Abstract
Background
Studies of insect-plant interactions have provided critical insights into the ecology and evolution of adaptive processes within and among species. Cactophilic Drosophila species have received much attention because larval development occurs in the necrotic tissues of cacti, and both larvae and adults feed on these tissues. Such Drosophila-cactus interactions include effects of the host plant on the physiology and behavior of the flies, especially so their nutritional status, mating condition and reproduction. The aim of this work was to compare the transcriptional responses of two species, Drosophila antonietae and Drosophila meridionalis, and identify genes potentially related to responses to odors released by their host cactus, Cereus hildmannianus. The two fly species are sympatric in most of their populations and use this same host cactus in nature.
Results
We obtained 47 unique sequences (USs) for D. antonietae in a suppression subtractive hybridization screen, 30 of these USs had matches with genes predicted for other Drosophila species. For D. meridionalis we obtained 81 USs, 46 of which were orthologous with genes from other Drosophila species. Functional information (Gene Ontology) revealed that these differentially expressed genes are related to metabolic processes, detoxification mechanisms, signaling, response to stimuli, and reproduction. The expression of 13 genes from D. meridionalis and 12 from D. antonietae were further analyzed by quantitative real time-PCR, showing that four genes were significantly overexpressed in D. antonietae and six in D. meridionalis.
Conclusions
Our results revealed the differential expression of genes related to responses to odor stimuli by a cactus, in two associated fly species. Although the majority of activated genes were similar between the two species, we also observed that certain metabolic pathways were specifically activated, especially those related to signaling pathways and detoxification mechanisms. The activation of these genes may reflect different metabolic pathways used by these flies in their interaction with this host cactus. Our findings provide insight into how the use of C. hildmannianus may have arisen independently in the two fly species, through genetic differentiation in metabolic pathways to effectively explore this cactus as a host.
Electronic supplementary material
The online version of this article (doi:10.1186/s12862-014-0191-2) contains supplementary material, which is available to authorized users.
Keywords: Drosophila-cactus interaction, Differential gene expression, Suppression subtractive hybridization, Adaptation
Background
Interactions between insects and their host plants are highly diverse and are an important area of study in evolutionary ecology [1-3]. Examples of these interactions include the effects of host plants on the physiology and behavior of insects, which in turn develop strategies to improve their nutritional status, mating conditions and reproductive success [4,5]. Interest into whether and how differentially expressed genes could be involved in this adaptive process has been a driver for an increasing number of studies aiming to understand the ecological and evolutionary mechanisms underlying these insect-plant interactions [6-9]. The extent and nature of these interactions can now be examined using hypothesis-driven transcriptome analyses based on an ecological perspective [10].
The substantial amount of publically available genomic information on drosophilids, including in-depth data on their evolutionary history and ecological niches, facilitates the study of these questions. Among the drosophilids, the cactophilic species belonging to the Repleta group breed and feed on cactus necrotic tissue [11]. The vast majority of these species are saprophytic, feeding as larvae and adults mainly on yeasts and bacteria growing in decaying tissue of cacti. While yeasts are a major source of nutrients for these drosophilids, these flies are then also exposed to xenobiotic compounds present on the host cactus [12]. Thus, cues for finding the appropriate cactus and necrotic tissue by drosophilids may be an important determinant for their feeding strategies and, ultimately, their reproductive fitness.
For many cactophilic Drosophila species the chemical composition of their cactus hosts is one of the major determinants of host-plant specificity [13,14], because of the presence of potentially toxic compounds, such as alkaloids, medium-chain fatty acids, sterol diols, and triterpene glycosides, especially so in columnar cacti [15]. The chemical composition of necrotic cactus tissues is largely dependent on the yeast and bacterial communities present, which metabolize these compounds and produce volatiles that are important cues for the flies to detect possible host plants [16-19].
Among the adaptive genetic changes presented by the Drosophila species that use these cacti, primary targets should be detoxification processes to remove or tolerate toxic compounds produced by their host cacti and their microorganism communities [13,20-22]. Furthermore, several studies have demonstrated an influence of these host cacti on fitness-related traits, such as developmental time and viability [23], and on cuticular hydrocarbons [24]. Nonetheless, there is little information on the genetic basis underlying the perception of host cacti necrotic tissue by different Drosophila species.
In this study, we used a suppression subtractive hybridization (SSH) approach to compare transcriptional responses of Drosophila antonietae and Drosophila meridionalis females and identify genes potentially related to responses to odor stimuli released by the columnar cactus Cereus hildmannianus. Both species are cactophilic and belong to the Repleta group. Within this group, Drosophila meridionalis belongs to the meridiana complex, a monophyletic basal subdivision of the Repleta group, and Drosophila antonieta belongs to the buzzatii complex, which is a monophyletic derived subdivision [25]. The two species have the same geographic range, which includes the core Missiones region (Paraná-Paraguay basin river), the South Athantic Seasonal Forest and the eastern part of the Chaco Biome in South America [26-29] and are sympatric in most of their populations. As these species belong to different lineages of the Repleta group and use the same host plant, an interesting question is to understand the genetic pathways and their evolution involved in the exploration of Cereus hildmaniannus as a host, which is considered a relevant factor in the diversification of the group [25,30].
Our study revealed the differential expression of a set of genes related to metabolic processes, detoxification mechanisms, signaling, responses to stimuli, and reproduction. We found that a portion of these genes are similar among the two species, while others are different, and we discuss possible functional roles of these genes in the interaction between Drosophila flies and their host cactus.
Materials and methods
Fly stocks
Both species of cactophilic drosophilids, D. antonietae (buzzatii cluster) and D. meridionalis (meridiana complex), were maintained in the Laboratory of Evolutionary Genetics of the University of São Paulo, Ribeirão Preto, Brazil. These flies were obtained by collecting decaying cladodes of the cactus Cereus hildmannianus from the Experimental Station at Sertãozinho of the Institute of Animal Science and Pastures (IZ), São Paulo, Brazil. The cladodes were placed in a terrarium to allow the flies to emerge. These were then collected, identified and transferred to glass bottles containing culture medium (Additional file 1). From these initial cultures, single females were placed into individual vials to establish isofemale lines. Species identification was refined by analysis of the genitalia (aedeagus) of the male progeny [31], and 10 isofemale lines from each species were mixed to establish the test population used in this study. Populations of both species were reared under identical conditions for 14 generations before the odor exposure tests were conducted.
Experimental design
Females of the two fly species that had previously been kept in the presence of conspecific males to enable mating were removed from their culture medium (Additional file 1) 24 h before the start of the experiment and deprived of food (they were placed in the middle of a dish containing 1% agar). From these flies, experimental units were formed consisting of 50 mature female flies (10 to 15 days old) kept in a sealed Erlenmeyer flask. The flasks were maintained in an incubator under controlled conditions (25 ± 1°C, R.H. of 70%) in the dark. Subsequently, the flies of the experimental units were either exposed or not exposed (control) to odor of the cactus C. hildmannianus for 6 hours (Figure 1). The cactus odor was produced by pieces of fresh cactus and liquid exudate of decaying cladodes from cacti previously collected at the same site as described above. These decaying cladodes had been kept for three days in the laboratory at −20°C, in plastic containers covered by gaze. After a 6 h exposure period to cactus odor, the female flies were immediately frozen in liquid nitrogen and stored at −80°C.
Figure 1.
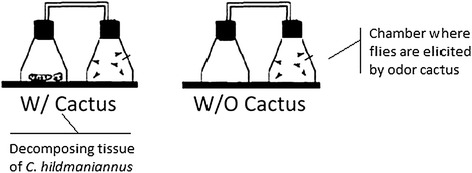
Schematic representation of the experimental set up. Flies were placed in 250 ml Erlenmeyer flasks connected by a hose to another Erlenmeyer flask containing decomposing tissue of C. hildmaniannus cactus. Flies were prevented by a small pore net from entering the flask containing cactus tissue.
Analysis of differentially expressed genes (DEGs)
mRNA extraction and cDNA synthesis
RNA was extracted from whole fly bodies using TRIzol reagent (Invitrogen) followed by purification with the Pure Link RNA Mini Kit (Invitrogen). The samples were treated with DNase I (Thermo Scientific) to remove genomic DNA from the samples. RNA quantity and purity were estimated by measuring the absorbance at 260 nm and the ratio of the absorbance at 260 and 280 nm, respectively, in a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). One microgram of total RNA was used to generate cDNA using the PCR cDNA Synthesis Kit SMARTer (Clontech) according to the manufacturer’s recommendations.
Suppressive subtraction protocol
Differentially expressed genes were identified by means of a suppressive subtraction protocol [32] using the PCR-Select kit (Clontech). This approach permits the enrichment of differentially expressed genes while removing genes that are shared across experimental groups. Briefly, tester cDNA (cDNA from flies that had been exposed to cactus odor) and driver cDNA (cDNA from flies that had not been exposed to cactus odor) was subjected to blunt-end digestion by RsaI restriction enzyme. Tester cDNA was split into two equimolar aliquots to which different PCR primer adapter sequences were ligated (adapter A for one pool of cDNA and adapter B for the other); the primer sequences are described in the PCR-Select kit manual (Clontech). No adapters were ligated to the respective driver cDNA samples. After ligation, the tester cDNAs were mixed with an excess (1:30) of driver cDNA for hybridization. Tester sequences with adapter A that remained single-stranded (did not hybridize to driver cDNA) after the first hybridization round were then used to hybridize with cDNA containing adapter B in the presence of driver cDNA (1:200). Adaptor A-B cDNA hybrids were then selectively amplified by PCR. This procedure allows for an enrichment of the population of cDNA that is specific to (differentially represented in) the respective tester cDNA sample (Figure 2).
Figure 2.
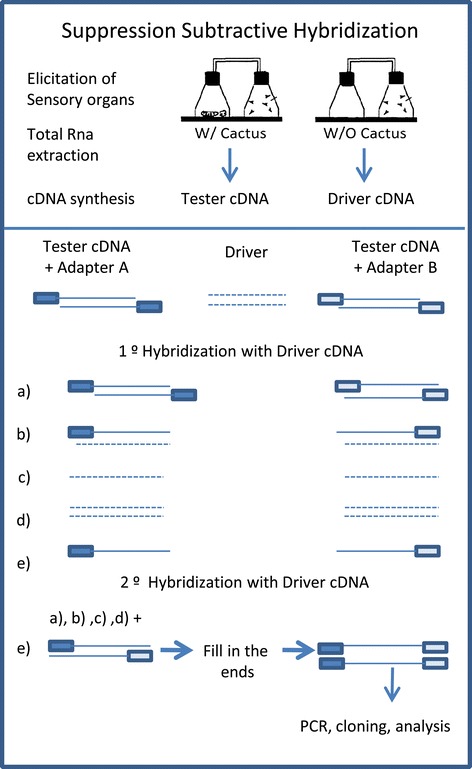
Schematic diagram of the Suppression Subtractive Hybridization protocol. Source: Clontech manual modified. Tester cDNA was divided into two aliquots, one for ligation with adapter A and the second with adapter B. Each half was then hybridized with driver cDNA that was not ligated to any adapter. Single-stranded (non-hybridized) sequences of the two independent hybridization steps were then mixed in a second hybridization step generating A-B hynrids. Primers to these adapters were then used for the selective amplification of these hybrids by PCR. This procedure allows for the enrichment of the cDNA population that is overrepresented (differentially represented) in the respective tester cDNA sample.
PCR products from the subtracted libraries were then ligated into pCR2.1 TOPO TA vector (Invitrogen) for transformation into electrocompetent TOP10 E. coli cells (Invitrogen); the transformants were grown on LB-agar media under ampicillin selection. Positive transformants detected by β-galactosidase expression were picked and individually grown in liquid LB-ampicillin medium. For sequencing of the cDNA inserts, the cells were lysed and the plasmids extracted and sequenced using a Big Dye Terminator v1.1 Cycle Sequencing kit (Applied Biosciences) in an automatic sequencer model ABI 3730 XL DNA Analyzer (Applied Biosystems, Foster City, CA).
Bioinformatics analysis
The sequencing reads were entered into an E-Gene annotation pipeline [33], where they were first filtered to detect and remove reads corresponding to ribosomal and mitochondrial genes. Subsequently, vector sequences were trimmed using the Crossmatch program module implemented in the E-Gene pipeline. Read quality was checked and the reads were assembled using the Phred-Phrap program module. Reads passing these quality checks were next submitted to a CAP3 assembly procedure [34] to obtain unique sequences (USs). These contigs or singlets were then dynamically translated and used in blastx queries against a non-redundant (nr) database (GenBank). The E-value cutoff was set to ≤10−5, which is a frequently used value in SSH approaches [35]. ESTs with similarities to genes of known function in other organisms were clustered using Gene Ontology (GO) terms attributed to their respective orthologs in D. melanogaster and D. mojavensis. Homology and Gene Ontology terms [36], enzyme classification codes (EC), and metabolic pathway analyses of the assembled sequences were retrieved using the BLAST2GO software [37].
Differential gene expression analysis by quantitative RT-PCR
SSH strategies enrich for differentially expressed genes (DEGs) and, therefore, are not strictly quantitative representations. Differences in expression among identified DEGs must, thus, be validated against independent biological samples. To this end, we performed real-time qRT-PCR analyses with gene-specific primers for a set of selected USs designed by Primer Express Version 3.0 software (Applied Biosystems). The predicted product length varied between 180 bp and 200 bp. An α-tubulin gene recommended for Drosophila real time qRT-PCR studies [38] was used as reference gene. In addition, we designed primers to a region of the α-tubulin gene at 84B (CG1913) that is conserved among Drosophila species. Primer specificity was tested by conventional PCR. The primer sequences are listed in Additional files 2 and 3.
Fly RNA was extracted with TRIzol followed by purification through a PureLink RNA Mini Kit (Invitrogen) and treated with DNase I (Thermo Scientific) to eliminate possible DNA contaminants. First-strand cDNA was produced from approximately 400 ng of total RNA using a SuperScript II kit (Invitrogen) in a protocol consisting of an incubation at 42°C for 50 min and 70°C for 15 min. The resulting cDNA was diluted five times for use in the real time qRT-PCR assays, which were conducted using SYBR Green PCR Master Mix according to the manufacturer’s protocol (Applied Biosystems). The reaction volume was 14 μl and contained 7 μl of SYBR Green reagent, 1.4 μl of the respective cDNA, 0.7 μl of forward and reverse primers (both at a concentration of 5 μM), and 4.2 μl of nuclease-free water. The amplification reactions were performed in a StepOnePlus system (Applied Biosystems) using the following protocol: 95°C for 20 s followed by 40 cycles of 95°C for 3 s and 60°C for 30 s. Subsequently, dissociation curves were acquired to evaluate amplification product quality by melting peak analysis. The assays were run with the three biological replicates, each as three technical replicates, and with assay controls consisting of non-template samples. The real time qRT-PCR results were analyzed using LinReg software [39]. For statistical analysis, Ct values were used as inputs for randomization tests implemented in the Relative Expression Software Tool (REST) [40], with P ≤ 0.05 considered as statistically significant.
Results
Differential expression library characteristics
The SSH libraries from the two Drosophila species contained genes enriched in expression in flies exposed to C. hildmannianus cactus odor relative to the non-exposed control flies. Among a total of 192 expressed sequence tag (EST) reads for D. antonietae, 149 were of high quality (Phred quality ≥20). After assembly, these ESTs resulted in 29 contigs and 18 singlets (47 Unique Sequences, USs). A blastx analysis of these USs showed that 30 out of 47 sequences (63.8%) had significant similarity with genes from the 12 Drosophila species with completely sequenced genomes [41], and among these, 24 (80%) were associated with known GO functional categories (Additional file 4).
A total of 336 sequencing reads were obtained in the D. meridionalis SSH library, 219 of which were of high quality (Phred quality ≥20). These sequences were assembled into 32 contigs and 49 remaining singlets (81 USs). A total of 46 out of 81 sequences (56.8%) had significant matches with genes from the 12 Drosophila species, and for these we could also retrieve GO terms. The remaining 35 genes (43.2%) had no significant matches in GenBank and consequently, also lack GO attributes (Additional file 5). The USs that showed significant matches with genes from Drosophila species were submitted to GenBank (JZ544508-JZ544575).
The functional characterization and grouping of DEGs for the two species are shown in the Tables 1 and 2. The results were inferred by using the BLAST2GO program [37]. The majority of the sequences for both D. antonietae and D. meridionalis were related to the following molecular function GO categories (level 3): ligation (GO:0005488), involving ligation to ions, nucleic acids, proteins, small molecules, cofactors, and transcription factors; catalytic activity (GO:0003824), such transferase, hydrolase,and oxidoreductase; structural molecules (GO:0005198), such as ribosome and nuclear pore constituents; and transporter activity (GO:0005215), including the transport of specific substrates and transmembrane transport (Table 1).
Table 1.
Classification of differentially expressed genes (DEGs) according to Gene Ontology (GO) molecular function attributes
| a). Drosophila antonietae | ||
|---|---|---|
| GO-id | GO-term | Score |
| GO:0000989 | Transcription factor binding transcription factor activity | 1.0 |
| GO:0036094 | Small molecule binding | 4.0 |
| GO:0097367 | Carbohydrate derivative binding | 3.0 |
| GO:0016787 | Hydrolase activity | 4.0 |
| GO:0030337 | DNA polymerase processivity factor activity | 1.0 |
| GO:0003735 | Structural constituent of ribosome | 1.0 |
| GO:0016491 | Oxidoreductase activity | 2.0 |
| GO:0061134 | Peptidase regulator activity | 1.0 |
| GO:0043167 | Ion binding | 8.0 |
| GO:0097159 | Organic cyclic compound binding | 7.0 |
| GO:0016853 | Isomerase activity | 1.0 |
| GO:0004857 | Enzyme inhibitor activity | 1.0 |
| GO:0048037 | Cofactor binding | 1.0 |
| GO:0016829 | Lyase activity | 1.0 |
| GO:0005515 | Protein binding | 3.0 |
| GO:0022892 | Substrate-specific transporter activity | 1.0 |
| GO:0016740 | Transferase activity | 5.0 |
| GO:0016874 | Ligase activity | 3.0 |
| GO:0022857 | Transmembrane transporter activity | 1.0 |
| GO:0017056 | Ttructural constituent of nuclear pore | 1.0 |
| GO:1901363 | Heterocyclic compound binding | 7.0 |
| b). Drosophila meridionalis | ||
| GO-id | GO-term | Score |
| GO:0008144 | Drug binding | 1.0 |
| GO:0000989 | Transcription factor binding transcription factor activity | 1.0 |
| GO:0097367 | Carbohydrate derivative binding | 6.0 |
| GO:0036094 | Small molecule binding | 7.0 |
| GO:0004871 | Signal transducer activity | 2.0 |
| GO:0003735 | Structural constituent of ribosome | 3.0 |
| GO:0016787 | Hydrolase activity | 4.0 |
| GO:0016491 | Oxidoreductase activity | 2.0 |
| GO:0061134 | Peptidase regulator activity | 1.0 |
| GO:0097159 | Organic cyclic compound binding | 10.0 |
| GO:0043167 | Ion binding | 11.0 |
| GO:0016853 | Isomerase activity | 1.0 |
| GO:0004857 | Enzyme inhibitor activity | 1.0 |
| GO:0048037 | Cofactor binding | 1.0 |
| GO:0005515 | Protein binding | 5.0 |
| GO:0003682 | Chromatin binding | 1.0 |
| GO:0022892 | Substrate-specific transporter activity | 3.0 |
| GO:0008289 | Lipid binding | 2.0 |
| GO:0016740 | Transferase activity | 5.0 |
| GO:0022857 | Transmembrane transporter activity | 3.0 |
| GO:0017056 | Structural constituent of nuclear pore | 1.0 |
| GO:1901363 | Heterocyclic compound binding | 10.0 |
Female flies were exposed to odor of the cactus Cereus hildmaniannus. The analyses were performed using BLAST2GO software [37].
Table 2.
Classification of differentially expressed genes (DEGs) according to Gene Ontology (GO) biological process attributes
| a). Drosophila antonietae | ||
|---|---|---|
| GO-id | GO-term | Score |
| GO: 0002376 | Immune system process | 1.0 |
| GO: 0044699 | Single-organism process | 13.0 |
| GO: 0050896 | Response to stimulus | 5.0 |
| GO: 0009987 | Cellular process | 20.0 |
| GO: 0008152 | Metabolic process | 17.0 |
| GO: 0048511 | Rhythmic process | 1.0 |
| GO: 0032502 | Developmental process | 7.0 |
| GO: 0071840 | Cellular component organization or biogenesis | 8.0 |
| GO: 0065007 | Biological regulation | 8.0 |
| GO: 0000003 | Reproduction | 4.0 |
| GO: 0040007 | Growth | 2.0 |
| GO: 0040011 | Locomotion | 2.0 |
| GO: 0051704 | Multi-organism process | 1.0 |
| GO: 0051179 | Localization | 7.0 |
| GO: 0032501 | Multicellular organismal process | 7.0 |
| GO: 0023052 | Signaling | 4.0 |
| b. Drosophila meridionalis | ||
| GO-id | GO-term | Score |
| GO: 0044699 | Single-organism process | 19.0 |
| GO: 0050896 | Response to stimulus | 8.0 |
| GO: 0008152 | Metabolic process | 16.0 |
| GO: 0009987 | Cellular process | 21.0 |
| GO:0071840 | Cellular component organization or biogenesis | 12.0 |
| GO: 0032502 | Developmental process | 6.0 |
| GO: 0065007 | Biological regulation | 9.0 |
| GO: 0000003 | Reproduction | 3.0 |
| GO: 0040007 | Growth | 2.0 |
| GO: 0051704 | Multi-organism process | 1.0 |
| GO: 0051179 | Localization | 10.0 |
| GO: 0032501 | Multicellular organismal process | 8.0 |
| GO: 0023052 | Signaling | 5.0 |
Female flies were exposed to odor of the cactus Cereus hildmaniannus. The analyses were performed using BLAST2GO software [37].
Some terms were species specific. For instance, the D. meridionalis SSH library contained sequences related to signaling receptor activity (GO:0038023), signal transducer activity (GO:0004871), drug binding (GO:0008144) and lipid binding (GO:0008289) (Table 1b), and in the D. antonietae library we found sequences related to DNA polymerase processivity factor activity (GO:0030337) (Table 1a).
In the biological processes category (set at level 2), genes differentially expressed in response to cactus odor were, for both species, related to the following GO terms: cellular process (GO:0009987), including processes such as cellular recognition, cell cycle, and cellular membrane organization; metabolic processes (GO:0008152), including the metabolism of macromolecules organic, nitrogen, and organophosphate metabolism, and oxidoreduction mechanisms; multicellular organismal process (GO:0032501), including mechanisms related to reproduction and cellular signaling; cellular component organization (GO:0071840); biological regulation processes (GO:0065007); localization (GO:0051179); response to stimulus (GO:0050896); and signaling (GO:0023052) (Table 2).
For D. meridionalis, cactus odor-activated genes were also related to neural precursor cell proliferation (GO: 0061351), interspecies interaction (GO: 0044419), establishment of cell polarity (GO: 0030010), and establishment of localization (GO: 0051234) (Additional file 5 and Table 2b). For D. antonietae, the DEG list also comprised genes related to cell death (GO: 0016265), immune response (GO: 0002376), response to other organisms (GO: 0051707), and rhythmic behavior (GO:0048511) (Additional file 4 and Table 2a). A statistical analysis (Fisher's exact test) revealed no significant difference for the two species with respect to the number of sequences associated with the GO categories.
Metabolic mapping of the DEGs by KEGG pathway analysis showed that D. antonietae females experienced an activation of the following pathways after cactus odor stimulation: inositol phosphate metabolism, lipoic acid metabolism, sphingolipid metabolism, pyrimidine metabolism, and purine metabolism (Additional file 4). In D. meridionalis, the activated pathways were glycerophospholipid metabolism; the metabolism of alanine, aspartate, and glutamate; nitrogen metabolism and oxidative phosphorylation (Additional file 5). The phosphatidylinositol signaling system was activated in both species (Additional files 4 and 5).
Real-time PCR validation
We employed real-time PCR analysis to validate the transcript levels of DEGs revealed in the subtractive libraries of D. antonietae and D. meridionalis females either exposed or not exposed to C. hildmannianus odor. The genes selected for quantitative real time-PCR analysis included those represented by high numbers of ESTs in the respective libraries, and genes with GO attributes related to reproduction, communication, oxidoreduction processes and mechanisms of detoxification. According to previous studies [10], genes involved in these processes are typically activated in Drosophila females when they are placed in contact with a host cactus or volatile compounds, such as ethanol, resulting from fermentation by yeast in decomposing cacti. We also included some genes that showed no significant matches to genes of other species, and thus would represent potentially novel genes involved in fly-cactus interaction.
We designed 12 and 13 primer pairs, respectively, for the USs from D. antonietae and D. meridionalis (Additional files 2 and 3) and used an α-tubulin gene to normalize the expression levels of these genes. In general, all of the genes analyzed by real-time PCR from the two species had a slightly higher expression in flies exposed to cactus odor when compared to non-exposed control flies. For four genes of D. antonietae and six of D. meridionalis overexpression in odor-exposed flies was denoted as statistically significant (P ≤ 0.05) by randomization tests using REST software (Figure 3) [40], this validating the SSH library results.
Figure 3.
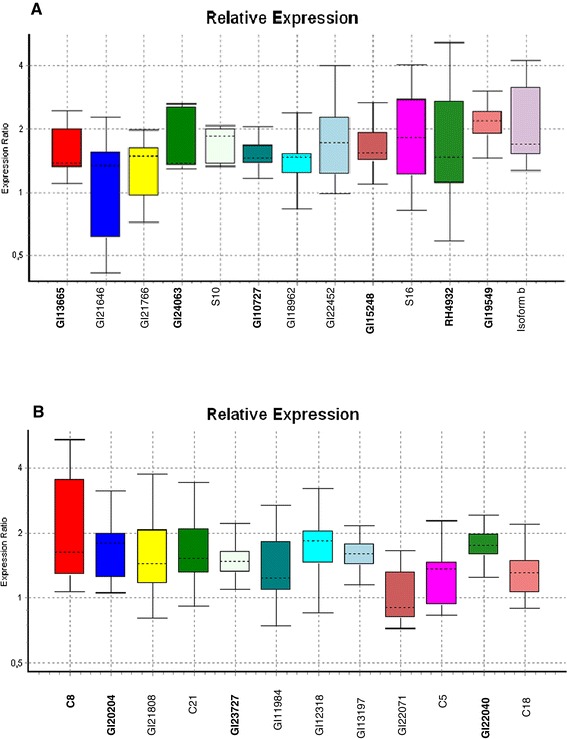
Relative expression levels in female flies exposed to odor cactus. Relative expression levels were assessed by quantitative RT-PCR for genes selected from the set of unique sequences of the SSH libraries for odor-exposed females of (A) Drosophila meridionalis and (B) Drosophila antonietae. Gene names in bold mark sequences that presented a statistical difference in relative expression (P ≤ 0.05) compared to control (flies not exposed to cactus odor). Statistical analyses were done using the randomization test implemented in REST 2008 [40].
The following genes were significantly overexpressed in female D. meridionalis flies exposed to cactus odor: Dmoj\GI24063, which is associated with biological reproduction of multicellular organisms; actin-binding protein ipp-like (GI10727), which is involved in the cytoplasmic sequestering of transcription factors; serine threonine-protein kinase 38 (GI13665), which is associated with protein phosphorylation and intracellular protein kinase cascades and the phosphatidylinositol signaling system; alanine aminotransferase (GI15248), which is involved in the metabolism of alanine, aspartate, and glutamate; RH49324p, which has unknown functions; and fk506 binding protein (GI19549), which is involved in protein folding (Figure 3a).
The following genes were found to be significantly overexpressed in D. antonietae: trna-dihydrouridine synthase (GI20204), a gene related to oxidoreduction; an f-box and wd-40 domain-containing protein (GI23727) involved in the regulation of signaling mechanisms, olfactory learning, and the ubiquitination of proteins; and a tnf receptor-associated factor (GI22040) related to defense responses, the regulation of the JNK cascade, Toll signaling pathway, receptor activity, protein ubiquitination; and C8 representing a no-match sequence (Figure 3b).
Discussion
By using an SSH approach we were able to identify genes overexpressed in D. antonietae and D. meridionalis females that were exposed to the odor of their host cactus C. hildmannianus. The majority of these genes showed significant similaritywith genes of D. melanogaster, but several had no known function associated with their sequence (Additional files 4 and 5).
A GO analysis revealed that the known-function genes can be grouped into several categories, including metabolic processes, signal transduction, response to stimulus, and reproduction. Previous studies on the cactophilic species D. mojavensis had also implicated the activation of genes belonging to these functional categories in flies reared on their native host cactus, as well as on chemically distinct host tissue [10,20,42].
The metabolic process category encompasses genes involved in the metabolism of carbohydrates, lipids, proteins, and amino acids (Table 2, Additional files 4 and 5). Interestingly, the activation of genes belonging to specific metabolic pathways was detected in each of the species studied herein. For example, genes related to the metabolism of inositol phosphate, lipoic acid, sphingolipids, and purines and pyrimidines were detected in D. antonietae, while genes associated with the metabolism of glycerophospholipids, alanine, aspartate, and glutamate were revealed in D. meridionalis. The activation of these genes could reflect differences in metabolic strategies employed by the two fly species to survive, develop, and reproduce on their host cactus C. hildmannianus.
Genes involved in catalytic activity were up-regulated in response to cactus odor in both species (Table 1). This category included genes encoding hydrolases, transferases, and oxidoreductases. These enzymes are directly linked to the metabolism of xenobiotic compounds typically present in cacti, and previous studies revealed the activation of members of these gene families, including glutathione S-transferases (Gst), cytochrome P450, and UDT-glucuronosyltransferase (UGT), in the detoxification of compounds present in necrotic cactus tissue [10,20]. In our study, a gene belonging to the Gst family was up-regulated in D. meridionalis females in response to cactus odor (Additional file 5). These genes are known to be involved in the detoxification of various compounds, particularly hydrophobic organic compounds [43]. In mammals, Gst genes are induced during exposure to carcinogens and in drug-resistance processes [44], and in insects they are associated with insecticide resistance [45]. Surprisingly, only the SSH library for D. meridionalis, but not D. antonietae, included ESTs representing genes involved in glutathione metabolism. It is possible that this difference resulted from the random selection of clones for sequencing; however, an alternative explanation is that D. antonietae employs another mechanism for the detoxification of such compounds. In fact, we found a gene in this species that is linked to lipoic acid metabolism, which is a mechanism employed by some fly species to detoxify insecticides. Lipoic acid acts as an antioxidant by quenching reactive oxygen species that can inflict damage to DNA, proteins and lipids. In Drosophila melanogaster, lipoic acid also is effective in counteracting the toxicity of paraquat, a quaternary nitrogen herbicide [46].
Another detoxification strategy used by insects is the excretion of toxic substances. This strategy could explain the activation of genes in both species related to transmembrane transporter activity (Table 1), which is a mechanism that mediates the transport of proteins, lipids, neurotransmitters, and other substrates across cell membranes. For this category, we found a somewhat larger number of genes activated in D. meridionalis than in D antonietae.
The exploitation of necrotic cactus tissue as a breeding site is an ecological characteristic of most species of the Repleta group. And this required genetic adjustments related to detoxification, as many cacti are rich in chemically complex secondary compounds. The current hypothesis is that the plesiomorphic fly-cactus relationship in the Repleta group is the use of Opuntia species as hosts, which are chemically less complex than columnar cacti [25]. Through independent processes, certain Drosophila species then evolved to explore chemically more complex columnar cacti that produce toxic compounds, such as alkaloids, medium-chain fatty acids, sterol diols, and triterpene glycosides [15].
Thus, the evolutionarily independent colonization of columnar cacti may have activated different transcriptional pathways in the two species studied here, this depending on the genetic characteristics of their respective ancestral populations. These pathways, when activated by the perception of cactus odors, may represent different metabolic strategies that allow flies to tolerate the toxicity of certain compounds present in tissue of columnar cacti, which, in the present case study, is C. hilmannianus..
In both species, the exposure of female flies to volatile cactus compounds, including ethanol, activated genes related to redox processes (Additional files 4 and 5 and Table 1). Ethanol produces oxidative stress by causing an imbalance in redox reactions and increases the formation of reactive oxygen species (ROS) [47,48]. The increase in ROS levels activates detoxification mechanisms that act as antioxidants and reduce the synthesis of ROS [49]. Glutathione metabolism is among the detoxification mechanisms involved in the response to ethanol and was shown to be activated in D. meridionalis in our study. Furthermore, oxidative stress is related to the apoptosis of neurons [50,51]. Interestingly, we found an increased expression of genes related to neurogenesis in both species in this study. At present, we cannot confirm whether this observation indicates a mechanism countering cell death, because most current evidence suggests that neurogenesis does not occur in the brain of adult flies [52] with the exception of a recent study that showed such events by lineage tracing [53].
Genes involved in the ubiquitination of proteins were differentially expressed in D. antonietae females exposed to cactus odor (Additional file 4). Oxidative stress increases protein damage and subsequent degradation, and in this process, ubiquitination marks proteins for proteasomal degradation and regulates the protein levels for a number of critical cellular functions, including gene expression [54,55]. In addition, protein ubiquitination plays a role in stress response and in the immune system in D. mojavensis [42].
Another important class of genes activated in both species is associated with signaling pathways (Table 2). This category includes genes related to signal transduction mechanisms, such as kinases, phosphatases, and transcription factors. We identified genes related to the Toll signaling pathway, which activates the expression of genes involved in the immune response of Drosophila [56]; and the phosphatidylinositol signaling pathway.
Other mechanisms were also specifically activated in each species. In D. antonietae, we found evidence for the activation of the Wnt pathway, which plays a central role in several developmental processes, including neurogenesis [57]. This pathway may also be linked to neuronal apoptosis, which can be a result of exposure to ethanol present in volatile compounds produced by cacti [49]. In contrast, the Notch pathway was activated in D. meridionalis. This pathway is a node in a conserved signaling mechanism that regulates development [58] and may contribute to plasticity-related processes, including changes in neurite structure [59] and the maintenance of neural stem cells [60]. The Notch pathway is also related to the learning and memory processes [61].
The activation of genes associated with the GO Biological process category “response to stimuli was also common for both species (Table 2), and this category includes G-protein-coupled receptors. These proteins represent a large class of cell-surface molecules that act as stimulus receptors, including receptors for neurotransmitters, hormones, growth factors, odor molecules, and light [62]. In general, these proteins act in response to extracellular signals and activate intracellular signal transduction mechanisms that culminate in the organism's response to the stimulus [63,42].
A noteworthy outcome of the SSH library analyses was the up-regulation of reproduction-related genes in female flies exposed to the odor of necrotic tissue from C. hildmannianus. For D. antonietae we found four genes involved with reproduction: CG3411, which is involved in oocyte localization in germarium-derived egg chamber formation; CG10262, involved in eggshell chorion formation; CG11989, associated with oogenesis; and CG2512, involved with embryo development ending in egg hatching (Additional file 4). For D. meridionalis we found a GI24063 gene associated with multicellular organism reproduction (Additional file 5). These findings are in accordance with those for D. sechellia [64], where flies placed in contact with their host cactus showed increased egg production, chorion formation, and oviposition. Thus, olfactory stimuli from necrotic cactus tissues may play an important role in the selection of an appropriate site for oviposition and larval development in Drosophila species.
The two fly species investigated herein, D. antonietae and D. meridionalis, belong to different complexes of the Repleta group. They are sympatric in certain localities along their species range and use the same host cactus as breeding and feeding site. Our data revealed the differential expression of genes related to metabolic processes, signaling pathways, detoxification mechanisms, reproduction, and response to stimuli for both species. While there was an overlap in some of the activated pathways for the two species, there were also specific mechanisms separating the two species. This data provides initial insights into how the use of the C. hildmannianus cactus as a host may have arisen by independent adaptation, involving species-specific genetic mechanisms and metabolic and signaling pathway activation in response to cactus odor in the two species.
Conclusions
In this study, we analyzed the transcriptional response to olfactory stimulation by necrotic C. hildmannianus cactus tissue in females of the cactophilic species D. antonietae and D. meridionalis, which use this cactus as an obligatory breeding site. We found several genes that were differentially expressed in response to cactus odor. The GO and KEGG pathway analysis of these genes showed that the cactus odor activated genes are related to metabolic processes, including the metabolism of carbohydrates, lipids, proteins, and amino acids, as well as to detoxification mechanisms, signaling, response to stimuli, and reproduction. These results provide a basic data set for future investigations into evolutionary mechanisms underlying adaptive diversification processes involved in Drosophila-host cactus interactions.
Acknowledgments
The project was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior), FAPESP (Fundação de Amparo à Pesquisa do Estado de São Paulo) process 10/10920-1, 11/51652-2., and CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico). CB is sponsored by CAPES, CC by FAPESP, and MS by a CNPq post-doctoral fellowship.
Additional files
Ingredients for the preparation of culture medium. This file show the ingredients for the preparation the culture medium used to maintain stocks of Drosophila flies.
qRT-PCR primer sequences and their respective target genes (when known) for Drosophila antonietae. This table show the sequence primer for D. antonietae used in qRT-PCR assay.
qRT-PCR primer sequences and their respective target genes (when known) for Drosophila meridionalis. This table show the sequence primer for D. meridionalis used in qRT-PCR assay.
ESTs sequenced from the Suppression-Subtractive Hybridization (SSH) library of Drosophila antonietae exposed to odor of the cactus Cereus hildmaniannus . This table shows the respective orthologs or predicted genes retrieved by blastx searches in GenBank for ESTs sequenced from library of Drosophila antonietae exposed to odor of the cactus Cereus hildmaniannus. Also included are the Gene Ontology terms and KEGG pathway information.
ESTs sequenced from the Suppression-Subtractive Hybridization (SSH) library of Drosophila meridionalis exposed to odor of the cactus Cereus hildmaniannus . This table shows the respective orthologs or predicted genes retrieved by blastx searches in GenBank for ESTs sequenced from library of Drosophila meridionalis exposed to odor of the cactus Cereus hildmaniannus. Also included are the Gene Ontology terms and KEGG pathway information.
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
CB conceived the study. CC and MM designed the study. CB and CC conducted experiments with female Drosophila and extracted the RNA. CC conducted the SSH. CB and RR cloned and sequenced the fragments. CB and MS conducted the bioinformatics analysis. CB designed the primers, conducted qRT-PCR, and analyzed the qRT-PCR results. CB, KH, MM, and JH wrote the manuscript. All authors read and approved the final manuscript.
Contributor Information
Camila M Borgonove, Email: cmborgonove@gmail.com.
Carla B Cavallari, Email: barsalobres@gmail.com.
Mateus H Santos, Email: mateushenrique@yahoo.com.br.
Rafaela Rossetti, Email: rafaelarossetti@hotmail.com.
Klaus Hartfelder, Email: klaus@fmrp.usp.br.
Maura H Manfrin, Email: maura.manfrin@usp.br.
References
- 1.Reznick D, Ricklefs RE. Darwin's bridge between microevolution and macroevolution. Nature. 2009;457:837–842. doi: 10.1038/nature07894. [DOI] [PubMed] [Google Scholar]
- 2.Thompson JN. The Coevolutionary Process. Chicago: University of Chicago Press; 1994. [Google Scholar]
- 3.Thompson JN. The Geographic Mosaic of Coevolution. Chicago: The University of Chicago Press; 2005. [Google Scholar]
- 4.Landolt PJ, Phillips TW. Host plant influences on sex pheromone behavior of phytophagous insects. Annu Rev Entomol. 1997;42:371–391. doi: 10.1146/annurev.ento.42.1.371. [DOI] [PubMed] [Google Scholar]
- 5.Bruce MJ, Heiling AM, Herberstein ME. Spider signals: are web decorations visible to birds and bees? Biol Lett. 2005;1:299–302. doi: 10.1098/rsbl.2005.0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuo T. Rapid evolution of two odorant-binding protein genes, Obp57d and Obp57e, in the Drosophila melanogaster species group. Genetics. 2008;178:1061–1072. doi: 10.1534/genetics.107.079046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Bruyne M, Smart R, Zammit E, Warr CG. Functional and molecular evolution of olfactory neurons and receptors for aliphatic esters across the Drosophila genus. J Comp Physiol A Neuroethol Sens Neural Behav Physiol. 2010;196:97–109. doi: 10.1007/s00359-009-0496-6. [DOI] [PubMed] [Google Scholar]
- 8.Govind G, Mittapalli O, Griebel T, Allmann S, Bocker S, Baldwin IT. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One. 2010;5:e8735. doi: 10.1371/journal.pone.0008735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogel H, Heidel AJ, Heckel DG, Groot AT. Transcriptome analysis of the sex pheromone gland of the noctuid moth Heliothis virescens. BMC Genomics. 2010;11:e29. doi: 10.1186/1471-2164-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Matzkin L. Population transcriptomics of cactus host shifts in Drosophila mojavensis. Mol Ecol. 2012;21:2428–2439. doi: 10.1111/j.1365-294X.2012.05549.x. [DOI] [PubMed] [Google Scholar]
- 11.Wasserman M. Cytological evolution of the Drosophila Repleta species group. In: Krimbas CB, Powell JR, editors. Drosophila inversion polymorphism. Boca Raton: CRC Press; 1992. pp. 455–555. [Google Scholar]
- 12.Matzikin LM. Ecological genomics of host shifts in Drosophila mojavensis. Adv Exp Med Biol. 2014;781:233–47. doi: 10.1007/978-94-007-7347-9_12. [DOI] [PubMed] [Google Scholar]
- 13.Fogleman JC, Abril JR. Ecological and evolutionary importance of host plant chemistry. In: Barker JSF, MacIntyre RJ, Starmer WT, editors. Ecological and evolutionary genetics of Drosophila. New York: Plenum Press; 1990. pp. 121–143. [Google Scholar]
- 14.Fogleman JC, Danielson PB. Chemical interactions in the cactus-microorganism-Drosophila model system of the Sonoran Desert. Am Zool. 2001;41:877–889. doi: 10.1668/0003-1569(2001)041[0877:CIITCM]2.0.CO;2. [DOI] [Google Scholar]
- 15.Fogleman JC, Heed WB. Columnar cacti and desert Drosophila: The chemistry of host plant specificity. In: Schmidt J, editor. Special biotic relationships in the arid southwest. Albuquerque: Univ. New Mexico Press; 1989. pp. 1–24. [Google Scholar]
- 16.Starmer WT, LaChance MA, Phaff HJ, Heed WB. The biogeography of yeasts associated with decaying cactus tissue in North America, the Caribbean, and northern Venezuela. Evol Biol. 1990;24:253–296. [Google Scholar]
- 17.Starmer WT, Schmedicke RA, Lachance MA. The origin of the cactus-yeast community. FEMS Yeast Res. 2003;3:441–448. doi: 10.1016/S1567-1356(03)00056-4. [DOI] [PubMed] [Google Scholar]
- 18.Moraes EM, Rosa CA, Sene FM. Preliminary notes on yeasts associated with necrotic cactus stems from different localities in Brazil. Braz J Biol. 2005;65:299–304. doi: 10.1590/s1519-69842005000200014. [DOI] [PubMed] [Google Scholar]
- 19.Date P, Dweck HKM, Stensmyr MC, Shann J, Hansson S, Rollmann SM. Divergence in olfactory host plant preference in D.mojavensis in response to cactus host use. PLoS One. 2013;8:e70027. doi: 10.1371/journal.pone.0070027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matzkin LM, Watts TD, Bitler BG, Machado CA, Markow TA. Functional genomics of cactus host shifts in Drosophila mojavensis. Mol Ecol. 2006;15:4635–4643. doi: 10.1111/j.1365-294X.2006.03102.x. [DOI] [PubMed] [Google Scholar]
- 21.Danielson PB, MacIntyre RJ, Fogleman JC. Molecular cloning of a family of xenobiotic inducible drosophilid cytochrome P450s: evidence for involvement in host plant allelochemical resistance. Proc Natl Acad Sci U S A. 1998;94:10797–10802. doi: 10.1073/pnas.94.20.10797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fogleman JC, Danielson PB, MacIntyre RJ. The molecular basis of adaptation in Drosophila: the role of cytochrome P450s. Evol Biol. 1998;30:15–77. doi: 10.1007/978-1-4899-1751-5_2. [DOI] [Google Scholar]
- 23.Soto IM, Carreira VP, Fanara JJ, Hasson E. Evolution of male genitalia: environmental and genetic factors affect genital morphology in two Drosophila sibling species and their hybrids. BMC Evol Biol. 2007;15:7–77. doi: 10.1186/1471-2148-7-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Etges WJ, de Oliveira CC, Ritchie MG, Noor MAF. Genetics of incipient speciation in Drosophila mojavensis. II. Host plants and mating status influence cuticular hydrocarbon QTL expression and G × E interactions. Evolution. 2009;63:1712–1730. doi: 10.1111/j.1558-5646.2009.00661.x. [DOI] [PubMed] [Google Scholar]
- 25.Oliveira DCSG, Almeida FC, O’Grady PM, Armella MA, DeSalle R, Etges WJ. Monophyly, divergence times and evolution of host plant use inferred form a revised phylogeny of the Drosophila repleta species group. Mol Phylogenet Evol. 2013;64:533–544. doi: 10.1016/j.ympev.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 26.Pereira MAQR, Vilela CR, Sene FM. Notes on breeding and feeding site of some of the repleta group of the genus Drosophila (Diptera, Drosophilidae) Cienc Cult. 1983;35:1313–1319. [Google Scholar]
- 27.Manfrin MH, Sene FS. Cactophilic Drosophila in South America: a model for evolutionary studies. Genetica. 2006;126:57–75. doi: 10.1007/s10709-005-1432-5. [DOI] [PubMed] [Google Scholar]
- 28.Leal DYB . Master thesis. São Paulo University: Genetics Department; 2013. História demográfica e estrutura de populações para a espécie cactófila Drosophila meridionalis. [Google Scholar]
- 29.Taylor N, Zappi D. Cacti of Eastern Brazil. Kew: Royal Botanic Gardens; 2004. Taxonomic Inventory; p. 281. [Google Scholar]
- 30.O’Grady PM, Markow TA. Rapid morphological, behavioral and ecological evolution in Drosophila: comparions beteween the endemic Hawaiian Drosophila and the cactophilic replete species group. In: Singh RS, Xu J, Kulathinal RJ, editors. Rapidly Evolving Genes and Genetic Systems. 1. Oxford: Oxford University Press; 2012. pp. 176–186. [Google Scholar]
- 31.Vilela CA. A revision of the Drosophila species group (Diptera-Drosophilidae) Rev Brasil Entomol. 1983;27:1–114. [Google Scholar]
- 32.Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996;93:6025–6030. doi: 10.1073/pnas.93.12.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Durham AM, Kashiwabara AY, Matsunaga FTG, Ahagon PH, Rainone F, Varuzza L, Gruber A. E-Gene: a configurable pipeline generation system for automated sequence analysis. Bioinformatics. 2005;21:2812–2813. doi: 10.1093/bioinformatics/bti424. [DOI] [PubMed] [Google Scholar]
- 34.Huang XQ, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Judice CC, Carazzole MF, Festa F, Sogayar MC, Hartfelder K, Pereira GAG. Gene expression profiles underlying alternative caste phenotypes in a highly eusocial bee, Melipona quadrifasciata. Insect Mol Biol. 2006;15:33–44. doi: 10.1111/j.1365-2583.2005.00605.x. [DOI] [PubMed] [Google Scholar]
- 36.Gene Ontology terms GO. [http://www.geneontology.com]
- 37.BLAST2GO. [http://www.blast2go.de]
- 38.Ponton F, Chapuis MP, Pernice M, Sword GA, Simpson SJ. Evaluation of potential reference genes for reverse transcription-qPCR studies of physiological responses in Drosophila melanogaster. J Insect Physiol. 2011;57:840–850. doi: 10.1016/j.jinsphys.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 39.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time PCR data. Neuroscience. 2003;339:62–66. doi: 10.1016/s0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 40.Pfaffl MW, Horgan GW, Dempfle L. Relative expression software tool (REST (c)) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:1–10. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drosophila 12 Genomes Consortium Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- 42.Smith G, Fang Y, Liu X, Kenny J, Cossins AR, Oliveira CC, Etges WJ, Ritchie MG. Transcriptome-wide expression variation associated with environmental plasticity and mating success in cactophilic Drosophila mojavensis. Evolution. 2013;67:1950–1963. doi: 10.1111/evo.12082. [DOI] [PubMed] [Google Scholar]
- 43.Atkins WM, Wang RW, Bird AW, Newton DJ, Lu AYH. The catalytic mechanism of glutathione-S-Transferase (gst): spectroscopic determination of the Pk (A) of Tyr-9 in rat alpha-1–1 gst. J Biol Chem. 1993;268:19188–19191. [PubMed] [Google Scholar]
- 44.Hayes JD, Pulford DJ. The glutathione S-Transferase supergene family: regulation of gst and the contribution of the isoenzymes to cancer chemoprotection and drug resistance. Crit Rev Biochem Mol Biol. 1995;30:445–600. doi: 10.3109/10409239509083491. [DOI] [PubMed] [Google Scholar]
- 45.Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14:3–8. doi: 10.1111/j.1365-2583.2004.00529.x. [DOI] [PubMed] [Google Scholar]
- 46.Bonilha E, Leendertz SM, Villalobos V, Molero L, Bohorquez A. Paraquat-induced oxidative stress in Drosophila melanogaster: effects of melatonin, glutathione, serotonin, minocycline, lipoic acid and ascorbic acid. Neurochem Res. 2006;31:1425–1432. doi: 10.1007/s11064-006-9194-8. [DOI] [PubMed] [Google Scholar]
- 47.González A, Pariente J, Salido GM. Ethanol stimulates ROS generation by mitochondria through Ca2+ mobilization and increases GFAP content in rat hippocampal astrocytes. Brain Res. 2007;1178:28–37. doi: 10.1016/j.brainres.2007.08.040. [DOI] [PubMed] [Google Scholar]
- 48.Morozova TV, Mackay TFC, Anholt RRH. Transcriptional networks for alcohol sensitivity in Drosophila melanogaster. Genetics. 2011;187:1193–1205. doi: 10.1534/genetics.110.125229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Awofala AA, Davies JA, Jones S. Functional roles for redox genes in ethanol sensitivity in Drosophila. Funct Integr Genomics. 2012;12:305–315. doi: 10.1007/s10142-012-0272-5. [DOI] [PubMed] [Google Scholar]
- 50.Ramachandran V, Watts LT, Maffi SK, Chen J, Schenker S, Henderson G. Ethanol-induced oxidative stress precedes mitochondrially mediated apoptotic death of cultured fetal cortical neurons. J Neurosci Res. 2003;74:577–588. doi: 10.1002/jnr.10767. [DOI] [PubMed] [Google Scholar]
- 51.Björk K, Saarikoski ST, Arlinde C, Kovanen L, Osei-Hyiaman D, Ubaldi M, Reimers M, Hyytiä P, Heilig M, Somme WH. Glutathione-S-transferase expression in the brain: possible role in ethanol preference and longevity. FASEB J. 2006;20:1826–1835. doi: 10.1096/fj.06-5896com. [DOI] [PubMed] [Google Scholar]
- 52.Kato M, Okugaw G, Wakeno M, Takekita Y, Nonen S, Tetsuo S, Nishida K, Azuma J, Kinoshita T, Serretti A. Effect of basic fibroblast growth factor (FGF2) gene polymorphisms on SSRIs treatment response and side effects. Eur Neuropsychopharmacol. 2009;19:718–725. doi: 10.1016/j.euroneuro.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 53.Fernández-Hernández I, Rhiner C, Moreno E. Adult neurogenesis in Drosophila. Cell Rep. 2013;3:1857–1865. doi: 10.1016/j.celrep.2013.05.034. [DOI] [PubMed] [Google Scholar]
- 54.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 55.Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- 56.Hoffmann JA, Reichhart JM. Drosophila innate immunity: an evolutionary perspective. Nat Immunol. 2002;3:121–126. doi: 10.1038/ni0202-121. [DOI] [PubMed] [Google Scholar]
- 57.Zhang L, Yang X, Yang S, Zhang J. The Wnt ⁄b-catenin signaling pathway in the adult neurogenesis. Eur J Neurosci. 2011;33:1–8. doi: 10.1111/j.1460-9568.2010.7483.x. [DOI] [PubMed] [Google Scholar]
- 58.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 59.Sestan N, Artavanis-Tsakonas S, Rakic P. Contact-dependent inhibition of cortical neurite growth mediated by notch signaling. Science. 1999;286:741–746. doi: 10.1126/science.286.5440.741. [DOI] [PubMed] [Google Scholar]
- 60.Hitoshi S, Alexson T, Tropepe V, Donoviel D, Elia AJ, Nye JS, Conlon RA, Mak TW, Bernstein A, van der Kooy D. Notch pathway molecules are essential for the maintenance, but not the generation, of mammalian neural stem cells. Genes Dev. 2002;16:846–858. doi: 10.1101/gad.975202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Costa RM, Honjo T, Silva AJ. Learning and memory deficits in Notch mutant mice. Curr Biol. 2003;13:1348–1354. doi: 10.1016/S0960-9822(03)00492-5. [DOI] [PubMed] [Google Scholar]
- 62.Marinissen MJ, Gutkind JS. G-protein-coupled receptors and signaling networks: emerging paradigms. Trends Pharmacol Sci. 2001;22:368–376. doi: 10.1016/S0165-6147(00)01678-3. [DOI] [PubMed] [Google Scholar]
- 63.Krieger J, Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- 64.Jones CD. Genetics of egg production in Drosophila sechellia. Heredity. 2004;92:235–241. doi: 10.1038/sj.hdy.6800401. [DOI] [PubMed] [Google Scholar]


