Abstract
Aim
Familial loading for alcohol dependence (AD) and variation in genes reported to be associated with AD or BMI were tested in a longitudinal study.
Materials & methods
Growth curve analyses of BMI data collected at approximately yearly intervals and obesity status (BMI > 30) were examined.
Results
High-risk males were found to have higher BMI than low-risk males, beginning at age 15 years (2.0 kg / m2 difference; p = 0.046), persisting through age 19 years (3.3 kg/m2 difference; p = 0.005). CHRM2 genotypic variance predicted longitudinal BMI and obesity status. Interactions with risk status and sex were also observed for DRD2 and FTO gene variation.
Conclusion
Variation at loci implicated in addiction may be influential in determining susceptibility to increased BMI in childhood and adolescence.
Keywords: addiction, BMI, familial risk for alcoholisml, multiplex families, obesity genes
Obesity is a prevalent medical condition that affects more than one third of adults in the United States and almost 17% of children and adolescents [1]. Obesity has been found to increase risk for a myriad of significant health problems, including Type 2 diabetes, coronary heart disease, stroke, hypertension, arthritis and cancer [2]. Altogether, it has been estimated that obese individuals are at 18% higher risk for all-cause mortality, compared with individuals with a healthy weight [3]. It is projected that by 2030, the treatment costs for preventable obesity-related diseases will reach between US$195 and US$276 billion, with economic productivity losses estimated to reach between US$390 and US$580 billion [2]. Thus, the societal burden of obesity is immense, both in terms of healthcare expenditures as well as premature morbidity and mortality.
Adiposity appears to be influenced by genetic factors with 40–70% of the variation in BMI attributable to genetic variation [4]. Recently, there has been increased focus on the possibility that those most susceptible to becoming obese also have a greater risk for addiction [5]. Neural circuits supporting the brain’s reward systems that are implicated in drug and alcohol abuse appear to also be involved in the urges to overeat observed in individuals with greater BMI and obesity. Specifically, the mesolimbic dopamine pathway appears to play an important role in mediating reinforcement of substance use as well as food consumption [6]. Administration of both drugs and palatable food activates the ventral tegmental area, nucleus accumbens and other striatal regions in both humans and animals [7,8]. Furthermore, individuals with substance use disorders or higher levels of BMI show greater activation of similar reward and attention regions, compared with healthy controls, when presented with cues related to abused substances or high-fat/high-sugar foods, respectively [9,10].
Abnormalities of the brain’s reward system are likely due to a complex interaction of genetic and environmental effects and recent research has hypothesized that addiction and obesity may have overlapping genetic liability [11]. Several genes have been identified as conferring risk for addiction, including BDNF [12], DRD2 [13], FTO [14], CHRM2 [15] and OPRM1 [16–19]. Interestingly, these genes have also been associated with BMI, obesity or overeating in subjects without family histories of alcohol dependence (AD) [20–24]. Thus, common genetic variants linked to a number of biological processes have been independently associated with both of these conditions. Two other genes that have been studied in association with obesity are LEP and SERPINF1, though findings in the literature have been mixed [25–27]. Whereas these genes have yet to be studied with regard to risk for addiction, the proteins they code for, leptin and PEDF, respectively, have each been implicated in alcoholism [28,29].
Because no studies have examined the genetic liability for addiction and obesity in the same sample, the current study sought to examine whether familial loading for AD, variation in genes associated with AD, as well as genes previously found to influence BMI would be associated with increased BMI and obesity in the current sample. Subjects with and without a high familial density of AD had their BMI assessed approximately annually from ages 8–19 years. Growth curve analysis was used to examine the main effect of seven candidate genes (18 SNPs) on BMI, as well as inter-actions between genotype, risk status and sex. BMI was analyzed both continuously and as a categorical obesity variable (i.e., BMI >30).
Materials & methods
Clinical sample & assessments High-risk group
A three generation study of multiplex AD families initiated in 1985 provided the participants for the present study. Third-generation offspring of second-generation probands were studied. All participants were of Caucasian background. These multiplex AD families were originally identified through the presence of two second-generation adult brothers (proband pair) with AD. The multiplex AD families were first identified through one member of the pair currently being in treatment at the time of identification. Probands were screened with the Diagnostic Interview Schedule (DIS) [30] for the presence of AD, and other Axis I (diagnostic criteria from the Diagnostic and Statistical Manual – Third Edition [DSM-III]) disorders (Feighner Criteria for AD was also obtained). Extensive in-person and family history information are available for these targeted families including the probands’ parents (first generation) and the second-generation siblings of the identified probands. The multiplex sampling strategy used in this study resulted in a high density of AD in the targeted pedigrees and increased familial loading for AD among third-generation offspring (an average of four first- and second-degree relatives for the present subsample) as previously described [31]. Therefore, third-generation offspring of the brother pairs and offspring of their siblings in these multiplex families provided the high-risk offspring for the present report. These offspring have a greater familial loading for AD than is commonly seen in studies designed to identify offspring of alcohol dependent parents. All existing third-generation offspring were recruited for annual follow-up during childhood as part of an ongoing longitudinal study. The study has ongoing approval from the University of Pittsburgh Institutional Review Board. Participants provided informed consent at each follow-up visit, while children provided assent with parental consent.
Low-risk group
Controls were recruited from the community through advertisements targeting families with children. To avoid bias among volunteers, no information was provided in the advertisements regarding the purpose of the study other than to indicate it was a large-scale family study of hereditary aspects of cognition and personality. Interested individuals contacted the research program and screening was initiated. Control families were included if two adult brothers without a lifetime history of alcohol or drug dependence were present and first- and second-degree relatives were without psychopathology by family history. The low-risk subjects of this report are the offspring of these identified index cases. Written informed consent was obtained from the parent and child after the study had been described and questions answered.
A total of 158 subjects were selected for inclusion in the current analyses for whom both genotyping data and a minimum of two BMI measurements between the ages 8 and 19 years were available. The sample included 96 subjects (51 male and 45 female) from multiplex for AD families and 62 (33 male and 29 female) low-risk control subjects. A total of 1002 BMI measurements were available for statistical modeling. Overall, the groups did not differ in mean age at entry into the study, age at last visit, or socioeconomic status (SES; Table 1).
Table 1.
Characteristics of study subjects by gender and familial-risk status.
| Male |
Female |
Significance | |||
|---|---|---|---|---|---|
| High | Low | High | Low | ||
| Subjects (n) | 51 | 33 | 45 | 29 | NS |
| BMI measurements (n) | 332 | 213 | 294 | 163 | NS |
| BMI† ≥30, n (%) | 15 (29.4 ) | 5 (15.2) | 7 (15.6) | 8 (27.6) | NS |
| Age at first visit, mean (SD) | 11. 2 (2.5 ) | 11.2 ( 2.4 ) | 10.8 (2.2) | 11.6 ( 2.4) | NS |
| Age at last visit, mean (SD) | 18.1 (1.3 ) | 18.0 (0.8) | 17. 8 (1. 3 ) | 17. 5 (1.9) | NS |
| BMI at first visit, mean (SD) | 20.8 (4.6) | 19.8 (4.8) | 20.3 (5.3) | 21.0 (4.1) | NS |
| BMI at last visit, mean (SD) | 26.4 (5.2) | 24.6 (4.4) | 24.5 (4.7) | 24.8 (5.5) | NS |
| SES‡, mean (SD) | 38.6 (10.2) | 44.3 (12.3) | 40.2 (10.3) | 44.0 (9.7) | NS |
BMI was calculated as weight in kilograms divided by height in meters squared
SES was calculated using occupation and education as previously described [31]
NS: Not significant; SD: Standard deviation; SES: Socioeconomic status.
BMI measurements
All third-generation offspring who were enrolled in a longitudinal follow-up initiated in 1989 were weighed and measured at the time of their yearly childhood (between the ages of 8–19 years) follow-up visits.
DNA extraction & genotyping
Genomic DNA was extracted from lymphocytes from whole blood or from Epstein–Barr virus-transformed lymphoblastic cell lines and assayed in our laboratory as previously described [32]. Genotyping was completed on a Biotage PSQ 96MA Pyrosequencer (Biotage AB, Uppsala, Sweden). Each polymorphism was analyzed by PCR amplification incorporating a biotinylated primer. Thermal cycling included 45 cycles at an annealing temperature of 60°C. The Biotage workstation was used to isolate the biotinylated single strand from the double-strand PCR products. The isolated product was then sequenced using the complementary sequencing primer. Seven candidate genes (18 SNPs) were chosen based on their previously demonstrated association with addiction (CHRM2, BDNF, OPRM1, DRD2) or obesity (FTO, LEP, SERPINF1). The following SNPs were assessed: BDNF rs6265; CHRM2 rs1424569, rs1424387, rs1824024, rs2061174, rs324650, rs8191992 and rs8191993; DRD2 rs6277; FTO rs9939609, rs17818902 and rs17820875; LEP rs7799039, rs2167270 and rs11763517; OPRM1 rs2281617; and SERPINF1 rs12150053 and rs12603825.
Quality control
SNP genotyping quality control involved ongoing monitoring of SNP signals provided by Qiagen software. Output is provided using three categories for each SNP: pass, fail and check. Data analysis was performed for only those signals meeting the ‘pass’ criterion. Signals that failed or were returned as needing further checking were rerun. If after three attempts the SNP did not meet the ‘pass’ criterion, it was eliminated from the analysis.
Data analysis
Mixed effects logistic and linear models were used to investigate the relationship between BMI development during childhood and adolescence and familial loading for AD along with potential modification by specific genes. The goal of the analyses was to determine if familial loading for AD was related to BMI growth curves and/or occurrence of obesity. A second goal was to determine if genes associated with addiction would modify the relationship between familial risk and BMI. A third goal was to determine if variation in genes previously reported to influence BMI would modify the relationship between risk status and body mass.
Genetic models
Due to the low frequency of individuals who were homozygous for the minor allele, we assumed a dominant genetic model with the minor allele as the effect allele to maximize statistical power. Each gene was tested for departure from Hardy–Weinberg equilibrium (HWE) using Haploview [33]. No SNP was found to exhibit statistically significant HWE departure.
Linear mixed-effects model
A linear mixed-effects model was used to test associations between the candidate genes or familial risk status with BMI development. The model was fit using xtmixed in Stata, specifying random effects to adjust for within subject correlations across time and family relatedness. Genetic effects were considered individually for each SNP, controlling for the linear effect of age and possible nonlinear effects of age (age2). Possible age-dependent genetic effects on BMI development were modelled, estimating a dominant minor allele effect for each of three age ranges: 8–13, 14–16 and 17–19 years. Potential age-dependent effects of familial risk on BMI were modelled with age as a continuous variable and tested for each of ages 8–19 years. Additionally, genetic effects were tested for possible differences by sex and familial risk. Risk was controlled for as an independent effect for SNPs showing significant interactions with sex. Similarly sex was controlled for as an independent effect for SNPs showing significant interactions with risk. Additionally, because of possible effects of smoking on BMI, number of cigarettes smoked per day was stratified into three groups: none, below the median of those who smoked (<one half pack) and above the median (>one half pack).
Mixed-effects logistic model
A mixed-effects logistic model was used to investigate the associations of obesity with SNP variation. Obesity was defined as an observed BMI >30 in the 8–19 years age range. The model was fit using xtmelogit in Stata, specifying random effects for family to adjust for family relatedness (siblings were present in some families). The interaction of genotypic variation with sex or risk was tested. Risk was controlled for as an independent effect for SNPs showing significant interactions with sex. Similarly sex was controlled for as an independent effect for SNPs showing significant interactions with risk.
Correction for multiple tests
Using a Bonferroni correction method, results would need to be <0.003, to account for multiple tests.
Results
Familial risk effects
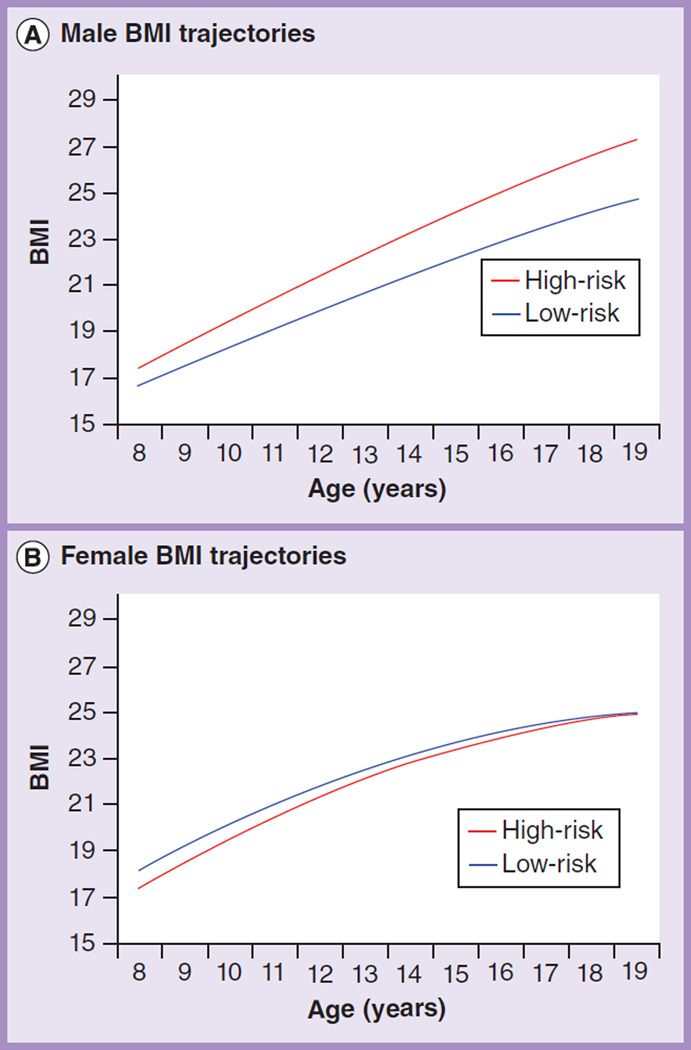
Results of the growth curve analysis of familial risk with BMI treated as a continuous variable are summarized in Table 2, and illustrated in Figure 1. High-risk males were found to have higher BMI than low-risk males, beginning at age 15 years (2.0 kg/m2 difference; p = 0.046). This group difference persisted through adolescence and increased in magnitude through age 19 years (3.3 kg/m2 difference; p = 0.005). High-risk females were not found to have higher BMI than low-risk females at any age.
Table 2.
Estimated difference in BMI between high and low familial risk males by age.
| Age (years) | Difference (kg/m2) | 95% CI | p-value |
|---|---|---|---|
| 8 | 0.48 | −1.88–2.84 | 0.69 |
| 9 | 0.74 | −1.59 –3. 07 | 0.53 |
| 10 | 1.00 | −1.31–3.30 | 0.40 |
| 11 | 1.25 | −1.03 – 3.54 | 0.28 |
| 12 | 1.51 | −0.75–3.78 | 0.19 |
| 13 | 1.77 | −0.49–4.02 | 0.12 |
| 14 | 2.03 | −0.22–4.27 | 0.077 |
| 15 | 2.28 | 0.04–4.53 | 0.046* |
| 16 | 2.54 | 0.29–4.79 | 0.027* |
| 17 | 2.80 | 0.54–5.06 | 0.015** |
| 18 | 3.05 | 0.78–5.33 | 0.009** |
| 19 | 3.31 | 1.01–5.61 | 0.005** |
Statistically significant values are in bold.
p < 0.05.
p < 0.01.
Figure 1. BMI trajectories for ages 8–19 years.
(A) High-risk males displayed significantly higher BMI than low-risk males from age 16 to 19 years. (B) No effect of familial risk on BMI was observed among the female participants at any age.
Longitudinal BMI: genetic & sex effects
Results of the growth curve analyses with BMI as a continuous variable are summarized in Table 3. Of the seven genes tested, only CHRM2 variation showed a significant main effect. Minor allele carriers for SNP rs8191993 (C>G) had greater BMIs in the 14–16 and 17–19 age ranges, regardless of sex or risk status. Additionally, two candidate genes demonstrated significant interactions with risk. Among high-risk participants with multiple relatives with alcohol dependence, DRD2 rs6277 minor allele (T>C) (C) carriers had higher BMIs in the 14–16 and 17–19 age ranges, and those who were homozygous for the major allele of the FTO rs17818902 (T>G) had greater BMI during late adolescence (17–19 years age range). No effect of DRD2 rs6277 or FTO rs17818902 was seen in the LR group at any age.
Table 3.
Linear regression analyses of BMI by familial risk status, gender, age range and the presence of a SNP minor allele.
| Age range (years)† |
CHRM2rs2061174 (T>C) |
DRD2rs6277 (T>C) |
FTOrs17818902 (T>G) |
CHRM2rs8191993 (C>G) |
|||||
|---|---|---|---|---|---|---|---|---|---|
| Regression coefficients (95% CI), kg/m2 |
p-value | Regression coefficients (95% CI), kg/m2 |
p-value | Regression coefficients (95% CI), kg/m2 |
p-value | Regression coefficients (95% CI), kg/m2 |
p-value | ||
| High risk | 8–13 | – | – | 1.6 (−0.2–3.5) | 0.088 | −1.5 (−3.3–0.4) | 0.12 | – | – |
| 14–16 | – | – | 1.9(0.1–3.8) | 0.040 | −1.4 (−3.2–0.5) | 0.14 | – | – | |
| 17–19 | – | – | 1.9(0.0–3.8) | 0.046 | −1.9 (−3.7–0.0) | 0.048 | – | – | |
| Low risk | 8–13 | – | – | −1.6 (−4.0–0.7) | 0.18 | 0.7 (−1.7–3.1) | 0.57 | – | – |
| 14–16 | – | – | −0.6 (−2.9–1.8) | 0.65 | 1.2 (−1.2–3.6) | 0.32 | – | – | |
| 17–19 | – | – | −0.7 (−3.0–1.7) | 0.58 | 0.8 (−1.6–3.2) | 0.53 | – | – | |
| Male | 8–13 | 2.3(0.5–4.2) | 0.014 | 1.7 (−0.3–3.7) | 0.088 | – | – | – | – |
| 14–16 | 2.6(0.7–4.5) | 0.006 | 2.6(0.6–4.6) | 0.010 | – | – | – | – | |
| 17–19 | 2.4(0.5–4.3) | 0.011 | 2.6(0.6–4.6) | 0.010 | – | – | – | – | |
| Female | 8–13 | −1.7 (−3.8–0.4) | 0.12 | −0.7 (−2.9–1.4) | 0.50 | – | – | – | – |
| 14–16 | −0.8 (−2.9–1.3) | 0.43 | −0.4 (−2.6–1.7) | 0.68 | – | – | – | – | |
| 17–19 | −0.5 (−2.6–1.6) | 0.63 | −0.6 (−2.7–1.5) | 0.58 | – | – | – | – | |
| All | 8–13 | 0.8 (−0.7–2.3) | 0.30 | ||||||
| 14–16 | – | – | – | – | – | – | 1.6(0.1–3.1) | 0.034 | |
| 17–19 | – | – | – | – | – | – | 1.7(0.2–3.2) | 0.026 | |
Age ranges were calculated to the midpoint of each year so that subjects who were 13.5 years or greater would be included in the next age range (14–16 years). Similarly, those aged 16.5 years or greater are included in the 17–19 years age range.
Two genes showed significant interactions with sex. Among males only, carriers of the CHRM2 rs2061174 minor allele (T>C) had significantly higher BMIs across all age ranges, and individuals with the C (T>C) minor allele of DRD2 rs6277 had greater BMIs in the 14–16 and 17–19 years age ranges. Significant inter-actions between sex and genetic variation may be seen in Table 3.
Obesity: genetic & sex effects
When obesity was analyzed as a categorical variable (i.e., BMI >30), many of the effects seen for the analyses using BMI as a continuous variable were replicated (Table 4). CHRM2 rs8191993 minor allele carriers (C>G) showed an increased odds ratio (OR) for being obese (OR: 2.5) independent of sex. There was also a significant interaction between this SNP and risk status, indicating that the relationship between minor allele status and obesity was stronger in the low-risk group. Consistent with the results obtained using the continuous BMI measure, using the binary (yes/no) obesity variable, the minor allele of DRD2 rs6277 (T>C) was significantly associated with greater odds of being obese (OR:5.0) but only in males. Also, the presence of the minor allele of CHRM2 rs2061174 (T> C) conferred greater risk for obesity, but only among males.
Table 4.
Odds ratio for obesity by familial risk status, gender, age range and SNP.
| Gene | SNP | Alleles | Odds ratio | 95% CI | p-value | |
|---|---|---|---|---|---|---|
| CHRM2 | rs8191993 | High risk | C/G | 1.2 | 0.4–3.5 | NS |
| Low risk | 11.4 | 1.3–103.3 | 0.03 | |||
| CHRM2 | rs2061174 | Male | T/C | 6.0 | 1.6–22.6 | 0.008 |
| Female | 0.8 | 0.3–2.6 | NS | |||
| DRD2 | rs6277 | Male | T/C | 5.0 | 1.1 – 23.7 | 0.04 |
| Female | 1.0 | 0.3–3.3 | NS | |||
| CHRM2 | rs8191993 | All | C/G | 2.5 | 1.0 – 6.1 | 0.05 |
NS: Nonsignificant.
Discussion
The current study supports the existence of a shared diathesis for addiction and obesity by demonstrating that males at ultra-high-risk for AD show significantly higher BMI between the ages of 16 and 19 than low-risk males. Emerging research suggests that abnormalities in reward circuitry may predispose individuals to both addiction and obesity [34]. Indeed, abnormalities in reward processing have been demonstrated among offspring with a family history of alcoholism [35], which are thought to be partially genetically determined. The current findings indicating that males at high risk for substance use problems also display a significant predisposition to weight gain in adolescence supports the notion that aberrant reward system function may represent a common etiological pathway to both conditions.
The current results also replicate previous reports of an association between BMI and genetic variation in CHRM2, DRD2 and FTO. Our findings demonstrate that CHRM2 rs2061174 accounted for 4.1% of the variance in BMI in our sample, DRD2 rs6277 explained 3.6% of BMI variance, and CHRM2 rs8191993 and FTO rs17818902 each accounted for 1.6% of the variance in BMI. The proportion of variance explained by each SNP was calculated using the R package MuMIn [36]. These estimates are comparable to earlier studies that have measured the effects of variation at these loci, which have reported that SNPs within the CHRM2, FTO and DRD2 genes explained approximately 1–3% of the variance in BMI [22,37–39].
The CHRM2 gene is a muscarinic acetylcholine receptor gene which has been linked to both substance use [40] and body mass [22]. There is evidence to suggest that increased disinhibition may represent a common mechanism supporting both of these associations, perhaps mediated by specific genetic variation. CHRM2 variation has been shown to influence indices of disinhibition [40], as well as externalizing behavior [41]. A robust literature has implicated disinhibitory traits in risk for addiction [42], and emerging data suggest that this factor may also play a critical role in weight status. A recent meta-analysis of 23 studies demonstrated significantly higher impulsivity among overweight and obese children, with the dimension of disinhibition showing the greatest effect [43]. Furthermore, obesity is also highly comorbid with childhood externalizing disorders [44]. Therefore, disinhibitory traits may predispose to both addiction and increased BMI with evidence that CHRM2 may be infuential in this process.
Congruently, prior work has demonstrated an association between CHRM2 rs1824024 and the P300 component of the event-related potential [45,46]. The relationships between reduced P300 amplitude, which is thought to be an indicator of disinhibitory conditions, familial risk for AD [47,48] and externalizing disorders of childhood [49] are well-documented. The presence of reduced amplitude P300 has also been related to the likelihood of developing obesity and nicotine dependence [50]. Additionally, reduced amplitude of P300 in childhood appears to predict the likelihood of developing substance use disorders many years later. The longest follow-up shows P300 amplitude recorded as early as age 9 years predicts substance use outcome by age 20 [51]. Follow-ups at 3 and 7 years, respectively, from other laboratories confirm this relationship [52,53]. Therefore, these data lend further support for the hypothesis that CHRM2 may influence risk for both addiction and obesity through the influence of the cholinergic system on behavioral disinhibition, particularly as it relates to electrophysiological indices of disinhibitory processes.
Additionally, interactions between familial risk for substance use and specific genes appeared to influence the observed BMI in childhood and adolescence. Among high-risk individuals, carriers of the minor allele (C) (T>C) of DRD2 SNP rs6277 displayed greater BMI in adolescence, whereas minor allele (G) (T>G) carriers of the FTO SNP rs17818902 were found to be protected, with the G carriers displaying lesser BMI between the ages of 17 and 19 years. Variation within the DRD2 gene has frequently been linked to hedonic response to drug rewards, and more recently has been implicated in binge eating and obesity [20]. Thus, our observation that DRD2 variation influences BMI among high-risk adolescents supports the contention that differences in the hedonic experience of reward may impact risk for obesity as well as substance abuse.
Some of the effects observed appear to be sex specific. DRD2 rs6277 and CHRM2 rs2061174 significantly influenced BMI, but only among males. Interestingly, prior research has also reported sex-specific effects of DRD2 variation at this locus on waist circumference [54]. Although the precise mechanism of the observed sex specificity is unclear, prior research has reported sex differences in mesocortical dopamine neurotransmission [55] as well as interactions between dopamine and estrogen in cognitive functioning [56]. Future research should explore how dopaminergic gene variation may differentially impact BMI in males and females.
Congruently, only male carriers of the minor allele (C) (T>C) of CHRM2 rs2061174 displayed greater BMI throughout childhood and adolescence. Indeed, earlier studies have also reported sex-specific effects of CHRM2 variation on nicotine dependence [57]. Future research is necessary to elucidate how CHRM2 variation leads to different effects in men and women. Nonetheless, these data implicate the minor C allele in risk for a number of pathological conditions.
Among the three obesity genes tested (FTO, LEP and SERPINF1), our data replicate the well-substantiated relationship between FTO and body mass, whereas associations with LEP and SERPINF1 were not observed in our sample. Variation in the FTO gene has been linked to BMI in a number of different populations, yet the exact physiological function of this gene is not well understood [23,24]. Similarly, the specific causal variants underlying this association remain unclear. Our data replicate prior findings that the minor allele of SNP rs17818902 (T>G) is associated with lesser BMI [24]. Together, these findings indicate that variation at this locus may be an important component of the relationship between the FTO gene and bodyweight.
Although prior studies have implicated SERPINF1 and LEP in obesity, the literature on these genes has been inconsistent. Animal research suggests that the SERPINF1 gene influences body mass [58], although results from human studies have been mixed. Böhm and colleagues [26] have reported an association between SERPINF1 genotype and total adipose tissue mass among 1974 middle-aged Caucasians at high risk for Type 2 diabetes. However, a second study of younger Mexican American individuals failed to detect an influence of genetic variation at this locus [27]. The discrepancy among existing studies of this gene may be attributable to differences in the selection criteria, age range or size of the samples studied. Future research is needed to clarify the role of SERPINF1 in the determination of bodyweight. Findings for the LEP gene and body mass have also been mixed with both positive and negative reports [59].
A number of studies have found that associations between particular genes and either increased BMI or obesity are highly dependent on the age of the subjects studied [60]. The present analysis focused on children and adolescents so the long-term effects of genes having greater influence in adulthood could not be known. Finally, the current sample was recruited based on risk for AD. The selection criteria did not include BMI. Therefore, the present sample would be expected to more closely match the general population with respect to BMI distribution than samples specifically selected for increased BMI or disease conditions such as Type 2 diabetes associated with increased BMI. These considerations may explain the lack of association between LEP, SERPINF1 and BMI in the current analysis. To our knowledge, this is the first study to find a main effect of CHRM2 SNP rs8191993 on BMI, although another SNP from this gene, rs12673281, was previously found in a large-scale BMI study [22]. Laramie et al. [22] found the relationship between the SNP rs12673281 to be highly significantly related to BMI in a family study (FBAT = 0.0041) and in the case–control analysis the p-value was reported at 2.2 × 10−6 Our SNP, rs8191993, was found to increase the odds of being obese (OR:2.5). The D´ for rs1267381 and rs819193 is reported at 0.82 in HapMap Release 22. Although r2 is 0.056 in HapMap, the distance of 57,000 bp suggests that these two SNPs are in reasonably high linkage disequilibrium.
Limitations of the study
One limitation of the current study is our reliance on BMI to measure adiposity. A broad range of adiposity may be associated with a given BMI value, as increases in both lean mass and body fat will inflate this index. Therefore, in some instances greater BMI may actually correspond to favorable changes in body composition. Such a bias towards increased BMI due to lean muscle may have impeded our ability to detect significant associations between genetic variation in the LEP and SERPINF1 genes and obesity. This could have explained our male-only findings for risk group differences in BMI as well. However, the male-limited effect on obesity suggests this was probably not the case.
The current analyses were conducted with a modest number of individual subjects although the total number of repeated observations was reasonably large. Consequently, a full genotypic analysis could not be performed. Rather, those both heterozygous and those homozygous for the minor allele were combined and contrasted with the homozygous major allele individuals (dominant genetic model).
Another possible limitation of our study is that most of the genes found in large-scale genome-wide association studies were not tested. Our goal was to test genes previously identified in studies of AD and related endophenotypes and to include genes previously associated with BMI. However, inclusion of genes previously associated with BMI was limited to just four genes (LEP, SERPINF1, BDNF and FTO). In contrast, a total of 32 loci have been found based in an analysis of 249,796 individuals for whom a BMI had been collected [61]. Although only one of these genes (FTO) appeared to map to BMI in our family study, this may not be unexpected. It is quite possible that genes identified through GWAS may not discriminate among individuals from multiplex for AD families if the diathesis for dependence and BMI are shared in common and genes discriminating those who carry increased risk do so because of highly penetrant loci.
A comment regarding the strength of our findings needs to be mentioned. Some of the findings would not reach the Bonferroni-adjusted threshold for significance (p < 0.003). However, the candidate genes were chosen based on a hypothesis-driven strategy for which Bonferroni correction may be too conservative. Also, to our knowledge no prior study has investigated the influence of familial loading for alcoholism on obesity and included the potential moderating effects of specific obesity and addiction genes. Therefore, the importance of directly assessing familial loading for AD to determine if this loading confers greater risk for higher BMI suggested the need to take advantage of this developmental data set, although of limited size. The presence of longitudinal follow-up assessments allowed us to show that risk status influences the trajectory of BMI across development, and the effect is not limited to specific time points.
Conclusion
A previous study from our laboratory demonstrated that individuals with a family history of alcoholism with prenatal exposure to cigarettes display a significantly greater susceptibility to weight gain in adolescence [62]. The current analysis found a main effect of familial risk on BMI and for the first time demonstrates that CHRM2, a gene that has been implicated in AD, has a role in the development of increased BMI across adolescence. Additionally, we found further support for the role of DRD2 and FTO in the determination of body mass across time. Our findings also highlight the importance of addressing developmental aspects of weight gain to better understand how genetic variation impacts obesity risk. Importantly, these data support the existence of a shared diathesis for addiction and obesity, which may be mediated by genetic influences on disinhibition and reward processing.
Future perspective
There is increasing evidence that liability for addiction and obesity may have overlapping neurobiological etiology. An improved understanding of the interactive effects of genetic and environmental variables has important implications for the development of personalized medicine, allowing preventative and treatment measures to be targeted to those at greatest risk.
Supplementary Material
Executive summary.
Clinical relevance of uncovering common pharmacogenetic underpinnings of addiction & obesity
Obesity is a prevalent condition affecting both children and adults with increased all-cause mortality.
Alcohol dependence (AD) is a prevalent condition especially among those with a family history of AD that substantially increases morbidity and mortality.
Successful treatment and prevention strategies uncovered in either obesity or addiction research, if tailored to specific genetic variation, could be utilized in both disorders.
Evidence that the dopaminergic reward circuit is involved in addictive behaviors
Dopaminergic signaling has been the most extensively studied system for understanding drugs of addiction including alcohol, nicotine and other drugs of abuse.
The outcome of treatment of nicotine dependence is better in those with the DRD2 A2 allele.
DRD2 C957T modulates striatal dopamine D2 receptor binding and mRNA stability and has been shown to vary with AD status within families with both affected and unaffected family members.
Evidence that the dopaminergic reward circuit is involved in obesity & normal feeding behavior
Dopamine neurons regulate feeding behaviors needed for survival. Dopamine-deficient mice die of starvation as result of reduced feeding behavior, but this behavior can be restored by replenishing striatal dopamine.
Exposure to food stimuli affects dopaminergic neurons in the ventral tegmental area and the nucleus accumbens, brain regions that express ghrelin, leptin and insulin receptors.
Evidence that CHRM2 is involved in addictive disorders
CHRM2 has been shown to be related to AD as well as being associated with a robust endophenotype for AD, the amplitude of the P300 component of the event-related potential.
Evidence that CHRM2 is involved in regulation of BMI & obesity
The present study, which was conducted to include those at ultra-high risk for AD, shows that major allele carriers of the CHRM2 SNP rs2061174 had a sixfold increase in risk for becoming obese.
Value of testing for genetic variation in multiplex families to uncover common diatheses for alcohol & drug dependence & increased BMI
A recent genome-wide association study of 249,796 individuals identified 32 loci associated with BMI, yet these loci explained only 1.5% of the variance.
Genome-wide association studies are designed to detect common variants of small effect and although important may not identify those genes of high penetrence affecting both vulnerability for developing drug dependence and AD, but also identify those genes likely to affect risk for increased BMI and obesity.
Acknowledgments
Funding for this study was provided by NIAAA grants AA018289, AA005909, AA008082 and AA015168 to SY Hill with support for manuscript preparation by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1247842 to SD Lichenstein.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Ethical conduct of research
The authors state that they have obtained appropriate institutional review board approval or have followed the principles outlined in the Declaration of Helsinki for all human or animal experimental investigations. In addition, for investigations involving human subjects, informed consent has been obtained from the participants involved.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Ogden CL, Carroll MD, Kit BK, Flegal KM. Prevalence of obesity in the United States, 2009–2010. NCHS Data Brief. 2012;82:1–8. [PubMed] [Google Scholar]
- 2.Voelker R. Escalating obesity rates pose health, budget threats. JAMA. 2012;308(15):1514. doi: 10.1001/jama.2012.13712. [DOI] [PubMed] [Google Scholar]
- 3.Flegal KM, Kit BK, Orpana H, Graubard BI. Association of all-cause mortality with overweight and obesity using standard body mass index categories: a systematic review and meta-analysis. JAMA. 2013;309(1):71–82. doi: 10.1001/jama.2012.113905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barsh GS, Farooqi IS, O’Rahilly S. Genetics of body-weight regulation. Nature. 2000;404(6778):644–651. doi: 10.1038/35007519. [DOI] [PubMed] [Google Scholar]
- 5. Volkow ND, Wang GJ, Tomasi D, Baler RD. The addictive dimensionality of obesity. Biol. Psychiatr. 2013;73(9):811–818. doi: 10.1016/j.biopsych.2012.12.020. • Review of evidence for common neural substrates for addiction and some forms of obesity.
- 6.Bruijnzeel AW, Repetto M, Gold MS. Neurobiological mechanisms in addictive and psychiatric disorders. Psychiatr. Clin. North Am. 2004;27(4):661–674. doi: 10.1016/j.psc.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 7.Kringelbach ML, O’Doherty J, Rolls ET, Andrews C. Activation of the human orbitofrontal cortex to a liquid food stimulus is correlated with its subjective pleasantness. Cereb. Cortex. 2003;13(10):1064–1071. doi: 10.1093/cercor/13.10.1064. [DOI] [PubMed] [Google Scholar]
- 8.Volkow ND, Fowler JS, Wang GJ, Goldstein RZ. Role of dopamine, the frontal cortex and memory circuits in drug addiction: insight from imaging studies. Neurobiol. Learn. Mem. 2002;78(3):610–624. doi: 10.1006/nlme.2002.4099. [DOI] [PubMed] [Google Scholar]
- 9. Filbey FM, Myers US, Dewitt S. Reward circuit function in high BMI individuals with compulsive overeating: similarities with addiction. Neuroimage. 2012;63(4):1800–1806. doi: 10.1016/j.neuroimage.2012.08.073. • Demonstrates that individuals with high BMI displayed hypersensitivity of the reward system in response to food cues.
- 10.Myrick H, Anton RF, Li X, et al. Differential brain activity in alcoholics and social drinkers to alcohol cues: relationship to craving. Neuropsychopharmacology. 2004;29(2):393–402. doi: 10.1038/sj.npp.1300295. [DOI] [PubMed] [Google Scholar]
- 11. Heber D, Carpenter CL. Addictive genes and the relationship to obesity and inflammation. Mol. Neurobiol. 2011;44(2):160–165. doi: 10.1007/s12035-011-8180-6. • Presents evidence that inflammation may represent a shared mechanism of genetic risk for addiction and obesity.
- 12.Janak PH, Wolf FW, Heberlein U, Pandey SC, Logrip ML, Ron D. BIG news in alcohol addiction: new findings on growth factor pathways BDNF, insulin, and GDNF. Alcohol Clin. Exp. Res. 2006;30(2):214–221. doi: 10.1111/j.1530-0277.2006.00026.x. [DOI] [PubMed] [Google Scholar]
- 13.Connor JP, Young RM, Lawford BR, Ritchie TL, Noble EP. D(2) dopamine receptor (DRD2) polymorphism is associated with severity of alcohol dependence. Eur. Psychiatr. 2002;17(1):17–23. doi: 10.1016/s0924-9338(02)00625-9. [DOI] [PubMed] [Google Scholar]
- 14. Sobczyk-Kopciol A, Broda G, Wojnar M, et al. Inverse association of the obesity predisposing FTO rs9939609 genotype with alcohol consumption and risk for alcohol dependence. Addiction. 2011;106(4):739–748. doi: 10.1111/j.1360-0443.2010.03248.x. • Demonstrates that a variation in the FTO gene that has been found to increase obesity risk is also an independent predictor of increased alcohol and tobacco use.
- 15.Wang JC, Hinrichs AL, Stock H, et al. Evidence of common and specific genetic effects: association of the muscarinic acetylcholine receptor M2 (CHRM2) gene with alcohol dependence and major depressive syndrome. Hum. Mol. Genet. 2004;13(17):1903–1911. doi: 10.1093/hmg/ddh194. [DOI] [PubMed] [Google Scholar]
- 16.Ray LA, Bujarski S, MacKillop J, Courtney KE, Monti PM, Miotto K. Subjective response to alcohol among alcohol-dependent individuals: effects of the µ-opioid receptor (OPRM1) gene and alcoholism severity. Alcohol Clin. Exp. Res. 2013;37(S1):E116–E124. doi: 10.1111/j.1530-0277.2012.01916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Setiawan E, Pihl RO, Benkelfat C, Leyton M. Influence of the OPRM1 A118G polymorphism on alcohol-induced euphoria, risk for alcoholism and the clinical efficacy of naltrexone. Pharmacogenomics. 2012;13(10):1161–1172. doi: 10.2217/pgs.12.99. [DOI] [PubMed] [Google Scholar]
- 18.Anton RF1, Oroszi G, O’Malley S, et al. An evaluation of mu-opioid receptor (OPRM1) as a predictor of naltrexone response in the treatment of alcohol dependence: results from the Combined Pharmacotherapies and Behavioral Interventions for Alcohol Dependence (COMBINE) study. Arch. Gen. Psychiatry. 2008;65(2):135–144. doi: 10.1001/archpsyc.65.2.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chamorro AJ, Marcos M, Mirón-Canelo JA, Pastor I, González-Sarmiento R, Laso FJ. Association of µ-opioid receptor (OPRM1) gene polymorphism with response to naltrexone in alcohol dependence: a systematic review and meta-analysis. Addict. Biol. 2012;17(3):505–512. doi: 10.1111/j.1369-1600.2012.00442.x. [DOI] [PubMed] [Google Scholar]
- 20.Davis CA, Levitan RD, Reid C, et al. Dopamine for “wanting” and opioids for “liking”: a comparison of obese adults with and without binge eating. Obesity (Silver Spring) 2009;17(6):1220–1225. doi: 10.1038/oby.2009.52. [DOI] [PubMed] [Google Scholar]
- 21.Gunstad J, Schofield P, Paul RH, et al. BDNF Val66Met polymorphism is associated with body mass index in healthy adults. Neuropsychobiology. 2006;53(3):153–156. doi: 10.1159/000093341. [DOI] [PubMed] [Google Scholar]
- 22.Laramie JM, Wilk JB, Williamson SL, et al. Multiple genes influence BMI on chromosome 7q31- 34: the NHLBI Family Heart Study. Obesity (Silver Spring) 2009;17(12):2182–2189. doi: 10.1038/oby.2009.141. [DOI] [PubMed] [Google Scholar]
- 23.Melka MG, Gillis J, Bernard M, et al. FTO, obesity and the adolescent brain. Hum. Mol. Genet. 2013;22(5):1050–1058. doi: 10.1093/hmg/dds504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonjes A, Zeggini E, Kovacs P, et al. Association of FTO variants with BMI and fat mass in the self-contained population of Sorbs in Germany. Eur. J. Hum. Genet. 2010;18(1):104–110. doi: 10.1038/ejhg.2009.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angeli CB, Kimura L, Auricchio MT, et al. Multilocus analyses of seven candidate genes suggest interacting pathways for obesity-related traits in Brazilian populations. Obesity (Silver Spring) 2011;19(6):1244–1251. doi: 10.1038/oby.2010.325. [DOI] [PubMed] [Google Scholar]
- 26.Böhm A, Ordelheide AM, Machann J, et al. Common genetic variation in the SERPINF1 locus determines overall adiposity, obesity-related insulin resistance, and circulating leptin levels. PLoS ONE. 2012;7(3):e34035. doi: 10.1371/journal.pone.0034035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Duran-Gonzalez J, Ortiz I, Gonzales E, et al. Association study of candidate gene polymorphisms and obesity in a young Mexican–American population from South Texas. Arch. Med. Res. 2011;42(6):523–531. doi: 10.1016/j.arcmed.2011.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiefer F, Jahn H, Otte C, Demiralay C, Wolf K, Wiedemann K. Increasing leptin precedes craving and relapse during pharmacological abstinence maintenance treatment of alcoholism. J. Psychiatr. Res. 2005;39(5):545–551. doi: 10.1016/j.jpsychires.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 29.Sogawa K, Kodera Y, Satoh M, et al. Increased serum levels of pigment epithelium-derived factor by excessive alcohol consumption-detection and identification by a three-step serum proteome analysis. Alcohol Clin. Exp. Res. 2011;35(2):211–217. doi: 10.1111/j.1530-0277.2010.01336.x. [DOI] [PubMed] [Google Scholar]
- 30.Robins LN, Helzer JE, Croughan J, Ratcliff KS. National Institute of Mental Health Diagnostic Interview Schedule: its history, characteristics and validity. Arch. Gen. Psychiatry. 1981;38(4):381–389. doi: 10.1001/archpsyc.1981.01780290015001. [DOI] [PubMed] [Google Scholar]
- 31.Hill SY, Shen S, Lowers L, Locke-Wellman J, Matthews AG, McDermott M. Psychopathology in offspring from multiplex alcohol dependence families with and without parental alcohol dependence: a prospective study during childhood and adolescence. Psychiatry Res. 2008;160(2):155–166. doi: 10.1016/j.psychres.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hill SY, Weeks DE, Jones BL, Zezza N, Stiffler S. ASTN1 and alcohol dependence: family-based association analysis in multiplex alcohol dependence families. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2012;159B(4):445–455. doi: 10.1002/ajmg.b.32048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 34. Volkow ND, Wang GJ, Fowler JS, Telang F. Overlapping neuronal circuits in addiction and obesity: evidence of systems pathology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008;363(1507):3191–3200. doi: 10.1098/rstb.2008.0107. •• Proposes a common model of dysfunctional dopaminergic neural circuitry in both addiction and obesity.
- 35.Herting MM, Schwartz D, Mitchell SH, Nagel BJ. Delay discounting behavior and white matter microstructure abnormalities in youth with a family history of alcoholism. Alcohol Clin. Exp. Res. 2010;34(9):1590–1602. doi: 10.1111/j.1530-0277.2010.01244.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakagawa S, Schielzeth H. A general and simple method for obtaining R2 from generalized liner mixed-effects models. Methods Ecol. Evol. 2013;4(2):133–142. [Google Scholar]
- 37.Cornes BK, Lind PA, Medland SE, Montgomery GW, Nyholt DR, Martin NG. Replication of the association of common rs9939609 variant of FTO with increased BMI in an Australian adult twin population but no evidence for gene by environment (G × E) interaction. Int. J. Obes. (Lond.) 2009;33(1):75–79. doi: 10.1038/ijo.2008.223. [DOI] [PubMed] [Google Scholar]
- 38.Frayling TM, Timpson NJ, Weedon MN, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scuteri A, Sanna S, Chen W-M. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:1200–1210. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hendershot CS, Bryan AD, Ewing SW, Claus ED, Hutchison KE. Preliminary evidence for associations of CHRM2 with substance use and disinhibition in adolescence. J. Abnorm. Child Psychol. 2011;39(5):671–681. doi: 10.1007/s10802-011-9511-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Latendresse SJ, Bates JE, Goodnight JA, et al. Differential susceptibility to adolescent externalizing trajectories: examining the interplay between CHRM2 and peer group antisocial behavior. Child Dev. 2011;82(6):1797–1814. doi: 10.1111/j.1467-8624.2011.01640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zucker RA, Heitzeg MM, Nigg JT. Parsing the undercontrol/disinhibition pathway to substance use disorders: a multilevel developmental problem. Child Dev. Perspect. 2011;5(4):248–255. doi: 10.1111/j.1750-8606.2011.00172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thamotharan S, Lange K, Zale EL, Huffhines L, Fields S. The role of impulsivity in pediatric obesity and weight status: a meta-analytic review. Clin. Psychol. Rev. 2013;33(2):253–262. doi: 10.1016/j.cpr.2012.12.001. [DOI] [PubMed] [Google Scholar]
- 44.Pulgaron ER. Childhood obesity: a review of increased risk for physical and psychological comorbidities. Clin. Ther. 2013;35(1):A18–A32. doi: 10.1016/j.clinthera.2012.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hill SY, Jones BL, Holmes B, Steinhauer SR, Zezza N, Stiffler S. 5Cholinergic receptor gene (CHRM2) variation and familial loading for alcohol dependence predict childhood developmental trajectories of P300. Psychiatry Res. 2013;209(3):504–511. doi: 10.1016/j.psychres.2013.04.027. •• Demonstrates that both familial loading for alcohol dependence and CHRM2 gene variation are associated with a low visual P300 trajectory. P300 amplitude is known to be associated with behavioral disinhibition, and may represent a shared mechanism of risk for substance use disorders and obesity.
- 46.Jones KA, Porjesz B, Almasy L, et al. A cholinergic receptor gene (CHRM2) affects event-related oscillations. Behav. Genet. 2006;36(5):627–639. doi: 10.1007/s10519-006-9075-6. [DOI] [PubMed] [Google Scholar]
- 47.Begleiter H, Porjesz B, Bihari B, Kissin B. Event-related brain potentials in boys at risk for alcoholism. Science. 1984;225(4669):1493–1496. doi: 10.1126/science.6474187. [DOI] [PubMed] [Google Scholar]
- 48.Hill SY, Steinhauer S, Park J, Zubin J. Event-related potential characteristics in children of alcoholics from high density families. Alcohol Clin. Exp. Res. 1990;14(1):6–16. doi: 10.1111/j.1530-0277.1990.tb00438.x. [DOI] [PubMed] [Google Scholar]
- 49.Iacono WG, Malone SM, McGue M. Substance use disorders, externalizing psychopathology, and P300 event-related potential amplitude. Int. J. Psychophysiol. 2003;48(2):147–178. doi: 10.1016/s0167-8760(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 50.Bauer L, Dick D, Bierut L, et al. Obesity, smoking, and frontal brain dysfunction. Am. J. Addict. 2010;19(5):391–400. doi: 10.1111/j.1521-0391.2010.00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hill SY, Steinhauer SR, Locke-Wellman J, Ulrich R. Childhood risk factors for young adult substance dependence outcome in offspring from multiplex alcohol dependence families: a prospective study. Biol. Psychiatry. 2009;66(8):750–757. doi: 10.1016/j.biopsych.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Habeych ME, Charles PJ, Sclabassi RJ, Kirisci L, Tarter RE. Direct and mediated associations between P300 amplitude in childhood and substance use disorders outcome in young adulthood. Biol. Psychiatry. 2005;57(1):76–82. doi: 10.1016/j.biopsych.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 53.Carlson SR, McLarmon ME, Iacono WG. P300 amplitude, externalizing psychopathology, and earlier- versus later-onset substance-use disorder. J. Abnorm. Psychol. 2007;116(3):565–577. doi: 10.1037/0021-843X.116.3.565. [DOI] [PubMed] [Google Scholar]
- 54.Kvaløy K, Kulle B, Romundstad P, Holmen TL. Sex-specific effects of weight-affecting gene variants in a life course perspective-The HUNT Study, Norway. Int. J. Obes. (Lond.) 2013;2013;37(9):1221–1229. doi: 10.1038/ijo.2012.220. [DOI] [PubMed] [Google Scholar]
- 55.Kritzer MF, Creutz LM. Region and sex differences in constituent dopamine neurons and immunoreactivity for intracellular estrogen and androgen receptors in mesocortical projections in rats. J. Neurosci. 2008;28(38):9525–9535. doi: 10.1523/JNEUROSCI.2637-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jacobs E, D’Esposito M. Estrogen shapes dopamine-dependent cognitive processes: implications for women’s health. J. Neurosci. 2011;31(14):5286–5293. doi: 10.1523/JNEUROSCI.6394-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mobascher A, Rujescu D, Mittelstrass K, et al. Association of a variant in the muscarinic acetylcholine receptor 2 gene (CHRM2) with nicotine addiction. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2010;153B(2):684–690. doi: 10.1002/ajmg.b.31011. [DOI] [PubMed] [Google Scholar]
- 58.Fontanesi L, Schiavo G, Galimberti G, et al. A genome wide association study for backfat thickness in Italian large white pigs highlights new regions affecting fat deposition including neuronal genes. BMC Genomics. 2012;13:583. doi: 10.1186/1471-2164-13-583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Labayen I, Ruiz JR, Moreno LA, et al. The effect of ponderal index at birth on the relationships between common LEP and LEPR polymorphisms and adiposity in adolescents. Obesity (Silver Spring) 2011;19(10):2038–2045. doi: 10.1038/oby.2011.74. [DOI] [PubMed] [Google Scholar]
- 60.Mei H, Chen W, Jiang F, et al. Longitudinal replication studies of GWAS risk SNPs influencing body mass index over the course of childhood and adulthood. PLoS ONE. 2012;7(2):e31470. doi: 10.1371/journal.pone.0031470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Speliotes EK, Willer CJ, Berndt SI, et al. Association analysis of 249,796 individuals reveal 18 new loci associated with body mass index. Nat. Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hill SY, Shen S, Locke-Wellman J, Rickin E, Lowers L. Offspring from families at high risk for alcohol dependence: increased body mass index in association with prenatal exposure to cigarettes but not alcohol. Psychiatry Res. 2005;135(3):203–216. doi: 10.1016/j.psychres.2005.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.