Abstract
MicroRNAs are small, non-coding RNAs that regulate gene expression post-transcriptionally. Here, we show that miR-210 is induced by Oct-2, a key transcriptional mediator of B-cell activation. Germline deletion of miR-210 results in the development of autoantibodies from 5 months of age. Overexpression of miR-210 in vivo resulted in cell autonomous expansion of the B1 lineage and impaired fitness of B2 cells. Mice over-expressing miR-210 exhibited impaired class-switched antibody responses, a finding confirmed in wild-type B-cells transfected with a miR-210 mimic. In vitro studies demonstrated a defect in cellular proliferation and cell-cycle entry, which was consistent with the transcriptomic analysis demonstrating down-regulation of genes involved in cellular proliferation and B cell activation. These findings indicate that Oct-2 induction of miR-210 provides a novel inhibitory mechanism for the control of B cells and autoantibody production.
Introduction
B-cells are activated upon antigenic stimulation to mediate a variety of effector functions including antibody production (1), and dysregulated B cell behaviour has been implicated in autoimmunity and malignancy(1, 2). Our understanding of the genetic control of B-cell maturation involving key transcriptional regulators such as Oct-2 remains incomplete (3). Oct-2 is a POU domain containing transcription factor required for normal humoral responses to T-dependent and T-independent antigens (4). Whilst recent studies have identified important targets including IL-6 and IL-5 receptor alpha chain(5, 6), the full extent of transcriptional complexity underlying Oct-2 mediated regulation of B-cell responses remains to be elucidated.
MicroRNAs (miRNAs) are small, non-coding RNAs that regulate gene expression post-transcriptionally. MiRNAs are processed by Dicer, and form miRNA-induced silencing complexes (miRISC) that base-pair imperfectly with target mRNAs at sites located mainly in their 3’UTR(7). In mammalian cells, target repression occurs by decreasing mRNA levels and translation (8). MiRNAs are subjected to regulation at several levels, including transcription, precursor processing and export, as well as by other miRNAs(9). Early studies have shown that miRNAs are important in B-cell development. Removal of Dicer at an early stage in B-cell development resulted in an almost complete block at the pro- to pre-B transition and reduced B-cell populations in the periphery (10). Dicer ablation in CD19+ B-cells resulted in skewing of B2 cellular subsets with increased transitional and marginal zone B-cells and reduced follicular B-cells(11).
MiRNAs are also important in the B-cell response to antigen (12). Dicer ablation at either the pro-B or CD19+ stage results in altered antibody repertoires (10, 11), and deletion of Dicer in murine antigen-activated B-cells results in impaired production of high-affinity class-switched antibodies, memory B-cells, and long-lived plasma cells (13). MiR-155, an activation-induced miRNA, enhances the formation of germinal centres in vivo and is essential for the generation of class-switched antibody-secreting cells via downregulation of Pu.1 (14, 15). Activation-induced cytidine deaminase (AID), an enzyme important in somatic hypermutation and class switch recombination, is targeted by both miR-155 and miR-181b to prevent AID-mediated Myc-IgH translocations and malignant transformation(16-18).
Previous studies have shown that most murine miRNAs are down-regulated upon B-cell activation (18, 19). In this study, we identify the highly conserved miR-210 as an Oct-2-regulated miRNA induced upon B-cell activation. MiR-210 is widely expressed and has been implicated in the hypoxic response (20), oncogenesis (21), and angiogenesis (22). It is expressed in haematopoietic stem cells, myeloid cells, and lymphocytes (19). In macrophages, miR-210 has been reported to negatively regulate production of pro-inflammatory cytokines by targeting NF-κB (23). Its function in lymphocytes has not been defined, and interestingly, miR-210 has been shown to be over-expressed in B-cell malignancies (24, 25). In this study, we demonstrate that miR-210 is a novel regulatory target of Oct-2 and has a physiologically important role in inhibiting the development of age-associated autoantibodies. Furthermore, over-expression of miR-210 can result in B-cell subset and functional abnormalities, by down-regulating genes involved in cellular proliferation and B-cell activation. Our results reveal a novel, miRNA-mediated mechanism for the control of B cell responses and autoantibody production.
Materials and Methods
Mouse Strains
C57BL/6 (B6), MRL, MRLlpr, and NZB mice were purchased from Harlan Europe. NOD mice were provided by Sarah Howlett (Juvenile Diabetes Research Foundation/Wellcome Trust Diabetes and Inflammation Laboratory, Cambridge Institute for Medical Research). All experiments were performed according to the regulations of the UK Home Office Scientific Procedures Act (1986). The animal experiments were approved by the UK Home Office.
Generation of miR-210 deficient mice
Heterozygous knockouts in the C57Black/6N ES cells were generated as part of the Wellcome Trust Sanger Institute’s mirKO initiative (Prosser et al, 2011). Full details of the targeting vector and ES cell reagents are described at http://www.knockoutmouse.org/martsearch/search?query=mir-210. Briefly, a targeting vector with a PuroΔtk selection cassette was generated by recombineering in E.coli. The AscI linearized vector was electroporated into JM8.F6 ES cells and correctly targeted clones identified by using long range PCR across both of the targeting arms. The primer sequences for the 5′ arm are TGAGAGTATCAGTCTTGGAGGAAGTAT and CCAGTGATAACTTCGTATAATGTATGCTAT with a product of 5,489bp and for the 3′ arm the primers are TCTAGAAAGTATAGGAACTTCCATGGTC and CCAAGTCCTCTGAAGAAGTAATAAATG with a product of 2,885bp. The PuroΔtk selection cassette was deleted from the targeted allele by transient transfection with a Cre recombinase expression plasmid followed by selection with FIAU (200nM). Surviving ES cells were pooled and microinjected into C57BL/6J-Tyr c-Brd blastocysts for the generation of chimaeric mice. Germline transmission was tested by breeding with albino C57BL/6J-Tyr c-Brd mice and the colony was maintained by backcrossing with C57BL/6N. Genotyping of the miR-120 knockout allele was by PCR across the deleted locus using primers aggtgaaatagaagggttacaaggtt and aaccctaatacacctaagaagaaagttcc annealing at 560C. The wild type product was 527bp and the mutant product was 443bp.
Generation of miR-210 overexpressing mice and mixed chimeras
A 260 bp region including the miR-210 precursor sequence was cloned into a construct containing the heavy chain (VH) promoter, IgH intronic enhancer (Eμ), and the Igκ 3′ enhancer (26, 27), and injected into CBA fertilized C57BL/6 eggs. Transgenic offspring were backcrossed onto C57BL/6 mice for at least five generations. The presence of the transgene was identified by tail DNA PCR assays using primers designed to amplify a 500 nt region spanning the Eμ-Vp junction (miR-210F: TCCAGCAGGAGTAGGTGCTT, miR-210R: GCTCCCATTCATCAGTTCCA). Fetal liver chimeras were generated as previously described (15). For μMT -chimeras, irradiated Rag2−/− Il2rγ−/− mice (5.0 Gy) received a total of 5 million fetal liver cells, 80% of μMT and 20% miR-210 TG or NTG cells. For mixed IgHa chimeras, the following combinations were generated: 50% IgHa/50% miR-210 TG, 90% IgHa/10% miR-210 TG, 50% IgHa/50% NTG, 100% miR-210 TG, 100% NTG. Mice were analysed 8 – 16 weeks after reconstitution.
Chromatin Immunoprecipitation (ChIP)
ChIP was carried out as previously described(24). Briefly, proteins were cross-linked to DNA using formaldehyde (0.4%), followed by cell lysis. Chromatin was fragmented using a Bioruptor sonicator (Diagenode SA) and separate immunoprecipitates were produced using polyclonal anti-Oct-2 serum, pre-immunised control serum(24), and antibodies to H3K4Me3, H3K9Ac (Millipore) and control IgG (Sigma-Aldrich). Primers A1 (TCAGGGTGGGCCTTACTAGA) and A2 (GATCAACTTCAGTGGCAGCA) were used to detect the miR-210 promoter and primers B1 (GCAGCTGCTCTTTTGCTTCT) and B2 (GAGCCAAGGACCAGGGTATC), C1 (TCATAGTTTCTCAACCCAAGAGG) and C2 (TATAGGCTACAGGGCCAGGA) were used to amplify regions 1 kb 5′ and 3′ of the Oct-2 binding site respectively. Data were normalised to genomic regions 1 kb upstream (5′ 1kb) and downstream (3′ 1kb) of the Oct-2 binding site to demonstrate localised enrichment, and the CD36 promoter was used as a positive control as previously described(5).
Luciferase assay and mutagenesis
The 2 kb miR-210 promoter was amplified from C57BL/6 genomic DNA and cloned into pGL4.14 and pGL4.26 hLuc vectors (Promega) using the primers D1 (AACTCGAGACACAGGGCAGTAAGGGTTG) and D2 (AAAAGATCTCTGAGCCTGGAGGGACTG). Deletion of the Oct-2 and HIF-1α consensus binding sites were generated using QuikChange mutagenesis (Agilent Technologies) with primers E1(CAGTTACCCCGCCCCATAGTCTATCTTGCAGC) and E2(GCTGCAAGATAGACTATGGGGCGGGGTAACTG), and E3(CCCGCCGCCGAGCAAGGCGCGTT) and E4(AACGCGCCTTGCTCGGCGGCGGG) respectively. WEHI231 cells were stimulated with LPS (10 μg/ml) and IL-4 (20 ng/ml) for 24 h before electroporating with different hLuc vectors and a pCMV Renilla vector control(25) using AMAXA (Lonza) according to manufacturer’s’ instructions. Luciferase and Renilla activity were analyzed after 24 h using the Dual-Glo assay system and Glomax reader (Promega). Relative luciferase activity was calculated by normalizing the luciferase to the Renilla OD.
Immunisations
Mice were immunized intraperitoneally with 100μg NP-Ficoll or NP-KLH (Biosearch Technologies) emulsified in alum (Thermo-Fisher) according to manufacturer’s instructions.
ELISA and ELISPOT assay
Serum anti-NP, anti-chromatin and anti-dsDNA ELISAs were carried out as previous described(25). Serum IgM, IgG, IgG2a, IgG2b, IgG1, and IgG3 were assayed using paired capture and HRP-conjugated antibodies (SouthernBiotech). For anti-dsDNA and anti-chromatin ELISAs, a positive result was determined as an O.D. of at least 1.5x above that of pooled sera from 8-12 wk old BL/6 mice, and pooled sera from NZB/W mice were used as a positive control. Plasma cells secreting anti-NP were detected by ELISPOT assay as previously described (25).
Immunofluorescence
Spleens were embedded in Tissue-Tek OCT compound (Bayer Healthcare), cut in 6-μm thick sections and fixed in acetone. Primary antibodies used were anti-mouse B220-biotin (clone RA3-6B2, Ebioscience), IgD-FITC (clone 11-26, Ebioscience), IgG1-AF633, IgM-Rhodamine Red (Jackson Immunoresearch) and MAdCAM1-biotin (clone MECA-367, Ebioscience). Biotinylated antibodies were detected with Steptavdin-AF555 (Invitrogen). FITC-conjugated antibodies were amplified with rabbit anti-FITC (Invitrogen), then goat anti-rabbit IgG FITC (Southern Biotech). Anti NP binding was detected with Rabbit IgG conjugated with NP, then goat anti-rabbit IgG FITC (Invitrogen). Sections were mounted using DABCO (Sigma-Aldrich). Confocal images were obtained using a LSM 510 Meta microscope (Zeiss) equipped with 405, 488, 543, and 633 nm lasers and image analysis used the Zeiss LSM software. Anti-nuclear antibodies were detected using a commercial indirect fluorescent antibody assay (ANA Hep-2 slide; Cambridge Life Sciences) with sera diluted at 1:50 as previously described (26).
MiR-210 mimics and Luciferase Assay
The target site of CD23 was amplified using primers F1 (AAACTCGAGGCACAGCAATGTGGGCTGGC) and F2 (AAAGCGGCCGCTCACTGCAGGCGAACCTGGC) and cloned into psiCheck-2 renilla luciferase reporter plasmid (Promega). HeLa cells cultured in 96-well plates were transfected with 2 ng of plasmid and miR-210 mimic or control (c =80nM, Dharmacon) using Lipofectamine 2000 (Invitrogen). Reporter renilla luciferase activity was measured after 24h with Dual-Luciferase Reporter Assay System (Promega) and normalised to firefly luciferase activity.
Cell separation and culture
B cells were isolated by magnetic cell purification using anti-CD19 beads (Miltenyi Biotech) according to manufacturer’s instructions, and routinely sorted to >95% purity. Cells were cultured in RPMI-1640 supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 10 000 units/ml penicillin, 10 mg/ml streptomycin, 1 mM sodium pyruvate, 10 mM HEPES pH7.5 and 0.1 mM non-essential amino acids. All reagents were from Sigma-Aldrich unless otherwise specified.
Cell stimulation and transfection
For miRNA expression profiling studies, cells were cultured at 2 × 106 cells/ml and stimulated with either goat anti–mouse IgM μ-chain specific F(ab′)2 (10 μg/ml: Jackson ImmunoResearch Laboratories) with recombinant murine CD40L (1 μg /ml: PeproTech) and IL-4 (20 ng/ml: PeproTech), or LPS from E.coli (10 μg /ml, Sigma). Oct2+/+ and Oct2−/− B cells were obtained from Rag1−/− reconstituted mice as the Oct2 mutation is lethal when homozygous, and stimulated with CpG for 48 h as previously described(6). For transfection, primary B cells were pre-stimulated with LPS and IL-4 overnight and electroporated with miR-210 mimic or control (10nM, Dharmacon) using the Mouse B cell Nucleofector kit (Amaxa) according to manufacturer’s instructions. For in vitro class-switching and plasma cell differentiation assays, cells were cultured at 1 × 105 cells/ml for 72 or 96 hours as previous described (15).
B cell proliferation and cell cycle analysis
For CFSE labeling, 5 × 107 purified B cells/ml were loaded with 5 μM CFSE (Invitrogen) in protein-free media or PBS by incubating for 10 mins at room temperature. Cells were then washed with complete media and the extent of CFSE labeling was analyzed by flow cytometry. Cell cycle analysis with propidium iodide was assayed as previously described (28).
RNA extraction and RT-PCR
Total RNA was extracted with TRIzol reagent (Invitrogen), and real-time quantitative PCR performed using TaqMan MiRNA Assays (Applied Biosystems, Foster City, CA) according to manufacturer’s instructions. Data was acquired on a 7900HT Fast Real-Time PCR System (Applied Biosystems) and normalised to snRNAU6. For mRNA expression assays, cDNA was generated using SuperRT reverse transcriptase (HT Biotechnology, Cambridge, UK) before quantification with Taqman probes (Applied Biosystems).
Gene Ontology Analysis
Gene Ontology (GO) overrepresentation analysis was performed using the Bioconductor topGO package, using a Fisher’s exact test in conjunction with the “elim” algorithm as described previously (26).
MicroRNA Target Prediction
MiR-210 predicted targets were generated from the mirWalk database (http://mirwalk.uni-hd.de/) using 5 different algorithms – mirWalk(27), RNA22 (28), TargetScan (29), Miranda (30), and RNAHybrid (31),with a minimum seed length of 7 nt, p value = 0.05.
Statistical Analysis
Statistical analysis was performed with two-tailed unpaired Student’s t test using GraphPad Prism 4 unless otherwise indicated.
MiRNA expression profiling
MiRNA expression profiling was carried out as previously described(29). Briefly, miRNAs were extracted from 5 μg of total RNA and adaptors were ligated at the 3′ and 5′ ends using T4 RNA ligase (Fermentas, Burlington, OT, CA). These bi-ligated products underwent reverse transcription using an adaptor specific primer and were amplified and labelled using PCR. PCR products were precipitated and hybridised overnight to oligonucleotide probes coupled to color-coded polystyrene beads. Unbound sample was removed from beads by washing and streptavidin-phycoerythrin (Invitrogen) was added to the beads to bind biotin moieties on the cDNA. Samples were processed on a Luminex 100 machine and median fluorescence intensity (MFI) values acquired using the StarStation software (ACS, Sheffield, UK).
Bioinformatic Analysis
For miRNA profiling of B cells across C57BL/6 and autoimmune strains of mice, well-to-well scaling was performed such that the total MFI of the post-labeling controls in each well was equal to the median value across all wells. Sample scaling was performed such that total MFI of the pre-labeling controls in each sample was equal to the median value across all samples. A threshold of 1 was applied and the data was log2-transformed. Filtering for ‘expressed’ miRNAs was performed by employing a cutoff value of three standard deviations above the median MFI of all readings. Differential gene expression between sample pairs was analysed by carrying out 1-way ANOVAs (1 way Analysis of Variance) for all combinations of sample pairs using Graphpad Prism. Combinations with p value <0.05 were then filtered to show only within-strain comparisons. Differential gene expression across all samples was analysed using GEPAS T-rex multi-class comparison (ANOVA) tool (30), which provided P-values adjusted for multiple testing, with control of family-wise error rate and false-discovery rate (31). All miRNA expression data have been submitted to the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with accession number GSE48186.
mRNA extraction, microarray hybridisation and data analysis
Total RNA was extracted using an RNeasy Mini kit (Qiagen), of which 200 ng was labelled using Affymetrix’s WT Sense Target labelling kit and hybridised to Mouse Gene 1.0 ST arrays (Affymetrix). Arrays were scanned using a GS 3000 scanner (Affymetrix), and raw data files were imported into R and subjected to variance stabilisation normalisation using the VSN package in BioConductor (32). Following normalisation, differentially expressed genes were identified using the Limma package (33, 34). Differential expression was defined as fold changes greater than 1.5 fold that were statistically significant following correction for multiple testing by setting the false discovery rate to 5%. Gene Set Enrichment Analysis (GSEA) was performed as previously described, with a false discovery rate of q < 0.25 indicating significant enrichment (35). All mRNA expression data have been submitted to ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) with accession number E-MTAB-1758.
Results
Mir-210 is an activation-induced miRNA regulated by Oct-2
Splenic B-cells from different mouse strains were stimulated with either anti-IgM, CD40L and IL-4 or LPS for 48 hours and miRNA expression determined using a bead-based microarray (29). In agreement with previous studies, most miRNAs are downregulated upon B-cell activation (Figure 1a). Strikingly, miR-210 was up-regulated 15-20 fold in both BCR-dependent and BCR-independent conditions across all strains examined (Figure 1a, b). Kinetic characterization revealed peak induction at 48 h, and high levels were maintained for at least 96 h after stimulation (Figure 1b).
Figure 1. Mir-210 is an activation-induced miRNA regulated by Oct-2.
A. B-cells from 8-12 wk old mice of various murine strains (top row) were stimulated with αIgM + CD40L + IL-4 or LPS for 48 hours, and their miRNA profiles compared to resting controls. Each row corresponds to a miRNA gene, and each column to an individual mouse. Red indicates increased, and blue reduced, expression. MiRNAs are ranked according to the degree of differential expression across samples. MiR-210 expression, which is up-regulated upon activation with both types of stimuli, is boxed in black.
B. Kinetics of miR-210 induction in C57BL/6 B-cells stimulated with αIgM + CD40L + IL-4 (top) or LPS (bottom) quantified by RT-PCR and normalised to levels of U6snRNA. Error bars represent SEM of 3 biological replicates.
C. Site-directed mutagenesis of Oct-2 and HIF-1 α binding sites in the miR-210 promoter. Firefly luciferase activity of a reporter vector was assayed, and normalised to control Renilla luciferase activity.
D. ChIP of endogenous Oct-2 bound to the miR-210 or CD36 (positive control) promoters in WEHI231 cells. Data is normalised to regions 1 kb 5′ and 3′ of the miR-210 promoter to demonstrate localised enrichment.
E. ChIP of H3K4Me3 (promoter-specific) and H3K9Ac (promoter/enhancer-enriched) histone modifications in WEHI231 cells, demonstrating enrichment of the Oct-2 binding site. Data is normalised to control IgG and input DNA.
F. Oct-2 deficient and wild-type B-cells were stimulated with CpG for 48 h and miR-210 induction quantified by RT-PCR, normalised to U6snRNA, and expressed as arbitrary units (AU).
Data is representative of 2-4 independent experiments for C-F, and error bars indicate SD of technical replicates.
As miR-210 induction occurs relatively late in B-cell activation, we were interested in its transcriptional regulation. Promoter analysis revealed the presence of putative binding sites for HIF-1α and Oct-2 upstream of the miR-210 transcription start site. HIF-1α has been shown to be a hypoxia-induced regulator of miR-210 in cancer cells (20) and whilst deletion of the HIF-1α binding site resulted in a 50% reduction in reporter luciferase activity, it did not completely abolish miR-210 induction in WEHI 231 cells (Figure 1c). The role of Oct-2 in miR-210 regulation on the other hand, has not been previously investigated. Oct-2 is predominantly expressed in lymphocytes (36), and its transcriptional activity is important in B-cell activation (37). Deletion of the Oct-2 consensus octamer (ATTTGCAT) resulted in a 25% reduction in reporter luciferase activity (Figure 1c). We demonstrated binding of Oct-2 to the miR-210 promoter by chromatin precipitation analysis (5) in WEHI231 B lymphoma cells (Figure 1d), and this same region was also highly enriched for the promoter-specific histone modification H3K4Me3 and H3K9Ac, which indicates likely promoter or enhancer activity (Figure 1e). In addition, we activated Oct-2 deficient and WT B-cells with CpG for 48 h (6) and measured miR-210 expression. Oct-2 deficient B-cells exhibited a 40% reduction in miR-210 levels compared to WT B-cells (Figure 1f), despite comparable levels of aicda induction (unpublished observation). These findings identify miR-210 as a novel regulatory target of Oct-2, with the latter being essential for the full induction of miR-210 during B-cell activation.
MiR-210 deficient mice develop increased autoantibodies with age
To investigate the physiological roles of miR-210, we generated mice deficient in miR-210 on a C57BL/6 background (38) (Figure S1a). MiR-210 KO mice were fertile and viable, and born at the expected Mendelian ratios to their littermates. These mice exhibited normal B, T and myeloid cell subsets (Table I, Table S1a) and there were no gross abnormalities in organ development.
Table I.
Cell populations in miR-210 KO mice1.
| Cell populations | Cell surface markers | miR-210 WT | miR-210 KO |
|---|---|---|---|
| Spleen | |||
| B cells ( × 106) | |||
| B1 cells | CD19+ CD5+/low CD43+ | 2.35 (0.94) | 1.94 (0.44) |
| B2 cells | |||
| Transitional B cells, T1 | B220+ CD93+ IgMhi CD23− | 2.94 (0.49) | 2.32 (0.88) |
| Transitional B cells, T2 | B220+ CD93+ IgMhi CD23+ | 4.32 (0.84) | 4.31 (2.14) |
| Transitional B cells, T3 | B220+ CD93+ IgMlo CD23+ | 4.16 (0.61) | 4.04 (2.04) |
| Marginal zone B cells | B220+ CD93− IgMhiCD21hi | 7.68 (2.71) | 7.98 (3.08) |
| Follicular B cells | B220+ CD93− IgM+CD21+ | 77.5 (27.7) | 90.5 (19.4) |
| T cells ( × 106) | |||
| CD4+ | CD3+ CD4+ | 3.49 (0.52) | 3.70 (0.96) |
| CD8+ | CD3+ CD8+ | 2.35 (0.30) | 2.25 (0.55) |
| Myeloid cells (× 105) | |||
| Macrophages | CD11b+ F4/80+ | 1.10 (0.28) | 0.87 (0.15) |
| Granulocytes | CD11b+ Gr-1+ | 4.88 (1.33) | 6.77 (2.00) |
| Peritoneal Cavity (× 106) | |||
| B1 cells | CD5+/low CD11b+ | 0.44 (0.28) | 0.58 (0.35) |
| Bone Marrow (× 104) | |||
| Pro-B cells | B220+ c-kit+ | 25.58 (5.70) | 25.83 (4.90) |
| Pre-B cells | B220+ CD25+ | 90.33 (24.9) | 117.8 (41.5) |
| Immature B cells | IgM+ B220+ | 108.3 (34.8) | 108.3 (37.2) |
| Mature B cells | IgM+ B220hi | 85.0 (32.5) | 101.5 (21.2) |
Cell counts derived from n= 4-6 mice (6-8 weeks old); Data shown are mean (SEM)
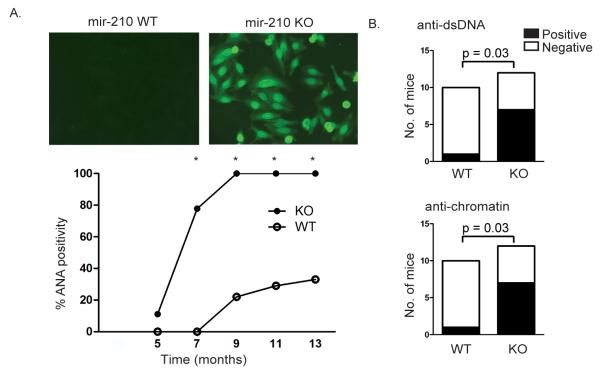
Strikingly, however, miR-210 KO mice developed spontaneous autoantibodies first detectable at 5 months of age (Figure 2a). MiR-210 KO mice had increased serum levels of anti-nuclear antibodies (ANA), and in particular anti-dsDNA and anti-chromatin antibodies classically associated with systemic lupus erythematosus (SLE), implying a role for miR-210 in the maintenance of B-cell tolerance (Figure 2b). Baseline serum IgM, IgG1, IgG3 and IgA levels were normal, and the ability to mount an antibody response to T-dependent and T-independent antigens was largely intact (unpublished observation). Interestingly, germinal centre B-cells were increased in aged miR-210 KO mice (Figure S2), suggesting that miR-210 may be induced in the later stages of B-cell activation to function as a negative feedback regulator in the prevention of age-associated development of autoimmunity.
Figure 2. MiR-210 deficient mice develop autoantibodies with age.
A. Sera from miR-210 KO and WT littermate controls were tested for the presence of anti-nuclear antibodies by immunofluorescence in 3 independent aged cohorts of n = 5-9 in each group, at 1:50 dilution. Representative slides from mice at 9 months old are shown in the top panel. Bottom panel illustrates percentage mice with ANA positivity in the largest aging cohort (n = 9 in each group) sampled at different time points. Fisher’s exact test was performed at each time point and * indicates p < 0.05.
B. Serum anti-dsDNA (top panel) and anti-chromatin antibodies (bottom panel) were measured by ELISA. Data is pooled from 2 independent aged cohorts of 10-11 month old miR-210 KO and littermate controls. Fisher’s exact test was performed with p values as indicated.
Overexpression of miR-210 in vivo results in B-cell subset abnormalities
To complement loss-of-function studies, we over-expressed miR-210 in vivo under the control of the heavy chain (VH) promoter, IgH intronic enhancer (Eμ), and the Igκ 3′ enhancer(26) (Figure S2b). Three lines of miR-210 transgenic (TG) mice were obtained in which expression of miR-210 in B-cells was moderately increased at 0.3, 1.2 and 12.3-fold that found in 48 h activated B-cells (Figure S2c). All three lines of miR-210 TG mice were fertile and viable. Mice from the highest expressing line exhibited a complex lymphoid phenotype, which were examined in more detail, and are henceforth referred to as miR-210 TG mice. Mice with low levels of expression did not exhibit obvious lymphoid abnormalities. MiR-210 TG mice were viable even when homozygous for the transgene, and karyotyping with fluorescence in-situ hybridisation (FISH) revealed no obvious karyotypic abnormalities (Figure S3a). The expression of other miRNAs (miR-24, miR-146b, let-7c, and miR-342) in miR-210 TG B-cells was unperturbed, indicating that microRNA processing pathways were not saturated (Figure S3b).
MiR-210 TG mice exhibited a 20-fold expanded splenic B1a compartment, and the proportion of B1a cells in the lymphocyte pool in the peritoneal cavity was also increased by a factor of 2.5 (Figure 3a). The B1a cells in both spleen and peritoneal cavity expressed markers consistent with the conventional phenotypic definition of B1a cells, that is CD19+B220loCD5+CD43+IgMhiIgDloCD23lo (39), and VH gene analysis(40) showed they were polyclonal (unpublished observation). Although there was no difference in splenic B2 cell numbers, immunohistochemistry revealed the absence of a well-defined marginal zone in miR-210 TG mice, and a reduced marginal zone B-cell population was verified by flow cytometry (Figure 3b, c). In the bone marrow, there was a decrease in the numbers of pro-B, pre-B and mature B-cells (Figure 3d), indicating impaired production, proliferation or survival of miR-210 TG B-cell progenitors.
Figure 3. B-cell subset abnormalities in miR-210 TG mice and impaired developmental fitness of B2 cells.
A. Representative FACS of B1a cell expansion in spleen and peritoneal cavity of 8–12 wk old miR-210 TG mice, with quantification shown in the right panel. Each dot represents one NTG (open circles) or TG (filled circles) mouse for A, C, and D. Student’s t-test; ** p<0.01.
B. The marginal zone B-cell population, seen here as IgMhiIgDlo marginal zone B-cells surrounding MAdCAM-1 expressing marginal zone macrophages, is not well-defined in the miR-210 TG spleen. Data is representative of n=3 mice in each group.
C. Quantification of follicular and marginal zone B-cells by flow cytometry. CD19+43-CD93- splenic B-cells are divided into follicular and marginal zone subsets based on higher expression of IgM and CD21 in the latter. Student’s t-test; ** p<0.01.
D. Quantification of B cell progenitors in the bone marrow of miR-210 TG mice by flow cytometry; Pro-B (B220+ c-kit+), Pre-B (B220+CD25+), Immature B (IgM+B220+) and Mature B (IgM+B220hi, with 6-8 biological replicates in each group, at 6-8 weeks old. Student’s t-test was performed and ** indicates p<0.01. The p-value for immature B cells in (b) is 0.0655.
E. Impaired developmental fitness of miR-210 TG B2 cells in the presence of wild-type competitors. Recipient mice were reconstituted with mixtures of WT IgHa and NTG or TG IgHb fetal liver cells and bar charts indicate the mean relative contribution of IgHb B-cells to each B-cell compartment. Striped gray columns represent the contribution of IgHb TG mice whilst plain gray columns represent the contribution of IgHb NTG mice. Student’s t-test was performed; ** p < 0.01
Impaired competitive fitness of miR-210 TG B2 cells
To elucidate the cellular basis of the increased B1a to B2 ratio in miR-210 TG mice, we investigated the behaviour of miR-210 TG B-cells in a competitive environment, using the allotypic markers IgHa (wild-type) and IgHb (TG/NTG). Mixed chimeras were generated by transferring miR-210 NTG or TG fetal liver cells mixed with wild-type B-cells in a 1:1 ratio into sublethally irradiated mice doubly deficient in Rag2 and the common cytokine receptor gamma chain (Rag2−/− Il2rγ−/−) (Figure 3d). Preservation of the 1:1 ratio in reconstituted B-cell compartments would reflect a similar competitive fitness between IgHa (wild-type) and IgHb (TG/NTG) B-cells. Surprisingly, splenic B2 cells in miR-210 TG/WT chimeras were derived mainly from WT progenitors, with an IgHa: IgHb ratio of 4:1. In contrast, the B1a compartment had an approximately equal contribution from TG and WT cells (Figure 3e, left panel). A similar pattern was observed in the peritoneal cavity (Figure 3e, middle panel). In addition, 80% of IgM+B220+ cells in the bone marrow of miR-210 TG/WT chimeras were derived from IgHa progenitors (Figure 3e, right panel), consistent with the decreased numbers of B2 cell progenitors in the bone marrow of non-chimeric mice (Figure 3d). Altogether, the data indicate that whilst the developmental fitness of miR-210 TG B1 cells is relatively unaffected, the ability of B2 cells to proliferate or survive is impaired in the presence of competing wild-type cells.
Overexpression of miR-210 results in impaired production of class-switched antibodies in a B cell-autonomous manner
MiR-210 TG mice exhibited a marked decrease in serum levels of class-switched antibodies, with a more than 10-fold decrease in IgG1, IgG2a, IgG2b, IgG3, and IgA isotypes (Figure 4a). Class-switched IgG1 antibody responses to the T-dependent antigen NP-KLH in miR-210 TG mice were also decreased, whereas the IgM response was intact (Figure 4b). This was also consistent with an ~8-fold decrease in the numbers of anti-NP IgG1 antibody-secreting cells in the spleen and bone marrow 14 days post-immunisation (Figure 4c). Similarly, when immunised with the T-independent antigen NP-Ficoll, IgM responses were preserved, with a non-significant trend towards lower IgG3 anti-NP antibodies 7 days post-immunisation (unpublished observation).
Figure 4. Over-expression of miR-210 results in impaired production of class-switched antibodies in a cell autonomous manner.
A. Levels of total serum Ig in miR-210 NTG (open circles) and TG (filled circles) mice.
B. MiR-210 TG mice were immunised with the T-dependent antigen NP-KLH, and anti-NP IgM (top panel) and IgG1 (bottom panel) titres were measured at day 0, 7, and 14 after immunisation with NP12-BSA.
C. ELISPOT analysis of anti-NP IgG1 antibody secreting cells in the spleen and bone marrow of miR-210 TG mice 14 days post-immunisation with NP-KLH.
D. Levels of total serum Ig in μMT-NTG (open circles) and μMT-TG (closed circles) chimeras.
E. Measurement of anti-NP antibody titres in μMT chimeras following immunization with the T-dependent antigen NP-KLH.
F. Anti-NP IgG1 antibody secreting cells were quantified by ELISPOT in μMT-NTG (open circles) or μMT-TG (closed circles) chimeras 7-days post immunisation with NP-KLH. Student’s t-test was performed for A, B, and C; * p<0.05 and ** p<0.01.
G. Immunohistochemistry of NP-specific B-cells in the spleen 7 days post-immunisation with NP-KLH. Data representative of n=3 mice in each group.
H. C57BL/6 B-cells transfected with miR-210 mimics exhibit increased class-switching to IgG1 upon activation with LPS (10 μg/ml) + IL-4 (40 ng/ml) for 96 hours. Data is pooled from two independent experiments, and a one-tailed Mann-Whitney test was performed, with p-value as indicated.
We investigated the cell autonomy of these B-cell abnormalities using μMT fetal liver chimeras, to exclude the effects of transgene expression in other lymphopoietic lineages(41). Mixed chimeras were created by transferring 80% of μMT-deficient fetal liver cells mixed with 20% of miR-210 TG or NTG fetal liver cells into Rag2−/−Il2rγ−/− mice, as previously described(15). As μMT-deficient fetal liver cells are unable to generate B-cells, the 80/20 ratio favours the reconstitution of non B-lineage haematopoietic cells from wild-type precursors, whilst the B-cell compartment would be reconstituted solely from miR-210 TG or NTG precursors. Consistent with our findings in germline mice, miR-210 TG chimeras exhibited a 8-fold increase in their splenic B1a population, as well as the absence of a well-defined marginal zone B-cell population in the spleen (unpublished observation). Serum levels of class-switched antibodies were also reduced, with a less pronounced reduction in IgM (Figure 4d). When immunised with the T-dependent antigen NP-KLH, μMT-miR-210 chimeras exhibited markedly decreased serum levels of anti-NP IgG1 antibodies, with a slight reduction in anti-NP IgM (Figure 4e). IgG1, but not IgM antibody-forming cells were reduced in the spleen 7 days post-immunisation (Figure 4f). Consistent with the above findings, immunohistochemical analysis revealed the absence of anti-NP IgG1 cells in the extrafollicular region of the μMT-TG spleen, whilst anti-NP IgM cells were present (Figure 4g). To further validate these observations in an independent system, given that only our highest expressing strain demonstrated this phenotype, we transfected activated WT B-cells with miR-210 mimics, and consistent with our in vivo findings, found impaired class-switching to IgG1 upon activation with LPS + IL-4 for 96 h (Figure 4h).
MiR-210 overexpression results in impaired B2 cellular proliferation, cell cycle entry, and class-switching in vitro
To investigate the underlying cause of the impaired production of class-switched antibodies and reduced “fitness” of miR-210 TG B2 cells observed in the IgHa/IgHb chimeras, we examined cellular proliferation and cell cycle entry in vitro. Splenic B2 cells were labelled with the intracellular dye carboxyfluorescein succinimidyl ester (CFSE) and stimulated with αIgM + CD40L + IL-4, in addition to activatory stimuli that induce class-switching to IgG1(42). MiR-210 TG B2 cells exhibited a marked defect in proliferation in response to different activatory conditions, and cell cycle analysis revealed a defect in G1 to S phase transition in miR-210 TG B2 cells (Figure 5a), indicating that a defect in cell cycle regulation contributes to the impaired proliferation of miR-210 TG B2 cells We further investigated the contribution of impaired B2 proliferation to reduced class-switching observed in vivo and in vitro. As class-switch recombination is a division-linked process(42, 43), the percentage of class-switched B2 cells was assessed in a specific subset of cells that has undergone at least 4 divisions (Figure 5b). There was a significant decrease in the proportion of IgG1 class-switched B-cells within this subset, suggesting a defect in class-switching that is independent of the proliferative defect. Quantification of aicda expression did not reveal significant changes between TG and NTG B2 cells (unpublished observation), indicating that other components of the class-switch recombination machinery are responsible for the impaired class-switching observed in miR-210 TG B2 cells.
Figure 5. Over-expression of miR-210 results in impaired proliferation and isotype switching in vitro, and downregulates genes involved in cell division and B cell activation.
A. Cell cycle analysis with propidium iodide of splenic B2 cells stimulated with αIgM + CD40L + IL-4 for 48 hours. Representative FACS plots are shown in the top panel and the percentage of cells in each phase of the cell cycle is quantified in the bottom panel. The average of three mice in each group ± SEM is shown.
B. Splenic B2 cells from miR-210 TG and NTG mice (8-12 weeks old) were exposed to various stimuli as indicated for 72 hours. Cellular proliferation was assessed by CFSE dilution, and the average number of divisions undergone by responding cells (division index) calculated. Top panel illustrates the gating strategy used to assess the percentage of IgG1 class-switched B-cells that have undergone 6 divisions. Histograms (bottom panel) are gated on this subset and the percentage of IgG1 class-switched B-cells quantified in the bottom right panel. Student’s t-test was performed for each condition, with n=3 in each group. * indicates p< 0.05 and ** indicates p< 0.01.
Influence of miR-210 on the B-cell transcriptome
As miR-210 is an activation-induced miRNA, we investigated the effect of miR-210 on the B-cell transcriptome by performing gene expression arrays of miR-210 mimic-transfected, miR-210 TG, and miR-210 KO activated B-cells. Overall, transcriptome changes with miR-210 overexpression were more striking than the effects of miR-210 deletion. Genes that were differentially expressed in mimic-transfected and miR-210 TG B-cells overlapped significantly (hypergeometric test, p < 2.2 × 10−16), with 1458 down-regulated and 115 up-regulated genes in common. When compared with a list of miR-210 targets predicted by 5 different algorithms(44-48), both the miR-210 mimic and miR-210 TG data sets showed significant overrepresentation of miR-210 target genes in their lists of down-regulated genes (hypergeometric p < 0.00021 and p < 0.0021 respectively), with no such enrichment in the lists of up-regulated genes. Genes that were upregulated in KO B-cells also overlapped with genes downregulated by miR-210 mimics with gene set enrichment analysis (GSEA, q = 0.22)(35). Though no single gene was strikingly dysregulated in miR-210 KO vs WT cells, there was a further overlap between genes upregulated in KO B-cells and miR-210 predicted targets (q = 0.25). Taken together, these data suggest that miR-210 functions by exerting a relatively subtle effect on a number of target genes, and this effect is amplified by over-expression.
Of the 169 predicted targets down-regulated in both miR-210 mimic and miR-210 TG B-cells (Figure 6a, Table S1b), gene ontology overrepresentation analysis demonstrated significant enrichment of genes involved in cell division, cell cycle and B-cell activation (Figure 6b), consistent with the impaired cellular proliferation observed in miR-210 over-expressing B-cells. As miRNAs are known to exert subtle effects on a large number of genes (49, 50) it is likely that the observed cellular phenotype is a consequence of the collective downregulation of several of these genes rather than a single target. Nevertheless, our array approach was validated by CD23 (FcεRII), which acts both as a low affinity IgE receptor and as an adhesion molecule that interacts with CD21 to regulate IgE production, survival of germinal centre B-cells, as well as the presentation of soluble protein antigen by B-cells to T cells (51). MiR-210 TG B2 cells exhibited lower surface expression of CD23 compared to controls (Figure 6c), and a closer examination of the CD23 mRNA sequence revealed the presence of a 7-mer miR-210 seed sequence target site CGCACAG located in exon 9 of the coding sequence. The ability of this site to mediate functional repression by miR-210 was confirmed using luciferase assays (Figure 6d).
Figure 6. Influence of miR-210 on the B-cell transcriptome.
A. 169 predicted miR-210 targets are downregulated in both miR-210 mimic transfected and miR-210 TG B cells 48 hours post-activation with LPS. Predicted targets were obtained from the MirWalk database (http://mirwalk.uni-hd.de/), and downregulated genes were defined as a fold change of >1.5, with a p-value cut-off of <0.05. Hypergeometric test p-values for overlap between miR-210 mimics and miR-210 TG B cells (p < 2.2 × 10-16); miR-210 mimic and miR-210 predicted targets (p < 0.00021); miR-210 TG and miR-210 predicted targets (p < 0.0021).
B. Gene ontology analysis demonstrates enrichment of genes involved in cell cycle, cell division and B-cell activation amongst miR-210 predicted targets downregulated in both miR-210 mimic-transfected and miR-210 TG activated B-cells.
C. CD23 expression is downregulated on miR-210 TG follicular B-cells (B220+ CD5− CD93−IgM+ CD21+).
D. Reporter luciferase assay of the miR-210 seed sequence within CD23 24 h after transfection into HeLa cells. Two-tailed Mann-Whitney test was performed, with p-values as indicated. Error bars indicate SD of technical replicates, and data representative of 2 independent experiments.
Discussion
We provide here the first description of a role for miR-210 in B-cells, as well as the first report of the effects of germline deletion of miR-210 in mice. As miR-210 was markedly up-regulated after B-cell activation, we investigated what controlled its expression, and demonstrated that miR-210 is a novel regulatory target of Oct-2. The peak of miR-210 induction occurs late in B-cell activation in comparison to other inducible miRNAs such as miR-155 (14), which is consistent with the kinetics of Oct-2 induction, as maximal levels of Oct-2 occur 8-12 hr post-stimulation with LPS (37). Oct-2 is essential in B-cells for progression through G1 phase of the cell cycle upon stimulation in vitro, whilst at the same time it contributes to the induction of miR-210 which has the opposite effect, suggesting an in-built regulatory mechanism to limit the activatory effects of Oct-2. Consistent with this, miR-210- deficient mice spontaneously develop autoantibodies with age, implying loss of an inhibitory mechanism to balance the immune response. Interestingly, mice with Dicer-deficient CD19+ B-cells also develop spontaneous autoantibodies, which was attributed to lower amounts of miR-185 resulting in accumulation of Btk and skewing of the B-cell repertoire towards more self-reactive MZB-cells (11). In contrast, miR-210 deficient mice displayed normal peripheral B-cell compartments including follicular and MZB-cells, suggesting a distinct mechanism for miRNA-mediated maintenance of tolerance, which is likely to contribute to the phenotype observed in the Dicer-deficient mice.
MiR-210 is expressed at low levels in bone marrow precursors and naïve peripheral B-cell subsets (52), but has been reported to be overexpressed in B-cell malignancies (24, 25). Here we show that ectopic over-expression of miR-210 in vivo can result in B-cell autonomous subset abnormalities including an increased number of B1a cells - a population thought to be of fetal origin that undergoes auto-renewal in the periphery(53-55). Our studies with IgHa/IgHb chimaeras suggest that their expansion in miR-210 transgenic mice may reflect a homeostatic response to the failure of division of B2 cells, as B1a cells were derived equally from WT and TG B-cell precursors, whilst the B2 cells were derived predominantly from WT precursors. B2 cells of miR-210 TG mice also had other abnormalities, including a decreased MZB-cell population and dysregulated expression of cell surface markers including CD23. Overexpression of miR-210 also resulted in cell-autonomous B-cell functional abnormalities, in particular reduced class-switched antibodies, both at baseline and in response to immunisation with a T-dependent antigen. Impaired isotype switching to IgG1 was confirmed in WT B-cells transfected with a miR-210 mimic. Our in vitro experiments demonstrate that the underlying basis of this defect is due at least in part, to reduced proliferation and cell cycle entry of miR-210 TG B2 cells, consistent with gene expression studies demonstrating the downregulation of genes involved in these processes.
That the effects of miR-210 overexpression and deletion do not result in exact inversely correlated phenotypes is not unexpected, as this has been observed for other miRNAs (56-58), and is consistent with the finding that miRNAs rarely regulate a single target, but instead are thought to exert modest effects on multiple genes in specific cellular contexts (49, 50). In our studies, the subtlety of these interactions is reflected at the transcriptome level; whilst there were no striking abnormalities in the expression of individual genes, genes upregulated in KO B-cells overlapped with genes that were downregulated by miR-210 mimics, and with miR-210 predicted target genes. Thus, in physiological contexts, the likely role of miR-210 is in fine-tuning cellular responses rather than acting as a “master regulator”, achieving the balance between pathogen clearance and autoimmunity. On the other hand, overexpression of miR-210, linked with malignancy and hypoxia, can clearly result in abnormal B-cell behaviour and impaired antibody responses. Our findings in both mouse models are consistent with miR-210 acting as an overall negative regulator of the B-cell immune response that is reminiscent of hypoxia-induced increase in miR-210 resulting in down-regulation of proliferation-related genes in xenografted cancer cell lines (20). Thus, the late induction of miR-210 may provide a more subtle, linked mechanism for control and inhibition of B-cell activation that accompanies the robust early activatory effects of Oct-2-driven transcription.
We have demonstrated that miR-210 is an Oct-2 regulated miRNA induced upon B-cell activation. Mice deficient in miR-210 develop increased spontaneous autoantibodies with age. In addition, overexpression of miR-210 expression causes abnormalities in B-cell subsets and function, in particular reducing B2 cell proliferation and isotype switching, as well as the down-regulation of genes involved in these processes. These results indicate that miR-210 provides a novel inhibitory mechanism that controls B-cells to prevent autoimmunity.
Supplementary Material
Acknowledgements
We thank members of the Smith lab for helpful discussions, Dr. A. Wood and Dr. B. Gottgens for assistance with ChIP studies, Dr. S.Howlett for providing NOD mice, and Dr. B. Fu and Dr. F. Yang from Team 70, Sanger Institute for FISH studies.
Abbreviations used in this article
- UTR
untranslated region
- AID
activation-induced cytidine deaminase
- HIF
hypoxia-inducible factor
- KO
knockout
- TG
transgenic
- NTG
non-transgenic
Footnotes
This work was supported by the Agency for Science, Technology and Research, Singapore (Y.M.), BRC Clinical Research Fellowship (V.S.), Wellcome Trust Programme Grant 06753AIA and NIHR Cambridge Biomedical Research Centre (K.G.C.S), BBSRC and MRC (E.V.), and Wellcome Trust WT098051 (A.B, H.M.P).
References
- 1.Allman D, Pillai S. Peripheral B cell subsets. Curr Opin Immunol. 2008;20:149–157. doi: 10.1016/j.coi.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol. 2008;8:22–33. doi: 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 3.Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol. 2005;5:497–508. doi: 10.1038/nri1633. [DOI] [PubMed] [Google Scholar]
- 4.Humbert PO, Corcoran LM. oct-2 gene disruption eliminates the peritoneal B-1 lymphocyte lineage and attenuates B-2 cell maturation and function. J Immunol. 1997;159:5273–5284. [PubMed] [Google Scholar]
- 5.Emslie D, D’Costa K, Hasbold J, Metcalf D, Takatsu K, Hodgkin PO, Corcoran LM. Oct2 enhances antibody-secreting cell differentiation through regulation of IL-5 receptor alpha chain expression on activated B cells. J Exp Med. 2008;205:409–421. doi: 10.1084/jem.20072049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karnowski A, Chevrier S, Belz GT, Mount A, Emslie D, D’Costa K, Tarlinton DM, Kallies A, Corcoran LM. B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1. J Exp Med. 2012;209:2049–2064. doi: 10.1084/jem.20111504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 10.Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- 11.Belver L, de Yebenes VG, Ramiro AR. MicroRNAs prevent the generation of autoreactive antibodies. Immunity. 2010;33:713–722. doi: 10.1016/j.immuni.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li J, Wan Y, Ji Q, Fang Y, Wu Y. The role of microRNAs in B-cell development and function. Cell Mol Immunol. 2013;10:107–112. doi: 10.1038/cmi.2012.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu S, Guo K, Zeng Q, Huo J, Lam KP. The RNase III enzyme Dicer is essential for germinal center B-cell formation. Blood. 2012;119:767–776. doi: 10.1182/blood-2011-05-355412. [DOI] [PubMed] [Google Scholar]
- 14.Thai TH, Calado DP, Casola S, Ansel KM, Xiao C, Xue Y, Murphy A, Frendewey D, Valenzuela D, Kutok JL, Schmidt-Supprian M, Rajewsky N, Yancopoulos G, Rao A, Rajewsky K. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 15.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KGC, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teng G, Hakimpour P, Landgraf P, Rice A, Tuschl T, Casellas R, Papavasiliou FN. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorsett Y, McBride KM, Jankovic M, Gazumyan A, Thai TH, Robbiani DF, Di Virgilio M, San-Martin BR, Heidkamp G, Schwickert TA, Eisenreich T, Rajewsky K, Nussenzweig MC. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Yebenes VG, Belver L, Pisano DG, Gonzalez S, Villasante A, Croce C, He L, Ramiro AR. miR-181b negatively regulates activation-induced cytidine deaminase in B cells. J Exp Med. 2008;205:2199–2206. doi: 10.1084/jem.20080579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuchen S, Resch W, Yamane A, Kuo N, Li Z, Chakraborty T, Wei L, Laurence A, Yasuda T, Peng S, Hu-Li J, Lu K, Dubois W, Kitamura Y, Charles N, Sun HW, Muljo S, Schwartzberg PL, Paul WE, O’Shea J, Rajewsky K, Casellas R. Regulation of MicroRNA Expression and Abundance during Lymphopoiesis. Immunity. 2010;32:828–839. doi: 10.1016/j.immuni.2010.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakada C, Tsukamoto Y, Matsuura K, Nguyen TL, Hijiya N, Uchida T, Sato F, Mimata H, Seto M, Moriyama M. Overexpression of miR-210, a downstream target of HIF1alpha, causes centrosome amplification in renal carcinoma cells. J Pathol. 2011;224:280–288. doi: 10.1002/path.2860. [DOI] [PubMed] [Google Scholar]
- 22.Fasanaro P, D’Alessandra Y, Di Stefano V, Melchionna R, Romani S, Pompilio G, Capogrossi MC, Martelli F. MicroRNA-210 modulates endothelial cell response to hypoxia and inhibits the receptor tyrosine kinase ligand Ephrin-A3. J Biol Chem. 2008;283:15878–15883. doi: 10.1074/jbc.M800731200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qi J, Qiao Y, Wang P, Li S, Zhao W, Gao C. microRNA-210 negatively regulates LPS-induced production of proinflammatory cytokines by targeting NF-kappaB1 in murine macrophages. FEBS Lett. 2012;586:1201–1207. doi: 10.1016/j.febslet.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 24.Roehle A, Hoefig KP, Repsilber D, Thorns C, Ziepert M, Wesche KO, Thiere M, Loeffler M, Klapper W, Pfreundschuh M, Matolcsy A, Bernd HW, Reiniger L, Merz H, Feller AC. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 25.Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS, Hatton CS, Harris AL. Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol. 2008;141:672–675. doi: 10.1111/j.1365-2141.2008.07077.x. [DOI] [PubMed] [Google Scholar]
- 26.Brownlie RJ, Lawlor KE, Niederer HA, Cutler AJ, Xiang Z, Clatworthy MR, Floto RA, Greaves DR, Lyons PA, Smith KGC. Distinct cell-specific control of autoimmunity and infection by FcgammaRIIb. J Exp Med. 2008;205:883–895. doi: 10.1084/jem.20072565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Higuchi T, Aiba Y, Nomura T, Matsuda J, Mochida K, Suzuki M, Kikutani H, Honjo T, Nishioka K, Tsubata T. Cutting Edge: Ectopic expression of CD40 ligand on B cells induces lupus-like autoimmune disease. J Immunol. 2002;168:9–12. doi: 10.4049/jimmunol.168.1.9. [DOI] [PubMed] [Google Scholar]
- 28.Xiao C, Calado DP, Galler G, Thai TH, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 29.Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, Barbosa-Morais NL, Teschendorff AE, Green AR, Ellis IO, Tavare S, Caldas C, Miska EA. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8:R214. doi: 10.1186/gb-2007-8-10-r214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Montaner D, Tarraga J, Huerta-Cepas J, Burguet J, Vaquerizas JM, Conde L, Minguez P, Vera J, Mukherjee S, Valls J, Pujana MA, Alloza E, Herrero J, Al-Shahrour F, Dopazo J. Next station in microarray data analysis: GEPAS. Nucleic Acids Res. 2006;34:W486–491. doi: 10.1093/nar/gkl197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Storey JD, Tibshirani R. Statistical methods for identifying differentially expressed genes in DNA microarrays. Methods Mol Biol. 2003;224:149–157. doi: 10.1385/1-59259-364-X:149. [DOI] [PubMed] [Google Scholar]
- 32.Huber W, von Heydebreck A, Sultmann H, Poustka A, Vingron M. Variance stabilization applied to microarray data calibration and to the quantification of differential expression. Bioinformatics. 2002;18(Suppl 1):S96–104. doi: 10.1093/bioinformatics/18.suppl_1.s96. [DOI] [PubMed] [Google Scholar]
- 33.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- 34.Smyth GK. Limma: linear models for microarray data. In: Gentleman VCR, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer; New York: 2005. pp. 397–420. [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubart K, Massa S, Schubart D, Corcoran LM, Rolink AG, Matthias P. B cell development and immunoglobulin gene transcription in the absence of Oct-2 and OBF-1. Nat Immunol. 2001;2:69–74. doi: 10.1038/83190. [DOI] [PubMed] [Google Scholar]
- 37.Corcoran LM, Karvelas M. Oct-2 is required early in T cell-independent B cell activation for G1 progression and for proliferation. Immunity. 1994;1:635–645. doi: 10.1016/1074-7613(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 38.Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berland R, Wortis HH. Origins and functions of B-1 cells with notes on the role of CD5. Annu Rev Immunol. 2002;20:253–300. doi: 10.1146/annurev.immunol.20.100301.064833. [DOI] [PubMed] [Google Scholar]
- 40.Keenan RA, De Riva A, Corleis B, Hepburn L, Licence S, Winkler TH, Martensson IL. Censoring of autoreactive B cell development by the pre-B cell receptor. Science. 2008;321:696–699. doi: 10.1126/science.1157533. [DOI] [PubMed] [Google Scholar]
- 41.Strasser A, Harris AW, Vaux DL, Webb E, Bath ML, Adams JM, Cory S. Abnormalities of the immune system induced by dysregulated bcl-2 expression in transgenic mice. Curr Top Microbiol Immunol. 1990;166:175–181. doi: 10.1007/978-3-642-75889-8_22. [DOI] [PubMed] [Google Scholar]
- 42.Hodgkin PD, Lee JH, Lyons AB. B cell differentiation and isotype switching is related to division cycle number. J Exp Med. 1996;184:277–281. doi: 10.1084/jem.184.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasbold J, Lyons AB, Kehry MR, Hodgkin PD. Cell division number regulates IgG1 and IgE switching of B cells following stimulation by CD40 ligand and IL-4. Eur J Immunol. 1998;28:1040–1051. doi: 10.1002/(SICI)1521-4141(199803)28:03<1040::AID-IMMU1040>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 44.Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 45.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, Thomson AM, Lim B, Rigoutsos I. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 46.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 51.Bonnefoy JY, Lecoanet-Henchoz S, Aubry JP, Gauchat JF, Graber P. CD23 and B-cell activation. Curr Opin Immunol. 1995;7:355–359. doi: 10.1016/0952-7915(95)80110-3. [DOI] [PubMed] [Google Scholar]
- 52.Spierings DC, McGoldrick D, Hamilton-Easton AM, Neale G, Murchison EP, Hannon GJ, Green DR, Withoff S. Ordered progression of stage-specific miRNA profiles in the mouse B2 B-cell lineage. Blood. 2011;117:5340–5349. doi: 10.1182/blood-2010-10-316034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hardy RR, Hayakawa K. A developmental switch in B lymphopoiesis. Proc Natl Acad Sci U S A. 1991;88:11550–11554. doi: 10.1073/pnas.88.24.11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hayakawa K, Hardy RR, Herzenberg LA, Herzenberg LA. Progenitors for Ly-1 B cells are distinct from progenitors for other B cells. J Exp Med. 1985;161:1554–1568. doi: 10.1084/jem.161.6.1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kantor AB, Stall AM, Adams S, Watanabe K, Herzenberg LA. De novo development and self-replenishment of B cells. Int Immunol. 1995;7:55–68. doi: 10.1093/intimm/7.1.55. [DOI] [PubMed] [Google Scholar]
- 56.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, Feldmann G, Yamakuchi M, Ferlito M, Lowenstein CJ, Arking DE, Beer MA, Maitra A, Mendell JT. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL, Linsley PS, Chen C, Lowe SW, Cleary MA, Hannon GJ. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Concepcion CP, Han YC, Mu P, Bonetti C, Yao E, D’Andrea A, Vidigal JA, Maughan WP, Ogrodowski P, Ventura A. Intact p53-dependent responses in miR-34-deficient mice. PLoS Genet. 2012;8:e1002797. doi: 10.1371/journal.pgen.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.