Abstract
Cervical cancer is responsible for 10–15% of cancer-related deaths in women worldwide1,2. The etiological role of infection with high-risk human papilloma viruses (HPV) in cervical carcinomas is well established3. Previous studies have implicated somatic mutations in PIK3CA, PTEN, TP53, STK11 and KRAS4–7 as well as several copy number alterations in the pathogenesis of cervical carcinomas8,9. Here, we report whole exome sequencing analysis of 115 cervical carcinoma-normal paired samples, transcriptome sequencing of 79 cases and whole genome sequencing of 14 tumor-normal pairs. Novel somatic mutations in 79 primary squamous cell carcinomas include recurrent E322K substitutions in the MAPK1 gene (8%), inactivating mutations in the HLA-B gene (9%), and mutations in EP300 (16%), FBXW7 (15%), NFE2L2 (4%) TP53 (5%) and ERBB2 (6%). We also observed somatic ELF3 (13%) and CBFB (8%) mutations in 24 adenocarcinomas. Squamous cell carcinomas had higher frequencies of somatic mutations in the Tp*C dinucleotide context than adenocarcinomas. Gene expression levels at HPV integration sites were significantly higher in tumors with HPV integration compared with expression of the same genes in tumors without viral integration at the same site. These data demonstrate several recurrent genomic alterations in cervical carcinomas that suggest novel strategies to combat this disease.
The prevention of cervical cancer by Pap smear-based screening and treatment programs has been largely successful in resource-rich countries. However, cervical cancer is the 2nd most common cause of cancer-related deaths in women in developing countries, where many patients are diagnosed at advanced stages of disease with limited treatment options and poor prognosis1. Recent advances in targeted therapy against specific somatic alterations have transformed the management of cancers in general10, and the discovery of novel therapeutic targets in cervical cancer could improve upon current strategies to combat cervical carcinomas.
To provide comprehensive data on the landscape of genomic aberrations that contribute to cervical cancer, we investigated a cohort that included 100 patients from Norway and 15 patients from Mexico (Supplementary Notes 1–7). We performed exome sequencing of 193,094 exons, covering a median of 34.2 Mb at a median of 89x (range: 56–122x) coverage for tumor samples and 88x (range: 69–122x) coverage for normal samples, followed by calling of somatic mutations using the Mutect algorithm11, and identified a total of 17,795 somatic mutations across the entire dataset, including 11,419 missense, 936 nonsense, 4,643 silent, 219 splice site, 29 translation start site mutations, as well as 401 deletions and 131 insertions.
The aggregate nonsilent mutation rate across the dataset was 3.7 per Mb. However, squamous cell carcinomas had a higher rate of nonsilent mutations (4.2 mutations/Mb) than adenocarcinomas (1.6 mutations/Mb) (Wilcoxon p =0.0095). The clinical, pathologic, epidemiologic and mutational characteristics of the tumors are summarized in Supplementary Figs. 1–6, Supplementary Tables 1–6 and Supplementary Notes 8 and 9.
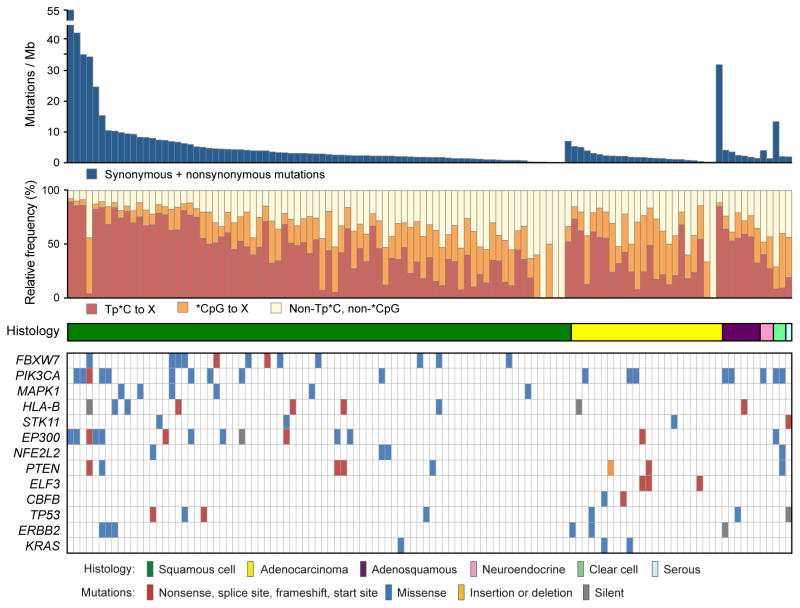
Hierarchical clustering of all 115 tumors based on the mutational context revealed that most tumors were characterized by previously described12 mutational signatures: with predominantly Tp*C to T/G mutations and *CpG to T mutations (Fig. 1, Supplementary Fig. 4). Tp*C mutations were present at a relative frequency of >0.5 in 53 (46%) tumors, and the relative frequency of Tp*C mutations was positively correlated with mutation rates, especially in squamous cell carcinomas (Fig. 1, Supplementary Notes 8, Supplementary Fig. 5). In addition, 5648 (54%) of the 10328 non-silent mutations observed in squamous cell carcinomas were Tp*C to T/G mutations.
Fig. 1. Relationship of mutational spectrum and rates with clinicopathological characteristics in cervical carcinoma.
All panels are aligned with vertical tracks representing 115 individuals. The data is sorted in order by histology (middle panel) and total mutational rate (top panel). The relative frequencies of nucleotide mutations occurring at cytosines preceeded by thymines (Tp*C) or at cytosines followed by guanines (*CpG) sites are depicted in red and orange respectively, on the second panel. The bottom heatmap shows the distribution of mutations in significantly mutated genes (q<0.1) in squamous cell carcinomas and adenocarcinomas in the order listed in Table 1. TP53, ERBB2 and KRAS were significant recurrence (q<0.1) among cancer driver genes reported in COSMIC.
We performed mutation significance analyses on 79 squamous cell carcinomas and 24 adenocarcinomas. Genes were determined to be significantly mutated if recurrent mutations were found in that gene at a false discovery rate of q<0.1 after correction for multiple hypothesis testing, as previously described13 (Supplementary Note 6). Details of candidate mutation validation are presented in Supplementary Figs 6 and 7.
As expected, recurrent mutations in PIK3CA, PTEN and STK11 were present in 14%, 6%, and 4%, respectively, of 79 squamous cell carcinomas (Table 1). In addition, we found significantly recurrent mutations in EP300 (16%), FBXW7 (15%), HLA-B (9%), MAPK1 (8%), and NFE2L2 (4%), here reported for the first time, to our knowledge, in primary squamous cell cervical carcinomas (Table 1, Fig. 1, Supplementary Table 7, Supplementary Fig. 8). In addition, TP53 (9%) and ERBB2 (5%) were found to be significantly mutated in analyses focused only on genes previously reported as mutated in the COSMIC database (http://cancer.sanger.ac.uk/cancergenome/projects/cosmic) (Supplementary Table 9a). Interestingly, 3 of the 6 ERBB2 mutations (S310F, S310Y and V842I; Supplementary Fig. 8) are known oncogenic driver mutations and in vitro therapeutic targets in lung 14 and breast cancer15.
Table 1.
Genes with Significantly Recurrent Somatic Mutations in Cervical Carcinomas
| Gene | Description | Nonsilent mutations | Relative frequency | Patients | Unique sites | Silent mutations | Indel + null | q |
|---|---|---|---|---|---|---|---|---|
| SQUAMOUS CELL CARCINOMA (N=79) | ||||||||
| FBXW7** | F-box and WD repeat domain containing 7 | 12 | 15% | 12 | 8 | 0 | 2 | 4.03E-12 |
| PIK3CA | phosphoinositide-3-kinase, catalytic, alpha polypeptide | 11 | 14% | 10 | 5 | 0 | 1 | <9.08e-12 |
| MAPK1** | mitogen-activated protein kinase 1 | 6 | 8% | 6 | 3 | 0 | 0 | 0.000671 |
| HLA-B+ | major histocompatibility complex, class I, B | 7 | 9% | 6 | 7 | 1 | 3 | 0.00169 |
| STK11 | serine/threonine kinase 11 | 3 | 4% | 2 | 2 | 0 | 1 | 0.012 |
| EP300+ | E1A binding protein p300 | 13 | 16% | 12 | 13 | 1 | 4 | 0.0354 |
| NFE2L2+ | nuclear factor (erythroid-derived 2)-like 2 | 3 | 4% | 3 | 2 | 0 | 0 | 0.0597 |
| PTEN | phosphatase and tensin homolog (mutated in multiple advanced cancers 1) | 5 | 6% | 5 | 5 | 0 | 3 | 0.0693 |
| ADENOCARCINOMA (N=24) | ||||||||
| ELF3* | E74-like factor 3 (ets domain transcription factor, epithelial-specific) | 3 | 13% | 3 | 3 | 0 | 3 | 0.03 |
| CBFB* | core-binding factor, beta subunit | 2 | 8% | 2 | 2 | 0 | 1 | 0.0342 |
Indel: insertions or deletions;
Null: nonsense, frameshft or splice-site mutations;
q: q value, false discovery rate (Benjamini-Hochberg procedure).
Genes with mutations observed in only squamous cell carcinomas
Genes with mutations observed in only adenocarcinomas
Genes with a majority of mutations occurring in squamous cell carcinomas
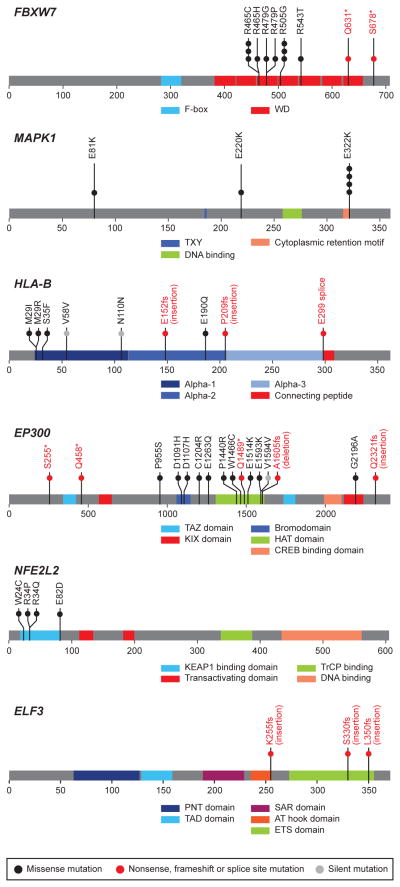
Somatic MAPK1 mutations were observed in 6/79 squamous cell carcinomas of the cervix (7%), each involving a G-to-A transition resulting in recurrent E322K mutations in 4 individuals, and E81K and E220K mutations in 1 individual each (Fig. 2). To our knowledge, this is the first report of recurrent mutations of MAPK1 in primary human cancer, although the MAPK1 E322K mutation has been reported in an oropharyngeal carcinoma cell line16 and scattered unpublished reports are summarized in COSMIC. The recurrent site-specific MAPK1 mutations and the known role of the MAPK signaling pathway in cancer17 suggest the possibility that mutant MAPK1 may exert oncogenic activity.
Fig. 2. Novel recurrent somatic mutations in cervical carcinoma.
The locations of somatic mutations in novel significantly mutated genes in 115 cervical carcinoma, FBXW7, MAPK1, HLA-B, EP300, NFE2L2 and ELF3 are shown in the context of protein domain models derived from UniProt and Pfam annotations. Numbers refer to amino acid residues. Each filled circle represents an individual mutated tumor sample: missense and silent mutations are represented by filled black and grey circles, respectively while nonsense, frameshift, and splice site mutations are represented by filled red circles and red text. Domains are depicted with various colors with an appropriate key located on the right hand of each domain model.
We observed EP300 and FBXW7 mutations in our dataset, similar to recent reports in endometrial and head and neck cancers18,19. Thirteen of 15 nonsilent EP300 mutations are novel in cancer, with 8 of these (including 2 nonsense) residing in the histone acetyltransferase and bromo- domains required for EP300 activity20 (Fig. 2). In addition, there were 2 truncating mutations at residues S255 and Q458 in EP300. The FBXW7 gene also had 2 novel truncating mutations at residues Q631 and R678, with 10 other mutations residing in the WD40 domains required to form the scaffold for the Skp1-Cul1-F-box protein complex21 (Supplementary Fig. 8). Furthermore, all four NFE2L2 mutations (W24C, R34P, R34Q and E82D) are in the domain required for interacting with its negative regulator, KEAP122 (Fig. 2), consistent with similar findings in lung squamous cancers23. Interestingly, mutations in these genes (FBXW7, EP300, MAPK1, NFE2L2) occur largely in a non-overlapping pattern in our dataset (Fig. 1, Supplementary Fig. 6b). These observations suggest that epigenetic regulation and the oxidative stress response may play important roles in cervical cancer pathogenesis.
We found 4 missense and 3 frameshift mutations in the HLA-B gene encoding the histocompatibility leukocyte antigen HLA-B (Table 1, Fig. 2). In addition, there were somatic mutations in other genes involved in antigen presentation, including splice site, nonsense and frameshift mutations in HLA-A, a gene previously reported as mutated in squamous cell carcinomas of the lung23, and in the beta-microglobulin (B2M) gene (Supplementary Fig. 8, Supplementary Table 7). All mutations in these 3 genes were within the antigen presenting domains of each respective protein24. Intriguingly, pathway analyses also revealed that the most significantly mutated geneset in squamous cell carcinomas involves immune response genes in the interferon gamma signaling pathway, including mutations in IFNG, IFNGR1, IKBKB, JAK2 and other genes (Supplementary Table 10a). Together, these data highlight the potential significance of the synergy between HPV infection and an altered immune response in the pathogenesis of squamous cell carcinomas of the cervix.
We also investigated a smaller subset of 24 adenocarcinomas. Our analysis revealed the ELF3 (13%) and CBFB (8%) genes as recurrently mutated at q< 0.1 (Table 1, Fig. 2, Supplementary Fig. 8, Supplementary Table 8). In addition, PIK3CA (16%) and KRAS (8%) were found to be significantly mutated in analyses focused only on genes previously reported as mutated in the COSMIC database (Supplementary Table 9b), consistent with previous reports6. Furthermore, geneset analyses revealed that the PIK3CA/PTEN pathway was significantly recurrently mutated across the adenocarcinoma subset (Supplementary Table 10b).
The ELF3 mutations in the adenocarcinomas involve frameshift insertional events at amino acid positions 255, 330 and 350. ELF3 (E74-like factor-3) encodes a member of the ETS transcription factor family which is expressed and upregulated in epithelial cancers, and is both a regulator and downstream effector of the ERBB2 signaling pathway25. Interestingly, ELF3-mutated tumors have higher gene expression levels than ELF3 wildtype tumors (Supplementary Fig. 9).
The spectrum of somatic copy number alterations, rearrangements, gene expression profiles, HPV integration and other genomic events observed in this cohort are documented in Supplementary Notes 10–14, Supplementary Tables 11–22, and Supplementary Figs. 10–30. HPV integration sites were defined by the presence of least six chimeric read pairs, derived from RNA sequencing (RNASeq) data, in which the pairmate of an HPV sequence read mapped to the human genome (Supplementary Note 7). As expected, HPV integration sites were found within or in close proximity to several fragile sites (Supplementary Note 11) as well as previously reported genes26–29 including MYC, ERBB2, TP63, FANCC, RAD51B and CEACAM5 (Supplementary Table 15).
HPV integration occurred closer to amplified regions than expected by chance (Mann Whitney p < 2.2 x 10−16; Fig. 3a), with 21 (41%) of 41 integration sites overlapping with amplified regions, supporting the hypothesis that viral integration may trigger genome amplification30. In general, viral integration was localized to one locus in most tumors investigated, and most of the integration sites were observed only in one tumor each (Supplementary Table 15). In addition, many of the genes involved in the integration events are members of cellular pathways known to play important roles in cancer (Supplementary Table 16). Similar to recent observations29, we observed recurrent HPV integration into the RAD51B locus in three different tumors; intriguingly, each involved a different HPV type: HPV16, HPV18, and HPV52 (Supplementary Figs. 20–22).
Fig. 3. Relationships between HPV integration, copy number amplifications and gene expression in cervical carcinoma.
Panel (a) shows comparative histograms of true and simulated genomic distances between HPV integration sites and the nearest copy number amplification (log segmean difference >0.5). Panel (b) shows boxplots of gene expression levels across 79 cervical tumors for 41 genes with chimeric human-HPV read pairs. The expression levels for tumors with HPV integration in the respective genes are highlighted in red circles. Panel (c) shows scatter plots comparing copy number alterations and gene expression levels across 79 tumors in selected integration site genes. The red circles represent data for the tumors with HPV integration events involving the respective genes.
We also observed that gene expression levels at sites of HPV integration were significantly higher in tumors with HPV integration compared with the expression levels of the same genes across the other tumors without integration at that site (p < 2.2 x 10−16; Fig. 3b; Supplementary Figs. 22–24). For some integration sites, including MYC, ERBB2, GLI2, TNIK, NR4A2, PROX1, EIF2C2, FAM179B, and SERPINB4, high gene expression levels were associated with copy number gains (Fig. 3c, Supplementary Fig. 25). Conversely, there were no copy number changes at several other highly expressed integration sites including RPS6KB1, MAFA, PARN, EGFL7, SNIP1, POC1B, and BCL11B (Fig. 3c, Supplementary Figs. 26, Supplementary Note 11E and 11F), supporting the hypothesis that the elevated expression of these genes may be driven in part by the integrated viral promoter27.
In summary, this study has demonstrated relationships between recurrent somatic mutations, copy number alterations, gene expression and HPV integration in cervical carcinomas. We report significantly recurrent somatic mutations in the MAPK1 gene in squamous cell cervical cancers, to our knowledge the first such report in human cancers. In addition, we found evidence of potential ERBB2 activation by somatic mutation, amplification, and HPV integration, suggesting that some cervical carcinoma patients could potentially be considered as candidates for clinical trials of ERBB2 inhibitors. Furthermore, our data suggest that alterations in immune response genes may synergize with HPV infection in the pathogenesis of squamous cell carcinomas. Finally, our data suggests that the association between HPV integration and increased expression of adjacent genes is a widespread phenomenon in primary cervical carcinomas.
METHODS
Sample preparation
All samples were obtained under institutional IRB approval and with documented informed consent. Surgically resected tumors or biopsies were snap frozen in liquid nitrogen and stored at −80° C. Genomic DNA and RNA were extracted from tumors found by frozen section investigations to have > 40% malignant epithelial cell component. A detailed description of sample collection is found in Supplementary Note 1. Nucleic acid was extracted using standard protocols described in Supplementary Note 2.
Sequence data generation
DNA from 115 tumor/normal paired samples was subjected to Agilent Sure-Select Human All Exon v2.0 based hybrid selection31 followed by exome library construction for Illumina sequencing, and 14 pairs for whole genome library construction; cDNA from 79 samples was subjected to transcriptome library construction, according to standard methods. Twelve tumor/normal pairs were sequenced with all three types of libraries. All libraries were sequenced with the Illumina HiSeq 2000 instrument (Supplementary Note 4). Exome sequencing was performed using a hybrid capture of 193,094 exon targets from 18,862 coding genes. Reads were aligned to human genome build GRCh37 using a Burrows-Wheeler aligner32. Data from the Illumina HiSeq were converted into BAM files 33 (http://samtools.sourceforge.net/SAM1.pdf)for each sample, using Picard (http://picard.sourceforge.net/) (Supplementary Note 6A).
Variant calling and significance analysis
All variant calls and significance analysis were obtained using the standard Cancer Genome Analysis Pipeline 13,34–37 with some modifications detailed in Supplementary Note 6B. Cross-individual contamination was estimated using ContEst 38 with both SNP Array (Supplementary Note 5) and sequencing data as input. SNV and Indel calls were generated using MuTect 11,13,34,35 and Indelocator 13,34,35, respectively, for all complete exome tumor-normal pairs. Variants were mapped to genes and transcripts using Oncotator, as well as being annotated with useful information such as overlapping COSMIC39 records. D-ToxoG (http://www.broadinstitute.org/cancer/cga/dtoxog) was used to filter mutations generated by a sequencing artifact that was discovered during this project. Significantly mutated genes and gene sets (Supplementary Note 9) were identified by MutSig2.0 based on the SNV and indel calls. Mutation rate calculations were also provided by MutSig2.0. Rearrangements were identified, using dRanger 36 running on 14 WGS tumor-normal pairs (Supplementary Note 13A), based on read pairings with unexpected distance or orientation. Somatic copy number alterations, both broad and focal, were identified with the GISTIC 2.013,35 tool, using segments identified from exome sequencing data (Supplementary Note 6B) as input (Supplementary Note 10A). Overlap between copy number alterations and somatic nonsilent mutations, for a subset of significantly mutated genes, are reported in Supplementary Note 10D and Supplementary Figure 14. UnifiedGenotyper 40,41 was used to identify germline mutations in genes in the Fanconi anemia pathway and TERC, as reported in Supplementary Note 14B and Supplementary Note 14C. ABSOLUTE 42 was used to generate purity and ploidy estimates for tumor samples where SNP Array data was available (Supplementary Table 3).
Variant validation
Two validation approaches were used in this study. The first was to resequence mutations in significant genes. Libraries were constructed with 200 bp flanks around key mutations in significant genes and sequenced using Illumina MiSeq. Manual validation was performed by examining mutations in the resulting BAM files. Mutations were considered validated if supported by five or more reads. The second validation approach was to compare exome SNV calls against the corresponding WGS and/or RNASeq calls where available. Mutations were considered validated if the alternate allele was seen in at least two reads and the calculated power was 80% or higher. These two approaches are detailed in Supplementary Note 6D. Due to the variable nature of human leukocyte antigen (HLA) genes43, mutations in HLA-A and HLA-B were validated manually using the procedure detailed in Supplementary Note 6E.
Hierarchical clustering of mutation signatures
Hierarchical clustering of all 115 samples, by nucleotide mutational context, using the heatmap.2 function from the gplots library (http://cran.r-project.org/web/packages/gplots/index.html) implemented in R 2.15.1 was performed. Mutation counts were scaled within each sample (i.e. converted to fraction of mutations corresponding to each category) and clustered using Ward’s minimum variance method44. Analysis of associations between mutation signature clusters and epidemiological factors (age, histology, geography, tumor grade and smoking status) was performed using Kruskal-Wallis, for continuous factors (age), and Fisher’s Exact Test, for discrete factors (histology, geography, tumor grade, and smoking status). A p-value threshold of 0.05 was used to decide association for all statistical testing. A detailed description and results can be found in Supplementary Note 8A. For display purposes in Figure 1, Tp*CpG mutations (which belong to both groups) are redistributed proportionately to each group, based on the relative frequencies of the other Tp*C and*CpG mutations in each tumor.
Hierarchical clustering of copy number variants
Copy number profiles, generated by GISTIC 2.0, were clustered into three categories. Each category was tested for association with tumor grade and histology using Fisher’s Exact Test and a p-value threshold of 0.05 (Supplementary Note 10B).
Gene expression analysis
Gene expression was measured using Cufflinks and cuffdiff 45 from the RNASeq data and fragments per kilobase of exon per million fragments mapped (FPKM) was obtained. Since non-transcribed genes tend to have more artifactual mutations 36,37,46,47, genes with low expression values (FPKM < 1) were filtered from significantly mutated gene lists produced by MutSig2.0 (Supplementary Note 6C). Genes with detected HPV integration sites were further analyzed for increased expression levels by ranking the expression levels of relevant genes for each sample (Supplementary Note 11E, Supplementary Note 11F, Supplementary Note 11G). Consensus clustering was performed on the RNASeq-derived gene expression data from 79 tumors, using ConsensusClusterPlus48. This analysis was run, using 1000 resampling iterations and a maximum of 25 clusters, on the 5000 genes with the largest deviation of FPKM scores across all patients. A final k value was chosen based on minimum threshold of change as k was increased (Supplementary Note 12). The correlative relationships between copy number variation and gene expression data were evaluated using Pearson correlation (Supplementary Note 10C).
Fusion analysis in transcriptome
RNASeq data was analyzed for fusion events by identifying inter-chromosomal chimeric read pairs or exon-exon read pairs separated by at least 1 Mb, with pairmates in the appropriate coding strand orientation. Unmapped reads spanning the putative fusion junction were also identified. High confidence fusion events are defined as having at least 3 reads mapped to a junction fusion (Supplementary Note 13B).
HPV typing
HPV typing was done by 2 multiplex HPV DNA PCR methods: the flourescent f-HPV assay49 and the mass spectrometry-based HPV PCR-MassArray50,51 (Supplementary Note 3A). Additionally, PathSeq 52 was used to generate HPV typing information from RNASeq data (Supplementary Note 3B).
HPV genome integration analysis
PathSeq 52 was used to identify sites of HPV genome integration in the cohort (Supplementary Note 7 and Supplementary Note 11A). Integration sites were identified using paired reads where one read aligned to the HPV genome (using NCBI viral databases) and the other to the human genome (Supplementary Note 11C). Validation of a recurrent integration site, in RAD51B, was performed by RT-PCR using primers targeting the junctions of specific HPV-RAD51B chimeric reads in three tumors (Supplementary Note 11B). In cases where the tumor with an integration site also had SNP array data, the distance between the integration site and the nearest copy number amplification was calculated. SNP Array data were used to identify amplifications. To evaluate co-occurrence of integration sites and copy number amplifications, data for a null model was created by simulating integration sites at uniformly distributed locations in the genome and assigning the simulated integration sites to random samples. The distribution of distances between the simulated integration sites and nearest copy number amplification compared to the true distribution of distances (Supplementary Note 11D). In Figure 3a, the true data (top histogram) was compared to distances generated from 100,000 permutations of randomly distributed integration sites across the genome with respect to the observed amplifications (bottom histogram). Overlapping amplification/integration sites have a distance of 0. Integration sites without amplification on the same chromosome were assigned a distance of the longest chromosome plus 1 (the bars on the right). MSigDB 53 was queried to evaluate whether the HPV integration sites co-occur in pathways with known roles in cancer (Supplementary Note 11H).
Epidemiological analysis for mutation rates
The statistical significance of mutation rate across histological types was corrected for the epidemiological factors: age (continuous), geography (discrete), tumor grade (discrete), and smoking status at diagnosis (discrete). The Fisher’s Exact Test, for discrete factors, and the Wilcoxon Test, for continuous factors, were run across histology (squamous cell and adenocarcinomas only). A p-value threshold of 0.05 was used to decide association for all statistical testing. For factors where an association was found with histology and mutation rate, we used a linear regression model to test whether histology was an independent predictor of mutation rate (Supplementary Note 8B).
Analysis of miscellaneous genes and pathways of interest
Additional analysis of notable genesets and pathways were performed using different techniques depending on the task. APOBEC mutations in the Tp*C context were counted and results reported in Supplementary Note 14A. The Fanconi anemia pathway has been implicated in suppressing HPV infection. Mutated genes in the Fanconi anemia pathway were identified from the somatic SNV, somatic Indel, and germline calls (Supplementary Note 14B). TERC has been identified as a marker for genomic instability and copy number gain. Therefore, we identified copy number variations encompassing the TERC gene as well as somatic and germline SNV and indel mutations (Supplementary Notes 14C)
Supplementary Material
Acknowledgments
This work was conducted as part of the Slim Initiative for Genomic Medicine in the Americas, a project funded by the Carlos Slim Health Institute in Mexico. This work was also partially supported by the Rebecca Ridley Kry Fellowship of the Damon Runyon Cancer Research Foundation (A.I.O.); MMRF Research Fellow Award (A.I.O.); Helse Vest, Research Council of Norway, Norwegian Cancer Society and Harald Andersens legat (H.B.S.); CONACyT grant SALUD-2008-C01-87625 and UANL PAICyT grant CS1038-1 (H.A.B.S); CONACyT grant 161619 (J M-Z). We also thank B. Edvardsen, K. Dahl-Michelsen, Å. Mokleiv, K. Madisso, T. Njølstad and E. Valen for technical and programmatic assistance; the staff of the Broad Institute Genomics Platform for their assistance in processing samples and generating the sequencing data used in the analyses; the Instituto Mexicano del Seguro Social (IMSS) for their Support; and L. Gaffney of Broad Institute Communications for figure layout and design.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature
Author Contributions
A.I.O., L.L., S.S.F., C.S.P., H.B.S. and M.M. wrote the manuscript with help from co-authors. A.I.O., L.L., K.C., C.S. and G.G performed whole exome and genome sequencing data analysis. A.I.O., I.I., V.T., K.V-S., A.S.G., S.R-C., C.R.E., S.S.F., and C.S.P. performed RNA sequencing data analysis. A.I.O., S.S.F., C.S.P and T.J.P. performed HPV integration analyses. A.I.O. and A.D.C. performed copy number analyses. A.I.O., F.D., B.K., R.W. and H.G. performed functional experiments on MAPK1. B.B., N.B.G., G.S.G-M., and C.P.C facilitated and performed pathology review. O.K.V., H.M.W., and T.E.C. performed HPV status determination. L.A., E.N. and M.L.C. facilitated project management. L.L., I.I., V.T., K.V-S., A.S.G., S.R-C., I.P.R.S. and C.R.E. performed sequencing data validation. M.E-C., M.K.H., E.W., E.A.H., C.K. and M.L.G-R. performed specimen processing, biobanking and data management. K.W., L.B., L.D.V-C., G.M., J.V., C.R., A.C. and H.B.S. collected patient materials and clinical information. A.I.O., L.L., and D.S.N. performed biostatistical and epidemiological analyses. A.I.O., L.L., S.F., C.S.P., V.T., A.A.W., T.J.P., M.W.R., M.S.L., C.S., S.L.C., A.M., H.B.S. and M.M. contributed text, figures (including supplementary information), and analytical tools. A.H.M., C.R.E., L.A.A., S.B.G., H.A.B-S., J.M-Z., G.G., H.B.S. and M.M. provided leadership for the project. All authors contributed to the final manuscript.
Sequence data used for this analysis are available in dbGaP under accession phs000600.
Reprints and permissions information is available at www.nature.com/reprints.
The authors declare no competing financial interests.
Readers are welcome to comment on the online version of this article at www.nature.com/nature.
References
- 1.Jemal A, et al. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.IARC. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 100B. International Agency for Research on Cancer; Lyon, France: 2012. A Review of Human Carcinogen: Biological Agents. [Google Scholar]
- 3.zur Hausen H. Papillomaviruses in the causation of human cancers - a brief historical account. Virology. 2009;384:260–5. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 4.Crook T, et al. Clonal p53 mutation in primary cervical cancer: association with human-papillomavirus-negative tumours. Lancet. 1992;339:1070–3. doi: 10.1016/0140-6736(92)90662-m. [DOI] [PubMed] [Google Scholar]
- 5.McIntyre JB, et al. PIK3CA mutational status and overall survival in patients with cervical cancer treated with radical chemoradiotherapy. Gynecol Oncol. 2013;128:409–14. doi: 10.1016/j.ygyno.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 6.Kang S, et al. Inverse correlation between RASSF1A hypermethylation, KRAS and BRAF mutations in cervical adenocarcinoma. Gynecol Oncol. 2007;105:662–6. doi: 10.1016/j.ygyno.2007.01.045. [DOI] [PubMed] [Google Scholar]
- 7.Wingo SN, et al. Somatic LKB1 mutations promote cervical cancer progression. PLoS One. 2009;4:e5137. doi: 10.1371/journal.pone.0005137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narayan G, Murty VV. Integrative genomic approaches in cervical cancer: implications for molecular pathogenesis. Future Oncol. 2010;6:1643–52. doi: 10.2217/fon.10.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vazquez-Mena O, et al. Amplified genes may be overexpressed, unchanged, or downregulated in cervical cancer cell lines. PLoS One. 2012;7:e32667. doi: 10.1371/journal.pone.0032667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arteaga CL, Baselga J. Impact of genomics on personalized cancer medicine. Clin Cancer Res. 2012;18:612–8. doi: 10.1158/1078-0432.CCR-11-2019. [DOI] [PubMed] [Google Scholar]
- 11.Cibulskis K, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–9. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrov LB, Nik-Zainal S, Wedge DC, Campbell PJ, Stratton MR. Deciphering signatures of mutational processes operative in human cancer. Cell Rep. 2013;3:246–59. doi: 10.1016/j.celrep.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Banerji S, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–9. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Greulich H, et al. Functional analysis of receptor tyrosine kinase mutations in lung cancer identifies oncogenic extracellular domain mutations of ERBB2. Proc Natl Acad Sci U S A. 2012;109:14476–81. doi: 10.1073/pnas.1203201109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bose R, et al. Activating HER2 Mutations in HER2 Gene Amplification Negative Breast Cancer. Cancer Discov. 2012 doi: 10.1158/2159-8290.CD-12-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arvind R, et al. A mutation in the common docking domain of ERK2 in a human cancer cell line, which was associated with its constitutive phosphorylation. Int J Oncol. 2005;27:1499–504. [PubMed] [Google Scholar]
- 17.De Luca A, Maiello MR, D’Alessio A, Pergameno M, Normanno N. The RAS/RAF/MEK/ERK and the PI3K/AKT signalling pathways: role in cancer pathogenesis and implications for therapeutic approaches. Expert Opin Ther Targets. 2012;16 (Suppl 2):S17–27. doi: 10.1517/14728222.2011.639361. [DOI] [PubMed] [Google Scholar]
- 18.Le Gallo M, et al. Exome sequencing of serous endometrial tumors identifies recurrent somatic mutations in chromatin-remodeling and ubiquitin ligase complex genes. Nat Genet. 2012;44:1310–5. doi: 10.1038/ng.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Agrawal N, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–7. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Ghazawi FM, Li Q. Interplay of bromodomain and histone acetylation in the regulation of p300-dependent genes. Epigenetics. 2010;5:509–15. doi: 10.4161/epi.5.6.12224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–5. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 22.Tong KI, et al. Keap1 recruits Neh2 through binding to ETGE and DLG motifs: characterization of the two-site molecular recognition model. Mol Cell Biol. 2006;26:2887–900. doi: 10.1128/MCB.26.8.2887-2900.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cancer Genome Atlas Research, N. et al. Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489:519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pamer E, Cresswell P. Mechanisms of MHC class I--restricted antigen processing. Annu Rev Immunol. 1998;16:323–58. doi: 10.1146/annurev.immunol.16.1.323. [DOI] [PubMed] [Google Scholar]
- 25.Neve RM, Ylstra B, Chang CH, Albertson DG, Benz CC. ErbB2 activation of ESX gene expression. Oncogene. 2002;21:3934–8. doi: 10.1038/sj.onc.1205503. [DOI] [PubMed] [Google Scholar]
- 26.Wentzensen N, Vinokurova S, von Knebel Doeberitz M. Systematic review of genomic integration sites of human papillomavirus genomes in epithelial dysplasia and invasive cancer of the female lower genital tract. Cancer Res. 2004;64:3878–84. doi: 10.1158/0008-5472.CAN-04-0009. [DOI] [PubMed] [Google Scholar]
- 27.Kraus I, et al. The majority of viral-cellular fusion transcripts in cervical carcinomas cotranscribe cellular sequences of known or predicted genes. Cancer Res. 2008;68:2514–22. doi: 10.1158/0008-5472.CAN-07-2776. [DOI] [PubMed] [Google Scholar]
- 28.Schmitz M, Driesch C, Jansen L, Runnebaum IB, Durst M. Non-random integration of the HPV genome in cervical cancer. PLoS One. 2012;7:e39632. doi: 10.1371/journal.pone.0039632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tang KW, Alaei-Mahabadi B, Samuelsson T, Lindh M, Larsson E. The landscape of viral expression and host gene fusion and adaptation in human cancer. Nat Commun. 2013;4:2513. doi: 10.1038/ncomms3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peter M, et al. Frequent genomic structural alterations at HPV insertion sites in cervical carcinoma. J Pathol. 2010;221:320–30. doi: 10.1002/path.2713. [DOI] [PubMed] [Google Scholar]
- 31.Gnirke A, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–9. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stransky N, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–60. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee RS, et al. A remarkably simple genome underlies highly malignant pediatric rhabdoid cancers. J Clin Invest. 2012;122:2983–8. doi: 10.1172/JCI64400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berger MF, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–20. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chapman MA, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cibulskis K, et al. ContEst: estimating cross-contamination of human samples in next-generation sequencing data. Bioinformatics. 2011;27:2601–2. doi: 10.1093/bioinformatics/btr446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Forbes SA, et al. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carter SL, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–21. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Erlich H. HLA DNA typing: past, present, and future. Tissue Antigens. 2012;80:1–11. doi: 10.1111/j.1399-0039.2012.01881.x. [DOI] [PubMed] [Google Scholar]
- 44.Ward JH., Jr Hierarchical Grouping to Optimize an Objective Function. Journal of the American Statistical Association. 1963;58:236–244. [Google Scholar]
- 45.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2012 doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bass AJ, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–8. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pleasance ED, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–6. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wilkerson MD, Hayes DN. ConsensusClusterPlus: a class discovery tool with confidence assessments and item tracking. Bioinformatics. 2010;26:1572–3. doi: 10.1093/bioinformatics/btq170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Canadas MP, et al. Comparison of the f-HPV typing and Hybrid Capture II(R) assays for detection of high-risk HPV genotypes in cervical samples. J Virol Methods. 2012;183:14–8. doi: 10.1016/j.jviromet.2012.03.005. [DOI] [PubMed] [Google Scholar]
- 50.Walline HM, et al. High-risk human papillomavirus detection in oropharyngeal, nasopharyngeal, and, oral cavity cancers: Comparison of multiple methods. JAMA-Otolaryngology. 2013 doi: 10.1001/jamaoto.2013.5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang H, et al. Sensitive detection of human papillomavirus in cervical, head/neck, and schistosomiasis-associated bladder malignancies. Proc Natl Acad Sci U S A. 2005;102:7683–8. doi: 10.1073/pnas.0406904102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kostic AD, et al. PathSeq: software to identify or discover microbes by deep sequencing of human tissue. Nat Biotechnol. 2011;29:393–6. doi: 10.1038/nbt.1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.