Abstract
In the arginylation branch of the N-end rule pathway, unacetylated N-terminal destabilizing residues function as essential determinants of protein degradation signals (N-degron). Here, we show that a neurostimulant, para-chloroamphetamine (PCA), specifically inhibits the Arg/N-end rule pathway, delaying the degradation of its artificial and physiological substrates, including regulators of G protein signaling 4 (RGS4), in vitro and in cultured cells. In silico computational analysis indicated that PCA strongly interacts with both UBR box and ClpS box, which bind to type 1 and type 2 N-degrons, respectively. Moreover, intraperitoneal injection of PCA significantly stabilized endogenous RGS4 proteins in the whole mouse brain and, particularly, in the frontal cortex and hippocampus. Consistent with the role of RGS4 in G protein signaling, treatment with PCA impaired the activations of GPCR downstream effectors in N2A cells, phenocopying ATE1-null mutants. In addition, levels of pathological C-terminal fragments of TDP43 bearing N-degrons (Arg208-TDP25) were significantly elevated in the presence of PCA. Thus, our study identifies PCA as a potential tool to understand and modulate various pathological processes regulated by the Arg/N-end rule pathway, including neurodegenerative processes in FTLD-U and ALS.
The N-end rule explains the correlation between a protein's N-terminal amino acid and its in vivo degradation rate1,2. It is the first identified constituent of the ubiquitin-proteasome system (UPS)-mediated proteolysis in eukaryotes3. However, the N-end rule pathway exists even in bacteria and archaea, which lack the bona fide UPS4,5. Eukaryotes have two branches of the pathway, the acetylation (Ac) branch and the arginylation (Arg) branch6. In the Arg/N-end rule pathway, Arg/N-degrons mainly include the primary “destabilizing” N-terminal amino acids, which are either positively charged residues (type 1), such as Arg, Lys, and His, or bulky, hydrophobic residues (type 2), such as Phe, Trp, Leu, Tyr, and Ile. Ac/N-degrons are generally created by cotranslational N-terminal acetylation, which are conditionally active only when they are spatially accessible to Ac/N-recognins7,8. The cleavages of protein by endopeptidases may generate not only Arg/N-degrons but also Ac/N-degrons by altering their sterically sequestered conformation.
The mammalian N-end rule pathway has been identified in various essential cellular processes, including cardiovascular development, neural tube formation, apoptosis, spermatogenesis, chromosomal stability and oxygen/heme sensing9,10,11,12,13,14. More recently, various neurodegenerative disease-implicated C-terminal fragments of proteins, such as, Tau, α-synuclein, and TDP43, were also identified as short-lived substrates of the Arg/N-end rule pathway15. Therefore, the Arg/N-end rule pathway might function to actively protect cells from detrimental effects of accumulated proteotoxic protein fragments. However, the physiological functions of the N-end rule pathway is far from completely elucidated, particularly given that the Ac/N-end rule pathway was only recently studied7,16 and more than 90% of human proteins are N-terminally acetylated in nascent proteins.
The UBR protein family are the N-recognins and specific E3 Ub ligases, which directly interact with the type 1 and type 2 “destabilizing” residues through the UBR box and the N-domain, respectively17. The UBR box mainly utilizes its acidic binding pocket for interaction with free α-amino groups (-NH3+) of type 1 destabilizing residues18,19. The N-domain, a eukaryotic sequelog of the E. coli ClpS domain, which also binds to type 2 residues20, appears to contain a hydrophobic pocket near its surface for type 2 interaction21. Cognate dipeptides have been widely used in vitro as competitive inhibitors of N-recognins, for example, Arg-Ala for type 1 and Phe-Ala for type 2, despite their low efficiency22. Simultaneously targeting these two binding sites of UBR proteins by heterovalent ligands significantly improved their inhibitory efficiency and druggable properties compared to dipeptides23. For examples, a small molecule having an N-terminal Arg and N-terminal Phe, which stretches ~15 Å and targets the UBR box and the N-domain, respectively, showed significant degradation inhibition on not only Arg/N-end rule model substrates, but also the physiological substrate RGS4 in cultured cells24. However, in mice, the multivalent inhibitors had little effects on the half-lives of RGS425. Therefore, it is becoming increasingly necessary to identify more potent in vivo inhibitors of the pathway, especially considering a wide range of developmental and pathological implications of the Arg/N-end rule pathway are being recognized.
In previous work, we identified the essential chemical components of N-degron, such as its l-conformation, protonated α-amine group, and hydrophobic side chains, required for direct interaction with N-recognins25. Several Phe-derived molecules with minimal interaction motifs for UBR proteins, including amphetamine and PCA, efficiently blocked the Arg/N-end rule pathway when model substrate degradation was biochemically tested in vitro25. PCA is a substituted amphetamine and serotonin (5-HT) releaser26, and was used as an antidepressant without significant psychotomimetic action until several animal studies identified neurotoxicity in various areas of the mouse brain after PCA injection27.
Here, we show that a neurostimulant PCA inhibits the Arg/N-end rule pathway in vivo and in the mouse brain. Treatment with PCA resulted in increased levels of endogenous RGS4 and, subsequently, impaired activity of downstream GPCR effectors and regulation of many GPCR target genes implicated in neuron development. PCA injection to mice produced similar phenotypes as observed in genetic studies of Arg/N-end rule utilizing knockout mice. In addition, pathologic C-terminal fragments of TDP43 (Arg208-TDP25) in cells, which are implicated in the sporadic and familial pathogenesis of frontotemporal lobar degeneration with Ub-positive, tau-negative inclusions (FTLD-U) and amyotrophic lateral sclerosis (ALS)28,29, formed cytosolic aggregation in cells with significantly increased rates in the presence of PCA, consistent with the counteractive functions of the Arg/N-end rule pathway in proteopathic protein accumulation. These results constitute the first evidence that the degradation of physiological substrates of the Arg/N-end rule pathway is pharmacologically inhibited in animals. Therefore, chemical inhibition of the Arg/N-end rule pathway using PCA may offer a novel strategy to understand and modulate the various pathological implications of the system in mice, including protein aggregation in the brain associated with various neurodegenerative diseases.
Results
In vitro biochemical assay and in silico computational analysis revealed that PCA effectively blocked the degradation of both type 1 and type 2 Arg/N-end rule substrates
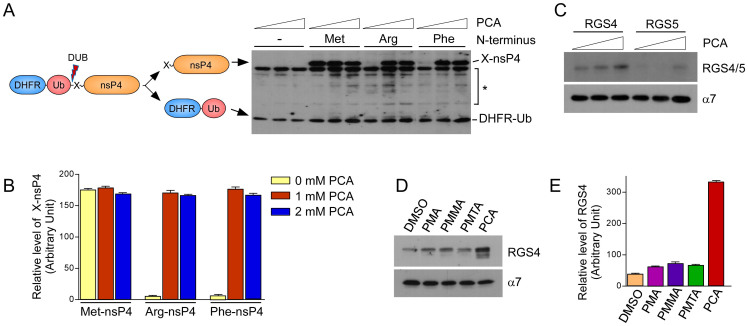
The inhibitory effect of PCA was first examined in vitro by artificial model substrates, DHFR-Ub-X-nsP4 fusion proteins, of the Arg/N-end rule pathway, which were expressed in rabbit reticulocyte lysates. The reticulocyte lysates were enriched with not only essential components for the cell-free expression of model substrates but also with a variety of enzymes involved in UPS-dependent proteolysis30,31. Under normal conditions, the type 1 substrate Arg-nsP4 and type 2 substrate Phe-nsP4 were short-lived, while Met-nsP4, a long-lived control of the model substrates, was long-lived (Fig. 1A). Stable DHFR-Ub proteins were used as references. Degradation of both of the N-end rule model substrates was completely blocked by PCA mainly through post-translational inhibition, considering the levels of DHFR-Ub. Furthermore, at the inhibitory concentrations, the levels of Met-nsP4 and Arg/Phe-nsP4 were virtually identical (Figs. 1A and B), suggesting that PCA is a potent inhibitor of both type 1 and type 2 Arg/N-end rule pathway. We also tested PCA for its ability to degrade RGS4 and RGS5, which are bona fide physiological substrates of the mammalian Arg/N-end rule pathway32. Similar to the artificial N-end rule substrates, the levels of these proteins were also significantly increased by PCA in a dose-dependent manner (Fig. 1C).
Figure 1. PCA inhibits the type 1 and type 2 Arg/N-end rule pathways in vitro.
(A) Artificial Arg/N-end rule substrates Arg-nsP4 and Phe-nsP4, which are type 1 and type 2 model substrates, respectively, were stabilized by PCA. Tripartite DHFR-Ub-X-nsP4 fusion proteins were expressed in vitro with biotin labels using rabbit reticulocyte lysates and biotinylated lysyl tRNA. Total reaction mixtures were subjected to SDS-PAGE followed by Western blot using horseradish peroxidase (HRP)-conjugated streptavidin. Cotranslational cleavage events by deubiquitinating enzymes (DUB) yields long-lived DHFR-Ub references and X-nsP4 substrates as indicated. Asterisk indicates nonspecific signals or cleaved forms of X-nsP4. The in vitro expression/degradation reactions were performed in the presence of 0, 1, 2 mM PCA for 90 min. (B) Relative amounts of remaining X-nsP4 proteins normalized by DHFR-Ub in the presence/absence of PCA. Multiple film images were quantified by densitometry. (C) Otherwise short-lived RGS4 and RGS5, physiological Arg/N-end rule substrates, were stabilized by PCA. As in Fig. 1A except that a proteasome core particle subunit α7 was used as loading control. (D) Compounds which are structurally similar to PCA did not inhibit the Arg/N-end rule pathway. Unlike PCA, PMMA, PMA, or PMTA exhibited little effects on RGS4 level compared to control (DMSO). Cropped gels/blots are used in (C) and (D). (E) Quantification of RGS4 levels, which are normalized to those of α7. Data represent mean ± SD (n = 3).
Small-molecules containing analogous structural scaffolds to those of PCA were examined to determine whether they possess inhibitory effects on the Arg/N-end rule pathway. Although PCA, para-methoxymethamphetamine (PMMA), para-methoxyamphetamine (PMA), and para-methylthioamphetamine (PMTA) have phenylisopropylamine backbones that are identical except for various chemical modifications at the para-position (Supplementary Fig. S1), only PCA showed a dramatic stabilization of RGS4 proteins (~8-fold higher level than vehicle control, Figs. 1D and E), suggesting the importance of a chlorine group in the phenyl core and a free amine at the isopropyl group for the intermolecular interaction with UBR proteins.
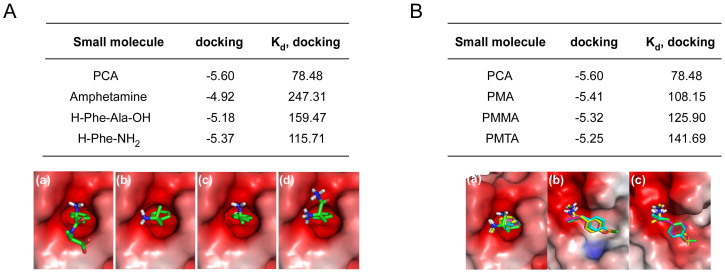
Consistent with the in vitro synthesis/degradation assay, in silico computational analysis using PCA docking to N-recognins revealed its strong and specific interactions with both the UBR box and the ClpS domain. PCA showed relatively stronger binding to the crystal structure of the type 2 N-degron-recognizing ClpS domain (PDB ID: 3DNJ) than other Phe-derived momomeric inhibitors (−5.60 kcal/mol of PCA vs. −3.90, −4.92, −5.18, and −5.37 kcal/mol of Phe-OH, amphetamine, Phe-Ala, and Phe-NH2, respectively, in binding affinity (Fig. 2A and Supplementary Fig. S2A). (See Methods section for detailed docking parameters and methodology) The dissociation constant (Kd) of PCA was also significantly lower than those of controls (78.5 μM of PCA vs. 1183.5 μM, 247 μM, 159.5 μM, and 115.7 μM of Phe-OH, amphetamine, Phe-Ala, and Phe-NH2, respectively in Kd). Blocking of the N-terminal free amino group such as acetylation substantially prevents the binding of the inhibitors to the ClpS domain (Supplementary Fig. S2B).
Figure 2. In silico analysis of PCA-UBR protein bindings.
(A) Binding affinities (docking, kcal/mol), dissociation constants (Kd, μM), and binding modes of (a) PCA, (b) amphetamine (c) Phe-Ala dipeptide, and (d) Phe-NH2 with the ClpS domain (PBD code: 3DNJ). (B) Binding parameters between PCA-like small molecules and the ClpS domain. The calculated binding modes of the chemicals and the (a) ClpS domain, (b) UBR1 box (3NY1), or (c) UBR2 box (3NY3) are shown in color: PCA, PMMA, PMA, and PMTA are cyan, yellow, magenta, and green, respectively.
In silico docking analysis indicated the stereospecific nature of the Arg/N-end rule pathway as anticipated (−8.03 μM of l-Arg-Ala vs. −6.27 μM of d-Arg-Ala in dissociation constant with the UBR box) (Supplementary Fig. S3A). The binding affinity of PCA toward the UBR2 box, which recognizes type 1 N-degrons, falls in micromolar ranges similar to the dissociation constants between the UBR box and type 1 peptides obtained from isothermal titration calorimetry (ITC)18,19 and was more than 10-fold stronger than the PCA-ClpS interaction (5.54 μM vs. 78.84 μM in dissociation constant). Consistent with the results of the biochemical degradation assay, in silico calculation substantiated that PCA had stronger and more specific interactions with both the UBR box and the ClpS domain than control molecules such as PMA, PMMA, and PMTA (Fig. 2B and Supplementary Fig. S3B). These data indicate that PCA is capable of inhibiting both the type 1 and type 2 Arg/N-end rule pathway in vitro and in silico. The para-positioned chlorine of PCA may function as an essential pharmacophore in the inhibition, which can be a target for further structure-activity relationship study.
PCA treatment inhibited the degradation of RGS4 and impaired the activation of GPCR signaling in mammalian cells
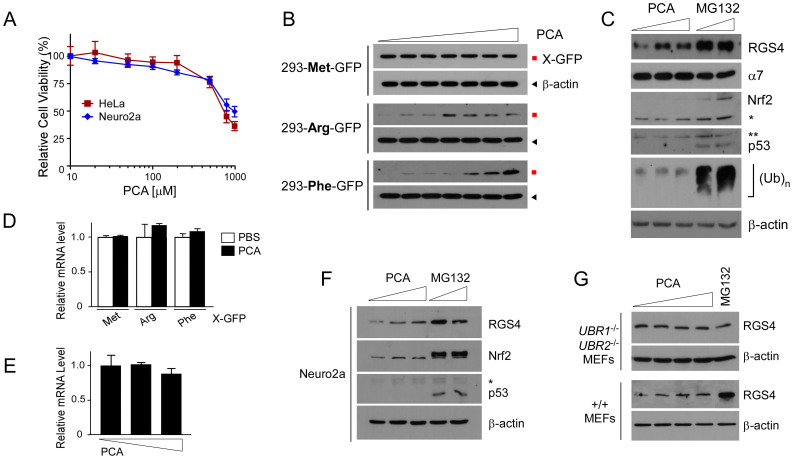
The chemical structure of PCA satisfies all of Lipinski's rules33, suggesting it is highly cell-permeable. Moreover, cytotoxicity of PCA on HeLa and Neuro2a cells was observed only at the concentration above 750 μM (Fig. 3A). The inhibitory effects of PCA on the Arg/N-end rule pathway in mammalian cells were first examined using stable HEK293 cell lines expressing Met-GFP (stable control), Arg-GFP (type 1), and Phe-GFP (type 2). Similar to their cognate in vitro model substrates, X-nsP4 (Fig. 1A), Arg-GFP and Phe-GFP, which have type 1 and type 2 primary destabilizing residues, respectively, were rapidly degraded under normal conditions while Met-GFP was highly stable (Fig. 3B and Supplementary Fig. S4). Proteasome inhibitors significantly enhanced levels of Arg- and Phe-GFP but did not affect Met-GFP levels (Supplementary Fig. S4). No additive or synergic effects were observed when PS341 (1 μM) was added to MG132 (10 μM). At 4 hr post-treatment of PCA, Arg-GFP and Phe-GFP proteins, which were typically undetectable, were prominently observed by immunoblotting starting at 50 μM and 100 μM PCA concentrations, respectively (Fig. 3B). While Phe-GFP levels increased with PCA in a dose-dependent manner up to 500 μM, stabilization of Arg-GFP appeared to be saturated at 50 μM, which might also reflect the low binding affinity of PCA to the UBR box than to the ClpS domain and stronger inhibition of type 1 substrates.
Figure 3. PCA specifically inhibits the Arg/N-end rule pathway in mammalian cells.
(A) The cytotoxicity of PCA on HeLa and Neuro2a cells was measured using the MTT assay. (B) HEK293-derived stable cell lines expressing Arg/N-end rule model substrates, including Ub-Arg-GFP (type 1), Ub-Phe-GFP (type 2), and Ub-Met-GFP (stable control), were generated and were treated with PCA at 0, 10, 20, 50, 100, 200, or 500 μM concentrations for 4 hr. Samples were analyzed by SDS-PAGE/Immunoblotting (IB) using indicated antibodies. β-actin, loading control. (C) A physiological substrate, RGS4 was transiently overexpressed in HeLa cells and, at two days posttransfection, PCA (0, 100, or 250 μM) and MG132 (10 or 20 μM) were used for treatment for 4 hr. Asterisks, nonspecific signals. (D & E) Total RNA was isolated after the experimental processes in (B) and (C) and used for quantitative RT-PCR of GFP and RGS4 mRNA levels, respectively. The values plotted are means ± SD of three independent experiments. (F) Endogenous RGS4 protein levels were examined in Neuro2a after treated with 0, 100, 250 μM of PCA or 10, 20 μM of MG132 for 4 hr. Nrf2 and p53 are endogenous UPS substrates. (G) As in (C) except that wild-type (+/+) and UBR1-/-UBR2-/- MEFs were used with 0, 50, 100, 250 μM of PCA or 10 μM of MG132. In UBR1-/-UBR2-/- MEFs, endogenous RGS4 proteins were detected. Cropped gels/blots are used in Western blot data.
To determine whether PCA could inhibit the degradation of physiological substrates of the Arg/N-end rule pathway, we transiently overexpressed RGS4 in HeLa cells and treated the cells with subcytotoxic doses of PCA or proteasome inhibitor MG132. PCA increased levels of RGS4 at the 100 μM concentration although the stabilization effect appears to be weaker than 10 μM MG132 (Fig. 3C). Future work requires to determine whether the weaker RGS4 stabilization of PCA than MG132 was due to its intrinsically limited inhibitory efficacy of PCA or other regulatory modification of RGS4. RGS4 stabilization by PCA treatment was specifically achieved without affecting other substrates or components of UPS. No significant changes were seen in proteasome levels, endogenous proteasome substrates such as Nrf2 and p53, or bulk cellular Ub conjugates (Fig. 3C). Moreover, PCA posttranscriptionally antagonized the Arg/N-end rule pathway because little change was detected in mRNA levels of X-GFPs or RGS4 (Figs. 3D and E).
Endogenous RGS4 was also significantly stabilized by PCA. In Neuro2a cells, in which we found that RGS4 was more abundant than in other cells, PCA treatment significantly increased levels of endogenous RGS4 (Fig. 3F). However, PCA showed still weaker RGS4 degradation inhibition than proteasomal inhibitor MG132 as observed when overexpressed RGS4 was compared (Fig. 3C). Unlike in wild-type MEFs, no inhibition of PCA was observed in UBR1-/-UBR2-/- or ATE1-/- MEFs (Fig. 3G and Supplementary Fig. S5), which lack key components of the Arg/N-end rule-dependent RGS4 degradation34,35. In UBR1-/-UBR2-/- MEFs, little changes of RGS4 were observed in the presence of MG132 (Fig. 3G), suggesting that proteasomal degradation of RGS4 is mainly mediated by the Arg/N-end rule pathway. These results together indicate that PCA may readily pass through the cellular membrane and specifically target the Arg/N-end rule pathway in mammalian cells.
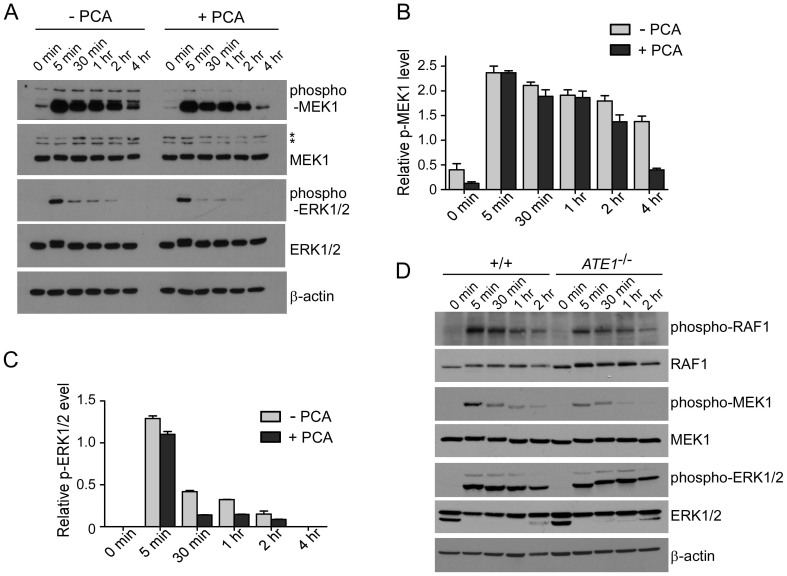
RGS4 protein is a GTPase activating protein that negatively regulates Gi- and Gq-mediated signaling pathways by accelerating the rate of Gα-GTP hydrolysis and re-association of the Gαβγ heterotrimer36,37. Considering the physiologic function of RGS4, we examined whether the activities of downstream G protein signaling were affected by PCA treatment. Time-course immunoblotting using HeLa cells after 12-hr serum starvation and subsequent serum activation in the presence and absence of PCA revealed that the activities of MEK1 were significantly attenuated in PCA-treated cells (Figs. 4A–C), similar to that which was observed in ATE1-/- MEFs (Fig. 4D). Therefore, chemical inhibition by PCA might produce phenocopies of the genetic mutations in the Arg/N-end rule pathways. Previous work has shown that the Arg/N-end rule pathway involves in a variety of essential cellular processes38. This may be reflected by the diverse functions of G protein signaling regulated by RGS proteins.
Figure 4. PCA impairs the downstream activations of GPCR signaling.
(A) Activation profiles of MAPK signaling proteins in HeLa cells in the presence or absence of 100 μM PCA. Cells were incubated with serum-free media for 12 hr, followed by serum-containing media. Samples were collected at the indicated time points and analyzed by SDS-PAGE/IB. (B & C) Quantification of phospho-MEK1 (B) and phospho-ERK1/2 (C) normalized by total MEK1 and ERK1/2, respectively, in the presence/absence of PCA. Multiple film images were quantified by densitometry and data represent mean ± SD (n = 3). (D) As in (A) except that the experiment was performed using wild-type (+/+) and ATE1-/- MEFs. Cropped gels/blots are used in Western blot data.
PCA inhibited RGS4 degradation and impaired GPCR signaling in the mouse brain
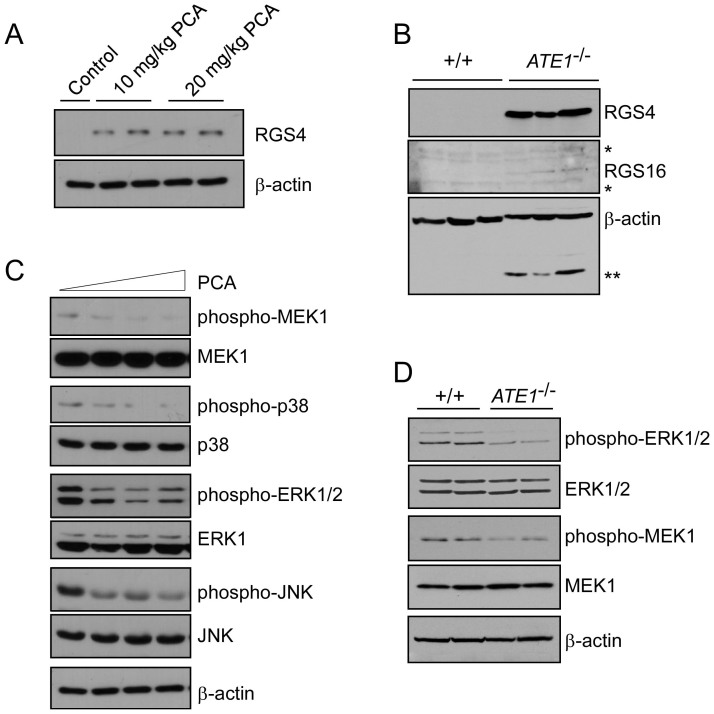
The physiological functions of the N-end rule pathway in the brain are largely undetermined. The mutant mice (or embryos) lacking key components of the pathway were reported to exhibit strong phenotypes in other organs such as heart and testis rather than the brain39. However, ATE1 gene showed intense expression in the brain and the spinal cord, as determined by X-gal staining for the reporter β-galactosidase, which was integrated into the targeted ATE1 allele (Supplementary Fig. S6), strongly suggesting an essential role of the Arg/N-end rule pathway in the dynamic regulation of GPCR signaling in the brain. Considering the blood-brain-barrier permeable nature of PCA, we tested whether PCA inhibits the degradation of RGS4 in the mouse brain and whether this degradation affects the downstream processes of the G protein signaling pathway. Mice were injected with PCA either at 10 mg/kg or 20 mg/kg for three days and sacrificed 6 hr after the final injection. First, whole brain extracts were prepared for immunoblotting. The levels of endogenous RGS4 in the whole brain were significantly increased by either dose of PCA administrations compared to those of the control, in which saline was injected and virtually no RGS4 was detected (Fig. 5A). The PCA effects on the brain were similar to when the arginylation branch of the N-end rule pathway was disrupted. In the ATE1-/- knockout mouse brain, both RGS4 and RGS16, which required to be arginylated at the N-termini for degradation, were strikingly stabilized compared to wild-type brains (Fig. 5B and Supplementary Fig. S7). These data identify that the PCA is the first in vivo N-end rule inhibitor targeting the Arg/N-end rule in mouse brains.
Figure 5. PCA inhibits the Arg/N-end rule pathway and impairs downstream effectors of G protein signaling in the mouse brain.
(A) RGS4 levels in the brain were significantly increased after intraperitoneal PCA injections at the indicated concentrations. Whole brain extracts were analyzed by SDS-PAGE and RGS4 IB. β-actin, loading control. (B) Immunoblotting of RGS4 and RGS16 in extracts of +/+ and ATE1-/- embryonic brains. *, non-specific. **, unstripped signals from RGS4 IB. (C) Activation of MAPK signaling molecules such as MEK1, p38, ERK1/2, and JNK in the mouse brain. Whole brain extracts after PCA treatments (0, 10, 20 mg/kg) were subjected to immunoblotting with the indicated antibodies. (D) MEFs from wild-type (+/+) or ATE1-/- were compared to examine ERK1/2 and MEF1 activities. Cropped gels/blots are used in Western blot data.
To determine the consequences of PCA-induced RGS4 stabilization in GPCR signaling in mice, we further examined the activities of downstream signaling transducers especially MAPKs12. PCA-treated mouse brains had reduced MEK1, p38, ERK1/2, and JNK activity (Fig. 5C), in agreement with their biochemical functions in cells and consistent with observations from ATE1-/- brains (Fig. 5D). These data indicate that chemical inhibition by PCA, a potent inhibitor of the Arg/N-end rule pathway, is sufficient to mimic the effects of GPCR-signaling inhibition and RGS hyperactivation, and that this effect is similar to genetic inhibition methods of the Arg/N-end rule components, which often resulted in embryonic lethality.
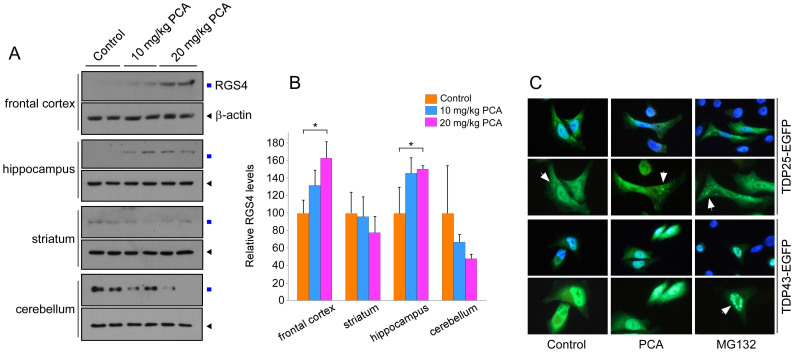
Next, the regional specificity of PCA was examined by dissecting the brain into frontal cortex, striatum, hippocampus, and cerebellum. Strong stabilization of RGS4 by intraperitoneal injection of PCA was observed in the frontal cortex and hippocampal areas (Figs. 6A and 6B), which are also known to express high levels of RGS4 mRNA40. Few effects were observed in the striatum and the cerebellum. It is to be determined whether the region-specific inhibition of PCA simply reflects differential levels of translated RGS4 or depends on characteristic distribution patterns of PCA perhaps through neuronal transport systems in brain capillary membranes. It is interesting that these regions are also known to be critically affected by various neurodegenerative diseases such as Alzheimer's disease (AD), ALS, and FTLD-U41,42. To identify putative GPCR target genes altered by PCA treatment in the brain, we profiled the expression of genes in the frontal cortex and hippocampus by microarray analysis. The microarray analysis results showed that 16 and 57 genes were downregulated by PCA in the frontal cortex and hippocampus, respectively (Supplementary Fig. S8). Many genes implicated in neuron differentiation and pattern specification processes were significantly decreased by PCA treatment in frontal cortex and hippocampus. It remains to be determined whether the altered expression of these genes are directly regulated by RGS4-dependent GPCR signaling and whether it underlies neurological phenotypes in Arg/N-end rule-related knockout mice.
Figure 6. PCA delays RGS4 degradation in the frontal cortex and hippocampus while facilitates aggregation formation of Arg208-TDP25.
(A) Mouse brains after control and PCA injection were dissected into the frontal cortex, striatum, hippocampus, and cerebellum. Then samples were extracted and used for RGS4 immunoblotting. Cropped gels/blots are used here. (B) Quantification of RGS4 levels in brains normalized by β-actin, which indicate significantly increased levels of RGS4 in the frontal cortex and hippocampus. Mean ± SD (n = 4/group). (C) TDP43-EGFP and Arg208-TDP25-EGFP was transfected into HeLa cells which were treated with MG132 (10 μM) or PCA (100 μM) for 4 hr after 36 hr post-transfection.
Bower et al. recently reported that the Arg/N-end rule pathway mediated the proteasomal degradation of many naturally occurring C-terminal fragments of tau, TDP43, and α-synuclein15. Among them, C-terminal TDP43 fragments Arg208-TDP43, produced by caspase-cleavage event, are highly pathologic in FTLD-U and ALS to form toxic, insoluble, and Ub- and phospho-positive cytoplasmic inclusions28,29. To determine whether PCA could inhibit the degradation of TDP25, we expressed Arg208-TDP25-EGFP in HeLa cells in the presence and absence of PCA or MG132. Transiently overexpressed TDP43-EGFP in HeLa cells was diffusively distributed mainly in nucleus. When treated with MG132, aggregated forms of TDP43 were readily observed in nucleus while PCA showed virtually no effects on TDP43 aggregation (Fig. 6C). Arg208-TDP25-EGFP, which lacks the NLS domain located in the N-terminus of TDP43, was mainly observed dispersive in whole cells, but forms cytoplasmic aggregation in PCA-treated cells, further supporting that the Arg/N-end rule branch has neuroprotective roles in TDP43-related neurodegenerative diseases.
Discussion
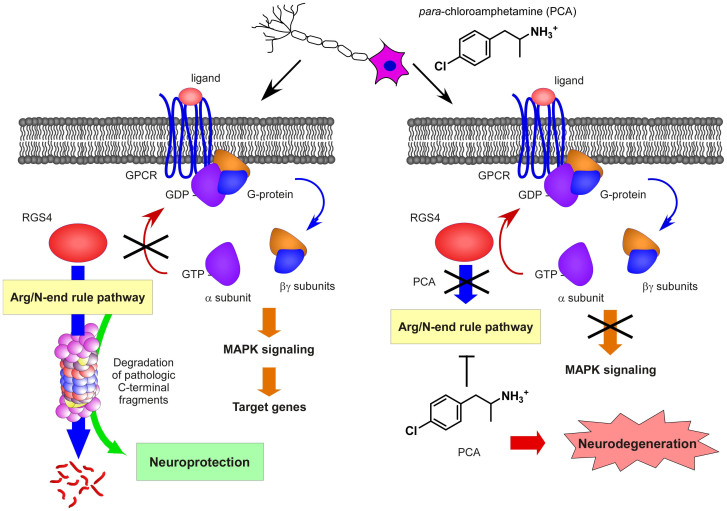
Here we report a small-molecule PCA for its capacity to inhibit the Arg/N-end rule pathway in vitro, in cultured cells, and in the mouse brain. PCA potently and specifically stabilized various artificial and physiological Arg/N-end rule substrates including RGS4. RGS4 is known to negatively regulate the G protein signaling pathway, and PCA-treated cells and brains indeed appeared to be significantly impaired in activation of downstream G protein signaling (Figs. 4 and 5). The detailed similarities observed between PCA treatments and the effects of genetic inactivation of the Arg/N-end rule pathway provide strong evidence that modulating the N-end rule pathway in the brain and in other organs is achievable through this chemical inhibition methods. Based on our results, we propose a model, in which the Arg/N-end rule mediated proteolysis of RGS4 is important for appropriate GPCR signaling in neuronal cells (Fig. 7).
Figure 7. A model for the effects of PCA in Arg/N-end rule pathway-dependent GPCR signaling in the brain and its implication in neurodegeneration.
In normal conditions, the protein levels of RGS4 are maintained in low concentrations in the neuronal cells by the Arg/N-end rule pathway, which facilitate various signal transductions mediated by GPCR. Moreover, the Arg/N-end rule pathway appears to have a neuroprotective role in neurons for the clearance of various proteotoxic C-terminal protein fragments implicated in neurodegenerative diseases. When the Arg/N-end rule pathway is genetically or chemically inhibited, RGS4 levels are elevated to hydrolyze GTP of G protein's α subunit, which results in inactivation of GPCR signaling.
This is the first evidence that a single molecule can inhibit both type 1 and type 2 Arg/N-end rule pathway in vivo. From our in vitro and in silico study, PCA was effectively bound to the UBR domain and the ClpS domain, which are responsible to recognize type 1 and type 2 Arg/N-end rule substrates, respectively (Figs. 1 and Fig 2). UBR-PCA interaction appeared to be much stronger than ClpS-PCA interaction. However, the underlying molecular mechanism is not yet clear. It was previously shown that the addition of a chloride moiety to unmodified amphetamine substantially increased its inhibitory efficiency43. Although the chloride atom substitution of PCA may facilitate the interaction between PCA and the active sites by allowing water molecules to occupy the space and mediate hydrogen bonds as seen in the UBR box and ClpS domain19,44, the structural basis of inhibitory effects of PCA is to be determined. The para-positioned chlorine of PCA may function as an essential pharmacophore in the inhibition, which can be a target for further structure-activity relationship study.
We demonstrate that PCA has the ability to specifically inhibit the Arg/N-end rule pathway in cultured cells using GFP-based model substrates and physiological substrate RGS4. The stabilizing effect of these substrates by PCA was significant, but weaker than proteasomal inhibitor MG132 (Fig. 3), indicating that the potency of PCA can be further improved by varying the structure of PCA. In contrast to other E3 Ub ligases that recognize substrates through protein-protein interfaces, UBR proteins recognize single amino acids as a degradation signal, which contributes to establish PCA as an attractive target for structure-activity relationship study. Interestingly, for Arg/N-end rule model substrates, no additive or synergic effects between PS341, which mainly targets chymotrypsin-like and caspase-like proteasomal activity, and MG132, which targets chymotrypsin-like and trypsin-like activity45, was observed (Supplementary Fig. 4). In addition, some of UBR proteins were identified to directly interact with proteasomes46, which may contribute the rapid turnover rates of Arg/N-end rule substrates. Taken together, it is highly possible that the components of Arg/N-end rule-dependent post-translational modificationand ubiquitination, such as ATE1 and UBR family, may allosterically interact with subunits of proteasomes and that the substrate degradation is highly sensitive to proteasomal degradation, which is perhaps commenced at the protein translation level.
A prominent pathway regulated by the Arg/N-end rule pathway is Gq-dependent signaling47, whose function is tightly controlled by RGS proteins. This might be related to the diverse physiological involvement of the Arg/N-end rule pathway in cellular processes. ATE1 gene encodes Arg-transferase, which is essential for the N-terminal modification and degradation of RGS412. Constitutional nullification of the ATE1 gene, resulted in embryonic lethality due to abnormal cardiovascular development9. However, a more recent study of ATE1-deficient adult mice, which were generated by a postnatal, tissue-specific knockout strategy, showed that the loss of ATE1 is implicated in a strikingly broad range of biological processes, which include abnormalities in metabolic rates, resistance to obesity, brain size, and male sterility38. Moreover, in the cultured cells, the delayed activation of ERK1/2 after PCA treatment was weaker than those of upstream targets such as RAF1 or MEK1 (Fig. 4), which might reflect complex crosstalk between GPCRs on downstream activators. It will be interesting to determine whether PCA can stabilize other R4 subfamily of RGS proteins such as RGS248. Overall, our data identify PCA as a potent and specific in vivo Arg/N-end rule inhibitor that results in phenocopies similar to the genetic inhibition of the Arg/N-end rule pathway (Fig. 5). Therefore, PCA has the potential to be used for determining the underlying regulatory circuits of the pathway and for identifying the hitherto unknown in vivo Arg/N-end rule substrates. Recently, it was reported that UBR proteins could recognize the N-terminal Met if the penultimate residues are large, hydrophobic amino acids such as Leu, Ile, Phe, Tyr, and Trp16, suggesting the dual targeting mechanisms of the two branches of the N-end rule pathway. PCA can be a useful tool to dissect the crosstalk between them and the conditional regulation of substrate degradation in vivo.
The scope of Arg/N-end rule-mediated proteolysis is not limited to the regulation of GPCR signaling. The Arg/N-end rule pathway may be a more general mechanism for protect cells, especially neuronal cells, to protect themselves from proteotoxic stress by degrading aggregation-prone proteins or protein fragments (Fig 7). For examples, PCA facilitated the aggregation formation of pathological C-terminal fragment of TDP43, which is degraded by the Arg/N-end rule pathway (Fig 6C). Therefore, the N-end rule pathway appears to have a neuroprotective role in neurons for the clearance of various proteotoxic C-terminal protein fragments implicated in neurodegenerative diseases. Using PCA, the biological scope of the Arg/N-end rule pathway can be extended to pathogenesis of more diverse human diseases. Moreover, pharmacological modulation of the Arg/N-end rule pathway by PCA has a potential to provide new insight into the pathological neurodegeneration controlled by the Arg/N-end rule pathway.
Methods
In vitro degradation assay
In vitro protein synthesis/degradation assays in the presence and absence of PCA or PCA-derivatives were performed using DHFR-Ub-X-nsP4, RGS4, and RGS5 plasmids as previously described12,25. Briefly, the plasmids were expressed with biotin labeling using biotinylated lysine-tRNA complex (Promega), and the steady-state levels of de novo synthesized proteins were detected by anti-biotin Western blotting using horseradish peroxidase-conjugated streptavidin (Pierce). Quantification of Western blotting was achieved by densitometry of multiple film images using ImageJ software (ver. 1.38e, NIH). Data are presented as mean ± SD and statistical analysis was performed by unpaired student's t-test or ANOVA. A value of P < 0.05 was accepted as statistically significant.
In silico docking analysis of protein-chemical interactions
Crystal structures of the UBR1 box, UBR2 box, and ClpS box were taken from a Protein Data Bank (PDB ID: 3NY1, 3NY3, and 3DNJ, respectively). The structure of the small molecules were optimized using the semi-empirical PM6 method of Gaussian 0949. AutoDockTools version 1.5.4 was used to add polar hydrogens and to assign Gasteiger charges for proteins and compounds50. For each protein, AutoGrid version 4.2 was used to create affinity grids centered on the active site51. The grids were large enough to contain all of the active sites. The binding affinities between the active sites and PCA or PCA-like molecules were calculated using a ligand-receptor docking computation with AutoDock version 4.2 with the Lamarckian genetic algorithm51. Important docking parameters were as follows: trials of 200 dockings, population size of 300, random starting position and conformation, and 50 million energy evaluations. Docked conformations were clustered using a tolerance of 0.5 Å RMSD. The lowest binding energy of the most populated cluster was used for ranking. Experimental binding free energies were calculated from the dissociation constants based on the equation: ΔG = RT lnKd, where R is a gas constant (1.987 cal K−1 mol−1) and T is temperature (298.15 K). The electrostatic potentials were calculated using the APBS package52. All structural figures were prepared using PyMOL package (http://sourceforge.net/projects/pymol/).
Assessment of cell viability
Cell viability was assessed using a modified MTT assay53. Briefly, HeLa cells and Neuro2a cells were treated with PCA at various concentrations (10 μM to 1000 μM) for 4 hr, followed by the addition of 10 μL of 5 mg/mL thiazolyl blue tetrazolium bromide (MTT, Sigma Aldrich) solution to the media and incubation for 2.5 h at 37°C in a humidified atmosphere of 95% air and 5% CO2. After discarding the media, 200 μL of DMSO was added to solubilize the blue MTT-formazan product and the cells were incubated for an additional 30 min at room temperature. The absorbance of the solution was read at 570 nm (test) and 630 nm (reference).
Mice
For the PCA experiment, male C57B6 mice weighing 20–25 g were given intraperitoneal injections of PCA at final doses of 10 mg/kg and 20 mg/kg once per day for three days. Control mice received an equivalent volume of saline. Mice were sacrificed 6 hr after the third PCA injection, and brains were extracted to prepare protein or mRNA samples. Homozygous ATE1-null mice have been previously generated by replacing exons 1 through 3 of the ATE1 gene with the NLS-lacZ marker in CJ7 embryonic stem cells34. ATE1-/- embryos were produced through heterozygous crosses in the mixed 129SvImJ/C57BL/6 genetic background. Animal studies were conducted according to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication no. 85-23, revised in 1996) and the protocols approved by the Institutional Animal Care and Use Committee at Kyung Hee University.
Cell cultures and transient expression
Mammalian cells used in this study were grown in DMEM supplemented with 10% FBS, 2 mM glutamine, and 100 units/ml penicillin/streptomycin. Stable HEK293 cell lines expressing X-GFP were generated largely as previously reported54. Cells were transfected with 2–4 μg of total plasmid DNA in a 6-well culture plate for 4 hr using LipofectAMINE 2000 (Invitrogen) when cells were >95% confluent or at a density of 106 cells/well. Cell lysates were prepared 36–48 hr post-transfection in RIPA buffer and used for immunoblotting. Ub-TDP43-EGFP and Ub-Arg208-TDP25-EGFP were cloned in the EGFP-N1 vector. After 36 hr post-transfection, HeLa cells were treated with 100 μM of PCA or 10 μM of MG132 for 4 hr and fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 10 min fluorescence studies. All of the fluorescence micrographic images are representatives of the total cell population.
Quantitative RT-PCR and microarray analysis
Total RNA from cultured cells was prepared using the TRIzol reagent (Invitrogen), followed by further purification through RNeasy mini-column (Qiagen) with on-column DNase I treatment. The cDNA samples were prepared by reverse transcription using Accupower® RT-pre mix (Bioneer). Real-time PCR reaction was then performed using StepOne Real-time PCR machine (ABI) with a diluted cDNA, SYBR Green qPCR master mixture (Life technologies, UK), and 10 pmole of gene-specific primers. Thermal cycling conditions were comprised of 95°C for 10 min for enzyme activation, 40 cycles at 95°C for 15 sec, 60°C for 1 min. After 40 cycles, perform the other 95°C for 15 sec, 60°C for 1 min and at last keep at 95°C for 15 sec. Each mRNA level was normalized to that of GAPDH and the values were plotted as means ± SD of three independent experiments. Primer sequences were as follows: for RGS4, forward (5′-GGCTGAATCGTTGGAAAACCT -3′) and reverse (5′-TCCTCGCTGTATTCCGACTTC-3′); for GFP, forward (5′-ACGTAAACGGCCACAAGTTC-3′) and reverse (5′-AAGTCGTGCTGCTTCATGTG-3′); for OTX2, forward (5′-TATCTAAAGCAACCGCCTTACG-3′) and reverse (5′- AAGTCCATACCCGAAGTGGTC-3′); for EGR2, forward (5′-ACTCAGCCTGAACTGGACCA-3′) and reverse (5′-TGGATGGCGGCGATAAGAATG-3′); for RASGRP1, forward (5′-GCCCCAAAGCAAGACTAGAGG-3′) and reverse (5′-CCCAGGGACACCATCATTCG-3′); for CIRBP, forward (5′-GGACTCAGCTTCGACACCAAC-3′) and reverse (5′-ATGGCGTCCTTAGCGTCATC-3′); for NME2, forward (5′-TGGCAGTGATTCAGTGGAGAG-3′) and reverse (5′-CTTGTAGTCGATCAGTTCTTCGG-3′); for HES5, forward (5′-AGTCCCAAGGAGAAAAACCGA-3′) and reverse (5′- GCTGTGTTTCAGGTAGCTGAC-3′); for GAPDH, forward (5′-AGGTCGGTGTGAACGGATTTG-3′) and reverse (5′-GGGGTCGTTGATGGCAACA-3′). Each mRNA level was normalized to that of GAPDH and the values were plotted as means ± SD of three independent experiments. For microarray analysis, 750 ng of labeled cRNA samples were hybridized to each Mouse Ref-8 expression v.2 bead array for 16-18 h at 58°C, according to the manufacturer's instructions (Illumina). Detection of the array signal was carried out using Amersham fluorolink streptavidin-Cy3 (GE Healthcare) following the bead array manual. Arrays were scanned with an Illumina bead array reader confocal scanner according to the manufacturer's instructions. All data analysis and visualization of differentially expressed genes was conducted using R 2.15.1 (www.r-project.org).
Analysis of GPCR pathways
For the serum-starvation/activation assay, wild-type and ATE1-/- MEFs or HeLa cells were starved in serum-free medium for 12 hr and then activated for time as indicated by adding DMEM containing 20% FBS. PCA was pretreated for 4 hr at 100 μM final concentration and the samples were harvested. Immunoblotting was performed to analyze the MAPK pathways as described.
LacZ staining
Whole embryos were fixed in 4% paraformaldehyde/PBS for 10 min, rinsed in PBS three times, and stained overnight at 37°C in X-gal solution (1.3 mg/mL potassium ferrocyanide, 1 mg/mL potassium ferricyanide, 0.3% Triton X-100, 1 mM MgCl2, 150 mM NaCl, and 1 mg/mL 4-choloro-5-bromo-3-indolyl-β-galactoside; X-gal, Roche Applied Science) in PBS (pH 7.4) followed by post-fixation.
Author Contributions
Y.J. carried out most in vitro studies and cell-based assays. W.H.C. performed mice experiments. J.H.L., D.H.H. and J.H.K. contributed to in silico analysis, cytotoxicity assay, and TDP43-related works. Y.-S.C. provided key reagents and intellectual input. S.H.K. and M.J.L. were responsible for the overall design and oversight of the project. Many authors contributed to preparation of the paper.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by grants of the Disease Oriented Translational Research and the Korea-UK Alzheimer's Disease Research funded by the KHIDI (HI14C0202 and A111227 to M.J.L.). This work was also supported by the Basic Science Research Program of NRF of Korea (2013R1A1A2059793 to J.H.L.).
References
- Sriram S. M., Kim B. Y. & Kwon Y. T. The N-end rule pathway: emerging functions and molecular principles of substrate recognition. Nat Rev Mol Cell Biol 12, 735–747 (2011). [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway and regulation by proteolysis. Protein Sci (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A., Finley D. & Varshavsky A. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234, 179–186 (1986). [DOI] [PubMed] [Google Scholar]
- Cerda-Maira F. A. et al. Molecular analysis of the prokaryotic ubiquitin-like protein (Pup) conjugation pathway in Mycobacterium tuberculosis. Mol Microbiol 77, 1123–1135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbard M. A. et al. Ubiquitin-like small archaeal modifier proteins (SAMPs) in Haloferax volcanii. Nature 463, 54–60 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The ubiquitin system, an immense realm. Annu Rev Biochem 81, 167–176 (2012). [DOI] [PubMed] [Google Scholar]
- Hwang C. S., Shemorry A. & Varshavsky A. N-terminal acetylation of cellular proteins creates specific degradation signals. Science 327, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shemorry A., Hwang C. S. & Varshavsky A. Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell 50, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. J. et al. Characterization of arginylation branch of N-end rule pathway in G-protein-mediated proliferation and signaling of cardiomyocytes. J Biol Chem 287, 24043–24052 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. Y. et al. UBR2 mediates transcriptional silencing during spermatogenesis via histone ubiquitination. Proc Natl Acad Sci U S A 107, 1912–1917 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J. Y. et al. UBR2 of the N-end rule pathway is required for chromosome stability via histone ubiquitylation in spermatocytes and somatic cells. PLoS One 7, e37414 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. J. et al. RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc Natl Acad Sci U S A 102, 15030–15035 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R. G., Wang H., Xia Z. & Varshavsky A. The N-end rule pathway is a sensor of heme. Proc Natl Acad Sci U S A 105, 76–81 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzel M. et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol 5, 467–473 (2003). [DOI] [PubMed] [Google Scholar]
- Brower C. S., Piatkov K. I. & Varshavsky A. Neurodegeneration-associated protein fragments as short-lived substrates of the N-end rule pathway. Mol Cell 50, 161–171 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. K. et al. The N-terminal methionine of cellular proteins as a degradation signal. Cell 156, 158–169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T. & Kwon Y. T. The mammalian N-end rule pathway: new insights into its components and physiological roles. Trends Biochem Sci 32, 520–528 (2007). [DOI] [PubMed] [Google Scholar]
- Choi W. S. et al. Structural basis for the recognition of N-end rule substrates by the UBR box of ubiquitin ligases. Nat Struct Mol Biol 17, 1175–1181 (2010). [DOI] [PubMed] [Google Scholar]
- Matta-Camacho E., Kozlov G., Li F. F. & Gehring K. Structural basis of substrate recognition and specificity in the N-end rule pathway. Nat Struct Mol Biol 17, 1182–1187 (2010). [DOI] [PubMed] [Google Scholar]
- Erbse A. et al. ClpS is an essential component of the N-end rule pathway in Escherichia coli. Nature 439, 753–756 (2006). [DOI] [PubMed] [Google Scholar]
- Roman-Hernandez G., Grant R. A., Sauer R. T. & Baker T. A. Molecular basis of substrate selection by the N-end rule adaptor protein ClpS. Proc Natl Acad Sci U S A 106, 8888–8893 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule: functions, mysteries, uses. Proc Natl Acad Sci U S A 93, 12142–12149 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M. J. et al. Synthetic heterovalent inhibitors targeting recognition E3 components of the N-end rule pathway. Proc Natl Acad Sci U S A 105, 100–105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sriram S. et al. Development and characterization of monomeric N-end rule inhibitors through in vitro model substrates. J Med Chem 56, 2540–2546 (2013). [DOI] [PubMed] [Google Scholar]
- Jiang Y. X. L. et al. Characterization of mammalian N-degrons and development of heterovalent inhibitors of the N-end rule pathway. Chem Sci 4, 3339–3346 (2013). [Google Scholar]
- Lassen J. B. The effect of p-chloroamphetamine on motility in rats after inhibition of monoamine synthesis, storage, uptake and receptor interaction. Psychopharmacologia 34, 243–254 (1974). [DOI] [PubMed] [Google Scholar]
- Harvey J. A., McMaster S. E. & Yunger L. M. P-Chloramphetamine: Selective neurotoxic action in brain. Science 187, 841–843 (1975). [DOI] [PubMed] [Google Scholar]
- Zhang Y. J. et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci U S A 106, 7607–7612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igaz L. M. et al. Expression of TDP-43 C-terminal Fragments in Vitro Recapitulates Pathological Features of TDP-43 Proteinopathies. J Biol Chem 284, 8516–8524 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonda D. K. et al. Universality and structure of the N-end rule. J Biol Chem 264, 16700–16712 (1989). [PubMed] [Google Scholar]
- Tasaki T. et al. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol 25, 7120–7136 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R. G. et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 (2005). [DOI] [PubMed] [Google Scholar]
- Lipinski C. A., Lombardo F., Dominy B. W. & Feeney P. J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 46, 3–26 (2001). [DOI] [PubMed] [Google Scholar]
- Kwon Y. T. et al. An essential role of N-terminal arginylation in cardiovascular development. Science 297, 96–99 (2002). [DOI] [PubMed] [Google Scholar]
- An J. Y. et al. Impaired neurogenesis and cardiovascular development in mice lacking the E3 ubiquitin ligases UBR1 and UBR2 of the N-end rule pathway. Proc Natl Acad Sci U S A 103, 6212–6217 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollinger S. & Hepler J. R. Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol Rev 54, 527–559 (2002). [DOI] [PubMed] [Google Scholar]
- Ross E. M. & Wilkie T. M. GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu Rev Biochem 69, 795–827 (2000). [DOI] [PubMed] [Google Scholar]
- Brower C. S. & Varshavsky A. Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougan D. A., Micevski D. & Truscott K. N. The N-end rule pathway: from recognition by N-recognins, to destruction by AAA + proteases. Biochim Biophys Acta 1823, 83–91 (2012). [DOI] [PubMed] [Google Scholar]
- Erdely H. A. et al. Regional expression of RGS4 mRNA in human brain. Eur J Neurosci 19, 3125–3128 (2004). [DOI] [PubMed] [Google Scholar]
- Krueger C. E. et al. Conflict monitoring in early frontotemporal dementia. Neurology 73, 349–355 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimidis I. et al. Frontal lobe dysfunction in amyotrophic lateral sclerosis. J Neurol Sci 195, 25–33 (2002). [DOI] [PubMed] [Google Scholar]
- Sriram S. et al. Development and Characterization of Monomeric N-End Rule Inhibitors through In Vitro Model Substrates. J Med Chem 56, 2540–2546 (2013). [DOI] [PubMed] [Google Scholar]
- Wang K. H., Roman-Hernandez G., Grant R. A., Sauer R. T. & Baker T. A. The molecular basis of N-end rule recognition. Mol Cell 32, 406–414 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev A. F., Callard A. & Goldberg A. L. Importance of the different proteolytic sites of the proteasome and the efficacy of inhibitors varies with the protein substrate. J Biol Chem 281, 8582–8590 (2006). [DOI] [PubMed] [Google Scholar]
- Besche H. C., Haas W., Gygi S. P. & Goldberg A. L. Isolation of mammalian 26S proteasomes and p97/VCP complexes using the ubiquitin-like domain from HHR23B reveals novel proteasome-associated proteins. Biochemistry 48, 2538–2549 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers J. H. et al. RGS4 causes increased mortality and reduced cardiac hypertrophy in response to pressure overload. J Clin Invest 104, 567–576 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodenstein J., Sunahara R. K. & Neubig R. R. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol Pharmacol 71, 1040–1050 (2007). [DOI] [PubMed] [Google Scholar]
- Stewart J. J. Optimization of parameters for semiempirical methods V: modification of NDDO approximations and application to 70 elements. J Mol Model 13, 1173–1213 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanner M. F. A component-based software environment for visualizing large macromolecular assemblies. Structure 13, 447–462 (2005). [DOI] [PubMed] [Google Scholar]
- Morris G. M. et al. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem 30, 2785–2791 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker N. A., Sept D., Joseph S., Holst M. J. & McCammon J. A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods 65, 55–63 (1983). [DOI] [PubMed] [Google Scholar]
- Dantuma N. P., Lindsten K., Glas R., Jellne M. & Masucci M. G. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat Biotechnol 18, 538–543 (2000). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information