. [DOI] [PubMed] [Google Scholar]
- 114.Malicdan MC, Noguchi S, Hayashi YK, Nishino I. Muscle weakness correlates with muscle atrophy and precedes the development of inclusion body or rimmed vacuoles in the mouse model of DMRV/hIBM. Physiol Genomics. 2008;35:106–115. doi: 10.1152/physiolgenomics.90219.2008. [DOI] [PubMed] [Google Scholar]
- 115.Malicdan MC, Noguchi S, Hayashi YK, Nonaka I, Nishino I. Prophylactic treatment with sialic acid metabolites precludes the development of the myopathic phenotype in the DMRV-hIBM mouse model. Nat Med. 2009;15:690–695. doi: 10.1038/nm.1956. [DOI] [PubMed] [Google Scholar]
- 116.Savelkoul PJ, Manoli I, Sparks SE, Ciccone C, Gahl WA, Krasnewich DM, Huizing M. Normal sialylation of serum N-linked and O-GalNAc-linked glycans in hereditary inclusion-body myopathy. Mol Genet Metab. 2006;88:389–390. doi: 10.1016/j.ymgme.2006.04.011. [DOI] [PubMed] [Google Scholar]
- 117.Huizing M, Rakocevic G, Sparks SE, Mamali I, Shatunov A, Goldfarb L, Krasnewich D, Gahl WA, Dalakas MC. Hypoglycosylation of alpha-dystroglycan in patients with hereditary IBM due to GNE mutations. Mol Genet Metab. 2004;81:196–202. doi: 10.1016/j.ymgme.2003.11.012. [DOI] [PubMed] [Google Scholar]
- 118.Ricci E, Broccolini A, Gidaro T, Morosetti R, Gliubizzi C, Frusciante R, Di Lella GM, Tonali PA, Mirabella M. NCAM is hyposialylated in hereditary inclusion body myopathy due to GNE mutations. Neurology. 2006;66:755–758. doi: 10.1212/01.wnl.0000200956.76449.3f. [DOI] [PubMed] [Google Scholar]
- 119.Broccolini A, Gidaro T, De Cristofaro R, Morosetti R, Gliubizzi C, Ricci E, Tonali PA, Mirabella M. Hyposialylation of neprilysin possibly affects its expression and enzymatic activity in hereditary inclusion-body myopathy muscle. J Neurochem. 2008;105:971–981. doi: 10.1111/j.1471-4159.2007.05208.x. [DOI] [PubMed] [Google Scholar]
- 120.Tajima Y, Uyama E, Go S, Sato C, Tao N, Kotani M, Hino H, Suzuki A, Sanai Y, Kitajima K, Sakuraba H. Distal myopathy with rimmed vacuoles: impaired O-glycan formation in muscular glycoproteins. Am J Pathol. 2005;166:1121–1130. doi: 10.1016/S0002-9440(10)62332-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Paccalet T, Coulombe Z, Tremblay JP. Ganglioside GM3 levels are altered in a mouse model of HIBM: GM3 as a cellular marker of the disease. PLoS One. 2010;5:e10055. doi: 10.1371/journal.pone.0010055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Krause S, Aleo A, Hinderlich S, Merlini L, Tournev I, Walter MC, Argov Z, Mitrani-Rosenbaum S, Lochmuller H. GNE protein expression and subcellular distribution are unaltered in HIBM. Neurology. 2007;69:655–659. doi: 10.1212/01.wnl.0000267426.97138.fd. [DOI] [PubMed] [Google Scholar]
- 123.Amsili S, Shlomai Z, Levitzki R, Krause S, Lochmuller H, Ben-Bassat H, Mitrani-Rosenbaum S. Characterization of hereditary inclusion body myopathy myoblasts: possible primary impairment of apoptotic events. Cell Death Differ. 2007;14:1916–1924. doi: 10.1038/sj.cdd.4402208. [DOI] [PubMed] [Google Scholar]
- 124.Darvish D. Magnesium may help patients with recessive hereditary inclusion body myopathy, a pathological review. Med Hypotheses. 2003;60:94–101. doi: 10.1016/s0306-9877(02)00339-0. [DOI] [PubMed] [Google Scholar]
- 125.Sparks S, Rakocevic G, Joe G, Manoli I, Shrader J, Harris-Love M, Sonies B, Ciccone C, Dorward H, Krasnewich D, Huizing M, Dalakas MC, Gahl WA. Intravenous immune globulin in hereditary inclusion body myopathy: a pilot study. BMC Neurol. 2007;7:3. doi: 10.1186/1471-2377-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Miyagoe-Suzuki Y, Takeda S. Gene therapy for muscle disease. Exp Cell Res. 2010;316:3087–3092. doi: 10.1016/j.yexcr.2010.05.022. [DOI] [PubMed] [Google Scholar]
- 127.Sohn RL, Gussoni E. Stem cell therapy for muscular dystrophy. Expert Opin Biol Ther. 2004;4:1–9. doi: 10.1517/14712598.4.1.1. [DOI] [PubMed] [Google Scholar]
- 128.Skuk D, Tremblay JP. Intramuscular cell transplantation as a potential treatment of myopathies: clinical and preclinical relevant data. Expert. 2011;11:359–374. doi: 10.1517/14712598.2011.548800. [DOI] [PubMed] [Google Scholar]
- 129.Nemunaitis G, Jay CM, Maples PB, Gahl WA, Huizing M, Yardeni T, Tong AW, Phadke AP, Pappen BO, Bedell C, Allen H, Hernandez C, Templeton NS, Kuhn J, Senzer N, Nemunaitis J. Hereditary Inclusion Body Myopathy: Single Patient Response to Intravenous Dosing of GNE Gene Lipoplex. Hum Gene Ther. 2011 doi: 10.1089/hum.2010.192. Epub Apr 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nemunaitis G, Maples PB, Jay C, Gahl WA, Huizing M, Poling J, Tong AW, Phadke AP, Pappen BO, Bedell C, Templeton NS, Kuhn J, Senzer N, Nemunaitis J. Hereditary inclusion body myopathy: single patient response to GNE gene Lipoplex therapy. J Gene Med. 2010;12:403–412. doi: 10.1002/jgm.1450. [DOI] [PubMed] [Google Scholar]
- 131.Varki A. Sialic acids in human health and disease. Trends Mol Med. 2008;14:351–360. doi: 10.1016/j.molmed.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Ivanov S, Gavazova E, Antonova M, Chelibonova-Lorer H. Studies on N-acetylneuraminic acid biosynthesis in chicken liver and hepatoma Mc-29 by using [14C]N-acetylmannosamine and [14C]glucosamine. Int J Biochem. 1985;17:1125–1128. doi: 10.1016/0020-711x(85)90047-3. [DOI] [PubMed] [Google Scholar]
- 133.Suzuki O, Tasaki K, Kusakabe T, Abe M. UDP-GlcNAc2-epimerase regulates cell surface sialylation and ceramide-induced cell death in human malignant lymphoma. Int J Mol Med. 2008;22:339–348. [PubMed] [Google Scholar]
- 134.Coll MJ, Serratosa J, Bachs O, Gahmberg CG, Enrich C. Calmodulin may decrease cell surface sialic acid and be involved in the expression of fibronectin during liver regeneration. FEBS Lett. 1986;208:418–422. doi: 10.1016/0014-5793(86)81060-2. [DOI] [PubMed] [Google Scholar]
- 135.Hughes RC, Sanford B, Jeanloz RW. Regeneration of the surface glycoproteins of a transplantable mouse tumor cell after treatment with neuraminidase. Proc Natl Acad Sci U S A. 1972;69:942–945. doi: 10.1073/pnas.69.4.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bekesi JG, Roboz JP, Holland JF. Therapeutic effectiveness of neuraminidase-treated tumor cells as an immunogen in man and experimental animals with leukemia. Ann N Y Acad Sci. 1976;277:313–331. doi: 10.1111/j.1749-6632.1976.tb41710.x. [DOI] [PubMed] [Google Scholar]
- 137.Grunholz HJ, Harms E, Opetz M, Reutter W, Cerny M. Inhibition of in vitro biosynthesis of N-acetylneuraminic acid by N-acyl- and N-alkyl-2-amino-2-deoxyhexoses. Carbohydr Res. 1981;96:259–270. doi: 10.1016/s0008-6215(00)81876-5. [DOI] [PubMed] [Google Scholar]
- 138.Blume A, Chen H, Reutter W, Schmidt RR, Hinderlich S. 2',3'-Dialdehydo-UDP-N-acetylglucosamine inhibits UDP-N-acetylglucosamine 2-epimerase, the key enzyme of sialic acid biosynthesis. FEBS Lett. 2002;521:127–132. doi: 10.1016/s0014-5793(02)02856-9. [DOI] [PubMed] [Google Scholar]
- 139.Zeitler R, Giannis A, Danneschewski S, Henk E, Henk T, Bauer C, Reutter W, Sandhoff K. Inhibition of N-acetylglucosamine kinase and N-acetylmannosamine kinase by 3-O-methyl-N-acetyl-D-glucosamine in vitro. Eur J Biochem. 1992;204:1165–1168. doi: 10.1111/j.1432-1033.1992.tb16743.x. [DOI] [PubMed] [Google Scholar]
- 140.Zhu X, Stolz F, Schmidt RR. Synthesis of thioglycoside-based UDP-sugar analogues. J Org Chem. 2004;69:7367–7370. doi: 10.1021/jo049077m. [DOI] [PubMed] [Google Scholar]
- 141.Stolz F, Reiner M, Blume A, Reutter W, Schmidt RR. Novel UDP-glycal derivatives as transition state analogue inhibitors of UDP-GlcNAc 2-epimerase. J Org Chem. 2004;69:665–679. doi: 10.1021/jo0353029. [DOI] [PubMed] [Google Scholar]
- 142.Al-Rawi S, Hinderlich S, Reutter W, Giannis A. Synthesis and biochemical properties of reversible inhibitors of UDP-N-acetylglucosamine 2-epimerase. Angew Chem Int Ed Engl. 2004;43:4366–4370. doi: 10.1002/anie.200453863. [DOI] [PubMed] [Google Scholar]
- 143.Miller ML, Blom N. Kinase-specific prediction of protein phosphorylation sites. Methods Mol Biol. 2009;527:299–310. x. doi: 10.1007/978-1-60327-834-8_22. [DOI] [PubMed] [Google Scholar]
- 144.Wilcken B, Don N, Greenaway R, Hammond J, Sosula L. Sialuria: a second case. J Inherit Metab Dis. 1987;10:97–102. doi: 10.1007/BF01800030. [DOI] [PubMed] [Google Scholar]
- 145.Saechao C, Valles-Ayoub Y, Esfandiarifard S, Haghighatgoo A, No D, Shook S, Mendell JR, Rosales-Quintero X, Felice KJ, Morel CF, Pietruska M, Darvish D. Novel GNE mutations in hereditary inclusion body myopathy patients of non-Middle Eastern descent. Genet Test Mol Biomarkers. 2010;14:157–162. doi: 10.1089/gtmb.2009.0157. [DOI] [PubMed] [Google Scholar]
- 146.Kim BJ, Ki CS, Kim JW, Sung DH, Choi YC, Kim SH. Mutation analysis of the GNE gene in Korean patients with distal myopathy with rimmed vacuoles. J Hum Genet. 2006;51:137–140. doi: 10.1007/s10038-005-0338-5. [DOI] [PubMed] [Google Scholar]
- 147.Weihl CC, Miller SE, Zaidman CM, Pestronk A, Baloh RH, Al-Lozi M. Novel GNE mutations in two phenotypically distinct HIBM2 patients. Neuromuscul Disord. 2011;21:102–105. doi: 10.1016/j.nmd.2010.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Broccolini A, Ricci E, Cassandrini D, Gliubizzi C, Bruno C, Tonoli E, Silvestri G, Pescatori M, Rodolico C, Sinicropi S, Servidei S, Zara F, Minetti C, Tonali PA, Mirabella M. Novel GNE mutations in Italian families with autosomal recessive hereditary inclusion-body myopathy. Hum Mutat. 2004;23:632. doi: 10.1002/humu.9252. [DOI] [PubMed] [Google Scholar]
- 149.Li H, Chen Q, Liu F, Zhang X, Liu T, Li W, Liu S, Zhao Y, Wen B, Dai T, Lin P, Gong Y, Yan C. Clinical and molecular genetic analysis in Chinese patients with distal myopathy with rimmed vacuoles. J Hum Genet. 2011;56:335–338. doi: 10.1038/jhg.2011.15. [DOI] [PubMed] [Google Scholar]
- 150.Liewluck T, Pho-Iam T, Limwongse C, Thongnoppakhun W, Boonyapisit K, Raksadawan N, Murayama K, Hayashi YK, Nishino I, Sangruchi T. Mutation analysis of the GNE gene in distal myopathy with rimmed vacuoles (DMRV) patients in Thailand. Muscle Nerve. 2006;34:775–778. doi: 10.1002/mus.20583. [DOI] [PubMed] [Google Scholar]
- 151.Del Bo R, Baron P, Prelle A, Serafini M, Moggio M, Fonzo AD, Castagni M, Bresolin N, Comi GP. Novel missense mutation and large deletion of GNE gene in autosomal-recessive inclusion-body myopathy. Muscle Nerve. 2003;28:113–117. doi: 10.1002/mus.10391. [DOI] [PubMed] [Google Scholar]
- 152.Broccolini A, Pescatori M, D'Amico A, Sabino A, Silvestri G, Ricci E, Servidei S, Tonali PA, Mirabella M. An Italian family with autosomal recessive inclusion-body myopathy and mutations in the GNE gene. Neurology. 2002;59:1808–1809. doi: 10.1212/01.wnl.0000031808.04545.e0. [DOI] [PubMed] [Google Scholar]
- 153.Grandis M, Gulli R, Cassandrini D, Gazzerro E, Benedetti L, Narciso E, Nobbio L, Bruno C, Minetti C, Bellone E, Reni L, Mancardi GL, Mandich P, Schenone A. The spectrum of GNE mutations: allelic heterogeneity for a common phenotype. Neurol Sci. 2010;31:377–380. doi: 10.1007/s10072-010-0248-y. [DOI] [PubMed] [Google Scholar]
- 154.Vasconcelos OM, Raju R, Dalakas MC. GNE mutations in an American family with quadriceps-sparing IBM and lack of mutations in s-IBM. Neurology. 2002;59:1776–1779. doi: 10.1212/01.wnl.0000039780.13681.ad. [DOI] [PubMed] [Google Scholar]
- 155.Darvish D, Vahedifar P, Huo Y. Four novel mutations associated with autosomal recessive inclusion body myopathy (MIM: 600737) Mol Genet Metab. 2002;77:252–256. doi: 10.1016/s1096-7192(02)00141-5. [DOI] [PubMed] [Google Scholar]
- 156.Chu CC, Kuo HC, Yeh TH, Ro LS, Chen SR, Huang CC. Heterozygous mutations affecting the epimerase domain of the GNE gene causing distal myopathy with rimmed vacuoles in a Taiwanese family. Clin Neurol Neurosurg. 2007;109:250–256. doi: 10.1016/j.clineuro.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 157.Ro LS, Lee-Chen GJ, Wu YR, Lee M, Hsu PY, Chen CM. Phenotypic variability in a Chinese family with rimmed vacuolar distal myopathy. J Neurol Neurosurg Psychiatry. 2005;76:752–755. doi: 10.1136/jnnp.2004.048876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Stober A, Aleo A, Kuhl V, Bornemann A, Walter MC, Lochmuller H, Lindner A, Krause S. Novel missense mutation p.A310P in the GNE gene in autosomal-recessive hereditary inclusion-body myopathy/distal myopathy with rimmed vacuoles in an Italian family. Neuromuscul Disord. 2010;20:335–336. doi: 10.1016/j.nmd.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 159.Behin A, Dubourg O, Laforet P, Pecheux C, Bernard R, Levy N, Eymard B. Distal myopathy due to mutations of GNE gene: clinical spectrum and diagnosis. Rev Neurol (Paris) 2008;164:434–443. doi: 10.1016/j.neurol.2008.02.040. [DOI] [PubMed] [Google Scholar]
- 160.Fisher J, Towfighi J, Darvish D, Simmons Z. A case of hereditary inclusion body myopathy: 1 patient, 2 novel mutations. J Clin Neuromuscul Dis. 2006;7:179–184. doi: 10.1097/01.cnd.0000211406.94445.f0. [DOI] [PubMed] [Google Scholar]
- 161.Krause S, Schlotter-Weigel B, Walter MC, Najmabadi H, Wiendl H, Muller-Hocker J, Muller-Felber W, Pongratz D, Lochmuller H. A novel homozygous missense mutation in the GNE gene of a patient with quadriceps-sparing hereditary inclusion body myopathy associated with muscle inflammation. Neuromuscul Disord. 2003;13:830–834. doi: 10.1016/s0960-8966(03)00140-8. [DOI] [PubMed] [Google Scholar]
- 162.Amouri R, Driss A, Murayama K, Kefi M, Nishino I, Hentati F. Allelic heterogeneity of GNE gene mutation in two Tunisian families with autosomal recessive inclusion body myopathy. Neuromuscul Disord. 2005;15:361–363. doi: 10.1016/j.nmd.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 163.Voermans NC, Guillard M, Doedee R, Lammens M, Huizing M, Padberg GW, Wevers RA, van Engelen BG, Lefeber DJ. Clinical features, lectin staining, and a novel GNE frameshift mutation in hereditary inclusion body myopathy. Clin Neuropathol. 2010;29:71–77. [PMC free article] [PubMed] [Google Scholar]
- 164.Kayashima T, Matsuo H, Satoh A, Ohta T, Yoshiura K, Matsumoto N, Nakane Y, Niikawa N, Kishino T. Nonaka myopathy is caused by mutations in the UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase gene (GNE) J Hum Genet. 2002;47:77–79. doi: 10.1007/s100380200004. [DOI] [PubMed] [Google Scholar]
- 165.Motozaki Y, Komai K, Hirohata M, Asaka T, Ono K, Yamada M. Hereditary inclusion body myopathy with a novel mutation in the GNE gene associated with proximal leg weakness and necrotizing myopathy. Eur J Neurol. 2007;14:e14–15. doi: 10.1111/j.1468-1331.2007.01905.x. [DOI] [PubMed] [Google Scholar]