Abstract
Human liver gene regulatory (Bayesian) network analysis was previously used to identify a cytochrome P450 (P450) gene subnetwork with Aldo-keto reductase 1D1 (AKR1D1) as a key regulatory driver of this subnetwork. This study assessed the biologic importance of AKR1D1 [a key enzyme in the synthesis of bile acids, ligand activators of farnesoid X receptor (FXR), pregnane X receptor (PXR), and constitutive androstane receptor (CAR), known transcriptional regulators of P450s] to hepatic P450 expression. Overexpression of AKR1D1 in primary human hepatocytes led to increased expression of CYP3A4, CYP2C8, CYP2C9, CYP2C19, and CYP2B6. Conversely, AKR1D1 knockdown decreased expression of these P450s. We resequenced AKR1D1 from 98 donor livers and identified a 3′-untranslated region (UTR) (rs1872930) single nucleotide polymorphism (SNP) significantly associated with higher AKR1D1 mRNA expression. AKR1D1 3′-UTR-luciferase reporter studies showed that the variant allele resulted in higher luciferase activity, suggesting that the SNP increases AKR1D1 mRNA stability and/or translation efficiency. Consistent with AKR1D1’s putative role as a driver of the P450 subnetwork, the AKR1D1 3′-UTR SNP was significantly associated with increased hepatic mRNA expression of multiple P450s (CYP3A4, CYP2C8, CYP2C9, CYP2C19, and CYP2B6) and CYP3A4, CYP2C8, CYP2C19, and CYP2B6 activities. After adjusting for multiple testing, the association remained significant for AKR1D1, CYP2C9, and CYP2C8 mRNA expression and CYP2C8 activity. These results provide new insights into the variation in expression and activity of P450s that can account for interindividual differences in drug metabolism/efficacy and adverse drug events. In conclusion, we provide the first experimental evidence supporting a role for AKR1D1 as a key genetic regulator of the P450 network.
Introduction
Pharmacogenetic efforts to date have largely been focused on identifying cis-coding and cis-regulatory polymorphisms affecting candidate gene expression and activity. This approach has been successful in identifying functional polymorphic alleles for CYP2C9, CYP2C19, CYP2B6, CYP2C8, and CYP3A4 (http://www.cypalleles.ki.se/). However, for many of these cytochrome P450 (P450) enzymes, a considerable percentage of variation in P450 activity cannot be explained by cis-genetic polymorphisms, and it is hypothesized that genetic variation in trans-regulators influences P450 variation (Schuetz, 2004). Indeed, we previously showed that polymorphisms in hepatic transcription factors that regulate P450s such as FoxA2, HNF4β, FoxA3, and PXR, together explained as much as 10% of the variation in hepatic CYP3A4 expression (Lamba et al., 2010).
One additional approach to identify P450 trans-regulators is to derive the P450 biologic coexpression networks using both a weighted gene coexpression network approach and a Bayesian network reconstruction approach. We previously (Yang et al., 2010) applied these integrative genomic approaches using mRNA expression from 466 human livers, to construct a probabilistic, causal P450 gene network. AKR1D1 (Δ4-3-oxosteroid, 5β-reductase) was predicted as one master regulator of the P450 subnetwork or module (Fig. 1). Expression of multiple P450s including CYP2C9, CYP3A4, and CYP2C19, was predicted to be influenced by Aldo-keto reductase 1D1 (AKR1D1).
Fig. 1.
Model for regulation of expression/activity of multiple P450s by AKR1D1. Cholesterol is converted to CA and CDCA via a classic pathway (starting with CYP7A1) and an alternative pathway (starting with CYP27A1). Both bile acid synthesis pathways include AKR1D1-mediated catalysis of the 5β-reduction of bile acid intermediates. These 5β-reduced intermediates are finally converted to the primary bile acids CDCA and CA, which can serve as endogenous ligands of nuclear receptors PXR and CAR that upon activation can regulate the transcription of multiple P450s including CYP3A, CYP2C9, and CYP2C19. CA, cholic acid.
AKR1D1 is a biologically plausible candidate gene regulating P450 expression because it participates in regulating bile acid concentrations in liver (Fig. 1). AKR1D1 catalyzes the NADPH-dependent reduction of the double bond in the steroid A-ring in bile acids and steroid hormones, and is a central enzyme in the synthesis of both chenodeoxycholic acid and cholic acid (Lee et al., 2009). Bile acids are now recognized not just as fat solubilizers, but as signaling hormones with endocrine effects that affect regulation of P450s as well as lipid and glucose metabolism (Houten and Auwerx, 2004; Thomas et al., 2008). Indeed, bile acids are known ligands/activators of the nuclear hormone receptors pregnane X receptor (PXR) and constitutive androstane receptor (CAR) (Schuetz et al., 2001; Congiu et al., 2009), which are transcriptional regulators of CYP3A4, CYP2C9, and CYP2C19. Bile acids also activate and regulate the nuclear hormone farnesoid X receptor (FXR), a known transcriptional regulator of bile acid homeostasis (Urquhart et al., 2007; Modica et al., 2009) and PXR (Jung et al., 2006), and CYP3A4 (Gnerre et al., 2004). AKR1D1 could also regulate P450s through reduction of steroid hormones. For example, AKR1D1-mediated 5β-reduction of progesterone to 5β-pregnane-3, 20-dione results in activation of PXR (Bertilsson et al., 1998).
This study was undertaken to validate whether AKR1D1 was indeed a regulator of hepatic P450 expression. In addition, we tested whether genetic variation in AKR1D1 was associated with P450 mRNA expression and activity. To date, genetic variation in AKR1D1 has not been explored in normal individuals, and rare genetic variants have been reported only in infants that lack cholic acid and chenodeoxycholic acid (Lemonde et al., 2003). In this study, we report that genetic variation in AKR1D1 contributes to the observed variation in P450 expression and activity of the P450 liver network.
Materials and Methods
Procurement of Human Liver Tissue.
The St. Jude Children’s Research Hospital and University of Pittsburgh Institutional Review Boards approved the human liver studies. Liver tissue from white donors was processed through St. Jude Liver Resource at St. Jude Children’s Research Hospital and was provided by the Liver Tissue Procurement and Distribution System (National Institutes of Health National Institute of Diabetes and Digestive and Kidney Diseases Contract N01-DK92310) and by the Cooperative Human Tissue Network. Liver tissue from 163 white donor livers (from the University of Pittsburgh and St. Jude Liver Resource) was previously used, together with livers from other resources, to isolate DNA for genotyping and RNA and microsomes for phenotyping mRNA expression and P450 activities, respectively (Schadt et al., 2008; Yang et al., 2010). A subset of 145 white donor livers from the University of Pittsburgh and St. Jude Liver Resource had sufficient tissue available for follow-up analysis in this study.
Plasmids.
The PXR expression plasmid (pSG5-hPXR) was generously provided by Dr. Steve Kliewer (Lehmann et al., 1998). The reporter plasmid [CYP3A4 +53 to 362(7836/7208)-Luciferase], hereafter called CYP3A4-PXRE2-LUC, was generously provided by Dr. Richard Kim (Zhang et al., 2001). The AKR1D1 3′-untranslated region (UTR)-luciferase reporter construct was purchased from Switchgear Genomics (Menlo Park, CA) and single mutants of rs1872929 and rs1872930 as well as different haplotypes [wild-type haplotype (WT-HAP) and variant haplotype (VAR-HAP)] were constructed by site directed mutagenesis. All nucleotide substitutions were confirmed by direct DNA sequencing.
Generation of Vectors, Adenoviruses, and Lentiviruses.
AKR1D1 was polymerase chain reaction (PCR) amplified from the human AKR1D1 expression plasmid (catalog no. SC116410; OriGene, Rockville, MD) and subcloned into pENTR/SD/D-TOPO (part no. 45-0219; Invitrogen, Grand Island, NY) for entry into the Adv-BGFP-Gateway destination vector (from Dr. John Gray, St. Jude) to make the Adv-BGFP-AKR1D1 overexpression vector. The pGIPz-AKR1D1-200099 lentiviral microRNA-adapted short hairpin RNA (shRNAmir) vector (catalog no. RHS-4430-98709631) to knockdown human AKR1D1 and pGIPz nonsilencing lentiviral shRNA control vector (catalog no. RHS4346) were purchased from Thermo Scientific Open Biosystems (Lafayette, CO). These vectors were used to make adenoviral or lentiviral particles by the Vector Development & Production shared resource at St. Jude Children’s Research Hospital.
RNA Profiling and Single Nucleotide Polymorphism Genotyping.
The microarray design, RNA sample preparation, amplification, hybridization, and expression analysis as well as single nucleotide polymorphism (SNP) genotyping were previously described in detail (Schadt et al., 2008; Yang et al., 2010). The AKR1D1 SNP rs1872930 was also genotyped with a Snapshot genotyping kit from Applied Biosystems (Grand Island, NY). The Snapshot primers for first-round PCR were as follows: forward primer, 5′-GCGCGATCATCCTGAATACCCATT-3′; and reverse primer, 5′-GACTGAGTCAAAGTCATTTACACTGCAC-3′. The extension primers used were as follows: forward, 5′-TCCTGAACAGATTTTTCACTCCC-3′; and reverse, 5′-CATTGCACCGTCTTGGCACTC-3′.
P450 Enzyme Activity Measurements in Human Liver Microsomes.
The P450 activities of CYP2B6 (bupropion to 1′-hydroxybupropion), CYP2C8 (paclitaxel to 6α-hydroxypaclitaxel), CYP2C9 (tolbutamide to hydroxytolbutamide), CYP2C19 ((S)-mephenytoin to 4′-hydroxymephenytoin), and CYP3A4 (midazolam to 1′-hydroxymidazolam and testosterone to 6β-hydroxytestosterone) were previously determined in microsomes isolated from human liver samples using probe substrate metabolism assays (Yang et al., 2010).
CYP3A4-PXRE2-LUC Reporter Assays and AKR1D1 3′-UTR-Luciferase Reporter Assays in HepG2 Cells.
HepG2 cells were cultured in minimum Eagle’s medium-α supplemented with 10% fetal bovine serum, 1% penicillin, and 1% streptomycin, and maintained in a humidified incubator at 37°C in an atmosphere of 5% CO2. For transfection studies, 3 × 105 cells per well were seeded into 24-well culture dishes. Twenty-four hours later, cells were transfected overnight by calcium phosphate precipitation with 300 ng PXRE2-CYP3A4-LUC reporter plasmid, 100 ng pSG5-hPXR, and 50 ng SV40-β-galactosidase along with either 100 ng pCMV-AKR1D1 expression plasmid or pCMV-XL5 empty vector plasmid as a control. The next day, cells were washed once with medium and incubated with fresh medium containing 10% charcoal dextran-treated fetal bovine serum (Hyclone Laboratories, Logan, UT) with either 50 µM chenodeoxycholic acid (CDCA) (Sigma-Aldrich, St. Louis, MO) or dimethylsulfoxide. For AKR1D1 3′-UTR-Luciferase reporter assays, HepG2 cells were transfected with 250 ng of different AKR1D1 3′-UTR- Luc reporter plasmid constructs along with 50 ng SV40-β-galactosidase. Twenty-four hours later, cells were harvested, lysed, and centrifuged at 1500g for 4 minutes, and luciferase activities were determined in an aliquot of supernatant according to the manufacturer’s instructions (Luciferase Assay System; Promega, Madison, WI) using Clarity luminescence microplate reader (BIO-TEK, Winooski, VT). β-galactosidase activities were determined in an aliquot of supernatant according to the manufacturer’s instructions (β-Galactosidase Enzyme Assay System; Promega). Luciferase activity was normalized to β-galactosidase activity to correct for transfection efficiency and expressed as fold change with respect to vector control. Data are reported as mean ± S.D. of three determinations and were representative of multiple experiments. Statistical difference between groups was determined by use of the t test. All experiments were performed in triplicate.
Adv-AKR1D1 and Lentiviral shRNAmir AKR1D1 Transduction of Primary Human Hepatocytes.
Freshly plated primary human hepatocytes isolated from four different male donor livers were purchased from BD Biosciences (catalog no. 454424; Woburn, MA). 4 × 105 cells per well in 24-well culture dishes were obtained from BD Biosciences and cultured in hepatocyte culture media supplemented with G418, amphotericin B, and l-glutamine at a final concentration of 50 mg/l, 750 µg/l, and 292 mg/l in the media, respectively. Hepatocytes were maintained in a humidified incubator at 37°C in an atmosphere of 5% CO2. Transfections were done after acclimatizing the cells for 24 hours. Cells were transfected with 20 m.o.i. (multiplicity of infection) of Adv-BGFP-AKR1D1 or control virus for overexpression of AKR1D1. Similarly, for AKR1D1 knockdown experiments, hepatocytes were transfected with 5 m.o.i. of either control (pGIPz-Lentiviral-RHS4346) or AKR1D1-specific shRNAmir lentiviruses (pGIPz-AKR1D1-200099). For bile acid treatment, cells were washed once with medium and incubated with fresh medium containing either 50 µM CDCA or 0.1% dimethylsulfoxide (vehicle). The CDCA-treated cells were harvested 24 hours after treatment, adenoviral infected cells were harvested 48 hours after infection, and lentiviral infected cells were harvested 72 hours after infection. Before harvesting, hepatocytes were washed once with phosphate-buffered saline (PBS) and were harvested by adding TRIzol reagent (Invitrogen) directly into the wells and passing a couple of times through the pipette tip.
Western Blotting for AKR1D1 in HepG2 Total Cell Lysate.
HepG2 cells were cultured as described above. Cells were infected with AKR1D1 lentiviral or adenoviral particles as described for human hepatocytes. After treatment cells were washed once with ice-cold PBS, harvested in 5 ml PBS, and centrifuged at 1500g for 10 minutes. The cell pellets were reconstituted in 50 μl microsome storage buffer (100 mM potassium phosphate, pH 7.4, 1.0 mM EDTA 20% glycerol with protease inhibitor cocktail) and sonicated to prepare total cell lysate. Cell lysates (100 µg protein) were subjected to SDS-PAGE and immunoblotted with anti-AKR1D1 (SC-67710; Santa Cruz Biotechnology, Dallas, TX) or anti-β-actin antibody, followed by appropriate secondary antibodies, and developed with SuperSignal West Dura Extended Duration Substrate (catalog no. 34076; Thermo Scientific, Waltham, MA).
mRNA Quantification by Quantitative Real-Time PCR.
Total RNA was isolated using TRIzol reagent. First-strand cDNA was prepared using oligo (dT) primers (catalog no. 11146-016, ThermoScript RT-PCR system; Invitrogen). Real-time PCR quantification of AKR1D1, PXR, CAR, CYP3A4, CYP2C8, CYP2C9, CYP2C19, CYP2B6, CYP7A1, small heterodimer partner (SHP), FXR, and GAPDH mRNAs was carried out using the SYBR GreenER quantitative PCR supermix (catalog no. 11760-100; Invitrogen) according to the manufacturer’s instructions. cDNA was analyzed in duplicate by quantitative real-time PCR on an ABI PRISM 7900HT Sequence Detection System (PE Applied Biosystems, Foster City, CA). Primers used for real-time quantification are provided in Table 1. Specificity of amplification was confirmed in each case by performing melt curve analysis and agarose gel electrophoresis. The averaged Ct values were analyzed by the comparative Ct method to obtain relative mRNA expression levels.
TABLE 1.
Primers used for real-time quantitative PCR
| S. No. | mRNA | Sequence (5′ → 3′) |
|---|---|---|
| 1 | GAPDH-FP | GGACCACCAGCCCCAGCAAGAG |
| 2 | GAPDH-RP | GAGGAGGGGAGATTCAGTGTGGTG |
| 3 | AKR1D1-FP | TCAGAACCTAAATCGACCCCT |
| 4 | AKR1D1-RP | CCCTGTGTCAATAGCAACCTTC |
| 5 | CYP3A4-FP | CCTTACACATACACACCCTTTGGAAGT |
| 6 | CYP3A4-RP | AGCTCAATGCATGTACAGAATCCCCGGTTA |
| 7 | CYP2C8-FP | GCAGGAAAAGGACAACCAAA |
| 8 | CYP2C8-RP | GTGTAAGGCATGTGGCTCCT |
| 9 | CYP2C9-FP | GATCTGCAATAATTTTTCTC |
| 10 | CYP2C9-RP | TCTCAGGGTTGTGCTTGTC |
| 11 | CYP2C19-FP | GAGGAGTTTTCTGGAAGAGGCC |
| 12 | CYP2C19-RP | CATTGCTGAAAACGATTCCAAA |
| 13 | CYP2B6-FP | CACCCTAACACCCATGACCG |
| 14 | CYP2B6-RP | GATCACACCATATCCCCGGA |
| 15 | FXR-FP | TATCTAGCCCAATATTTACAGTT |
| 16 | FXR-RP | GTATAACTTTCCTTTATTTCAC |
| 17 | CYP7A1-FP | AAACTGGATGGAAATAGCCATTTG |
| 18 | CYP7A1-RP | TTTCATCTGCAAGTCATTTAGCG |
| 19 | SHP-FP | GCTGTCTGGAGTCCTTCTGG |
| 20 | SHP-RP | CCAATGATAGGGCGAAAGAA |
| 21 | CAR-FP | CCAGCTCATCTGTTCATCCA |
| 22 | CAR-RP | GGTAACTCCAGGTCGGTCAG |
| 23 | PXR-FP | CAAGCGGAAGAAAAGTGAAC |
| 24 | PXR-RP | CACAGATCTTTCCGGACCTG |
AKR1D1 cDNA Amplification for Sequencing and To Screen for Alternative AKR1D1 mRNAs Associated with Polymorphisms.
RNA was extracted from 98 donor livers with tissue available using TRIzol and was used to prepare cDNA. In the first PCR, a forward primer (AKR-FP) (Table 2) that annealed in the 5′-UTR and a reverse primer (AKR-RP) (Table 2) that annealed in 3′-UTR were used to amplify all 9 exons of AKR1D1 from 12 samples heterozygous for rs1872930. To detect any low abundance alternatively spliced transcripts that might undergo nonsense-mediated decay, the first-round PCR product was used as template for PCR with a second primer pair (AKR-FPN and AKR-RPN) (Table 2). This PCR amplified a 989 bp fragment that still included all of the 9 exons of AKR1D1. Because amplification of a long PCR product might fail to detect alternatively spliced products with small insertions or deletions, and because rs1872930 was in linkage disequilibrium (LD) with two intron 8 SNPs, DNA from subjects with rs1872930 TT, TC, and CC genotypes was used as a template to specifically amplify a region around exon 8/intron8 using primers AKR-FP1 and AKR-RP (Table 2). Again to detect any low abundance alternatively spliced transcripts that might undergo nonsense-mediated decay, the first round PCR product was used as template for PCR with a second primer pair (AKR-FP1 and AKR-RPN) to amplify a 416 bp normal or any low abundance transcripts that might have been associated with the rs1872930 allele. All of the amplified PCR products were analyzed by agarose gel electrophoresis to look for differential banding patterns among genotypes caused by polymorphic splicing events. The first PCR products from 98 subjects (54 TT wild type, 42 TC heterozygous, and 2 CC variant homozygous) were sequenced directly to look for any coding region SNPs.
TABLE 2.
Primers used to test for alternative AKR1D1 mRNA transcripts
| Primer | Sequence (5′ → 3′) | Location in mRNA | Length |
|---|---|---|---|
| bp | |||
| AKR-FP | CCCTAGGACACCTTTCTA | 5′ UTR | 1353 |
| AKR-RP | TCATATGTTGCCTTTGATG | 3′ UTR | |
| AKR-FPN | ATGGATCTCAGTGCTGCAAGT | Start of Exon 1 | 989 |
| AKR-RPN | CCCTGCAGTCAGTATTCATCA | Exon 9/3′ UTR | |
| AKR-FP1 | ACCAGGTTGAGTGCCAT | Exon 5/Exon 6 | 711 |
| AKR-RP | TCATATGTTGCCTTTGATG | 3′ UTR | |
| AKR-FP1 | AACCAGGTTGAGTGCCAT | Exon 5/Exon 6 | 416 |
| AKR-RPN | CCCTGCAGTCAGTATTCATCA | Exon 9/3′ UTR |
RNA Secondary Structure Analysis.
The RNA secondary structure of AKR1D1 wild-type and variant haplotypes was determined using the Mfold web server (Zuker, 2003). For this analysis, a 370 nucleotide region surrounding the SNPs (from 170 nucleotides upstream of rs1872929 to 170 nucleotides downstream of rs1872930) was used to create wild-type and variant haplotypes and these sequences were used to predict the effect on secondary structure of RNA.
microRNA (miRNA) Recognition Sites Using miRanda.
To define the potential for microRNAs to be differentially bound to AKR1D1 wild-type or AKR1D1 variant (rs1872930), miRanda software (version 3.0; http://www.microrna.org/microrna/getDownloads.do) was used with the default settings and with a decreased energy reporting threshold (-en -10). Human microRNA sequences were obtained from the miRBase database (version 18; http://www.mirbase.org/index.shtml). The full-length mRNA coding sequence of AKR1D1 with and without the rs1872930 variant was used as the reference sequence. The best energy and heuristic score for each pairing was compared between wild-type and rs1872930 variant AKR1D1 sequences to find the maximum difference in either of these parameters.
Statistical Analysis.
The chi-squared test for deviation from Hardy–Weinberg equilibrium was used to calculate the observed versus expected distribution of genotypes. Because the phenotypic markers were not normally distributed, group differences were analyzed nonparametrically using the Wilcoxon rank sum test, which is more robust against outliers, to compare binary groups (e.g., TT versus TC+CC). The Kruskal–Wallis test was used to compare three groups of genotype for each polymorphism (e.g., TT versus TC versus CC) with the phenotype. To account for multiple testing, we adjusted the statistical significance threshold using the permutation method in consideration of the correlation among phenotypes. For the association between genotype and P450 activity levels or mRNA expression, we considered P < 0.0034 as statistically significant after adjusting for multiple testing.
Results
The Hepatic Expression of Multiple P450 mRNAs Is Significantly Correlated.
We first examined P450 expression (previously profiled by Agilent custom expression arrays) in a cohort of 163 human livers from white donors (Schadt et al., 2008) and found a significant degree of correlation between expression of mRNAs for CYP3A4 and CYP2C9 (r = 0.76, P < 2.2e−16), CYP3A4 and CYP2C19 (r = 0.76, P < 2.2e−16), and CYP3A4 and CYP2B6 (r = 0.76, P < 2.2e−16) (Fig. 2). A second cohort of 206 human liver tissues that had been separately profiled with a different Agilent custom expression array (Innocenti et al., 2011) was used to confirm correlation between P450s mRNA expression, and the results from the second analysis demonstrated virtually identical results. These P450s are located on different chromosomes (CYP3A4, CYP2C19, and CYP2B6 on chromosomes 7, 10, and 19, respectively). These results are consistent with the membership of these P450s in the P450 coexpression module (Yang et al., 2010).
Fig. 2.
Significant human liver P450-P450 mRNA correlation indicates that these P450s share trans-regulation. Correlation between CYP3A4 versus CYP2C9 mRNA (A), CYP3A4 versus CYP2C19 mRNA (B), and CYP3A4 versus CYP2B6 mRNA (C). P450 mRNA expression data from 163 human liver samples were taken from an earlier study (Schadt et al., 2008; Yang et al., 2010) and were utilized to study this correlation. The microarray design, RNA sample preparation, amplification, hybridization, and expression analysis were previously described in detail (Schadt et al., 2008; Yang et al., 2010). The Pearson's product-moment correlation r values are indicated.
AKR1D1 Overexpression in HepG2 Cells Induces CYP3A4 Transcription.
In our initial experiment, we used a mechanistic gain-of-expression study for experimental validation that AKR1D1 is a regulator of CYP3A transcription. Human hepatoblastoma HepG2 cells were transiently cotransfected with a CYP3A4-luc reporter plasmid (containing multiple PXR binding elements) and a PXR expression plasmid, with and without an AKR1D1 expression plasmid. As expected, CDCA treatment induced CYP3A4 transcription (Fig. 3A). In the presence of transfected PXR, cotransfected AKR1D1 significantly stimulated CYP3A4 expression (P = 0.002) in HepG2 cells to the same extent as the addition of 50 µM CDCA alone (P = 0.05). We speculate that transfected AKR1D1 increases endogenous bile acids that ligand activate PXR to induce CYP3A4 transcription.
Fig. 3.
AKR1D1 overexpression increases CYP3A4 promoter activity, and modulating AKR1D1 expression in HepG2 cells. HepG2 cells were transfected with PXRE2-CYP3A4-LUC and pSG5-hPXR along with either the pCMV-AKR1D1 expression plasmid or pCMV-XL5 empty vector control plasmid. Cotransfected pSV40-LacZ served as a transfection efficiency control. The next day, empty vector transfected cells were treated with either 50 µM CDCA or dimethylsulfoxide. Twenty-four hours later, luciferase and β-galactosidase activities were determined. Results are presented as fold change with respect to vector control (A). HepG2 cells cultured for 24 hours were infected with AKR1D1 adenovirus (B) or with AKR1D1 shRNA lentiviral particles (C). Cell lysates were prepared and 100 µg protein was subjected to SDS-PAGE and immunoblotted with anti-AKR1D1 or anti-β-actin antibody.
Successful Lentiviral shRNA Mediated AKR1D1 Knockdown and Adenoviral Mediated Overexpression of AKR1D1 in Hepatic Cells.
Since transient transfection is inefficient in introducing plasmid DNA into 100% of cells, we used adenovirus-mediated overexpression and lentiviral-mediated shRNA knockdown approaches to demonstrate that we could experimentally modulate AKR1D1 expression in human hepatic cells. HepG2 cells transduced for 48 hours with an AKR1D1 adenovirus showed an approximately 443% increase in AKR1D1 immunoreactive protein (Fig. 3B). Conversely, HepG2 cells transduced for 72 hours with the AKR1D1 shRNA lentivirus exhibited approximately 90% cell transduction, and an approximately 64% decrease in AKR1D1 protein compared with cells transduced with the scrambled shRNA (Fig. 3C).
AKR1D1 Overexpression Induces P450 mRNA Expression in Primary Human Hepatocytes.
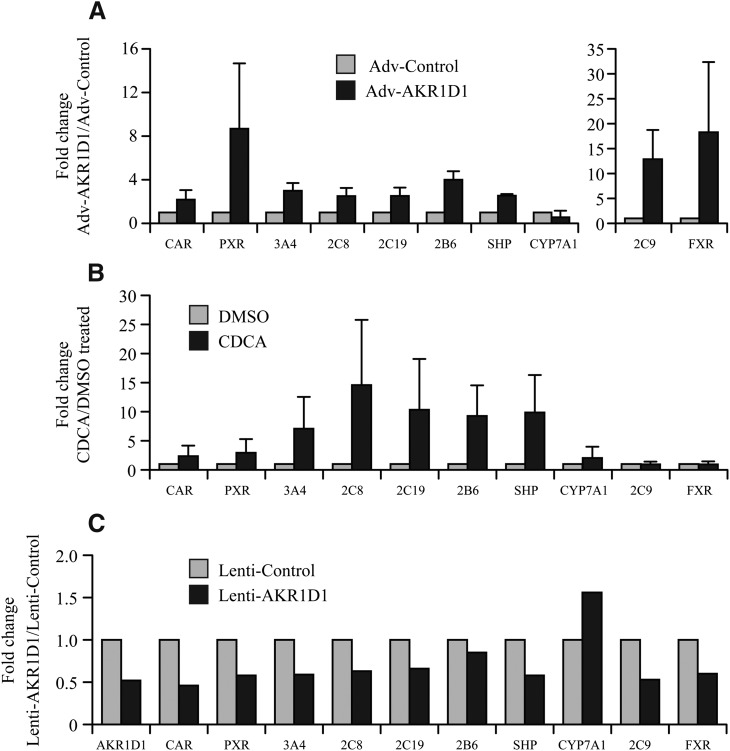
To test whether AKR1D1 is a key driver of the P450 network, presumably through elevation in bile acid levels, we used adenovirus-mediated overexpression of AKR1D1 in primary human hepatocytes and quantified mRNA expression of bile acid regulated target genes. Adenoviral-mediated AKR1D1 overexpression increased the mRNA expression of a variety of bile acid regulated genes including the nuclear receptors CAR and PXR and multiple P450s (CYP2C8, CYP2C9, CYP2C19, CYP2B6, and CYP3A4) (Fig. 4A). AKR1D1 overexpression, and presumably increased bile acids, engaged the FXR feedback loop to maintain bile acid homeostasis. Specifically, AKR1D1 overexpression induced expression of the bile acid sensor farnesoid X receptor (FXR), resulting in feedback inhibition of CYP7A1 (the rate limiting enzyme in bile acid synthesis) through induction of small heterodimer partner (SHP) (Fig. 4A). Treatment of hepatocytes with the primary bile acid CDCA elicited a P450, PXR, and CAR induction profile (Fig. 4B) similar to that observed with AKR1D1 overexpression. Although the fold induction of mRNA changes did not reach statistical significance for either CDCA treatment or AKR1D1 overexpression, this is likely due to the large human variation in fold induction between the different hepatocyte preparations.
Fig. 4.
AKR1D1 overexpression increased and AKR1D1 silencing decreased expression of multiple P450s in primary human hepatocytes. Primary human hepatocytes were transduced with Adv-BGFP-AKR1D1 or control virus, cells were harvested after 48 hours, and mRNA quantification was performed using real-time PCR. Results from four donors are presented as mean ± S.E.M. (A). As a positive control, hepatocytes were treated with either dimethylsulfoxide or 50 µM CDCA. Results from three donors are presented as mean ± S.D. (B). For AKR1D1 knockdown experiments, hepatocytes were transfected with either control or AKR1D1 lentiviruses. Cells were harvested 72 hours after infection for real-time PCR experiments. Results are presented as the average of two livers (C). The expression of genes involved in bile acid homeostasis FXR, SHP, and CYP7A1 was also studied and served as positive controls. The results did not reach statistical significance, likely due to the small number of donor hepatocytes tested and the wide human interindividual variability in the observations.
AKR1D1 Silencing Decreases Multiple P450s mRNA Expression in Primary Human Hepatocytes.
We next tested whether lentivirus shRNA-mediated AKR1D1 knockdown would lead to downregulation of CAR, PXR, and P450 mRNA expression. Primary hepatocytes transduced with the AKR1D1 shRNA lentiviruses (but not the control shRNA scrambled lentivirus) had decreased expression of AKR1D1 (50%) and a similar decrease in expression of CAR, PXR, and multiple P450s (CYP2C8, CYP2C9, CYP2C19, CYP2B6 and CYP3A4) (Fig. 4C). ShRNA-mediated AKR1D1 knockdown led to a decrease in FXR, a decrease in SHP (the CYP7A1 repressor), and a corresponding induction of CYP7A1 (Fig. 4C), although these changes did not reach statistical significance
The AKR1D1 3′-UTR rs1872930 Variant Allele Is Associated with Increased Hepatic AKR1D1 Expression.
Having established that AKR1D1 can act as a trans-regulator of multiple P450s, we hypothesized that functional SNPs in AKR1D1 could affect its expression. To identify AKR1D1 functional polymorphisms, we first surveyed AKR1D1 [Chromosome 7: 137761178 - 137803050 (+)] genotype information from 90 unrelated HapMap (http://gvs.gs.washington.edu/GVS137/index.jsp) white subjects. There were 22 AKR1D1 SNPs reported and the Genome Variation server binned the AKR1D1 SNPs into 12 linkage groups with the tag SNPs for each group (Table 3). Tag SNP genotypes for nine of the groups were already available in our St. Jude human liver cohort (n = 145 livers) (Schadt et al., 2008). The AKR1D1 3′-UTR SNP rs1872930 was most significantly associated with higher hepatic AKR1D1 mRNA expression quantified by both microarray analysis (P = 0.003) (Fig. 5A) and by quantitative PCR (P = 0.005) (Fig. 5B; Table 3). This 3′-UTR SNP (rs1872930) was in LD with three other SNPs [an additional 3′-UTR SNP (rs1872929) and two SNPs in intron 8 (rs2035647 and rs2306847)] in either a WT-HAP or VAR-HAP.
TABLE 3.
Tag SNPs from Caucasian HapMap genotype data and the association of SNP genotypes with liver AKR1D1 mRNA expression
The r2 threshold for tag SNPs analysis was 0.8. A dash indicates that the SNP was not genotyped.
| Group | Minor Allele Frequency | TAG SNPs | Location in Gene | Hepatic AKR1D1 mRNA Association |
|---|---|---|---|---|
| % | P | |||
| 1 | 23 | rs2120846 | Intron 1 | 0.045 |
| rs6977075 | Intron 2 | — | ||
| rs6980334 | Intron 4 | — | ||
| rs7795946 | Intron 4 | — | ||
| 2 | 43 | rs8180809 | Intron 3 | 0.14 |
| 3 | 21 | rs1872929 | 3′ UTR | 0.003 |
| rs1872930 | 3′ UTR | 0.003 | ||
| rs2035647 | Intron 8 | — | ||
| rs2306847 | Intron 8 | — | ||
| 4 | 18 | rs2306846 | Intron 6 | — |
| rs3805362 | Intron 7 | 0.17 | ||
| rs12668157 | Intron 1 | — | ||
| 5 | 7 | rs6943935 | Intron 2 | — |
| rs17169518 | Intron 7 | 0.53 | ||
| rs17169521 | Intron 8 | 0.22 | ||
| 6 | 37 | rs2035648 | Intron 8 | 0.1 |
| 7 | 3 | rs2306845 | Intron 4 | 0.9 |
| 8 | 3 | rs2882947 | Intron 7 | — |
| 9 | 17 | rs3735023 | 3′ UTR | 0.87 |
| 10 | 13 | rs6467735 | Intron 4 | 0.26 |
| 11 | 1 | rs17169523 | 3′ UTR | — |
| 12 | 11 | rs17543126 | Intron 4 | — |
Fig. 5.
AKR1D1 SNP (rs1872930) variant is associated with increased human liver AKR1D1 mRNA expression as well as increased transcript stability and/or translation efficiency. AKR1D1 microarray expression data taken from an earlier study (Schadt et al., 2008) (A) or analyzed in this study by quantitative PCR analysis (B) were significantly higher in livers with the AKR1D1 3′-UTR SNPs (P = 0.003 and P = 0.005, respectively). HepG2 cells were transfected with wild-type or variant AKR1D1 3′-UTR-Luc reporter plasmid constructs (C and D) and luciferase assays were performed 24 hours later. Results are expressed as fold change with respect to vector control. Statistical difference between groups was determined using the t test (D).
AKR1D1 cDNA Resequencing.
The tag SNPs may not represent the functional SNPs but could be in LD with other variants with functional significance. Hence, the AKR1D1 cDNA was resequenced in 196 alleles from 98 human livers for which tissue was available and with the following known AKR1D1 rs1872930 genotypes: 54 TT wild type, 42 TC heterozygous, and 2 CC variant homozygous. AKR1D1 resequencing identified no coding region polymorphism.
AKR1D1 Polymorphisms Did Not Influence Expression of Individual Alternatively Spliced mRNA Transcripts.
Since four SNPs in LD [two in the 3′-UTR (rs1872929 and rs1872930) and two in intron 8 (rs2035647 and rs2306847)] were significantly associated with AKR1D1 mRNA expression, we first determined whether the intron 8 SNPs (or any other intronic SNPs in partial LD) were affecting AKR1D1 mRNA splicing, particularly since there are two alternative AKR1D1 mRNAs in the UCSC Genome Browser (http://genome.ucsc.edu) that skip individual exons [exon 5 (125 bp), or exon 8 (83 bp)]. The full-length AKR1D1 cDNA and UTR regions (1353 bp) were amplified from livers wild type, heterozygous, and homozygous for the 3′-UTR SNPs. No alternative mRNAs were observed in any of the livers. However, this amplification strategy would fail to detect the consequences of SNPs that lead to the following: 1) alternative AKR1D1 transcripts differing from the wild-type mRNA by only a few nucleotides, and 2) alternative transcripts with premature termination codons that undergo accelerated degradation by nonsense-mediated decay. Hence, the 1353 bp AKR1D1 first-round PCR product was used as template for a second round of nested PCR amplification of various portions of AKR1D1 (Table 2) including amplification that would have detected any skipping of exon 8. AKR1D1 alternative mRNAs were detected on second- round PCR amplification, but the pattern of amplified bands was the same for all samples regardless of intron 8 (rs2035647 and rs2306847), or any other AKR1D1 genotypes. Hence, we unequivocally determined by PCR analysis of AKR1D1 mRNA transcripts in human livers that none of the AKR1D1 alternative mRNAs was related to a polymorphism.
The AKR1D1 VAR-HAP Has Increased Transcript Stability and/or Translation Efficiency Compared with the WT-HAP.
We conducted AKR1D1 3′-UTR-luciferase reporter assays (the entire 3′UTR is cloned downstream of a luciferase cDNA) to address whether either of the 3′-UTR SNPs (rs1872929 and rs1872930) was affecting mRNA abundance (by affecting mRNA translational efficiency or mRNA stability). Reporter plasmids containing the WT-HAP, VAR-HAP (rs1872929 and rs1872930), and single 3′-UTR variants were constructed by site directed mutagenesis, and the plasmids transiently transfected into HepG2 cells (Fig. 5, C and D). The AKR1D1 VAR-HAP and the rs1872930 single 3′-UTR variant construct had similar reporter activity that was significantly higher compared with the WT-HAP and the rs1872929 single 3′UTR variant. These AKR1D1 3′-UTR-luciferase reporter assay results (VAR-HAP > WT-HAP) (Fig. 5, C and D) corresponded well with the increased expression of AKR1D1 in livers with the VAR-HAP compared with the WT-HAP (Fig. 5, A and B). These results suggest that the rs1872930 3′-UTR SNP is increasing AKR1D1 mRNA transcript stability and/or translational efficiency.
The AKR1D1 3′-UTR rs1872930 Polymorphism Is Associated with Increased Expression of Many mRNAs in the P450 Subnetwork.
Having established that AKR1D1 may act as a master regulator of mRNA expression of multiple P450s in primary human hepatocytes (Fig. 4), and that the rs1872930 is a functional SNP affecting hepatic AKR1D1 mRNA expression, our next goal was to study the association of this polymorphism with the mRNA expression of other members of the P450 subnetwork (Fig. 6). The AKR1D1 functional SNP (rs1872930) was most significantly associated with expression of the nuclear hormone receptor CAR (NR1I3) and the bile acid uptake carrier Na+-taurocholate cotransporting polypeptide (NTCP) or solute carrier family 10 (sodium/bile acid cotransporter family) member (SLC10A1), but was also significantly associated with expression of multiple P450s involved in bile acid synthesis including CYP27A1, CYP7A1, and CYP8B1 (Fig. 6). The AKR1D1 polymorphism was also significantly associated with mRNA expression of multiple drug-metabolizing P450s including CYP2C19, CYP2C9, CYP2C8, and CYP3A4 (Fig. 6). To account for multiple testing, we adjusted the statistical significance threshold using the permutation method in consideration of the correlation among phenotypes. After adjusting for multiple testing, the association remained statistically significant for AKR1D1, SLC10A1, CYP2C9, CYP2C8, CYP27A1, CYP7A1, CYP8B1, and CAR mRNA expression (Fig. 6). Similarly, the AKR1D1 genotype was significantly associated with P450 enzyme activities measured in liver microsomes from the same livers. Livers with the AKR1D1 variant allele had significantly higher activity of CYP2C8 (P = 0.002) using paclitaxel as substrate, CYP3A4 (P = 0.03) using testosterone as substrate, CYP2B6 (P = 0.037) using bupropion as substrate, and CYP2C19 (P = 0.055) using S-mephenytoin as substrate (Fig. 7). The association for CYP2C8 remained statistically significant even after adjusting for multiple testing, whereas the association for CYP3A4, CYP2C19, and CYP2B6 could not escape the stringent P value threshold (P < 0.0034) of multiple testing.
Fig. 6.
The AKR1D1 3′-UTR (rs1872930) polymorphism is associated with increased liver mRNA expression of downstream targets including PXR, CAR, and multiple P450s, A pruned P450 subnetwork showing the master regulator AKR1D1 (dark gray oval) and downstream target P450s (light gray ovals) and other transcription factors including the nuclear receptors PXR and CAR (boxes) is shown along with the P values of association of the variant allele with increased liver mRNA expression. The mRNA expression data used for calculating associations are from previous microarray studies (Schadt et al., 2008; Yang et al., 2010). mRNA expression was compared between the 83 livers available with the AKR1D1 rs1872930 wild-type TT genotype versus livers with one (n = 57) or two (n = 5) variant alleles (TC+CC genotypes). To account for multiple testing, we adjusted the statistical significance threshold using the permutation method in consideration of the correlation among phenotypes. For the association between genotype and mRNA expression, we consider P < 0.0034 as statistically significant after adjusting for multiple testing. The statistically significant values after multiple testing are marked with an asterisk.
Fig. 7.
The AKR1D1 3′-UTR variant (rs1872930) is associated with increased activity of multiple P450s in human liver microsomes. P450 activities were compared between the 83 livers with the AKR1D1 rs1872930 wild-type TT genotype versus livers with one (n = 57) or two (n = 5) variant alleles (indicated as TC genotype). The activities of P450s were measured using probe substrate metabolism assays for CYP3A4 (testosterone), CYP2C8 (paclitaxel), CYP2C9 (tolbutamide), CYP2C19 (S-mephenytoin), and CYP2B6 (bupropion). P values of the association between AKR1D1 genotypes and P450 activities are shown on the top of each panel. For the association between genotype and P450 activity, we considered P < 0.0034 statistically significant after adjusting for multiple testing. The statistically significant value after multiple testing is marked with an asterisk.
To determine whether association of the AKR1D1 rs1872930 genotype with expression of other members of the P450 subnetwork was specific, and not simply because of the higher degree of correlation in mRNA expression between subnetwork members, we examined the relationship between several P450 polymorphisms with AKR1D1 and P450 mRNA expression. Livers heterozygous for either the CYP2C9*2 or 2C9*3 allele had significantly lower CYP2C9 activity compared with livers with the CYP2C9*1 genotype (P = 0.004), but there was no effect of the same genotypes on mRNA expression of CYP3A4 (P = 0.43) or AKR1D1 (P = 0.75). Similarly, the CYP2C19 diplotypes (2C19*1/*1, *1/*17, *1/*2, *17/*17, *2/*17, or *2/*2) were not associated with mRNA expression of either CYP3A4 (P = 0.3229) or CYP2C9 (P = 0.9716).
Mfold Analysis of AKR1D1 Wild-Type and Variant mRNA Structure.
To test whether the two 3′-UTR SNPs change AKR1D1’s mRNA secondary structure, we performed Mfold analysis of AKR1D1 mRNA. The local sequences of AKR1D1 mRNA (370 nucleotide) surrounding the two 3′UTR SNPs were used to predict mRNA secondary structure. Since RNAs can potentially adopt multiple conformations, Mfold calculated 13 alternative structures for the variant allele and 12 for the wild type. The average minimum free energy was −80.01 for the variant allele and −78.1 for the wild-type allele. The more negative, thermodynamically favorable, free energy of the variant versus wild-type allele suggests that this is one reason the variant allele has a higher level of AKR1D1 mRNA. The 3′UTR polymorphism has altered the thermodynamic folding landscape of AKR1D1’s mRNA to a more thermodynamically favorable conformation, which may be affecting its mRNA expression (Fig. 5).
In Silico Analysis of AKR1D1 Wild-Type and Variant 3′-UTR Sequences.
Both the full-length wild-type and variant AKR1D1 mRNA (3′-UTR) were investigated for any differential miRNA recognition/binding sites (using miRanda) that might affect the expression of AKR1D1 mRNA. There was no significant difference (P < 0.05) in the binding scores or energy of any known miRNAs between the wild-type and variant alleles demonstrating the absence of miRNA-mediated regulation of AKR1D1 via the rs1872930 SNP.
Discussion
This study was prompted by a recent report that expression profiled 466 human liver samples and constructed a P450 gene regulatory (Bayesian) subnetwork that showed striking correlation between expression of many drug-metabolizing P450s (Yang et al., 2010) (Fig. 2). This network modeling approach identified AKR1D1 as one master regulatory node for the human liver P450 subnetwork (Yang et al., 2010). In this report, we validated that AKR1D1 is a key regulator of the P450 subnetwork. Overexpression of AKR1D1 induced expression of P450s, whereas lentiviral-mediated AKR1D1 knockdown caused a decrease in P450 mRNA expression in hepatic cells. Our results show that an AKR1D1 3′-UTR polymorphism causes a perturbation in AKR1D1 mRNA expression that ripples through the network, ultimately affecting mRNA expression of multiple P450s. This result makes mechanistic sense since, presumably, alterations in AKR1D1 expression (an enzyme critical in both the classic and alternative bile acid synthesis pathways) would be expected to result in altered production of cholic acid and chenodeoxycholic acid, known activators of nuclear hormone receptors, such as PXR and CAR, that transcriptionally regulate a number of drug-metabolizing P450s. The fact that the association of the AKR1D1 SNP is greater with the bile acid pathway genes than with the genes involved in drug metabolism is further support for the notion that the functional consequence of the AKR1D1 polymorphism is on hepatic bile acid homeostasis. The fact that AKR1D1 overexpression also resulted in induction of the FXR target gene SHP and feedback repression of the CYP7A1 gene further suggests that AKR1D1 was modifying primary bile acid synthesis in hepatocytes.
Our AKR1D1 resequencing study failed to find any AKR1D1 coding variation. The absence of AKR1D1 coding variation likely reflects the important function of this gene in regulating bile acid levels. Human AKR1D1 deficiency is rare and patients with AKR1D1 deficiency accumulate bile acid intermediates and lack downstream functional bile acids, and leads to hepatotoxicity (Drury et al., 2010). For example, AKR1D1 sequence variation was reported in infant patients presenting with primary bile acid deficiency (Gonzales et al., 2004) who had a compound heterozygous status represented by two nonconservative missense mutations, P133R (C467G) in exon 4 and R261C (C850T) in exon 7. However, these variations were not detected in 100 chromosomes of control individuals of the same ethnicity. AKR1D1 genetic variations were also reported in three patients with neonatal onset cholestatic liver disease. These patients had rare disease causing AKR1D1 genetic variation that was not detected in 100 chromosomes of control individuals indicating that these are disease-specific mutations (Lemonde et al., 2003). Our study did identify a common 3′-UTR SNP (rs1872930) genotype that correlated significantly with total hepatic AKR1D1 and P450 mRNA expression. To our knowledge, this is the first study to report the occurrence of a functional genetic variation (rs1872930) in AKR1D1 in healthy livers. The minor allele frequency of rs1872930 varies among populations (e.g., Caucasians, 21%; Chinese, 39%; Japanese, 23%; African Americans, 12%; and Mexicans, 39%) and one could imagine the population-specific pharmacogenetic implications associated with this SNP. To rule out the possibility of another linked coding SNP responsible for the higher activity, we directly sequenced the cDNA from these samples and found no coding region polymorphism in AKR1D1. This data are further substantiated by the DNA sequencing data (Caucasians) from the 1000 Genome Project, which also revealed the absence of any AKR1D1 coding region SNP in LD to rs1872930.
The P450 coexpression subnetwork (Yang et al., 2010) included a significant number of drug-metabolizing P450s and nuclear hormone (PXR, CAR) regulators of these P450s. Importantly, the interconnected genes in the P450 subnetwork can sense perturbations from genetic loci (Wang et al., 2012) and identify the key intermediates that contribute to the perturbations. It was thus informative that the subnetwork was also enriched for a number of genes important for the bile acid biologic pathway, including AKR1D1 which was identified as a key regulator of the network. Indeed, the confirmation that the P450 subnetwork senses perturbations in AKR1D1 (driven by its genetic variation) supports the regulatory role for AKR1D1 in the P450 network.
The finding that AKR1D1 sequence variation was predictive of mRNA expression for AKR1D1 and multiple P450s also validates the notion that identification of regulatory nodes in transcription networks is a reasonable approach to identify new candidate genes whose genetic variation is predictive of expression of other genes in regulatory networks. This approach represents a radical departure from the traditional pharmacogenetic reductionist candidate gene strategy, and it represents an elegant approach that harnesses the ability of systems biology to identify the nodes that, when individually perturbed, regulate the entire network. This is particularly useful since the common P450 cis-variation does not explain the majority of human variation in P450 activity. Moreover, genetic variation in P450 (e.g., CYP3A4) trans-regulatory factors such as PXR, FoxA2 and HNF4a, can only explain another 10% of P450 phenotypic variation (Lamba et al., 2010).
This study demonstrates that genetic variation in genes (AKR1D1) that control the amount of ligands that activate P450 transcriptional regulators such as PXR, CAR, and FXR can be another important source of phenotypic variation in drug-metabolizing P450s. This is an interesting finding since we also recently found that variation in intestinal CYP3A4 expression was directly related to genetic variation in both its transcriptional regulator VDR, and probably in the level of its ligand 1,25-dihydroxyvitamin D3 because CYP3A4 intestinal expression varied seasonally, correlating with the documented levels of ultraviolet sunlight and reported seasonal levels of vitamin D (Thirumaran et al., 2012).
Supplementary Material
Acknowledgments
The authors thank the Hartwell Center and the Vector Development and Production Shared Resource at St. Jude Children’s Research Hospital for DNA sequencing and oligonucleotide synthesis and for development and production of adenoviruses and lentiviruses, respectively, used in this study. The authors also thank Dr. Steven Paugh for analyzing miRNA recognition sites using miRanda and Wenjian Yang for multiple testing and statistical analysis of the data.
Abbreviations
- AKR1D1
Aldo-keto reductase 1D1
- CAR
constitutive androstane receptor
- CDCA
chenodeoxycholic acid
- FP
forward primer
- FXR
farnesoid X receptor
- HAP
haplotype
- LD
linkage disequilibrium
- miRNA
microRNA
- P450
cytochrome P450
- PCR
polymerase chain reaction
- PXR
pregnane X receptor
- SHP
small heterodimer partner
- shRNA
short hairpin RNA
- shRNAmir
microRNA-adapted short hairpin RNA
- RP
reverse primer
- SNP
single nucleotide polymorphism
- UTR
untranslated region
- VAR-HAP
variant haplotype
- WT-HAP
wild-type haplotype
Authorship Contributions
Participated in research design: Chaudhry, Thirumaran, Yasuda, Schuetz.
Conducted experiments: Chaudhry, Yasuda.
Contributed new reagents or analytic tools: Thirumaran, Strom.
Performed data analysis: Chaudhry, Thirumaran, Yasuda, Fan, Schuetz.
Wrote or contributed to the writing of the manuscript: Chaudhry, Thirumaran, Yasuda, Yang, Fan, Strom, Schuetz.
Footnotes
References
- Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Bäckman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. (1998) Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc Natl Acad Sci USA 95:12208–12213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Congiu M, Mashford ML, Slavin JL, Desmond PV. (2009) Coordinate regulation of metabolic enzymes and transporters by nuclear transcription factors in human liver disease. J Gastroenterol Hepatol 24:1038–1044 [DOI] [PubMed] [Google Scholar]
- Drury JE, Mindnich R, Penning TM. (2010) Characterization of disease-related 5beta-reductase (AKR1D1) mutations reveals their potential to cause bile acid deficiency. J Biol Chem 285:24529–24537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnerre C, Blättler S, Kaufmann MR, Looser R, Meyer UA. (2004) Regulation of CYP3A4 by the bile acid receptor FXR: evidence for functional binding sites in the CYP3A4 gene. Pharmacogenetics 14:635–645 [DOI] [PubMed] [Google Scholar]
- Gonzales E, Cresteil D, Baussan C, Dabadie A, Gerhardt MF, Jacquemin E. (2004) SRD5B1 (AKR1D1) gene analysis in delta(4)-3-oxosteroid 5beta-reductase deficiency: evidence for primary genetic defect. J Hepatol 40:716–718 [DOI] [PubMed] [Google Scholar]
- Houten SM, Auwerx J. (2004) The enterohepatic nuclear receptors are major regulators of the enterohepatic circulation of bile salts. Ann Med 36:482–491 [DOI] [PubMed] [Google Scholar]
- Innocenti F, Cooper GM, Stanaway IB, Gamazon ER, Smith JD, Mirkov S, Ramirez J, Liu W, Lin YS, Moloney C, et al. (2011) Identification, replication, and functional fine-mapping of expression quantitative trait loci in primary human liver tissue. PLoS Genet 7:e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D, Mangelsdorf DJ, Meyer UA. (2006) Pregnane X receptor is a target of farnesoid X receptor. J Biol Chem 281:19081–19091 [DOI] [PubMed] [Google Scholar]
- Lamba V, Panetta JC, Strom S, Schuetz EG. (2010) Genetic predictors of interindividual variability in hepatic CYP3A4 expression. J Pharmacol Exp Ther 332:1088–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee WH, Lukacik P, Guo K, Ugochukwu E, Kavanagh KL, Marsden B, Oppermann U. (2009) Structure-activity relationships of human AKR-type oxidoreductases involved in bile acid synthesis: AKR1D1 and AKR1C4. Mol Cell Endocrinol 301:199–204 [DOI] [PubMed] [Google Scholar]
- Lehmann JM, McKee DD, Watson MA, Willson TM, Moore JT, Kliewer SA. (1998) The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J Clin Invest 102:1016–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemonde HA, Custard EJ, Bouquet J, Duran M, Overmars H, Scambler PJ, Clayton PT. (2003) Mutations in SRD5B1 (AKR1D1), the gene encoding delta(4)-3-oxosteroid 5beta-reductase, in hepatitis and liver failure in infancy. Gut 52:1494–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modica S, Bellafante E, and Moschetta A (2009) Master regulation of bile acid and xenobiotic metabolism via the FXR, PXR and CAR trio. Front Biosci 14:4719–4745. [DOI] [PubMed]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. (2008) Mapping the genetic architecture of gene expression in human liver. PLoS Biol 6:e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuetz EG. (2004) Lessons from the CYP3A4 promoter. Mol Pharmacol 65:279–281 [DOI] [PubMed] [Google Scholar]
- Schuetz EG, Strom S, Yasuda K, Lecureur V, Assem M, Brimer C, Lamba J, Kim RB, Ramachandran V, Komoroski BJ, et al. (2001) Disrupted bile acid homeostasis reveals an unexpected interaction among nuclear hormone receptors, transporters, and cytochrome P450. J Biol Chem 276:39411–39418 [DOI] [PubMed] [Google Scholar]
- Thirumaran RK, Lamba JK, Kim RB, Urquhart BL, Gregor JC, Chande N, Fan Y, Qi A, Cheng C, Thummel KE, et al. (2012) Intestinal CYP3A4 and midazolam disposition in vivo associate with VDR polymorphisms and show seasonal variation. Biochem Pharmacol 84:104–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Pellicciari R, Pruzanski M, Auwerx J, Schoonjans K. (2008) Targeting bile-acid signalling for metabolic diseases. Nat Rev Drug Discov 7:678–693 [DOI] [PubMed] [Google Scholar]
- Urquhart BL, Tirona RG, Kim RB. (2007) Nuclear receptors and the regulation of drug-metabolizing enzymes and drug transporters: implications for interindividual variability in response to drugs. J Clin Pharmacol 47:566–578 [DOI] [PubMed] [Google Scholar]
- Wang IM, Zhang B, Yang X, Zhu J, Stepaniants S, Zhang C, Meng Q, Peters M, He Y, Ni C, et al. (2012) Systems analysis of eleven rodent disease models reveals an inflammatome signature and key drivers. Mol Syst Biol 8:594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Zhang B, Molony C, Chudin E, Hao K, Zhu J, Gaedigk A, Suver C, Zhong H, Leeder JS, et al. (2010) Systematic genetic and genomic analysis of cytochrome P450 enzyme activities in human liver. Genome Res 20:1020–1036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Kuehl P, Green ED, Touchman JW, Watkins PB, Daly A, Hall SD, Maurel P, Relling M, Brimer C, et al. (2001) The human pregnane X receptor: genomic structure and identification and functional characterization of natural allelic variants. Pharmacogenetics 11:555–572 [DOI] [PubMed] [Google Scholar]
- Zuker M. (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.