Abstract
Background
Glypican 3 (GPC3) is a member of the family of glypican heparan sulfate proteoglycans (HSPGs). The GPC3 gene may play a role in controlling cell migration, negatively regulating cell growth and inducing apoptosis. GPC3 is downregulated in several cancers, which can result in uncontrolled cell growth and can also contribute to the malignant phenotype of some tumors. The purpose of this study was to analyze the mechanism of action of the GPC3 gene in clear cell renal cell carcinoma.
Methods
Five clear cell renal cell carcinoma cell lines and carcinoma samples were used to analyze GPC3 mRNA expression (qRT-PCR). Then, representative cell lines, one primary renal carcinoma (786-O) and one metastatic renal carcinoma (ACHN), were chosen to carry out functional studies. We constructed a GPC3 expression vector and transfected the renal carcinoma cell lines, 786-O and ACHN. GPC3 overexpression was analyzed using qRT-PCR and immunocytochemistry. We evaluated cell proliferation using MTT and colony formation assays. Flow cytometry was used to evaluate apoptosis and perform cell cycle analyses.
Results
We observed that GPC3 is downregulated in clear cell renal cell carcinoma samples and cell lines compared with normal renal samples. GPC3 mRNA expression and protein levels in 786-O and ACHN cell lines increased after transfection with the GPC3 expression construct, and the cell proliferation rate decreased in both cell lines following overexpression of GPC3. Further, apoptosis was not induced in the renal cell carcinoma cell lines overexpressing GPC3, and there was an increase in the cell population during the G1 phase in the cell cycle.
Conclusion
We suggest that the GPC3 gene reduces the rate of cell proliferation through cell cycle arrest during the G1 phase in renal cell carcinoma.
Keywords: GPC3, Cell lines, Cell proliferation, Renal carcinoma, Transfection
Background
Renal cell carcinoma (RCC) is the most lethal urological disease [1] and is responsible for 3% of all malignant neoplasms [2]. The incidence of RCC has been increasing over the last few decades [3] due to advances in early detection of renal tumors provided by ultrasound, computed tomography and magnetic resonance imaging [4–6].
RCC is a heterogeneous histological disease, and clear cell renal cell carcinoma (CCRCC) is the most common histological subtype, making up approximately 75-80% of the cases of renal tumors [1, 7]. Renal cell carcinoma is diagnosed in the advanced stage of the disease in 25% of patients [8]. Although nephrectomy and radiotherapy are effective, 30% of patients develop metastatic disease after treatment, with a median survival period of one year [7, 9].
The occurrence of RCC is usually sporadic, although genetic syndromes can cause a familial pattern of inheritance. For example, Von–Hippel Lindau disease, which is associated with mutations and inactivation of the VHL gene [10], is correlated with the occurrence of clear cell renal cell carcinoma [11]. Therefore, it is important to identify genes associated with CCRCC and to better understand their possible mechanisms of action in renal tumor cells. Several studies have identified genes differentially expressed in clear cell renal cell carcinoma and normal renal samples [9, 12]. One of these genes is GPC3, which is decreased in clear cell renal cell carcinoma [9].
Glypican 3 (GPC3), which is located on the human X chromosome (Xq26), is a member of the heparan sulfate proteoglycan (HSPG) family [13, 14]. This protein can bind to the surface of the cell membrane via glycosylphosphatidylinositol (GPI) anchorage [15]. GPC3 plays important roles in cell growth regulation, proliferation, differentiation, migration and apoptosis [16, 17]. It is differentially expressed in some tumor types – in hepatocellular carcinoma and melanoma, GPC3 is highly expressed [18]; however, its expression is reduced in ovarian and breast cancer [19, 20], a finding which suggests that GPC3 may be involved in tumor development [21]. The GPC3 gene is considered a potential molecular marker in hepatocellular carcinoma [22] and may act as a tumor suppressor in the ovary [19].
In the present study, we investigated the mechanisms of action of GPC3 in renal cell carcinoma using colony formation, cell proliferation, cell cycle progression and apoptosis assays to assess the potential role of GPC3 in this type of cancer.
Methods
Clear cell renal cell carcinoma samples
Thirty-five clear cell renal cell carcinoma samples and two normal renal fresh-frozen tissue samples were obtained from the Tumor Bank from the Pio XII Foundation/IBILCE-UNESP, Sao Paulo, Brazil. The use of patient-derived material was approved by the Research Ethics Committee of the Tumor Bank from the Pio XII Foundation/IBILCE-UNESP, Sao Paulo, Brazil, and written consent was obtained from all patients. Tissues were obtained during surgery on patients undergoing tumor resection, and the diagnosis of clear cell renal cell carcinoma was verified post-operatively using histopathology. The samples were classified according to the criteria provided by the International Union against Cancer [23].
Cell lines
The cell lines ACHN, 786-O, A-498, CaKi-1 and CaKi-2 were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). ACHN and A-498 cells were cultured in a MEM Alpha medium (Gibco by Life Technologies, Grand Island, NY, USA), CaKi-1 and CaKi-2 cells were cultured in a McCoy’s 5A medium (Gibco by Life Technologies, Grand Island, NY, USA) and 786-O cells were cultured in a RPMI1640 medium (Gibco by Life Technologies, Grand Island, NY, USA). Cell lines were supplemented with 10% FBS (Cultilab, SP, Brazil), 100 U/mL penicillin (Invitrogen, Grand Island, NY, USA) and 100 μg/mL streptomycin (Invitrogen, Grand Island, NY, USA) and were grown in a 37°C, 5% CO2 atmosphere.
GPC3 mRNA expression was analyzed in all cell lines. Then, representative cell lines, one primary renal carcinoma (786-O) and one metastatic renal carcinoma (ACHN), were chosen to carry out the functional studies.
Plasmid construction
DNA oligonucleotides were chemically synthesized, and appropriate restriction sites were introduced via PCR amplification with the following primers: CATCGGTACCATGGCCGGGACCGTGCG (Forward) and TCGACTCGAGCACCAGGAAGAAGAAGCACACCACCG (Reverse). After PCR purification, products and the pcDNA3.1/V5-HisB vector were digested by the restriction enzymes KpnI and XhoI (Uniscience, New England Biolabs, Hitchin, UK). The products were ligated by T4 DNA ligase (Uniscience, New England Biolabs, Hitchin, UK). The construct was confirmed using DNA sequencing.
Transfection
The pcDNA3.1/GPC3 expression vector and pcDNA3.1 (empty vector) were transfected into ACHN and 786-O cell lines using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s manual.
RNA extraction and qRT-PCR
Total RNA was extracted using TRIzol reagent (Life Technologies, Grand Island, NY, USA) according to the manufacturer’s instructions. Approximately 5 μg of total RNA from each sample was used to synthesize cDNA, using the High Capacity cDNA Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer’s instructions. Real Time PCR was performed using an ABI Prism 7300 Real Time PCR system and SYBR Green PCR Core Reagent (Applied Biosystems, Warrington, UK) following the manufacturer’s protocol. The primer sequences were designed using Primer 3 software: GPC3: GTGCTTTGCCTGGCTACATC (Forward) and TCCACGAGTTCTTGTCCATTC (Reverse), and GAPDH (endogenous control): ACCCACTCCTCCACCTTTGA (Forward) and CTGTTGCTGTAGCCAAATTCGT (Reverse). In brief, the reaction mixture (20 μL total volume) contained 25 ng of cDNA, gene-specific forward and reverse primers for each gene and 10 μL of 2× Quantitative SYBR Green PCR Master Mix. The samples were tested in triplicate.
The relative expression of each specific gene was calculated using the following formula: R = (E target)ΔCt target (control - sample)/(E endogenous)ΔCt endogenous (control - sample), which had been published previously [24]; a cutoff higher than a 2-fold change was used. The expression of the gene GPC3 was analyzed in thirty-five clear cell renal cell carcinoma samples and the cell lines ACHN, 786-O, A-498, CaKi-1 and CaKi-2. Two normal renal fresh-frozen tissue samples were used as the normal reference (control group). All samples were collected from the renal cortex.
Immunocytochemistry
ACHN and 786-O cells were seeded on coverslips in 24-well plates. The cells were washed with PBS twice and fixed with 4% paraformaldehyde for 30 min. Endogenous peroxidase activity was blocked with 3% hydrogen peroxide in methanol for 30 min in the dark and, after washing in PBS, the non-specific proteins were blocked in 1% bovine serum albumin (BSA) for 1 h. The cells were incubated at 4°C overnight with rabbit polyclonal anti-GPC3 (5 μg/mL) (ABCAM, Cambridge, UK) diluted in 1% BSA. After washing, cells were incubated with the biotinylated secondary antibody (1:200) (Santa Cruz Biotechnology, California, USA), diluted in 1% BSA for 45 min at 37°C and then exposed to an HRP-conjugated streptavidin complex (Santa Cruz Biotechnology, CA, USA). The reactions were visualized using DAB substrate (Dako, Cambridge, UK) and the slides were counterstained with hematoxylin. Densitometric analyses of GPC3 were performed with an Axioshop II Microscope (Zeiss, Germany) using the Software Axiovision (Zeiss). For the analyses, eleven different fields from the coverslips were used and 15 points were analyzed. The values were obtained on an arbitrary scale.
Colony formation assay
ACHN and 786-O cells transfected with pcDNA3.1/GPC3 and pcDNA3.1 were plated in 6-well plates (300 μL cell per well) containing 700 μg/mL geneticin (G418, Sigma Aldrich, St Louis, MO, USA). After 14 days, the colonies were stained with 0.01% crystal violet. Each experiment was performed in triplicate and in two independent assays.
Proliferation assay
ACHN and 786-O were seeded into 96-well plates. After the transfection, 1 mg/mL 3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) (Sigma Aldrich, St Louis, MO, USA) was added to the wells and incubated for 30 min at 37°C. Then, the MTT was removed, 100 μL of 100% DMSO (Sigma Aldrich, St Louis, MO, USA) was added to each well and the absorbance was measured at 562 nm. Each experiment was performed in triplicate and in two independent assays.
Apoptosis assay
Apoptotic cells were analyzed using a FITC Annexin V Apoptosis Detection Kit II (BD Biosciences, San Diego, CA, USA) according to the manufacturer’s instructions. After transfection, cells were washed twice with PBS and then resuspended in binding buffer. Next, 5 μL FITC-Annexin V and 5 μL Propidium Iodide (PI) were added and the cells were incubated for 15 min in the dark at room temperature. The cells were analyzed using an easyCyte 5-HT flow cytometry (Millipore Guava Technologies, Hayward, USA). Data are from two independent experiments.
Cell cycle analysis
ACHN and 786-O cells were analyzed 24 h, 48 h and 72 h after the transfection. The cells were washed twice with PBS and then fixed with ice-cold ethanol (70%). Next, the samples were stained with 200 μL of Guava Cell Cycle Reagent (EMD Millipore Corporation, Hayward, CA, USA), incubated for 30 min at room temperature, and the analysis was conducted by using the easyCyte 5-HT flow cytometry (Millipore Guava Technologies, Hayward, USA). Two independent experiments were performed.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 5 Software. The Mann–Whitney U Test and Wilcoxon Single Ranks Test were used to compare the protein expression levels detected through immunohistochemistry. The comparisons of protein expression levels in cells overexpressing GPC3 to cells lacking GPC3 were performed using analysis of variance (ANOVA), with the appropriate post-hoc test. Group comparisons in the MTT assay were performed with two-tailed paired Student’s t test. In all analyses, the differences were considered statistically significant when p < 0.05.
Results
Analysis of GPC3gene expression in clear cell renal cell carcinoma samples and renal carcinoma cell lines
The GPC3 gene was downregulated in all clear cell renal cell carcinoma samples, with the exception of one (Figure 1A). There was no association between GPC3 gene expression and the clinical data of the clear cell renal cell carcinoma patients (Table 1). The renal carcinoma cell lines were also compared with normal renal samples, with fold-change values for gene expression ranging from -1 to -10.3 in primary clear cell renal cell carcinoma samples and -7.8 to -14.4 in primary renal carcinoma cell lines (CaKi-2, A-498 and 786-O) and metastatic renal carcinoma cell lines (CaKi-1 and ACHN) (Figure 1B). GPC3 expression was lower in metastatic cell lines than in primary cell lines. The same observation was made in the case of clear cell renal cell carcinoma samples, in which GPC3 gene expression was lower in metastatic samples when compared with non-metastatic samples. Unfortunately, the number of metastatic clear cell renal cell carcinoma samples used in this study was too small to perform a statistical test. To assess the potential role of GPC3 in this type of cancer, we performed assays in the 786-O and ACHN renal carcinoma cell lines.
Figure 1.
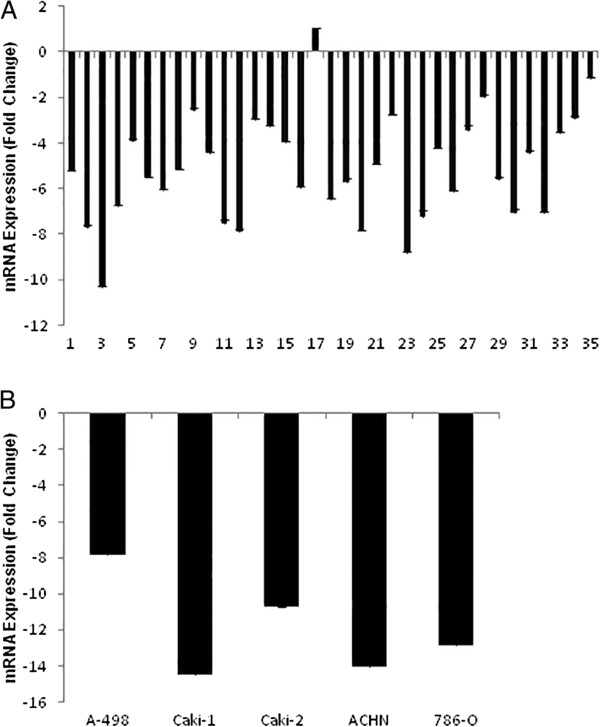
Endogenous GPC3 expression in clear cell renal cell carcinoma. Quantitative mRNA expression of GPC3 was determined via qRT-PCR in clear cell renal cell carcinoma and renal carcinoma cell lines and shown as fold change (log2) relative to expression in normal renal tissue. A) Expression (mRNA) of GPC3 in 35 clear cell renal cell carcinoma samples. B) Expression (mRNA) of GPC3 in five clear cell renal cell carcinoma cell lines.
Table 1.
Description of clinical data of patients with clear cell renal cell carcinoma
| Variable | Number of patients |
|---|---|
| Gender | |
| Male | 19 |
| Female | 16 |
| T stage | |
| T1 | 13 |
| T2 | 14 |
| T3 | 7 |
| T4 | 1 |
| N stage | |
| N0 | 31 |
| N1 | 2 |
| N2 | 1 |
| N3 | 1 |
| M stage | |
| M0 | 28 |
| M1 | 7 |
| Average age in years | |
| Males | 57.8 |
| Females | 57.1 |
| Smoker | |
| No | 30 |
| Yes | 5 |
| Alcoholic | |
| No | 27 |
| Yes | 8 |
We evaluated GPC3 mRNA and protein expression in the ACHN and 786-O cell lines before and after transfection with the pcDNA3.1/GPC3 expression vector or an empty vector using qRT-PCR and immunohistochemistry, respectively. GPC3 was upregulated in both cell lines 48 h after transfection with the pcDNA3.1/GPC3 vector (Figure 2A). GPC3 protein expression was increased in the cells transfected with pcDNA3.1/GPC3 vector compared with cells transfected with an empty vector in both cell lines. GPC3 immunostaining increased significantly in the membranes of ACHN and 786-O cells transfected with the pcDNA3.1/GPC3 vector (p < 0.0001) (Figures 2B and C).
Figure 2.
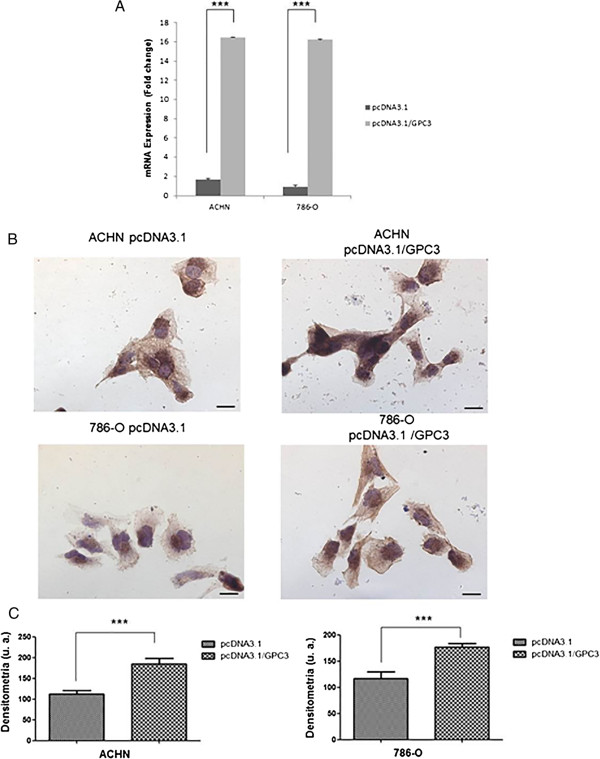
GPC3 expression in renal cell carcinoma cell lines. Restoration of GPC3 expression after transfection with the plasmid pcDNA3.1/GPC3. ACHN and 786-O cells were transiently transfected with pcDNA3.1 (empty vector) or pcDNA3.1/GPC3 and re-expression of GPC3 was confirmed 48 h post-transfection by qRT-PCR and immunocytochemistry. A) Quantitative mRNA expression of the GPC3 gene in renal cell carcinoma cell lines after 48 h of transfection with the pcDNA3.1/GPC3 plasmid is shown as fold change (log2) relative to expression of GPC3 in cell lines after 48 h of transfection with pcDNA3.1 (empty vector). B) Immunolocalization of the GPC3 protein in ACHN and 786-O cell lines after 48 h of transfection with the plasmids pcDNA3.1-GPC3 and pcDNA3.1 (empty vector). Bars = 20 μm. C) Densitometry graphic of GPC3 in ACHN and 786-O cell lines (***p < 0.0001, Mann Whitney test).
GPC3 suppresses colony formation
The pcDNA3.1/GPC3 and pcDNA3.1 vectors were transfected in ACHN and 786-O cell lines, and colony formation ability was assessed after 14 days. Cell lines overexpressing GPC3 had suppressed growth in the colony formation assay. GPC3-transfected cells grew significantly fewer colonies than cells transfected with an empty vector in both cell lines (p < 0.01) (Figure 3).
Figure 3.
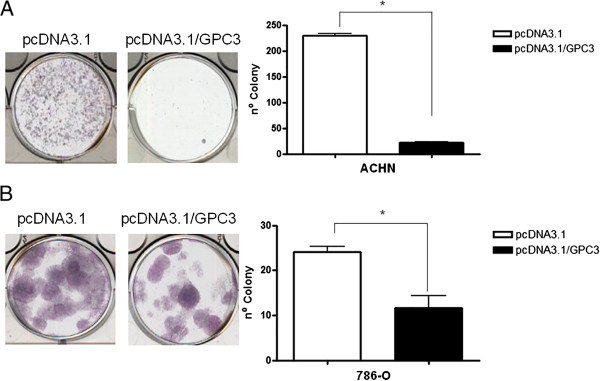
Effect of GPC3 on colony formation. The GPC3 gene suppresses the colony formation in ACHN (A) and 786-O (B) cell lines. Tumor cell proliferation was assessed in vitro. Cells were transiently transfected with pcDNA3.1 (empty vector) or pcDNA3.1/GPC3 and replated 24 h post-transfection for selection with Geneticin/G418. After 14 days of selection, colonies were stained with Giemsa and counted. Data are presented as the mean of two independent experiments ± SEM. Group comparisons were carried out using Student’s t test, *p < 0.01.
Effect of GPC3 on proliferation in cell lines
Cell proliferation was determined by an MTT assay in ACHN and 786-O cells overexpressing GPC3 at 24 h, 48 h, 72 h, 96 h and 120 h after transfection. ACHN cells overexpressing GPC3 experienced a significant reduction in cell proliferation compared with ACHN cells lacking GPC3 expression after 48 h (p < 0.01), 72 h, 96 h and 120 h (p < 0.0001) (Figure 4A). 786-O cells overexpressing GPC3 also had reduced proliferation after 48 h, 72 h, 96 h and 120 h (p < 0.0001) (Figure 4B).
Figure 4.
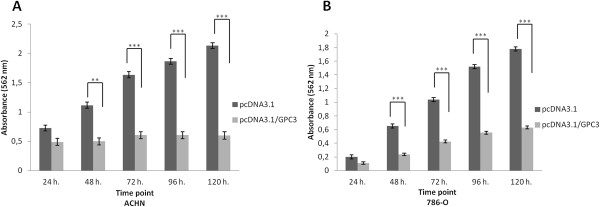
Effect of GPC3 on cell proliferation. Cell viability of ACHN and 786-O cells was measured 24 h, 48 h, 72 h, 96 h and 120 h post-transfection with pcDNA3.1 (empty vector) and pcDNA3.1/GPC3 by MTT assay. Data are presented as the mean of two independent experiments ± SEM. Group comparisons were carried out using Student’s t test. A) ACHN cells overexpressing GPC3 experienced a significant reduction in proliferation rate after 48 h (**p < 0.01), 72, 96 and 120 h (***p < 0.0001). B) 786-O overexpressing GPC3 experienced a significant reduction in proliferation rate after 48 h, 72 h, 96 h and 120 h (*** p <0.0001).
Effect of GPC3 on apoptosis in cell lines
The ability of GPC3 to induce apoptosis was also evaluated. The rate of apoptosis was analyzed in cell lines after 24 h, 48 h and 72 h of transfection with pcDNA3.1/GPC3 and pcDNA3.1 vectors using FITC-Annexin V/PI. No difference in apoptosis was observed between ACHN and 786-O cells overexpressing or lacking GPC3 at any analyzed period of time (p > 0.05) (Figure 5).
Figure 5.
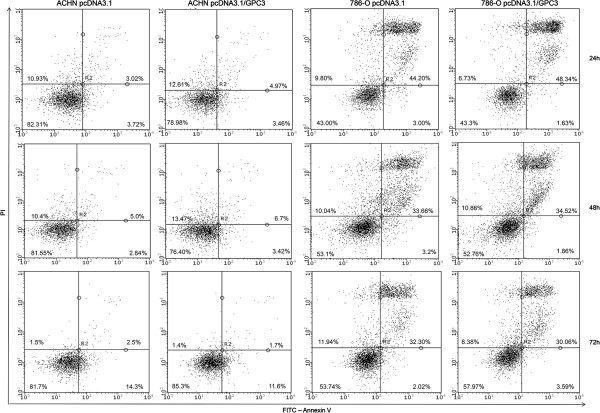
The effect of GPC3 on apoptosis. Apoptosis in ACHN and 786-O cell lines was analyzed using flow cytometry 24 h, 48 h and 72 h post-transfection. No difference in apoptosis was observed between ACHN and 786-O cell overexpressing and lacking GPC3 at any of the time points analyzed. Viable cells (Annexin V-/PI-) are located in the bottom left, early apoptotic cells (Annexin V+/PI-) in the bottom right, late apoptotic cells (Annexin V+/PI+) in the top right and necrotic cells (Annexin V-/ PI+) in the top left quadrants, respectively.
GPC3 alters cell cycle progression
We then performed cell cycle analysis on ACHN and 786-O cell lines, using flow cytometry 24 h, 48 h and 72 h after transfection with pcDNA3.1/GPC3. The number of ACHN and 786-O cells overexpressing GPC3 was evaluated in each phase of the cell cycle and was compared with the number of ACHN and 786-O cells lacking GPC3. There were more ACHN cells overexpressing GPC3 in the G1 phase (24 h and 48 h, p < 0.001; 72 h, p < 0.01) and fewer cells overexpressing GPC3 in the S phase after transfection (48 h and 72 h, p < 0.001) (Figure 6A). In the 786-O cell line, a higher number of cells overexpressing GPC3 was observed in the G1 phase after transfection (48 h, p < 0.01; 72 h, p < 0.05) compared with 786-O cells lacking GPC3 (Figure 6B).
Figure 6.
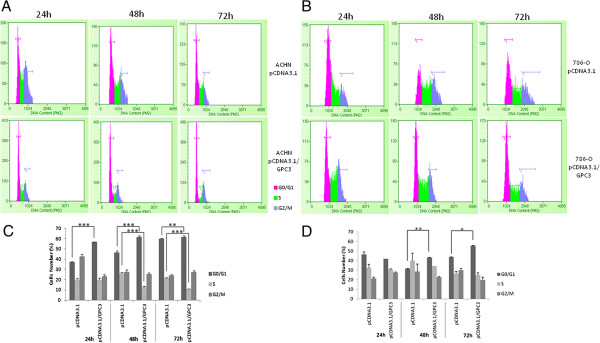
Effect of GPC3 on cell cycle in cell lines. A) ACHN cell cycle histogram. B) 786-O cell cycle histogram. C) The number of ACHN pcDNA3.1/GPC3 cells was significantly increased during the G1 phase (24 h and 48 h ***p < 0.001, 72 h **p < 0.01, ANOVA) and decreased during the S phase (48 h and 72 h ***p < 0.001, ANOVA). D) The number of 786-O pcDNA3.1/GPC3 cells was significantly increased in the G1 phase 48 h (**p < 0.01, ANOVA) and 72 h (*p < 0.05, ANOVA) post-transfection.
Discussion
Glypican-3 (GPC3) is a member of the family of heparan sulfate proteoglycans [25, 26]. Heparan sulfate proteoglycans (HSPGs) have profound effects on both tumor cell growth kinetics and metastasis formation [27]. HSPGs have the functional capacity to regulate a myriad of molecular interactions that induce tumor cell proliferation and metastasis [28]. Glypican-3 expression is higher in several tissues, such as the gastrointestinal tract, in human embryos [29].
In different types of tumors, GPC3 expression and its carcinogenic role are variable. In some tissues, such as ovary [19, 30], breast [20, 31] and lung (adenocarcinoma) [32], this glypican is downregulated, acting as a tumor suppressor, whereas in other tumors, it is overexpressed and functions as an oncoprotein, as observed in liver [33–35], lung (squamous cell carcinoma) [32, 36], melanoma [18] and embryonal tumors [37]. In a recent study, Gailey and Bellizzi [38] analyzed GPC3 protein expression in squamous cell carcinoma (SCCs) of diverse anatomic sites and in invasive urothelial carcinomas from the urinary bladder. They observed GPC3 staining was present in 17.3% of 156 tumors, including those of the anus (10.0%), cervix (27.3%), esophagus (28.6%), larynx (30.0%), lung (50.0%), tongue base/tonsil (12.5%), urinary bladder (12.2%), ventral tongue/floor of mouth (12.5%) and vagina (40.0%).
In the present study, GPC3 expression was downregulated in primary clear cell renal cell carcinoma samples and cell lines. To the best of our knowledge, this report is the first that has detected downregulation of the GPC3 gene in clear cell renal cell carcinoma cell lines. Okon et al. [39] have shown a point of distinction between GPC3 expression in chromophobe carcinoma and clear cell renal cell carcinoma; GPC3 expression was upregulated in chromophobe carcinoma and downregulated in clear cell carcinoma, agreeing with our expression results in tissue samples and cell lines.
Gailey and Bellizzi [38] also observed an absence of GPC3 expression in squamous cell carcinomas (SCCs) from the penis, skin and vulva. As opposed to the results of this study in clear cell renal cell carcinoma, GPC3 overexpression has emerged as a positive marker in liver cancer because it is highly expressed in 70-100% of hepatocellular carcinomas (HCCs) but not in normal adult liver tissue [33–35]. In addition to being proposed as a marker for liver tumor diagnosis, GPC3 has also been evaluated as a target for antibody- and cell-based therapies of HCC [18].
Cell growth was reduced in cells overexpressing GPC3 protein, as measured by colony formation and proliferation rates 48 h post-transfection in both the 786-O and ACHN cell lines used in this study. Lin et al. [19] and Murthy et al. [40] found that GPC3 re-expression in ovarian cancer cell lines resulted in inhibition of the efficiency of colony formation. Some studies have shown that GPC3 plays an important role in cell growth and differentiation [41, 42], such as in the case of hepatocellular carcinoma [43]. The data obtained in the present study suggest that GPC3 inhibits cell proliferation in clear cell renal cell carcinoma.
Therefore, we studied whether the reduced proliferation rate in cells overexpressing GPC3 occurs through the induction of apoptosis or through cell cycle arrest. Apoptosis, or programmed cell death, is a crucial point in the carcinogenic process. Cancer cells can overcome the apoptosis mechanism, and tumor progression continues [26, 44]. We observed that most cells, whether overexpressing or lacking GPC3, were viable at all time points in ACHN and 786-O cell lines. Our results suggest that cells overexpressing GPC3 in renal cell carcinoma do not induce apoptosis; therefore, we hypothesized that the inhibition of cell proliferation in renal carcinoma cells might occur due to cell cycle arrest.
To verify whether GPC3 overexpression arrests the cell cycle in renal cell carcinoma, we used flow cytometry to perform cell cycle analysis. The observed growth-repressive effect of GPC3 overexpressing cells was reflected by their arrest in the G1 cell cycle phase, which caused these cells to proliferate less in both cell lines. In this study, we observed changes in cell proliferation through the use of MTT assays in the ACHN cell line 48 h post-transfection. Using flow cytometry, we were able to identify changes in the cell cycle and, consequently, in cell proliferation 24 h after transfection for the same cell line. This difference is likely due to the increased sensitivity of the flow cytometry methodology to detecting changes in cell cycle and proliferation compared with the MTT assay. One previous study demonstrated that inhibiting GPC3 expression released hepatocyte carcinoma cells from G1 arrest and thus modulated cell cycle progression in this type of cancer [45].
Moreover, cell surface HSPGs inhibit invasion by promoting tight cell–cell and cell–extracellular matrix (ECM) adhesion. Previous studies documented diminished quantity and quality of heparan sulfate isolated from transformed cells compared with normal cells [46]. This alteration in heparan sulfate accompanies a reduction in the adhesive capacity of transformed cells. Low levels of cell surface heparan sulfates also correlate with high metastatic activity in melanomas [47, 48]. Furthermore, GPC3 downregulation was more significant in invasive areas, a result which further supports an inhibitory role for GPC3 in tumor progression in hepatocellular carcinomas [49]. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Conversely, glypican-3 expression is decreased in human breast cancers, and ectopic expression of GPC3 inhibits growth of breast cancer cell lines [20]. Additionally, glypican-3 was associated with the inhibition of invasion and metastasis of a mammary carcinoma cell line in vivo [50]. Thus, cells with normal epithelial morphology exhibit retention of cell surface HSPGs and tight attachment to the extracellular matrix. Cells that are beginning to invade exhibit reduced adhesion to the ECM, loss of epithelial morphology, and diminished levels of HSPG expression, whereas deeply invading cells completely lose HSPG expression [28].
Conclusions
GPC3 gene expression is downregulated in primary clear cell renal cell carcinoma, and GPC3 protein overexpression in clear cell renal cell carcinoma cell lines arrests cells during the G1 phase of the cell cycle, consequently reducing the proliferation rate. These results suggest that GPC3 acts as a tumor suppressor in clear cell renal cell carcinoma.
Acknowledgements
We thank Dr. Daniel Seabra and Dr. Eliney Ferreria Faria for their collaboration in collecting the samples and Dr. Lígia Maria Kerr for pathological review.
Abbreviations
- GPC3
Glypican 3
- HSPG
Heparan sulfate proteoglycan
- qRT-PCR
Real time quantitative polymerase chain reaction
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- mRNA
Messenger ribonucleic acid
- RCC
Renal cell carcinoma
- CCRCC
Clear cell renal cell carcinoma
- VHL
Von Hippel-Lindau tumor suppressor
- GPI
Glycosylphosphatidylinositol
- ATCC
American Type Culture Collection
- FBS
Fetal bovine serum
- PCR
Polymerase chain reaction
- RNA
Ribonucleic acid
- cDNA
Complementary deoxyribonucleic acid
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- HRP
Horseradish peroxidase
- 3
3′-diaminobenzidine
- FITC
Fluorescein isothiocyanate
- PI
Propidium iodide
- PBS
Phosphate buffered saline
- ECM
Extracellular matrix.
Footnotes
Competing interests
The authors declare they have no competing interests.
Authors’ contributions
MFC, PH and WASJr designed the research, supervised all experiments and drafted this paper. PJSP constructed the plasmid used in this study. ABBO and NMC performed qRT-PCR experiments. MCV executed the colony formation assay, proliferation assay and statistical analysis. LFA performed immunocytochemistry. ALGC executed the apoptosis assay. BS performed cell cycle analysis. All authors read and approved the final manuscript.
Contributor Information
Marina Curado Valsechi, Email: mcvalsechi@yahoo.com.br.
Ana Beatriz Bortolozo Oliveira, Email: anabbortolozo@gmail.com.
André Luis Giacometti Conceição, Email: andre4487@gmail.com.
Bruna Stuqui, Email: bru.stuqui@gmail.com.
Natalia Maria Candido, Email: na_candido@hotmail.com.
Paola Jocelan Scarin Provazzi, Email: paolaprovazzi@gmail.com.
Luiza Ferreira de Araújo, Email: luizaferreiraaraujo@gmail.com.
Wilson Araújo Silva, Jr, Email: wilsonjr@usp.br.
Marilia de Freitas Calmon, Email: macal131@gmail.com.
Paula Rahal, Email: rahalp@yahoo.com.br.
References
- 1.Xue YJ, Xiao RH, Long DZ, Zou XF, Wang XN, Zhang GX, Yuan YH, Wu GQ, Yang J, Wu YT, Xu H, Liu FL, Liu M. Overexpression of FoxM1 is associated with tumor progression in patients with clear cell renal cell carcinoma. J Transl Med. 2012;10:200. doi: 10.1186/1479-5876-10-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xu L, Zhu Y, Xu J, Wu K, Li J, Xu W, Liu H, Wang S, Yin H, Chen L, Wang G, Lin Z. Notch1 activation promotes renal cell carcinoma growth via PI3K/Akt signaling. Cancer Sci. 2012;103(7):1253–1258. doi: 10.1111/j.1349-7006.2012.02291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Poppel H, Becker F, Cadeddu JA, Gill IS, Janetschek G, Jewett MA, Laguna MP, Marberger M, Montorsi F, Polascik TJ, Ukimura O, Zhu G. Treatment of localised renal cell carcinoma. Eur Urol. 2011;60(4):662–672. doi: 10.1016/j.eururo.2011.06.040. [DOI] [PubMed] [Google Scholar]
- 4.Drucker BJ. Renal cell carcinoma: current status and future prospects. Cancer Treat Rev. 2005;31(7):536–545. doi: 10.1016/j.ctrv.2005.07.009. [DOI] [PubMed] [Google Scholar]
- 5.Martignoni G, Brunelli M, Gobbo S, Remo A, Ficarra V, Cossu-Rocca P, Pea M, Chilosi M, Menestrina F, Cheng L. Role of molecular markers in diagnosis and prognosis of renal cell carcinoma. Anal Quant Cytol Histol. 2007;29(1):41–49. [PubMed] [Google Scholar]
- 6.Lang K, Danchenko N, Gondek K, Schwartz B, Thompson D. The burden of illness associated with renal cell carcinoma in the United States. Urol Oncol. 2007;25(5):368–375. doi: 10.1016/j.urolonc.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Girgis AH, Iakovlev VV, Beheshti B, Bayani J, Squire JA, Bui A, Mankaruos M, Youssef Y, Khalil B, Khella H, Pasic M, Yousef GM. Multilevel whole-genome analysis reveals candidate biomarkers in clear cell renal cell carcinoma. Cancer Res. 2012;72(20):5273–5284. doi: 10.1158/0008-5472.CAN-12-0656. [DOI] [PubMed] [Google Scholar]
- 8.Wood LS. Spotlight on Symposia from the ONS 32nd Annual Congress: 2007; Las Vegas, NV. 2007. Managing the Complex Journey of Renal Cell Carcinoma. [Google Scholar]
- 9.Takahashi M, Rhodes DR, Furge KA, Kanayama H, Kagawa S, Haab BB, Teh BT. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci U S A. 2001;98(17):9754–9759. doi: 10.1073/pnas.171209998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moch H. An overview of renal cell cancer: pathology and genetics. Semin Cancer Biol. 2013;23(1):3–9. doi: 10.1016/j.semcancer.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 11.Nickerson ML, Jaeger E, Shi Y, Durocher JA, Mahurkar S, Zaridze D, Matveev V, Janout V, Kollarova H, Bencko V, Navratilova M, Szeszenia-Dabrowska N, Mates D, Mukeria A, Holcatova I, Schmidt LS, Toro JR, Karami S, Hung R, Gerard GF, Linehan WM, Merino M, Zbar B, Boffetta P, Brennan P, Rothman N, Chow WH, Waldman FM, Moore LE. Improved identification of von Hippel-Lindau gene alterations in clear cell renal tumors. Clin Cancer Res. 2008;14(15):4726–4734. doi: 10.1158/1078-0432.CCR-07-4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirota E, Yan L, Tsunoda T, Ashida S, Fujime M, Shuin T, Miki T, Nakamura Y, Katagiri T. Genome-wide gene expression profiles of clear cell renal cell carcinoma: identification of molecular targets for treatment of renal cell carcinoma. Int J Oncol. 2006;29(4):799–827. [PubMed] [Google Scholar]
- 13.Yu G, Yao W, Wang J, Ma X, Xiao W, Li H, Xia D, Yang Y, Deng K, Xiao H, Wang B, Guo X, Guan W, Hu Z, Bai Y, Xu H, Liu J, Zhang X, Ye Z. LncRNAs expression signatures of renal clear cell carcinoma revealed by microarray. PLoS ONE. 2012;7(8):e42377. doi: 10.1371/journal.pone.0042377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu B, Bell AW, Paranjpe S, Bowen WC, Khillan JS, Luo JH, Mars WM, Michalopoulos GK. Suppression of liver regeneration and hepatocyte proliferation in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2010;52(3):1060–1067. doi: 10.1002/hep.23794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sawada Y, Sakai M, Yoshikawa T, Ofuji K, Nakatsura T. A glypican-3-derived peptide vaccine against hepatocellular carcinoma. Oncoimmunology. 2012;1(8):1448–1450. doi: 10.4161/onci.21351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kohashi K, Nakatsura T, Kinoshita Y, Yamamoto H, Yamada Y, Tajiri T, Taguchi T, Iwamoto Y, Oda Y. Glypican 3 expression in tumors with loss of SMARCB1/INI1 protein expression. Hum Pathol. 2013;44(4):526–533. doi: 10.1016/j.humpath.2012.06.014. [DOI] [PubMed] [Google Scholar]
- 17.Wang YL, Zhu ZJ, Teng DH, Yao Z, Gao W, Shen ZY. Glypican-3 expression and its relationship with recurrence of HCC after liver transplantation. World J Gastroenterol. 2012;18(19):2408–2414. doi: 10.3748/wjg.v18.i19.2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho M, Kim H. Glypican-3: a new target for cancer immunotherapy. Eur J Cancer. 2011;47(3):333–338. doi: 10.1016/j.ejca.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin H, Huber R, Schlessinger D, Morin PJ. Frequent silencing of the GPC3 gene in ovarian cancer cell lines. Cancer Res. 1999;59(4):807–810. [PubMed] [Google Scholar]
- 20.Xiang YY, Ladeda V, Filmus J. Glypican-3 expression is silenced in human breast cancer. Oncogene. 2001;20(50):7408–7412. doi: 10.1038/sj.onc.1204925. [DOI] [PubMed] [Google Scholar]
- 21.Li L, Jin R, Zhang X, Lv F, Liu L, Liu D, Liu K, Li N, Chen D. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology. 2012;56(4):1380–1390. doi: 10.1002/hep.25891. [DOI] [PubMed] [Google Scholar]
- 22.Sun CK, Chua MS, He J, So SK. Suppression of glypican 3 inhibits growth of hepatocellular carcinoma cells through up-regulation of TGF-beta2. Neoplasia. 2011;13(8):735–747. doi: 10.1593/neo.11664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.International Union againt cancer . TNM Classification of Malignant Tumors. 7 2009. [Google Scholar]
- 24.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Umezu T, Shibata K, Shimaoka M, Kajiyama H, Yamamoto E, Ino K, Nawa A, Senga T, Kikkawa F. Gene silencing of glypican-3 in clear cell carcinoma of the ovary renders it more sensitive to the apoptotic agent paclitaxel in vitro and in vivo. Cancer Sci. 2010;101(1):143–148. doi: 10.1111/j.1349-7006.2009.01382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu S, Li Y, Chen W, Zheng P, Liu T, He W, Zhang J, Zeng X. Silencing glypican-3 expression induces apoptosis in human hepatocellular carcinoma cells. Biochem Biophys Res Commun. 2012;419(4):656–661. doi: 10.1016/j.bbrc.2012.02.069. [DOI] [PubMed] [Google Scholar]
- 27.Liu D, Shriver Z, Qi Y, Venkataraman G, Sasisekharan R. Dynamic regulation of tumor growth and metastasis by heparan sulfate glycosaminoglycans. Semin Thromb Hemost. 2002;28(1):67–78. doi: 10.1055/s-2002-20565. [DOI] [PubMed] [Google Scholar]
- 28.Sanderson RD. Heparan sulfate proteoglycans in invasion and metastasis. Semin Cell Dev Biol. 2001;12(2):89–98. doi: 10.1006/scdb.2000.0241. [DOI] [PubMed] [Google Scholar]
- 29.Lin CW, Mars WM, Paranjpe S, Donthamsetty S, Bhave VS, Kang LI, Orr A, Bowen WC, Bell AW, Michalopoulos GK. Hepatocyte proliferation and hepatomegaly induced by phenobarbital and 1,4-bis [2-(3,5-dichloropyridyloxy)] benzene is suppressed in hepatocyte-targeted glypican 3 transgenic mice. Hepatology. 2011;54(2):620–630. doi: 10.1002/hep.24417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esheba GE, Pate LL, Longacre TA. Oncofetal protein glypican-3 distinguishes yolk sac tumor from clear cell carcinoma of the ovary. Am J Surg Pathol. 2008;32(4):600–607. doi: 10.1097/PAS.0b013e31815a565a. [DOI] [PubMed] [Google Scholar]
- 31.Buchanan C, Stigliano I, Garay-Malpartida HM, Rodrigues Gomes L, Puricelli L, Sogayar MC, Bal De Kier Joffe E, Peters MG. Glypican-3 reexpression regulates apoptosis in murine adenocarcinoma mammary cells modulating PI3K/Akt and p38MAPK signaling pathways. Breast Cancer Res Treat. 2010;119(3):559–574. doi: 10.1007/s10549-009-0362-9. [DOI] [PubMed] [Google Scholar]
- 32.Tsuta K, Tanabe Y, Yoshida A, Takahashi F, Maeshima AM, Asamura H, Tsuda H. Utility of 10 immunohistochemical markers including novel markers (desmocollin-3, glypican 3, S100A2, S100A7, and Sox-2) for differential diagnosis of squamous cell carcinoma from adenocarcinoma of the Lung. J Thorac Oncol. 2011;6(7):1190–1199. doi: 10.1097/JTO.0b013e318219ac78. [DOI] [PubMed] [Google Scholar]
- 33.Capurro M, Wanless IR, Sherman M, Deboer G, Shi W, Miyoshi E, Filmus J. Glypican-3: a novel serum and histochemical marker for hepatocellular carcinoma. Gastroenterology. 2003;125(1):89–97. doi: 10.1016/S0016-5085(03)00689-9. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Li P, Zhai Y, Qu CF, Zhang LJ, Tan YF, Li N, Ding HG. Diagnostic value of glypican-3 in serum and liver for primary hepatocellular carcinoma. World J Gastroenterol. 2010;16(35):4410–4415. doi: 10.3748/wjg.v16.i35.4410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L, Liu H, Sun L, Li N, Ding H, Zheng J. Glypican-3 as a potential differential diagnosis marker for hepatocellular carcinoma: a tissue microarray-based study. Acta Histochem. 2012;114(6):547–552. doi: 10.1016/j.acthis.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 36.Aviel-Ronen S, Lau SK, Pintilie M, Lau D, Liu N, Tsao MS, Jothy S. Glypican-3 is overexpressed in lung squamous cell carcinoma, but not in adenocarcinoma. Mod Pathol. 2008;21(7):817–825. doi: 10.1038/modpathol.2008.37. [DOI] [PubMed] [Google Scholar]
- 37.Chan ES, Pawel BR, Corao DA, Venneti S, Russo P, Santi M, Sullivan LM. Immunohistochemical expression of glypican-3 in pediatric tumors: an analysis of 414 cases. Pediatr Dev Pathol. 2013;16(4):272–277. doi: 10.2350/12-06-1216-OA.1. [DOI] [PubMed] [Google Scholar]
- 38.Gailey MP, Bellizzi AM. Immunohistochemistry for the novel markers glypican 3, PAX8, and p40 (DeltaNp63) in squamous cell and urothelial carcinoma. Am J Clin Pathol. 2013;140(6):872–880. doi: 10.1309/AJCP4NSKW5TLGTDS. [DOI] [PubMed] [Google Scholar]
- 39.Okon K. Glypican-3 is expressed in chromophobe renal cell carcinomas. Pol J Pathol. 2008;59(1):15–20. [PubMed] [Google Scholar]
- 40.Murthy SS, Shen T, De Rienzo A, Lee WC, Ferriola PC, Jhanwar SC, Mossman BT, Filmus J, Testa JR. Expression of GPC3, an X-linked recessive overgrowth gene, is silenced in malignant mesothelioma. Oncogene. 2000;19(3):410–416. doi: 10.1038/sj.onc.1203322. [DOI] [PubMed] [Google Scholar]
- 41.Gao W, Ho M. The role of glypican-3 in regulating Wnt in hepatocellular carcinomas. Cancer Rep. 2011;1(1):14–19. [PMC free article] [PubMed] [Google Scholar]
- 42.Midorikawa Y, Ishikawa S, Iwanari H, Imamura T, Sakamoto H, Miyazono K, Kodama T, Makuuchi M, Aburatani H. Glypican-3, overexpressed in hepatocellular carcinoma, modulates FGF2 and BMP-7 signaling. Int J Cancer. 2003;103(4):455–465. doi: 10.1002/ijc.10856. [DOI] [PubMed] [Google Scholar]
- 43.Miao HL, Pan ZJ, Lei CJ, Wen JY, Li MY, Liu ZK, Qiu ZD, Lin MZ, Chen NP, Chen M. Knockdown of GPC3 inhibits the proliferation of Huh7 hepatocellular carcinoma cells through down-regulation of YAP. J Cell Biochem. 2013;114(3):625–631. doi: 10.1002/jcb.24404. [DOI] [PubMed] [Google Scholar]
- 44.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 45.Farooq M, Hwang SY, Park MK, Kim JC, Kim MK, Sung YK. Blocking endogenous glypican-3 expression releases Hep 3B cells from G1 arrest. Mol Cells. 2003;15(3):356–360. [PubMed] [Google Scholar]
- 46.Robinson J, Viti M, Hook M. Structure and properties of an under-sulfated heparan sulfate proteoglycan synthesized by a rat hepatoma cell line. J Cell Biol. 1984;98(3):946–953. doi: 10.1083/jcb.98.3.946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kure S, Yoshie O, Aso H. Metastatic potential of murine B16 melanoma correlates with reduced surface heparan sulfate glycosaminoglycan. Jpn J Cancer Res. 1987;78(11):1238–1245. [PubMed] [Google Scholar]
- 48.Moczar M, Caux F, Bailly M, Berthier O, Dore JF. Accumulation of heparan sulfate in the culture of human melanoma cells with different metastatic ability. Clin Exp Metastasis. 1993;11(6):462–471. doi: 10.1007/BF00054937. [DOI] [PubMed] [Google Scholar]
- 49.Zhu ZW, Friess H, Wang L, Abou-Shady M, Zimmermann A, Lander AD, Korc M, Kleeff J, Buchler MW. Enhanced glypican-3 expression differentiates the majority of hepatocellular carcinomas from benign hepatic disorders. Gut. 2001;48(4):558–564. doi: 10.1136/gut.48.4.558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters MG, Farias E, Colombo L, Filmus J, Puricelli L, Bal De Kier Joffe E. Inhibition of invasion and metastasis by glypican-3 in a syngeneic breast cancer model. Breast Cancer Res Treat. 2003;80(2):221–232. doi: 10.1023/A:1024549729256. [DOI] [PubMed] [Google Scholar]
Pre-publication history
- The pre-publication history for this paper can be accessed here:http://www.biomedcentral.com/1471-2407/14/631/prepub


