Abstract
Background
The methylotrophic yeast Pichia pastoris is frequently used for the production of recombinant proteins. However, expression levels can vary depending on the target protein. Allowing for simultaneous regulation of many genes, which may elicit a desired phenotype like increased protein production, overexpression of transcription factors can be used to overcome expression bottlenecks. Here, we present a novel P. pastoris transcription factor currently annotated as Aft1, activator of ferrous transport.
Results
The promoter regions of key secretory P. pastoris genes were screened for fungal transcription factor binding sites, revealing Aft1 as an interesting candidate for improving secretion. Genome wide analysis of transcription factor binding sites suggested Aft1 to be involved in the regulation of many secretory genes, but also indicated possible novel functions in carbohydrate metabolism. No Aft binding sites were found in promoters of characteristic iron homeostasis genes in P. pastoris. Microarrays were used to study the Aft1 regulon in detail, confirming Aft1 involvement in the regulation of carbon-responsive genes, and showing that iron regulation is dependent on FEP1, but not AFT1 expression levels. The positive effect of AFT1 overexpression on recombinant protein secretion was demonstrated for a carboxylesterase from Sphingopyxis sp. MTA144, for which secretion was improved 2.5-fold in fed batch bioreactor cultivations.
Conclusion
This study demonstrates that the transcription factor Aft1 can be used to improve recombinant protein secretion in P. pastoris. Furthermore, we discovered possible novel functions of Aft1 in carbohydrate metabolism and provide evidence arguing against a direct role of Aft1 in P. pastoris iron regulation.
Electronic supplementary material
The online version of this article (doi:10.1186/s12934-014-0120-5) contains supplementary material, which is available to authorized users.
Keywords: Pichia pastoris, Aft1, Transcription factor, Novel functions, Enhanced secretion
Background
The methylotrophic yeast Pichia pastoris (syn. Komagataella phaffii) is among today’s most frequently used yeast systems for the production of recombinant proteins [1]. Benefits of this yeast are the capability of high cell density cultivations, eukaryotic posttranslational modifications and good secretion capacity. A low level of endogenously secreted proteins allows for the production of relatively pure, recombinant secretory proteins. The recent availability of the genomic sequence boosted the generation of a versatile P. pastoris toolbox, including various strains, plasmids and promoters of different strength. To overcome individual bottlenecks during protein folding and secretion, a variety of helper factors such as the ER foldases Pdi1 or BiP (Kar2) have been studied in recent years [2,3]. The capability of transcription factors (TFs) as expression helpers was demonstrated by Guerfal et al. and Gasser et al. [4,5], who improved the secretion of the mIL-10 protein and antibody fragments by overexpression of the UPR (unfolded protein response) transcription factor HAC1. Also overexpression of the gene encoding the TF Nrg1 was shown to positively influence the secretion of recombinant porcine and human trypsinogen as well as the antibody Fab fragment 2 F5 [6].
Allowing for the simultaneous regulation of different proteins involved in e.g. folding and secretion, TFs have huge potential to overcome bottlenecks in the cellular protein production machinery.
Here, we present a novel P. pastoris TF which was annotated as Aft1 (Activator of ferrous transport) by sequence homology to Saccharomyces cerevisiae Aft1/2. While no information is currently available on the function of P. pastoris Aft1, its two S. cerevisiae homologs have been studied extensively. In S. cerevisiae the transcriptional activators Aft1/2 are responsible for iron uptake and homeostasis fulfilling overlapping but non-redundant roles [7,8]. It was shown that iron homeostasis is primarily maintained by Aft1, while Aft2 is the weaker transcriptional activator [8,9]. Still, both TFs can interact with the same iron-responsive element (FeRE) found within promoters of the genes of the iron regulon such as the iron reductases FRE1–6 or the multicopper oxidases FET3/FET5 [8]. Under iron limiting conditions, Aft1 binds to promoters of the iron regulon genes and increases their expression [10,11]. Under iron repletion, the monothiol glutaredoxins (Grx3/4) attached to an iron-sulfur cluster bind Aft1 and initiate its dissociation from target promoters [12]. Export of Aft1 from the nucleus is mediated by the nuclear export receptor Msn5 [13]. Several amino acid (aa) residues have been shown to be important for the iron-responsive regulation of Aft1. While phosphorylation of Ser210/Ser224 and an intermolecular interaction are essential for recognition by Msn5 [13], the residues Leu99, Leu102, Cys291, Cys293 are involved in the interaction with Grx3/4 and iron dependent regulation [12].
In contrast to S. cerevisiae, in most fungi such as Candida albicans, Pichia stipitis or Schizosaccharomyces pombe, the iron-regulatory pathway is controlled by a conserved zinc finger GATA-type repressor [14,15]. Also in P. pastoris iron uptake was shown to be under control of a GATA-factor, named Fep1. Fep1 was shown to bind to DNA only under iron repletion and disruption of FEP1 led to constitutively high expression of FET3, independent of the availability of iron [16]. Interestingly, several species such as C. albicans, P. stipitis and also P. pastoris have been found to possess an Aft-type regulator in addition to the GATA-type repressor, leading to the question on the function of Aft in these species [16,17].
In this study, we investigated the functions of the P. pastoris Aft1 regulator based on the prediction of putative Aft binding sites in promoters, focusing on the secretion of recombinant proteins. We provide evidence that Aft1 is not directly involved in P. pastoris iron regulation, but rather in carbon-responsive regulation. Furthermore, we show that AFT1, when overexpressed under its natural promoter, increased the secretion of a model protein up to 2.5-fold in fed batch bioreactor cultivations.
Results and discussion
Analysis of transcription factor binding sites in selected genes shown to enhance protein secretion
To identify novel TFs useful for improving protein secretion in P. pastoris, the promoter regions, i.e. 1000 bp upstream of the ATG start codon, of key secretory genes involved in folding, transport and exocytosis were studied for putative fungal TF binding sites using the program MatInspector (Genomatix, [18]). The following genes, which were previously identified to improve secretion if modified [4,5,19,20], and/or differentially regulated upon expression of a human sialic acid transporter [21], were studied: ERO1, HAC1, KAR2, PDI1, YDJ1, CNE1, SSE1, SSA4, SSB1, IRE1, UBI4, KIN2, SSO2, CUP5, CDS1, PGS1, ERG1, ERG3, ERG25, NCP1 and INO1.
Obtained TF hits were analysed according to frequency, function (link to the secretory machinery or stress response), matrix similarity (similarity of the input sequence to TF matrixes stored in the database, cut-off 0.9) and distance to the start codon (proximal more weighty than distal).
S. cerevisiae Aft1/2 binding sites were found enriched in the P. pastoris genes PDI1, KAR2, SSA4, KIN2 and NCP1 (Table 1). By sequence homology, the gene product of PAS_chr1-4_0361/PP7435_Chr1-1146 [NCBI] was identified as the single P. pastoris Aft protein, therefore called Aft1.
Table 1.
Putative P. pastoris Aft1 binding sites found in promoters of selected genes known to enhance protein secretion
| Gene | Functions a | Aft1 UBS (bp) | Sequence |
|---|---|---|---|
| PDI1 | Protein disulfide isomerase, ER chaperone, formation of disulfide bonds | -423b to -409 | gatcacaCACCctct |
| KAR2 | ER chaperone, mediating protein folding, UPR regulation | -870b to -856 | tcgtataCACCctca |
| -194b to -180 | tgtaataCACCcttg | ||
| SSA4 | HSP70 protein, co-translational protein-membrane targeting and translocation of nascent proteins into the ER | -608c to -594 | actcatgCACCctta |
| -342b to -328 | catggaaCACCccat | ||
| KIN2 | Serine/threonine kinase involved in exocytosis | -177c to -163 | ataactgCACCcaga |
| NCP1 | P450 reductase, involved in lipid metabolism (ergosterol biosynthesis) | -539c to -525 | ggttttgCACCcagg |
aFunctions are derived for the S. cerevisiae homologs of the P. pastoris genes in the Saccharomyces Genome Database.
bAft2 matrix.
cAft1 matrix.
Upstream binding sites (UBS): 5′ → 3′, -1000 to -1 A(0)TG. The core binding sequences, representing the highest conserved, consecutive positions are highlighted. TFBS analysis: MatInspector [18] using the search groups fungi and general core promoter elements.
P. pastoris Aft1 binding sites were predicted for the regulatory regions of Pdi1 and BiP, two important ER chaperones, indicating that Aft1 is involved in oxidative protein folding. Consistently, Blaiseau et al. [7] showed S. cerevisiae Aft2 involved in oxidative stress resistance. Also, the chaperone Ssa4 and the serine/threonine kinase Kin2, both recently identified as secretion enhancing factors [3], were found to have an upstream Aft1 binding site, confirming Aft1 as an interesting candidate for improving recombinant protein secretion in P. pastoris. In addition, Aft1 binding was predicted for the promoter region of NCP1, suggesting that Aft1 is also involved in lipid metabolism [22].
Prediction of Aft1 binding sites in the P. pastoris genome
To elucidate the function of Aft1 in P. pastoris, Regulatory Sequence Analysis Tools [23] was used to search for putative Aft1 binding sites in P. pastoris promoters. Using the binding motifs known for S. cerevisiae Aft1/2, ANTGCACCC and BRCACCCB, resulted in 972 genes with a putative Aft binding site. Thereof, 561 were found annotated [24] and mapped to broader parent terms, GO slim terms, using AmiGO GO Slimmer [25]. Aft1 was found involved in the regulation of approximately 100 different biological processes (Additional file 1), including nucleobase-containing small molecule metabolic processes (57 hits), carbohydrate metabolic processes (50 hits), transcription from RNA polymerase II promoter (47 hits) or amino acids metabolic processes (47 hits). Interestingly, within the categories ion transport (20 hits) and cellular ion homeostasis (11 hits) only 6 proteins with a possible connection to iron regulation were found: Ccc1, mediating transfer of iron from cytosol to vacuole [26]; Gef1, a chloride channel localized to the Golgi or endomembrane system, which has also been reported to be governing iron-limited growth [27]; Hmx1, a heme oxygenase, required for the reutilization of iron from heme, also involved in oxidative stress resistance [28,29] and Nfu1, a protein involved in mitochondrial iron sulfur cluster assembly [30]. Notably, none of these are characteristic iron regulatory genes such as the iron reductases FRE1-6 or the multicopper oxidases FET3/5. However, a putative Aft1 binding site was found within the upstream region of FEP1, the GATA type repressor, which has been shown to negatively control iron uptake/homeostasis in P. pastoris [16]. Thus, though no Aft1 binding site was found within the upstream region of characteristic iron regulators, the presence of such within the upstream region of FEP1 raises the possibility of a latent, indirect involvement of Aft1 in P. pastoris iron regulation.
Analysing possible other functions of P. pastoris Aft1, a high number of biological processes related to the cellular secretory pathway machinery was found (Table 2). 23 gene hits were found for protein targeting, 19 for Golgi to vesicle transport, 12 for response to oxidative stress, 11 for folding, 10 for glycosylation and vesicle organization, 9 for vacuole organization and regulation of transport, and 6 for exocytosis. Additionally to the already discussed ER chaperones Pdi1 and BiP, several important secretory regulators were identified. Sec12 and Sec23, both involved in COPII vesicle formation and ER to Golgi transport [31], Sec61, forming a channel for protein translocation into and out of the ER [32], Gos1, a v-SNARE protein found to be involved in ER to Golgi and/or intra-Golgi transport [33], Yap1, a transcription factor involved in oxidative stress response [34], Och1, a Golgi resident mannosyltransferase initiating the hypermannosylation of glycoproteins [35] and Kin1, a protein kinase involved in exocytosis [36], were found to possess a putative upstream Aft1 binding site. In accordance with the above-mentioned data, an Aft1 binding site was found in the upstream region of several genes conferring resistance to oxidative stress, such as YAP1 or HMX1, supporting the hypothesis of a role of Aft1 in oxidative protein folding and/or stress response.
Table 2.
P. pastoris genes with putative Aft1 binding site(s) related to the cellular secretory pathway machinery
| GO slim term - Biological process | Number (Genes) |
|---|---|
| GO:0006605 protein targeting | 23 (ATG1 ATG12 ATG18 GET3 GET4 IMP2 KAR2 LHS1 NPL6 NUP188 NUP84 PAM17 PEX14 PEX7 PEX8 SEC61 SPC1 SPC3 TAM41 VPS21 VPS64 VPS68 VPS8) |
| GO:0048193 Golgi vesicle transport | 19 (AGE2 APL4 CHS5 DRS2 EMP24 GET3 GOS1 HRR25 KES1 SEC12 SEC23 SFB3 SNC2 SVP26 TED1 TLG1 TRS31 VTI1 YPT32) |
| GO:0006979 response to oxidative stress | 12 (AFT1 CTA1 EOS1 GCY1 GSH1 HMX1 HSP104 MXR1 POS5 TSA1 YAP1 YBL055C) |
| GO:0006457 protein folding | 11 (CAJ1 CCT2 CCT6 CPR6 EGD2 ERV2 HLJ1 HSP104 PDI1 SIS1 TSA1) |
| GO:0006486 protein glycosylation | 10 (ALG7 EOS1 KRE5 MNN11 OCH1 OST6 PMT1 PMT5 STT3 SVP26) |
| GO:0016050 vesicle organization | 10 (EMP24 GOS1 SEC12 SEC23 SFB3 SNC2 TLG1 UBC1 VPS4 VTI1) |
| GO:0007033 vacuole organization | 9 (ATG1 ATG12 ENO1 GYP7 RDI1 TPM2 VPS21 VTI1 YHC3) |
| GO:0051049 regulation of transport | 9 (AKL1 ARG81 CUP9 FPK1 GEF1 SEC12 SEC23 SLG1 TUP1) |
| GO:0006887 exocytosis | 6 (ARG81 KES1 KIN1 SNC2 TPM2 YPT32) |
Search field: -1000 bp upstream of the P. pastoris GS115 coding sequences. Binding motifs: ScAft1 (ANTGCACCC) and ScAft2 (BRCACCCB). Gene hits were categorized into biological function GO terms using AmiGO GO Slimmer (Yeast GO slim set, [25]).
P. pastoris Aft1 has a conserved Aft-domain, but does not contain the iron-responsive motif
The amino acid sequence of P. pastoris (Pp) Aft1 was compared to S. cerevisiae (Sc) Aft1/2 and Kluyveromyces lactis (Kl) Aft using ClustalW2 [37]; all of these proteins display the positive mode of regulation and the characteristic residues Leu99, Leu102, Cys291 and Cys293 conferring iron sensitivity (numbering based on ScAft1, [14]). Interestingly, the P. pastoris N-terminal sequence is 41-98 amino acids shorter, lacking the conserved residues Leu99 and Leu102 (PpAft1: Met1, Ile4; Figure 1). Also the residues Cys291 and Cys293 were not found conserved in the P. pastoris sequence (PpAft1: Ser180, Ser182), suggesting a role different from iron regulation for PpAft1. Still, also regions of high homology were found in the N-terminal part of the P. pastoris protein, particularly the Aft-domain (supposedly the DNA binding domain [38]) between the amino acids 10-130. Especially high conservation was found for the residues 10-45 (identity 53%) and 102-130 (identity 45%), including the two conserved cysteines and histidines, which have been suggested to be part of a WRKY-motif involved in zinc binding [14,39]. Regarding nuclear export, no conservation was found for Ser210 in P. pastoris (PpAft1: Arg97) and K. lactis (KlAft: Gly224), but Ser224 is conserved in all 4 sequences (PpAft1: Ser111, KlAft: Ser238). However, as Ueta et al. [13] showed that individual serine mutations did not affect ScAft1 localization, export via Msn5p seems still possible for PpAft1 and KlAft. In contrast to ScAft1 and KlAft, a glutamine-rich region was found not at the C-terminal end, but between the residues 164-177. This glutamine-rich region is followed by serine (aa 180-213) and asparagine repeats (aa 216-269), maybe as part of a protein-protein interaction domain [40]. No homology was found for the C-terminal part of the PpAft1 protein (identity <1% ), in particular the residues 162 to 363. Interestingly, with only 363 amino acids PpAft1 is considerably shorter than ScAft1 (690 aa), ScAft2 (416 aa) and KlAft (823 aa).
Figure 1.

Aft1 protein sequence comparison. Amino acid sequences: S. cerevisiae Aft1 [NCBI protein: NP_011444] and Aft2 [NCBI protein:NP_015122], K. lactis Aft [NCBI protein:CAH00307], and P. pastoris Aft1 [NCBI protein:CCA37276] were aligned using ClustalW2 [37]. asterisk: “indicates positions which have a single, fully conserved residue”, colon: “indicates conservation between groups of strongly similar properties”; period: “indicates conservation between groups of weakly similar properties”; shaded grey: conserved residues (Leu99*, Leu102, Cys291 and Cys293 conferring iron sensitivity. Ser210 and Ser224 involved in Msn5 recognition and nuclear export. Cys143, Cys215, His239 and His241, which are suggested to be part of a WRKY-motif involved in zinc binding [39]). Differences are boxed and/or highlighted red; *numbering is based on ScAft1.
PpAft1 was further compared to C. albicans Aft [NCBI protein: XP_714862], which also lacks the characteristic residues Leu99, Leu102, Cys291 and Cys293. Similar to P. pastoris, in C. albicans iron regulation was shown to be under control of a GATA-type repressor [17]. Sequence comparison of these two proteins revealed only one short N-terminal region of high homology between the residues 3-47 (identity 67%, data not shown). This region was also found to be highly similar in ScAft1/2 and KlAft. Additionally, we searched the genome of the genetically and biochemically close methylotrophic yeast Hansenula polymorpha for a homolog of PpAft1. The protein encoded by HPODL_04658 in H. polymorpha (NCBI protein: ESX01890) has high N-terminal sequence homology including the two conserved cysteines und histidines (aa 3-132: identity 55%, Additional file 2). A region of high similarity was also found between the residues 244-332 (identity 31%), suggesting that HPODL_04658 functions similar to PpAft1. As PpAft1 and CaAft, also HPODL_04658 lacks the characteristic iron sensitivity residues. Accordingly, we also found a homolog of the GATA-type repressor in the H. polymorpha genome (HPODL_03720; 48% sequence identity to PpFep1). In summary, Aft homologs from species that are known to contain also a Fep1-like GATA type repressor (C. albicans, Pichia stipitis, Debaryomyces hansenii according to [14] and H. polymorpha) share high sequence homology to PpAft1 in the N-terminal DNA binding region, but lack the characteristic residues that are conferring iron sensitvity in ScAft1/2 (data not shown). Interestingly, all these species possess another protein of unknown function having a domain with sequence similarity to parts of the Aft DNA-binding domain, but low similarity to ScAft1/2 (i.e. less than 15% overall identity).
The P. pastoris Aft1 regulon
To investigate if P. pastoris Aft1 is involved in iron regulation, we generated an AFT1 overexpression (AFT1-OE) and an Δaft1 disruption mutant, and tested their growth in low and high iron containing media. AFT1-OE was achieved by expressing an additional copy of AFT1 under control of its native promoter (see below), while the Δaft1 strain was generated by exchanging parts of the gene for the KanMX marker cassette using the split marker approach as described by Heiss et al. [41]. Low iron media was prepared by addition of the iron chelator BPS (bathophenanthroline disulphonate) to YPD or YNB-Glucose agar plates as described by Miele et al. [16]. In order to exclude the possibility to be outside the sensitivity range we tried several concentrations of BPS in the iron sensitivity assay. Miele et al. [16] reported the use of YPD containing 80–160 μM BPS for P. pastoris. We tested concentrations ranging from 80-200 μM BPS in YP and minimal medium (YNB or M2) using either glucose or methanol as carbon source. We did not observe growth impairment of P. pastoris CBS7435 wild type using 80 μM BPS in YPD or YPM (data not shown). When using higher BPS concentrations, growth of all strains was significantly delayed. However, we did not observe differences between the Δaft1 strain, the AFT1-OE and the wild type control on any of the media tested (Figure 2). Thus, disruption of AFT1 does not render P. pastoris sensitive to iron-limited conditions. This behaviour is contrary to the phenotype observed for Δaft1/2 in S. cerevisiae and Δaft1 in K. lactis, which grow only poorly or are unable to grow in the same low iron conditions [7,14], suggesting that P. pastoris Aft1 is not involved in iron regulation. Additionally, the P. pastoris Δaft1 mutant did not show impaired growth on the cell wall disturbing agent Calcofluor White, which is again in contrast to what is reported for S. cerevisiae Δaft1 (not shown).
Figure 2.
Spotting assay on iron deficient media. Serial 1:10 dilutions of P. pastoris CBS7435 wild type, AFT1-OE and Δaft1 cell suspensions were spotted on YP plates containing either 2% glucose or 1% methanol as carbon source and on YNB agar plates containing 2% glucose as carbon source. Iron-deficient conditions were generated by addition of the iron chelator BPS (200 μM). Plates were incubated at 28°C for 70 h. While addition of BPS resulted in generally delayed growth on both substrates, no difference between the strains was seen.
In order to identify the regulon of Aft1 in P. pastoris, we analysed the transcription patterns of the Δaft1 strain and control using DNA microarrays. All strains were cultivated in synthetic minimal medium with glucose feed beads for 5 h in three biological replicates. Interestingly, although Aft-like proteins are described as transcriptional activators, an even higher number of genes was up-regulated (54 genes, 33 thereof more than 1.5-fold) than down-regulated (34 genes, 13 thereof more than 1.5-fold) in the Δaft1 mutant compared to the wild type (an adjusted p-value of < 0.05 was applied as cut-off to identify significantly regulated genes, Additional file 3). 42% of the up-regulated and 38% of the down-regulated genes contain at least one putative Aft1 binding site in their promoters (compared to 18% of total P. pastoris genes), suggesting that both, up- and down-regulation, are a direct consequence of AFT1 disruption. Table 3 summarizes differentially regulated genes in the Δaft1 mutant compared to the wild type strain according to their GO term category.
Table 3.
Differentially expressed and annotated genes in the Δ aft1 vs. control (wild type) strain
| Up-regulated genes (Δ aft1 vs. control) | |
|---|---|
| GO slim term - Biological process | Number (Genes) |
| GO:0006811 ion transport | 7 ( AQR1 ATX1 GEF1 MEP2 TPO3 YHL008C ZRT1 ) |
| GO:0008150 biological_process* | 6 ( ECM13 GPM3 YEL023C YLR156C-A YLR278C YOR292C ) |
| GO:0055085 transmembrane transport | 6 ( AQR1 GEF1 MEP2 STL1 TPO3 ZRT1 ) |
| GO:0006366 transcription from RNA polymerase II promoter | 5 ( GCR1 PHD1 RPM2 UGA3 YLR278C ) |
| GO:0042221 response to chemical | 5 ( AQR1 ATX1 GCR1 SLI1 UGA3 ) |
| GO:0006520 cellular amino acid metabolic process | 4 ( ADH2 ECM4 SFA1 UGA3 ) |
| GO:0007124 pseudohyphal growth | 4 ( HMS1 MEP2 PHD1 PTP1 ) |
| GO:0051186 cofactor metabolic process | 4 ( ADH2 ALD4 BIO2 THI21 ) |
| GO:0005975 carbohydrate metabolic process | 3 ( DOG1 GCR1 GPM3 ) |
| GO:0006091 generation of precursor metabolites and energy | 2 ( GCR1 GPM3 ) |
| GO:0006766 vitamin metabolic process | 2 ( BIO2 THI21 ) |
| GO:0006873 cellular ion homeostasis | 2 ( ATX1 GEF1 ) |
| GO:0055086 nucleobase-containing small molecule metabolic process | 2 ( ADH2 ALD4 ) |
| GO:0001403 invasive growth in response to glucose limitation | 1 ( PTP1 ) |
| GO:0002181 cytoplasmic translation | 1 ( RPM2 ) |
| GO:0006325 chromatin organization | 1 ( GCR1 ) |
| GO:0006397 mRNA processing | 1 ( RPM2 ) |
| GO:0006470 protein dephosphorylation | 1 ( PTP1 ) |
| GO:0006629 lipid metabolic process | 1 ( YJU3 ) |
| GO:0006865 amino acid transport | 1 ( AQR1 ) |
| GO:0006979 response to oxidative stress | 1 ( ATX1 ) |
| GO:0007005 mitochondrion organization | 1 ( RPM2 ) |
| GO:0008033 tRNA processing | 1 ( RPM2 ) |
| GO:0008643 carbohydrate transport | 1 ( STL1 ) |
| GO:0070647 protein modification by small protein conjugation or removal | 1 ( PCI8 ) |
| Down-regulated genes (Δ aft1 vs. control) | |
| GO slim term - Biological process | Number (Genes) |
| GO:0006811 ion transport | 4 ( ATO2 JEN1 TAT2 VMA8 ) |
| GO:0055085 transmembrane transport | 4 ( ITR1 JEN1 PEX13 TAT2 ) |
| GO:0006281 DNA repair | 3 ( PCD1 RVB1 YRA1 ) |
| GO:0006325 chromatin organization | 3 ( ACS1 RLF2 RVB1 ) |
| GO:0006366 transcription from RNA polymerase II promoter | 3 ( MIG1 RVB1 YHP1 ) |
| GO:0006974 cellular response to DNA damage stimulus | 3 ( PCD1 RVB1 YRA1 ) |
| GO:0008150 biological_process* | 3 ( NBA1 TOS8 YHR177W ) |
| GO:0005975 carbohydrate metabolic process | 2 ( CTS1 MIG1 ) |
| GO:0006766 vitamin metabolic process | 2 ( SNO1 SNZ3 ) |
| GO:0051186 cofactor metabolic process | 2 ( ACS1 SNZ3 ) |
| GO:0055086 nucleobase-containing small molecule metabolic process | 2 ( PCD1 YRA1 ) |
| GO:0070271 protein complex biogenesis | 2 ( PEX13 RLF2 ) |
| GO:0000278 mitotic cell cycle | 1 ( YHP1 ) |
| GO:0000910 cytokinesis | 1 ( CTS1 ) |
| GO:0002181 cytoplasmic translation | 1 ( RPL4A ) |
| GO:0006091 generation of precursor metabolites and energy | 1 ( ACS1 ) |
| GO:0006520 cellular amino acid metabolic process | 1 ( SNO1 ) |
| GO:0006605 protein targeting | 1 ( PEX13 ) |
| GO:0006865 amino acid transport | 1 ( TAT2 ) |
| GO:0006873 cellular ion homeostasis | 1 ( VMA8 ) |
| GO:0007031 peroxisome organization | 1 ( PEX13 ) |
| GO:0008643 carbohydrate transport | 1 ( JEN1 ) |
| GO:0015931 nucleobase-containing compound transport | 1 ( YRA1 ) |
| GO:0016570 histone modification | 1 ( ACS1 ) |
| GO:0018193 peptidyl-amino acid modification | 1 ( ACS1 ) |
| GO:0042221 response to chemical | 1 ( MIG1 ) |
| GO:0042594 response to starvation | 1 ( MIG1 ) |
| GO:0043543 protein acylation | 1 ( ACS1 ) |
| GO:0048193 Golgi vesicle transport | 1 ( BRE5 ) |
| GO:0051049 regulation of transport | 1 ( BRE5 ) |
| GO:0051169 nuclear transport | 1 ( YRA1 ) |
| GO:0070647 protein modification by small protein conjugation or removal | 1 ( BRE5 ) |
*Biological process is unkown.
Genes were categorized into biological function GO terms using AmiGO GO Slimmer (Yeast GO slim set, [25]).
In the Δaft1 strain, 9 putative transcriptional regulators are among the regulated genes, seven of them carrying putative Aft1 binding sites in their promoters. PAS_chr4_0324, a fungal specific transcription factor of unknown function with a Zn2/Cys6 DNA-binding domain is the highest up-regulated gene, while MIG1-1, encoding a transcription factor connected to glucose repression, has lower expression levels in the Δaft1 mutant. Most other regulated transcription factors are also of the fungal-specific Zn2/Cys6 type, but lack annotated function and defined target genes. Furthermore, 23 of the 54 up-regulated and 9 of the 34 down-regulated genes encode hypothetical proteins of unknown function. The second largest group of regulated genes comprises transport proteins, mainly transmembrane transporters, however, there is no clear preference for transported substrates, which range from ions to polyamines and sugars.
Regarding iron regulation, no changes in transcript levels were observed for the GATA-type repressor FEP1, in line with the unaltered expression levels of genes involved in iron uptake and homeostasis (i.e. FRE1–6 or FET3/FET5). Re-analysis of our previous microarray data (obtained in different environmental conditions, [42,43]) for expression changes of iron regulatory genes indeed revealed that induction of the iron regulon is dependent on the levels of FEP1, but not AFT1 in P. pastoris.
Notably, Atx1 and Gef1, which were described to be required for the correct assembly of the high affinity iron transporter Fet3 in S. cerevisiae, were found among the up-regulated genes in the Δaft1 mutant, pointing towards a different regulation of iron acquisition genes in P. pastoris.
Among the up-regulated genes several genes which are described to be repressed by glucose in S. cerevisiae are found, including alcohol dehydrogenase ADH2 and the two putative mitochondrial aldehyde dehydrogenases ALD4-1 and ALD5. All of these contain putative Aft1 binding sites in their promoter regions. Interestingly, deletion of these gene functions has recently been predicted by the genome scale metabolic model of P. pastoris to enhance recombinant protein production [44]. Moreover, carbon-source responsive transporters of the multifacilitator superfamily are among the regulated genes in the Δaft1 mutant. While the genes encoding the high affinity glucose transporter Ght1 and the putative glycerol transporter PAS_c034_0021 are up-regulated in the Δaft1 mutant, the second high affinity glucose transporter of P. pastoris, encoded by GHT2 as well as a homolog of the S. cerevisiae myo-inositol transporter ITR1, are repressed in this strain. This implicates that Aft1 is involved in the regulation of glucose-repressed genes, at least in an indirect manner by regulating MIG1-1 expression, but maybe also directly as some of these genes also contain Aft1 binding sites in their promoters. In this respect, it should be noted that AFT1 expression levels are significantly higher not only in glucose-limited conditions, but also in cells grown on methanol as compared to glucose or glycerol surplus (see below and own unpublished data). A correlation of AFT1/2 expression levels and the carbon source has also been observed in S. cerevisiae, where differences between the fermentable carbon source glucose and the non-fermentable carbon source glycerol were reported [38].
Regulation of the P. pastoris AFT1 promoter
To study the expression strength of the natural AFT1 promoter and to test if it is applicable for overexpression studies, intracellular expression of green fluorescent protein was analysed (cycle-3-GFP, [45]). 96-well deep-well plate screening, which has been applied for P. pastoris promoter studies before [46], was used for cultivation. However, the protocol was adjusted for GAP promoter driven expression, which was used for the expression of our model protein carboxylesterase.
P. pastoris was transformed with plasmid pPpKan-S-GFP. Clones expressing GFP from the AFT1 promoter were studied for intracellular fluorescence levels in comparison to clones expressing GFP from the constitutive GAP promoter. On average, the tested PAFT1 clones reached 724 relative fluorescence units (RFU) after 69 h of cultivation, which was 60% in comparison to the respective PGAP clones. In terms of average yields, the PAFT1 clones reached 343 RFU OD-1, corresponding to 50% of the yield of the PGAP clones, characterising PAFT1 as fairly strong promoter under the conditions tested. Analysing expression at 24, 48 and 69 h of cultivation, differences in regulation were observed. While the PGAP clones showed the expected constitutive expression profile with stable titers of 1201 ± 64 RFU at the different time points, PAFT1 controlled expression significantly increased with longer cultivation times, reaching a maximum at 69 h of cultivation (Figure 3A). Assuming that the observed effect was related to a decrease in glucose concentration, a batch series starting with 1, 2 and 4% glucose was studied for about 48 h (data not shown). The results confirmed enhanced PAFT1 activity at lower glucose concentrations, showing average yields of 104, 81 and only 16 RFU OD-1, for the 1, 2 and 4% glucose batches, respectively.
Figure 3.
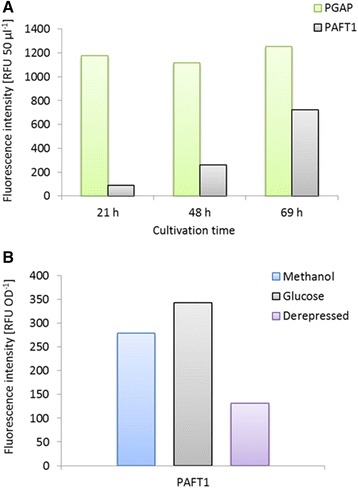
Characterization of the AFT1 promoter. (A) GFP expression levels under control of PGAP and PAFT1. Cultivation in 96-well plate. RFU: relative fluorescence units, average of 87 P. pastoris clones. (B) GFP expression levels under control of PAFT1 using different cultivation conditions. Glucose: cultivation time 69 h, 2% glucose batch, 3 times addition of glucose to 0.5%. Methanol: cultivation time 92 h, 48 h glucose batch (1%), 3 times addition of methanol to 0.5%. Derepressed: cultivation time 69 h, 2% glucose batch, no additional supplementation.
Interestingly and matching above data, the AFT1 promoter was also found inducible by methanol, reaching 80% (280 RFU OD-1) of the yield of glucose cultivated cells (Figure 3B). In comparison, using a 2% glucose batch without supplementation (derepressed protocol), a yield of only 40% was reached. While PAFT1 is obviously a rather strong and also methanol-inducible promoter, unlimited access of glucose negatively affects PAFT1 activity and presumably decreases overexpression effects.
The effect of AFT1 overexpression on the secretion of recombinant proteins
Although none of the secretion-related genes was differentially expressed in the Δaft1 mutant under the analysed conditions, our next step was to analyse whether AFT1 overexpression influences the secretion of recombinant proteins. Our model protein of choice was a carboxylesterase from Sphingopyxis sp. MTA144. This carboxylesterase is under development for use as a feed additive enzyme, because it hydrolyses an antinutritive substance that may be naturally contained in animal feed [47]. P. pastoris has recently been shown to secrete active carboxylesterase [48]. However, for a technological application of this carboxylesterase as feed enzyme for gastrointestinal detoxification in animals, a high yield recombinant production process is required.
Generation of carboxylesterase secreting strains
Carboxylesterase secreting strains were generated by transformation of P. pastoris CBS7435 with plasmid pPM2dZ30-PGAPα-CE expressing carboxylesterase under control of the GAP promoter and using the S. cerevisiae alpha mating factor signal sequence as secretion leader. Carboxylesterase represented the major fraction of total secreted protein of each transformant, yielding a strong band at the calculated size of 52 kDa. However, due to varying numbers of integrated expression cassettes, strains secreting lower and higher levels of carboxylesterase were observed (Additional file 4). The best-secreting strain CE#18 contained 6 copies of the expression cassette, while the average strain CE#12 had only one expression cassette integrated in its genome, confirming a positive correlation between secretion level and gene copy number. Using ELISA, an expression level of 80 μg mL-1 was determined for CE#18, whereas a titer of approximately 20 μg mL-1 was determined for strain CE#12.
AFT1 co-expression studies
The strains CE#12 and CE#18 were chosen to study whether overexpression of AFT1 can positively influence recombinant protein secretion. Both strains were transformed with linearized pPM2aK21-AFT1, comprising the AFT1 gene under control of its natural promoter and terminator. Supernatants of 8 transformants of each strain were studied for carboxylesterase secretion after 48 h of cultivation in shake flask. ELISA was used to reveal small, but significant differences in carboxylesterase secretion levels, showing improved secreting clones for both strain backgrounds. Figure 4 shows the results of the best two clones of each strain. Interestingly, while the best CE#12-AFT1 clones yielded a strong improvement of 62 ± 5%, the best two CE#18-AFT1 strains showed considerably less improvement, reaching an increase of only 19 ± 6%. However, it has to be considered that strain CE#18 produced four times more carboxylesterase (80 μg mL-1) than strain CE#12 (20 μg mL-1). Assuming that Aft1 is involved in oxidative protein folding, high overproduction of carboxylesterase could have led to an overload of recombinant protein in the ER and subsequent induction of ERAD. Newly elicited upstream or downstream bottlenecks could also be the reason for the reduced influence of AFT1 overexpression and the only modest improvement for strain CE#18.
Figure 4.
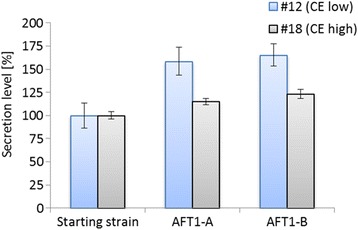
Screening results of improved carboxylesterase-AFT1 P. pastoris strains. Secretion levels: determined by ELISA and normalized to the respective starting strain. Starting strains: CE#12 (blue, low level expressing strain), CE#18 (grey, high level expressing strain). AFT1-A and AFT1-B: AFT1 overexpressing clones.
The growth behaviour of AFT1 overexpressing strains was studied in addition. In contrast to secretion levels, growth under these conditions was seemingly unaffected by overexpression of AFT1, showing neither significant changes in final OD levels (<15% , 48 h) nor any detectable alteration in the growth curve (data not shown).
Bioreactor cultivations
To confirm the positive effect of AFT1 overexpression on carboxylesterase secretion, the best strains from screening were studied in bioreactor cultivations. First, the performance of the AFT1 overexpressing strain CE#12-AFT1-A was compared to the starting strain (CE#12). Cultivations were performed using aerobic and hypoxic production conditions as preceding bioreactor experiments had indicated that a low oxygen supply is beneficial for carboxylesterase secretion (data not shown), an observation that had also been made for the production of antibody Fab fragments and trypsinogen [49]. While under aerobic production conditions a dissolved oxygen (DO) concentration of 20% was maintained throughout the whole process, a DO level of 5% was applied during the feed phase for hypoxic conditions. Bioreactor cultivations were monitored for 100 h. In addition to the quantification of secreted recombinant carboxylesterase by SDS-PAGE, also the functional quality of the enzyme was assessed by an activity assay.
The positive effect of AFT1 overexpression on the secretion rate of strain CE#12 was indeed confirmed in bioreactor. A maximum of 141 U L-1 was reached for strain CE#12-AFT1-A after 100 h of cultivation under hypoxic conditions, while the starting strain reached only a level of 76 U L-1 at this time point (Figure 5A). Confirming the beneficial effect of hypoxic production conditions, carboxylesterase activity levels were 3-4 fold lower under aerobic conditions. Applying 20% DO, a maximum of only 37 U L-1 was reached for strain CE#12-AFT1-A after 100 h of cultivation. However, comparing to the starting strain under the same conditions a 2.5-fold higher activity level was achieved for the AFT1 overexpressing strain. It seems that the amount of dissolved oxygen does not abrogate the beneficial impact of AFT1 overexpression on carboxylesterase secretion. As observed previously [43], lower amounts of biomass were reached in hypoxic compared to normoxic conditions due to the production of ethanol at the low DO setpoint. Consistently, about 2-fold higher carboxylesterase yields (U g-1 biomass) were obtained for the AFT1 overexpressing strain under both conditions. The results of the activity assays were confirmed by SDS-PAGE (Figure 5B). Comparing to a bovine serum albumin (BSA) standard, a carboxylesterase protein level of about 0.75 g L-1 was estimated for strain CE#12-AFT1-A after 100 h of cultivation under hypoxic conditions, while the starting strain produced below 0.5 g L-1 until this time point.
Figure 5.
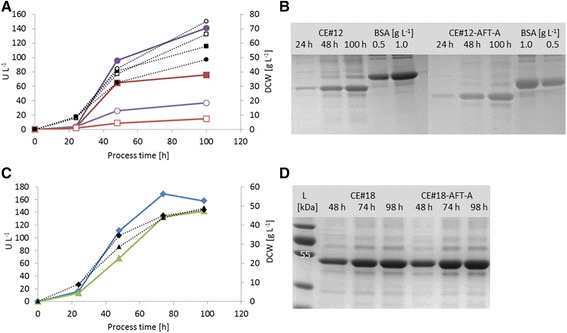
Bioreactor cultivations of improved carboxylesterase-AFT1 P. pastoris strains. (A) Carboxylesterase activity levels (U L-1) of strain CE#12 (red squares) and strain CE#12-AFT1-A (purple circles). Closed symbols: hypoxic conditions; open symbols: aerobic conditions. (C) Carboxylesterase activity levels (U L-1) of strain CE#18 (green triangles) and strain CE#18-AFT1-A (blue rhomboids). Dashed lines: biomass values (DCW, g L-1). (B) and (D) Reducing SDS-PAGE of supernatant samples from different cultivation time points (hypoxic conditions). BSA: bovine serum albumin standard (66.5 kDa).
Also the performance of the multi-copy carboxylesterase AFT1 overexpressing strain CE#18-AFT1-A was investigated in bioreactor under hypoxic conditions (Figure 5C). Again the positive effect of AFT1 overexpression was confirmed. Strain CE#18-AFT1-A reached 169 U L-1 after 74 h of cultivation, while the starting strain (CE#18) only reached 133 U L-1 at the same time point. As before, results were confirmed by SDS-PAGE, showing increased protein levels for strain CE#18-AFT1-A after 74 and 98 h of cultivation (Figure 5D). Comparing yields, secretion per biomass was increased 24% for strain CE#18-AFT1-A. The AFT1 overexpressing strain also yielded 25% higher ethanol levels, reconfirming the involvement of P. pastoris Aft1 in carbon-responsive regulation.
The AFT1 copy number of the overexpressing strains was also investigated. RT-PCR revealed one AFT1 overexpression cassette for strain CE#12-AFT1-A and three for strain CE#18-AFT1-A. Notably, strain CE#18-AFT1-B, which had only one AFT1 overexpression cassette integrated, did not show superior behaviour in bioreactor (data not shown). Seemingly, several AFT1 expression cassettes are necessary to positively influence the secretion of the high-level carboxylesterase producer #18, which might allow for further improvement by targeted AFT1 copy number amplification or the use of stronger promoters for AFT1 overexpression.
Both carboxylesterase-strains, CE#12 and CE#18, were also transformed with an empty vector control plasmid (pPM2aK21-empty). However, carboxylesterase secretion levels were not influenced, neither in shake flask (CE#12-empty) nor in bioreactor cultivations (CE#18-empty).
Conclusions
By analysing the promoter regions of secretion enhancing P. pastoris genes, the ortholog of the S. cerevisiae iron regulators Aft1/2 was selected as a novel factor to improve recombinant protein secretion. A genome-wide analysis of putative Aft1 binding sites in P. pastoris showed Aft1 to be involved in the regulation of many secretory genes, in addition to genes involved in carbohydrate metabolism. The absence of Aft1 binding sites in iron regulatory genes, i.e. FRE1–6 or FET3/FET5, led us to assume iron-independent functions of Aft1 in P. pastoris. These findings were supported by primary amino acid sequence analysis, showing that the DNA binding domain, but not the iron-responsive motif is conserved in the P. pastoris protein. Using DNA microarrays we unveiled further evidence that Aft1 might not directly be required for iron regulation, but is rather involved in regulatory mechanisms in response to carbon source availability, showing e.g. higher transcriptional activation at low glucose concentrations. We also discovered an involvement of P. pastoris Aft1 in the expression of glucose-repressible genes, which needs to be analysed in more detail in future studies. Finally, though microarray data did not reveal differential regulation of any secretion related genes, the secretion enhancing effect of AFT1 was confirmed in overexpressing strains, yielding up to 2.5-fold more secreted carboxylesterase.
Materials and methods
Strains and plasmids
P. pastoris wild type CBS7435 (Centraalbureau voor Schimmelcultures, NL) was used as host strain [50]. The plasmid pPpKan-S-GFP for GFP expression was described by Näätsaari et al. ([GenBank:JQ519694], [51]). Plasmid pPM2dZ30-PGAPα, a derivative of pPUZZLE [6], was used for the expression of a carboxylesterase from Sphingopyxis macrogoltabida (aa residues 48 to 540, [GenBank: ACS27056]) under control of the P. pastoris GAP promoter, with the S. cerevisiae α-MF leader sequence for secretion and a Zeocin resistance marker cassette. Prior to transformation, the expression vector was linearized within the GAP promoter using the restriction enzyme BlnI for homologous integration into the native GAP promoter locus of the P. pastoris genome.
For overexpression of P. pastoris AFT1 the pPUZZLE derived plasmid pPM2aK21 was used, which contains the KanMX4 cassette conferring resistance to Kanamycin/Geneticin (G418), and an AOX1 terminator sequence, which, when linearized with AscI, provides the homologous stretches for integration into the native P. pastoris AOX1 terminator locus. The AFT1 expression cassette, including the AFT1 gene [P. pastoris gene identifier:PAS_chr1-4_0361] and 1000 bp up- and 400 bp downstream sequences, was amplified from P. pastoris genomic DNA using following primers: ApaI-AFT-fw (AAAGGGCCCCCAGGTGAATGTACGTAATGGAG) and AgeI-AFT-rv (TTTACCGGTGGGGAGAAGCCGAATTGGAAG). After ApaI/AgeI digestion, the PCR product was cloned into pPM2aK21, creating pPM2aK21-AFT.
AFT1 gene knock out
A split marker cassette approach was used as described by Heiss et al. [41] to generate transformants with a disrupted AFT1 gene locus. The AFT flanking regions (A upstream, D downstream) were fused to the G418 resistance cassette fragments B and C, respectively, by overlap PCR. Then, equal amounts of both split marker fragments (AB and CD) were pooled and simultaneously transformed into P. pastoris. The G418 resistance gene is reconstituted when both split marker fragments integrate at the correct locus. Verification of positive Δaft1 transformant strains was done by PCR using a primer pair designed to bind on the native AFT1 locus up- and downstream of the split-marker cassette on genomic DNA of Geneticin-resistant transformants. Primers and split marker fragment sizes are shown in Additional file 5.
Transformation
Electrocompetent P. pastoris cells were transformed using the following parameters: 1.5-2.0 kV, 25 μF and 200 Ω. After two hours of regeneration on YP medium, containing 20 g L-1 glucose, selection of positive transformants was done by incubation for 48 h and 28°C on YPD agar plates (20 g L-1 yeast extract, 10 g L-1 peptone, 20 g L-1 glucose, 20 g L-1 agar-agar) supplemented with antibiotic, 50 μg mL-1 Zeocin and/or 450 μg mL-1 Geneticin, respectively.
Media and cultivation
If not stated otherwise chemicals were purchased from BD, Carl Roth, Merck and Sigma Aldrich. YP medium contained 20 g peptone and 10 g yeast extract per liter. Buffered minimal (BM)-medium contained 10 g yeast extract, 10 g peptone, 100 mM potassium phosphate buffer (pH 6.0), 13.4 g yeast nitrogen base without amino acids and 0.4 mg biotin per liter.
Microscale cultivations for GFP expression were performed in 96-well deep-well plates. 300 μL of BM-medium, containing 2% glucose, were inoculated using a toothpick and incubated at 25°C and 360 rpm. The culture was supplemented with glucose to 0.5% at 24 and 36 h of cultivation. Cells were finally harvested after 69 h of cultivation. Methanol induced cultivations were carried out based on the protocol of Weis et al. [52]. Cells were grown for 48 h in 300 μL of BM-medium, containing 1% glucose. Cells were induced by addition of methanol (0.5%) after 48, 56 and 72 h of cultivation. Finally, cells were harvested after 92 h of cultivation.
Shake flask cultivations were performed in 100 mL shake flasks without baffles. As pre-culture, 2.5 mL YP medium, containing 2% glycerol and the respective antibiotic(s), were inoculated in a 50 mL falcon tube and incubated for a minimum of 24 h at 25°C and 180 rpm. The main cultures, containing 10 mL BM-medium with 2% glucose, were inoculated to an OD600 of 0.1. Cultures were then incubated at 25°C and 180 rpm, and supplemented three times with glucose to 0.5% in 12 h intervals. Cells were harvested after 48 h by centrifugation at 4000 rpm. Subsequently, supernatants were analyzed for carboxylesterase production and cell pellets were used for cell weight analysis. Alternatively, OD600 was measured.
Pre-cultures for bioreactor cultivations were performed in 500 mL baffled shake flasks and 50 mL YP medium, containing 2% glucose and 0.01 g L-1 Glanapon DG160 (Bussetti). The main medium used for fed batch cultivations was used as described by Zhao et al. [53], supplemented with biotin and Glanapon DG160. The batch medium consisted of 4 g L-1 KH2PO4, 4 g L-1 (NH4)2SO4, 0.38 g L-1 CaCl2, 18.2 g L-1 K2SO4, 9.4 g L-1 MgSO4.7H2O, 40 g L-1 glucose monohydrate (Tereos Syral), 1 g L-1 Glanapon DG160 and 1 mL L-1 trace element solution. The trace element solution contained 2.50 g L-1 MnSO4.H2O (Riedel-de-Haën), 54.17 g L-1 FeSO4.7H2O (Merck), 16.67 gL-1 ZnCl2.2H2O (Riedel-de-Haën) and 0.17 gL-1 Na2MoO4.2H2O. The batch medium was supplemented with 2 mL of 0.2 g L-1 biotin stock solution per liter medium. The feed medium consisted of 600 g L-1 glucose monohydrate, 2 g L-1 (NH4)2PO4 and 1 g L-1 Glanapon DG160 and was supplemented with 2 mL of biotin stock solution per liter medium.
High cell-density fermentations were carried out in 1 L bioreactors (DASGIP). A starting volume of 500 mL batch medium was inoculated to an OD600 of 0.3. The pH was measured using a glass electrode (Mettler Toledo) and maintained at 5.0 ± 0.1 by automatic addition of 25% ammonium hydroxide (AppliChem). Dissolved oxygen (DO) was monitored using an optical, dissolved oxygen electrode (Hamilton) and maintained at 20% of saturation during the batch phase by a DO cascade of agitation and aeration. Hypoxic conditions were applied during the feed phase. Maximum agitation was 1200 rpm (corresponding to a maximum tip speed of 2.89 m s-1); maximum aeration was set to 1.3 vvm. Temperature was maintained at 25°C.
SDS-PAGE and western blot
10 μL of culture supernatant (containing secreted carboxylesterase enzyme) were run on a reducing sodium dodecyl sulfate (SDS) NuPAGE® 12% Bis-Tris polyacrylamide gel (Life technologies™) with NuPAGE® morpholinepropanesulfonic acid (MOPS) buffer at 180 V for 60 min. Protein bands were visualized using Coomassie staining solution. For Western blotting, SDS-PAGE separated proteins were transferred to a nitrocellulose membrane using the XCell II™ Blot Module for wet (tank) transfer (Life technologies™) according to the manufacturer’s instructions. Carboxylesterase was detected using anti-carboxylesterase antiserum as described by Heinl et al. [48].
Gene copy number determination using real time PCR
Copy numbers were determined by Real time (RT)-PCR as described by Abad et al. [54]. The ABI PRISM 7300 Real Time PCR System and Power SYBR® Green PCR Master Mix were used (Life technologies™). Normalization of the data was achieved using the P. pastoris ARG4 gene as reference. The number of copies per μL was calculated using Avogadro’s number. Following primers were used for amplification: ARG4-RTfw (TCCTCCGGTGGCAGTTCTT), ARG4-RTrv (TCCATTGACTCCCGTTTGAG), AFT-RTfw (GGGCAATATCCAATAGGGCTAA), AFT-RTrv (GGGTGCGCCAAGACTAACA), ZEO-RTfw (CGGCCTGGACGAGCTGTA), ZEO-RTrv (GGCTGCTCGCCGATCTC). Genomic DNA was prepared according to Hoffman and Winston [55].
Fluorescence measurements
GFP fluorescence levels were determined per 50 μL of cell culture. Measurements were performed in microplate using the SynergyMX plate reader (Biotek) applying the following settings: excitation 395 nm and emission 507 nm.
Carboxylesterase activity assay
Enzymatic activity was determined photometrically at 405 nm by hydrolysis of p-Nitrophenyl 2-(trimethylsilyl) ethyl carbonate (pNSi) to p-Nitrophenol + 2-(trimethylsilyl) ethanol + carbon dioxide. The reaction was performed at 37°C in microtiter plates and followed over time using a Tecan Sunrise™ plate reader (XFLUOR4 version: V 4.51). Clarified fermentation supernatants were used undiluted and in several dilutions (10-1 - 10-5) using 1 × FCE buffer. 10 × FCE buffer consisted of 200 mM Tris-Cl, pH 8.0 and 1 mg mL-1 BSA. 100 mM pNSi stock solution was prepared by dissolving 141.68 mg pNSi in 5 mL of 96% ethanol. 1 mM pNSi reaction solution was made by mixing 100 mM pNSi stock solution with 1× FCE buffer. 20 μL of the test samples were provided in a microtiter plate and the reaction was started by addition of 180 μL reaction solution. Absorbance was measured every 30 s for 2 h and activity was determined according to Lambert-Beer’s law.
Iron-dependent growth analysis: spotting assay
The ferrous iron chelator Bathophenanthrolinedisulfonate disodium salt (BPS; Sigma) reduces the amount of free iron in the medium. YP agar plates containing 80 - 200 μM BPS were prepared by addition of BPS to the melted YP or YPD (YP + 2% glucose) agar right before pouring the plates. Methanol (1%) was applied directly in 250 μL volumes onto YP agar plates and spread using a sterile spatula. YNB agar plates containing 200 μM BPS were prepared by addition of BPS to the melted YNB agar right before pouring the plates. YNB agar contained 3.4 g L-1 Yeast Nitrogen Base (Becton Dickinson, NJ), 10 g L-1 ammonium sulfate, 20 g L-1 glucose and 100 mM potassium phosphate buffer (pH 6.0). Cells from a YPD agar plate were resuspended in 1 mL sterile PBS, the optical density (OD600) was determined and set to OD600 = 0.3. Five μL of serial 1:10 dilutions (in sterile PBS) were applied on each agar plate. The plates were then incubated at 30°C for 72 h.
Regulatory sequence analysis tools (RSAT)
RSAT subcategory pattern matching and string genome-scale dna-pattern was used to search for the S. cerevisiae Aft1 and Aft2 binding sites within -1000 bp upstream of the P. pastoris GS115 coding sequences. Default settings were applied.
AmiGO Go Slimmer
AmiGO Go Slimmer (version 1.8) was used to map genes into GO slim terms according to their biological process. SGD was used as database filter (Evidence Code: all) and Yeast GO slim as pre-existing GO slim set (GO database release: 27.10.2012 (analysis of P. pastoris Aft1 binding sites) and 15.07.2013 (analysis of microarray data: Δaft1 vs. control). Advanced results option was used to display gene products and counts for each slim term.
Microarray hybridization and data analysis
For the generation of samples for microarray analysis the Δaft1 strain and the wild type control were cultivated in three biological replicates. Pre-cultures were cultivated as described above. The main cultures, containing 2 mL M2 medium, were inoculated to an optical density OD600 of 4.0 and a 12 mm glucose FeedBead (Kuhner Shaker) was added. The synthetic medium M2 contained per liter: 22.0 g Citric acid monohydrate, 3.15 g (NH4)2PO4, 0.49 g MgSO4*7H2O, 0.80 g KCl, 0.0268 g CaCl2*2H2O, 1.47 mL of PTM1 trace metals and 4 mg Biotin; pH was set to 5 with KOH (solid). Cultures were shaken at 180 rpm and 25°C. Slow release of glucose ensured glucose limited growth. Samples were taken after five h of main culture (estimated specific growth rate: 0.08 h-1), fixed in phenol/ethanol (5% phenol (v/v) in pure ethanol, ice-cold), and stored at -80°C until total RNA extraction.
Total RNA extraction was performed using Trizol as described in Graf et al. [42]. cDNA synthesis and labelling as well as the microarray hybridizations (in-house designed P. pastoris specific oligonucleotide arrays, AMAD-ID 034821, 8x15K custom arrays, Agilent) were carried out according to the Agilent protocols Quick Amp Labelling Kit (Cat. No. 5190-0444) and Gene Expression Hybridisation Kit (Cat. No. 5188-5242) using a reference design. Therefore, each sample was labelled in a dye-swap manner and hybridized against a reference cDNA, which was generated from a pool of cells grown under different culture conditions. Normalization steps and statistical analysis of microarray data included removal of color bias using locally weighted MA-scatterplot smoothing (LOESS) followed by between array normalization using the “Aquantile” method. For identifying differentially expressed genes and calculating p-values a linear model fit with an eBayes correction was used. P-values were adjusted for multiple testing with the false discovery method (FDR) by Benjamini & Yekutieli. For identifying differentially expressed genes, a fold change cut-off of at least 1.5 > FC >1/1.5 was applied. All steps were done using the R software (http://www.rproject.org) and the limma package. The expression changes of some genes selected based on their regulation pattern was confirmed using quantitative real time PCR (Additional file 6).
Acknowledgements
This work has been supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT - Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG. Further support by Biomin Research Center, Boehringer-Ingelheim RCV, Lonza AG, Biocrates Life Sciences AG, VTU Technology GmbH, and Sandoz GmbH is acknowledged. The carboxylesterase activity assay with pNSi as substrate originated at evocatal GmbH, Monheim am Rhein, Germany. The authors thank Dr. Alexandra B. Graf (University of Applied Sciences - FH Campus Vienna) for the processing of the microarray data, Dr. Minoska Valli (ACIB) for help with P. pastoris gene annotation, and Gabriele Wilt (ACIB) and Elena Pietrich (ACIB) for their excellent technical support.
Additional files
P. pastoris genes with putative Aft1 binding site(s). Search field: -1000 bp upstream of the P. pastoris GS115 coding sequences. Binding motifs: ScAft1 (ANTGCACCC) and ScAft2 (BRCACCCB). Genes were categorized into biological function GO-Terms using AmiGO GO Slimmer (Yeast GO slim set, [25]). Genes not found in the database and removed from the calculation: FLO104, ATG30, SHL23 and FEP1.
Aft1 protein sequence comparison (ClustalW2, [ 37 ]). Amino acid sequences: P. pastoris Aft1 [NCBI protein:CCA37276] and H. polymorpha HPODL_04658 [NCBI protein: ESX01890]; asterisk: “indicates positions which have a single, fully conserved residue”, colon: “indicates conservation between groups of strongly similar properties”; period: “indicates conservation between groups of weakly similar properties”; red boxed: conserved residues (Cys45*, Cys102, His126 and His128), shaded grey: Aft domain (pfam_ls:AFT, Motif Scan-My Hits-SIB), *numbering is based on PpAft1.
List of significantly regulated genes in Δ aft1 compared to wild type. GO term enrichment was analysed using Amigo GO Slimmer [25]. Significantly regulated GO terms were determined by GO term Finder. List of homologs of the S. cerevisiae iron regulon which are not regulated in Δaft1 in P. pastoris.
SDS-PAGE of carboxylesterase secreting P. pastoris strains. Clones #8-20: CBS7435 transformed with plasmid pPM2dZ30-PGAP-CE; negative control: CBS7435.
Primers used for the construction of the split marker cassette for AFT1 disruption in P. pastoris.
Primers used for the verification of gene regulation in Δ aft1 and wild type. Comparison of gene regulation patterns from microarrays and qPCR for genes selected based on their regulatory behaviour.
Footnotes
Claudia Ruth and Markus Buchetics contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CR, SN, DM, HP and BG participated in the design of the study. CR and VV carried out the sequence analysis, promoter and overexpression studies. Bioreactor cultivations were performed by DK. MB and BG carried out the DNA microarrays and data analysis. CR and MB drafted the manuscript. DK, SN, DM, HP and BG revised the manuscript. All authors read and approved the final manuscript.
Contributor Information
Claudia Ruth, Email: claudia.ruth@acib.at.
Markus Buchetics, Email: markus.buchetics@acib.at.
Viktorija Vidimce, Email: viktorija.vidimce@acib.at.
Daniela Kotz, Email: daniela.kotz@biomin.net.
Stefan Naschberger, Email: stefan.naschberger@biomin.net.
Diethard Mattanovich, Email: diethard.mattanovich@boku.ac.at.
Harald Pichler, Email: harald.pichler@tugraz.at.
Brigitte Gasser, Email: brigitte.gasser@boku.ac.at.
References
- 1.Gasser B, Prielhofer R, Marx H, Maurer M, Nocon J, Steiger M, Puxbaum V, Sauer M, Mattanovich D. Pichia pastoris: protein production host and model organism for biomedical research. Future Microbiol. 2013;8:191–208. doi: 10.2217/fmb.12.133. [DOI] [PubMed] [Google Scholar]
- 2.Damasceno LM, Anderson KA, Ritter G, Cregg JM, Old LJ, Batt CA. Cooverexpression of chaperones for enhanced secretion of a single-chain antibody fragment in Pichia pastoris. Appl Microbiol Biotechnol. 2007;74:381–389. doi: 10.1007/s00253-006-0652-7. [DOI] [PubMed] [Google Scholar]
- 3.Gasser B, Sauer M, Maurer M, Stadlmayr G, Mattanovich D. Transcriptomics-based identification of novel factors enhancing heterologous protein secretion in yeasts. Appl Environ Microbiol. 2007;73:6499–6507. doi: 10.1128/AEM.01196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guerfal M, Ryckaert S, Jacobs PP, Ameloot P, Van Craenenbroeck K, Derycke R, Callewaert N. The HAC1 gene from Pichia pastoris: characterization and effect of its overexpression on the production of secreted, surface displayed and membrane proteins. Microb Cell Fact. 2010;9:49. doi: 10.1186/1475-2859-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasser B, Maurer M, Gach J, Kunert R, Mattanovich D. Engineering of Pichia pastoris for improved production of antibody fragments. Biotechnol Bioeng. 2006;94:353–361. doi: 10.1002/bit.20851. [DOI] [PubMed] [Google Scholar]
- 6.Stadlmayr G, Benakovitsch K, Gasser B, Mattanovich D, Sauer M. Genome-scale analysis of library sorting (GALibSo): isolation of secretion enhancing factors for recombinant protein production in Pichia pastoris. Biotechnol Bioeng. 2010;105:543–555. doi: 10.1002/bit.22573. [DOI] [PubMed] [Google Scholar]
- 7.Blaiseau PL, Lesuisse E, Camadro JM. Aft2p, a novel iron-regulated transcription activator that modulates, with Aft1p, intracellular iron use and resistance to oxidative stress in yeast. J Biol Chem. 2001;276:34221–34226. doi: 10.1074/jbc.M104987200. [DOI] [PubMed] [Google Scholar]
- 8.Rutherford JC, Jaron S, Winge DR. Aft1p and Aft2p mediate iron-responsive gene expression in yeast through related promoter elements. J Biol Chem. 2003;278:27636–27643. doi: 10.1074/jbc.M300076200. [DOI] [PubMed] [Google Scholar]
- 9.Rutherford JC, Ojeda L, Balk J, Mühlenhoff U, Lill R, Winge DR. Activation of the iron regulon by the yeast Aft1/Aft2 transcription factors depends on mitochondrial but not cytosolic iron-sulfur protein biogenesis. J Biol Chem. 2005;280:10135–10140. doi: 10.1074/jbc.M413731200. [DOI] [PubMed] [Google Scholar]
- 10.Yamaguchi-lwai Y, Dancis A, Klausner RD. AFT1: a mediator of iron regulated transcriptional control in Saccharomyces cerevisiae. EMBO J. 1995;14:1231–1239. doi: 10.1002/j.1460-2075.1995.tb07106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yamaguchi-Iwai Y, Stearman R, Dancis A, Klausner RD. Iron-regulated DNA binding by the AFT1 protein controls the iron regulon in yeast. EMBO J. 1996;15:3377–3384. [PMC free article] [PubMed] [Google Scholar]
- 12.Ueta R, Fujiwara N, Iwai K, Yamaguchi-Iwai Y. Iron-induced dissociation of the Aft1p transcriptional regulator from target gene promoters is an initial event in iron-dependent gene suppression. Mol Cell Biol. 2012;32:4998–5008. doi: 10.1128/MCB.00726-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ueta R, Fujiwara N, Iwai K, Yamaguchi-iwai Y. Mechanism underlying the iron-dependent nuclear export of the iron-responsive transcription factor Aft1p in Saccharomyces cerevisiae. Mol Cell Biol. 2007;18:2980–2990. doi: 10.1091/mbc.E06-11-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Conde e Silva N, Gonçalves IR, Lemaire M, Lesuisse E, Camadro JM, Blaiseau PL. KlAft, the Kluyveromyces lactis ortholog of Aft1 and Aft2, mediates activation of iron-responsive transcription through the PuCACCC Aft-type sequence. Genetics. 2009;183:93–106. doi: 10.1534/genetics.109.104364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haas H, Eisendle M, Turgeon BG. Siderophores in fungal physiology and virulence. Annu Rev Phytopathol. 2008;46:149–187. doi: 10.1146/annurev.phyto.45.062806.094338. [DOI] [PubMed] [Google Scholar]
- 16.Miele R, Barra D, di Patti MC B. A GATA-type transcription factor regulates expression of the high-affinity iron uptake system in the methylotrophic yeast Pichia pastoris. Arch Biochem Biophys. 2007;465:172–179. doi: 10.1016/j.abb.2007.05.020. [DOI] [PubMed] [Google Scholar]
- 17.Lan C-Y, Rodarte G, Murillo LA, Jones T, Davis RW, Dungan J, Newport G, Agabian N. Regulatory networks affected by iron availability in Candida albicans. Mol Microbiol. 2004;53:1451–1469. doi: 10.1111/j.1365-2958.2004.04214.x. [DOI] [PubMed] [Google Scholar]
- 18.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 19.Gasser B, Saloheimo M, Rinas U, Dragosits M, Rodríguez-Carmona E, Baumann K, Giuliani M, Parrilli E, Branduardi P, Lang C, Porro D, Ferrer P, Tutino ML, Mattanovich D, Villaverde A. Protein folding and conformational stress in microbial cells producing recombinant proteins: a host comparative overview. Microb Cell Fact. 2008;7:11. doi: 10.1186/1475-2859-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Idiris A, Tohda H, Kumagai H, Takegawa K. Engineering of protein secretion in yeast: strategies and impact on protein production. Appl Microbiol Biotechnol. 2010;86:403–417. doi: 10.1007/s00253-010-2447-0. [DOI] [PubMed] [Google Scholar]
- 21.Vogl T, Thallinger GG, Zellnig G, Drew D, Cregg JM, Glieder A, Freigassner M. Towards improved membrane protein production in Pichia pastoris: general and specific transcriptional response to membrane protein overexpression. N Biotechnol. 2014;ᅟ:ᅟ. doi: 10.1016/j.nbt.2014.02.009. [DOI] [PubMed] [Google Scholar]
- 22.Daum G, Tuller G, Nemec T, Hrastnik C, Balliano G, Cattel L, Milla P, Rocco F, Conzelmann A, Vionnet C, Kelly DE, Kelly S, Schweizer E, Schüller HJ, Hojad U, Greiner E, Finger K. Systematic analysis of yeast strains with possible defects in lipid metabolism. Yeast. 1999;15:601–614. doi: 10.1002/(SICI)1097-0061(199905)15:7<601::AID-YEA390>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 23.Thomas-Chollier M, Defrance M, Medina-Rivera A, Sand O, Herrmann C, Thieffry D, van Helden J. RSAT 2011: regulatory sequence analysis tools. Nucleic Acids Res. 2011;39(Web Server issue):W86–W91. doi: 10.1093/nar/gkr377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mattanovich D, Callewaert N, Rouzé P, Lin Y-C, Graf A, Redl A, Tiels P, Gasser B, De Schutter K. Open access to sequence: browsing the Pichia pastoris genome. Microb Cell Fact. 2009;8:53. doi: 10.1186/1475-2859-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. the Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li L, Chen OS, McVey Ward D, Kaplan J. CCC1 is a transporter that mediates vacuolar iron storage in yeast. J Biol Chem. 2001;276:29515–29519. doi: 10.1074/jbc.M103944200. [DOI] [PubMed] [Google Scholar]
- 27.Greene JR, Brown NH, DiDomenico BJ, Kaplan J, Eide DJ. The GEF1 gene of Saccharomyces cerevisiae encodes an integral membrane protein; mutations in which have effects on respiration and iron-limited growth. Mol Gen Genet. 1993;241:542–553. doi: 10.1007/BF00279896. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Yukl ET, Moënne-Loccoz P, Montellano PR. Fungal heme oxygenases: functional expression and characterization of Hmx1 from Saccharomyces cerevisiae and CaHmx1 from Candida albicans. Biochemistry. 2006;45:14772–14780. doi: 10.1021/bi061429r. [DOI] [PubMed] [Google Scholar]
- 29.Collinson EJ, Wimmer-Kleikamp S, Gerega SK, Yang YH, Parish CR, Dawes IW, Stocker R. The yeast homolog of heme oxygenase-1 affords cellular antioxidant protection via the transcriptional regulation of known antioxidant genes. J Biol Chem. 2011;286:2205–2214. doi: 10.1074/jbc.M110.187062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schilke B, Voisine C, Beinert H, Craig E. Evidence for a conserved system for iron metabolism in the mitochondria of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1999;96:10206–10211. doi: 10.1073/pnas.96.18.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee MCS, Miller EA. Molecular mechanisms of COPII vesicle formation. Semin Cell Dev Biol. 2007;18:424–434. doi: 10.1016/j.semcdb.2007.06.007. [DOI] [PubMed] [Google Scholar]
- 32.Römisch K. Surfing the Sec61 channel: bidirectional protein translocation across the ER membrane. J Cell Sci. 1999;112(Pt 2):4185–4191. doi: 10.1242/jcs.112.23.4185. [DOI] [PubMed] [Google Scholar]
- 33.McNew JA, Coe JG, Søgaard M, Zemelman BV, Wimmer C, Hong W, Söllner TH. Gos1p, a Saccharomyces cerevisiae SNARE protein involved in Golgi transport. FEBS Lett. 1998;435:89–95. doi: 10.1016/S0014-5793(98)01044-8. [DOI] [PubMed] [Google Scholar]
- 34.Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vervecken W, Kaigorodov V, Callewaert N, Geysens S, De Vusser K, Contreras R. In vivo synthesis of mammalian-like, hybrid-type N-glycans in pichia pastoris. ᅟ. 2004;70:2639–2646. doi: 10.1128/AEM.70.5.2639-2646.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Elbert M, Rossi G, Brennwald P. The yeast par-1 homologs kin1 and kin2 show genetic and physical interactions with components of the exocytic machinery. Mol Biol Cell. 2005;16:532–549. doi: 10.1091/mbc.E04-07-0549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 38.Rutherford JC, Jaron S, Ray E, Brown PO, Winge DR. A second iron-regulatory system in yeast independent of Aft1p. Proc Natl Acad Sci U S A. 2001;98:14322–14327. doi: 10.1073/pnas.261381198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Babu MM, Iyer LM, Balaji S, Aravind L. The natural history of the WRKY-GCM1 zinc fingers and the relationship between transcription factors and transposons. Nucleic Acids Res. 2006;34:6505–6520. doi: 10.1093/nar/gkl888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson JD. Molecular Biology of the Cell, 3rd Addition. New York: Garland Science; 1994. [Google Scholar]
- 41.Heiss S, Maurer M, Hahn R, Mattanovich D, Gasser B. Identification and deletion of the major secreted protein of Pichia pastoris. Appl Microbiol Biotechnol. 2013;97:1241–1249. doi: 10.1007/s00253-012-4260-4. [DOI] [PubMed] [Google Scholar]
- 42.Graf A, Gasser B, Dragosits M, Sauer M, Leparc GG, Tüchler T, Kreil DP, Mattanovich D. Novel insights into the unfolded protein response using Pichia pastoris specific DNA microarrays. BMC Genomics. 2008;9:390. doi: 10.1186/1471-2164-9-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baumann K, Carnicer M, Dragosits M, Graf AB, Stadlmann J, Jouhten P, Maaheimo H, Gasser B, Albiol J, Mattanovich D, Ferrer P. A multi-level study of recombinant Pichia pastoris in different oxygen conditions. BMC Syst Biol. 2010;4:141. doi: 10.1186/1752-0509-4-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nocon J, Steiger MG, Pfeffer M, Sohn SB, Kim TY, Maurer M, Rußmayer H, Pflügl S, Ask M, Haberhauer-Troyer C, Ortmayr K, Hann S, Koellensperger G, Gasser B, Lee SY, Mattanovich D. Model based engineering of Pichia pastoris central metabolism enhances recombinant protein production. Metab Eng. 2014;24:129–138. doi: 10.1016/j.ymben.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 46.Hartner FS, Ruth C, Langenegger D, Johnson SN, Hyka P, Lin-Cereghino GP, Lin-Cereghino J, Kovar K, Cregg JM, Glieder A. Promoter library designed for fine-tuned gene expression in Pichia pastoris. Nucleic Acids Res. 2008;36:e76. doi: 10.1093/nar/gkn369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartinger D, Moll W-D. Fumonisin elimination and prospects for detoxification by enzymatic transformation. World Mycotoxin J. 2011;4:271–283. doi: 10.3920/WMJ2011.1285. [DOI] [Google Scholar]
- 48.Heinl S, Hartinger D, Thamhesl M, Vekiru E, Krska R, Schatzmayr G, Moll W-D, Grabherr R. Degradation of fumonisin B1 by the consecutive action of two bacterial enzymes. J Biotechnol. 2010;145:120–129. doi: 10.1016/j.jbiotec.2009.11.004. [DOI] [PubMed] [Google Scholar]
- 49.Baumann K, Maurer M, Dragosits M, Cos O, Ferrer P, Mattanovich D. Hypoxic fed-batch cultivation ofPichia pastoris increases specific and volumetric productivity of recombinant proteins. Biotechnol Bioeng. 2008;100:177–183. doi: 10.1002/bit.21763. [DOI] [PubMed] [Google Scholar]
- 50.Küberl A, Schneider J, Thallinger GG, Anderl I, Wibberg D, Hajek T, Jaenicke S, Brinkrolf K, Goesmann A, Szczepanowski R, Pühler A, Schwab H, Glieder A, Pichler H. High-quality genome sequence of Pichia pastoris CBS7435. J Biotechnol. 2011;154:312–320. doi: 10.1016/j.jbiotec.2011.04.014. [DOI] [PubMed] [Google Scholar]
- 51.Näätsaari L, Mistlberger B, Ruth C, Hajek T, Hartner FS, Glieder A. Deletion of the Pichia pastoris KU70 homologue facilitates platform strain generation for gene expression and synthetic biology. PLoS One. 2012;7:e39720. doi: 10.1371/journal.pone.0039720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Weis R, Luiten R, Skranc W, Schwab H, Wubbolts M, Glieder A. Reliable high-throughput screening with Pichia pastoris by limiting yeast cell death phenomena. FEMS Yeast Res. 2004;5:179–189. doi: 10.1016/j.femsyr.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 53.Zhao W, Wang J, Deng R, Wang X. Scale-up fermentation of recombinant Candida rugosa lipase expressed in Pichia pastoris using the GAP promoter. J Ind Microbiol Biotechnol. 2008;35:189–195. doi: 10.1007/s10295-007-0283-8. [DOI] [PubMed] [Google Scholar]
- 54.Abad S, Kitz K, Hörmann A, Schreiner U, Hartner FS, Glieder A. Real-time PCR-based determination of gene copy numbers in Pichia pastoris. Biotechnol J. 2010;5:413–420. doi: 10.1002/biot.200900233. [DOI] [PubMed] [Google Scholar]
- 55.Hoffman CS, Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]


