Abstract
Background:
Pancreatic cancer has poor prognosis by surgical and chemotherapy when it is diagnosed, so other anti-cancerous assistant therapeutic drugs are suggested e.g. epigenetic reversal of tumor-suppressor genes on promoter hypermethylation. 5-Aza-CdR is a nucleoside analog of DNMTi but it has long-term cytotoxicity effects. This study compares the anticancer effect of 5-Aza-CdR and Disulfiram potencies on PANC-1 cell line and up-regulation of p21.
Materials and Methods:
PANC-1 cell line was cultured in DMEM high glucose and treated by 5-Aza-CdR with 5 and 10 μM concentration for four days and 13 μM DSF (Diulfiram) for 24 hours. MS-PCR and RT-PCR were carried out to detect the methylation pattern and estimate the mRNA expression of RASSF1A and p21 in PANC-1.
Result:
MS-PCR demonstrated partial unmethylation after treatment with 5-Aza-CdR while there was no unmethylated band after DSF treatment. RT-PCR showed significant differences between re-expression of RASSF1A before and after treatment with 10 μM 5-Aza-CdR (P < 0.01) but not after treatment with 13 μM DSF (P > 0.05). The significant correlation was observed between RASSF1A re-expression and p21 up-regulation before and after treatment with 10 μM 5-Aza-CdR (P < 0.01) but not after treatment with 13 μM DSF (P > 0.05), while p21 up-regulation was significantly higher after DSF treatment (P < 0.01).
Conclusion:
Our findings indicated that 5-Aza-CdR induces the re-expression of RASSF1A and p21 up-regulation in PANC-1. DSF showed no epigenetic reversion while it affected p21 up-regulation.
Keywords: 5-Aza-CdR, Disulfiram, DNMT inhibitor, epigenetic, p21, PANC-1, RASSF1A
INTRODUCTION
Pancreatic cancer is the 4th leading cause of cancer death in the US.[1,2] It is called “silent killer” as it has no clear symptoms neither at the early stage nor the late. Therefore, pancreatic cancer is often not diagnosed until it is advanced.[3,4] Surgery represents the only curative treatment, but due to late diagnosis, only minorities (10-20%) of patients are willing to surgical intervention. Owing to the high pancreatic cancer recurrence, chemotherapy with or without radiotherapy is performed but unfortunately they have a poor prognosis and less than 5% of them live more than 5 years.[5]
Research in the past 10 years indicates that epigenetic alterations play a critical role in tumor genesis. Epigenetic gene silencing through promoter hypermethylation is a common and early event in the pathogenesis of many solid tumors.[6,7] The process of DNA methylation involves the transfer a methyl group from S-adenosyl-L-methionine (SAM) to C5 position in CpG dinucleotides. The catalyzing enzymes for this procedure are known as DNMTs which DNMT1 is considered to maintain DNMT by copying DNA methylation from parental strand to newly synthesized strand.[8,9] Methylated gene detection can be used for early diagnosis of cancer and to monitor the response to chemotherapy, epigenetic events can re-exchange so the DNA methyltransferase inhibitors can be used as assistant therapeutic anti-cancerous drugs in low doses and have clinical activity against some tumors.[10,11,12]
There are two kinds of DNMT inhibitors, the nucleoside and non-nucleoside analogous demethylating agents. The nucleoside analogues such as 5-Aza-CR (5-Azacytidine) and 5-Aza-CdR (5-aza-deoxycytidine) are produced from cytosine by a nitrogen substitution at the fifth carbon. During replication, they are replaced instead of cytosine and trap DNMT enzyme, so there is decrease in cytosine methylation by reducing the cell DNMTs enzymes after treatment.[9] 5-Aza-CdR is approved by FDA for MDS (myelodysplastic syndromes) treatment, but it has some cytotoxic effects as a result of high concentration required for cytosine demethylation. Furthermore, the nucleoside analogues perform their function by trapping DNMT (DNA methyl- transferases enzyme) and it doesn’t effect on consequences of demethylation. They also become inactivated by cytosine deamination in neutral situation and are unstable both in-vitro and in-vivo.[8]
DSF has a long history of alcoholic abuse treatment.[13] It contains strong thiol reaction functional group which attacks the thiol group of cytosine and limits the catalytic mechanism of DNMTs on the C6 position of cytosine. A recent screen of more than 3,000 patients revealed that DSF can induce prostate cancer cell death. Furthermore, DSF shows mild side-effects and is well-tolerated in alcohol abuse therapy so makes it a new candidate for repurposing.[14]
RASSF1A is one of the tumor suppressor genes, which is a key cell cycle-related gene at the G-S checkpoint by modulation of cycline D1 levels and inhibition of JNK pathway. Its inactivation is occurred by gene deletion, point mutation and inappropriate promoter hypermethylation, but epigenetic inactivation is most common in different solid tumors and cell lines such as lung, thyroid, liver, kidney, breast cancers, cervix adenocarcinoma, prostate carcinomas, pancreatic adenocarcinomas, pancreatic neuroendocrine tumor, pancreatitis and pancreatic cell line.[15,16,17] Furthermore, RASSF1A may upgrade CDKI (cycline-dependent kinase inhibitor) P21.[18] P21 Cip1/Waf1 is a significant production of Ras/Raf activation. P21 expression is occurred at the transcriptional and posttranscriptional level, for instance at transcription level, it is expressed by p53-dependent and independent mechanisms while its expression during senescence is induced by Ras/Raf and make cell cycle arrest in primary fibroblasts and keratinocytes.[19]
As pancreatic cancer is incurable at the time of diagnosis, other anti-cancer assistant therapeutic are necessary. On the other hand little is known about the anti-tumor mechanisms of DSF, so the present study investigates the comparison of DSF and 5-Aza-CdR effects on re-expression of RASSF1A in pancreatic cancer cell line. Subsequently up regulation of p21 gene was considered.
MATERIALS AND METHODS
Cell line and cell culture
Human PANC-1 cell line was purchased from national cell bank Pasteur Institute of Iran. The cells were cultured in high glucose DMEM (Sigma USA) supplemented with 10% fetal bovine serum (FBS) (Sigma USA) and 1% penicillin-streptomycin (Sigma USA) at 37oC and in humidified atmosphere containing 5% CO2. After 80% confluence in T25 culture flask, 105 cells were transferred to 24 well cultural plates. Drug treatment was done one day after seeding.
Drug treatment
5-Aza-CdR treatment
5-Aza-CdR was purchased from Sigma, USA. Its Stock solution (50 mM) was prepared by dissolving in DMSO, and stored at –80°C. The stock was diluted in cell culture media at different concentration (0, 1, 5, 10, 15, 20, 25, and 30 μM) at experience day. 5 and 10 μm for 4 days were chosen according to Stresemann 2006. MTT survival assay was carried out for evaluating cell viability (group 1. Untreated, 5 and 10 μM).[20]
DSF treatment
The DSF was purchased (Sigma USA). DSF stock solution (50 mM) was prepared by dissolving in DMSO and stored at –20°C. The final concentration of DSF drug was obtained by the IC50 assay. 105 PANC-1 cells were transferred in each well of 24 -well cell culture microplate and treated with different concentrations of DSF (0, 2.5, 5, 7.5, 10, 15, 20, 25, 30 μM) for 24 hours. Then MTT survival assay was carried out for evaluating cell viability (group 2: Untreated, 13 μm).[21]
MTT assay
To examine viability of PANC-1 cells in the presence of DSF, MTT (3-[4,5-dimethythiaziazol-2yl]-2,5-diphenyl tetrazoliumbromide), was dissolved in PBS at 5 mg/ml. The stock solution was added to the culture medium at a dilution 1:10. The plates were incubated at 370C for 4 hours. The medium was then aspirated and 400 μl of DMSO was added to extract the MTT formazan and the absorbance of each well was detected by microplate reader (Hiperion MPR 4, Germany) at the wavelength of 540 nm.[21]
DNA extraction, bisulfite treatment, and methylation-specific PCR (MSP)
Genomic DNA was isolated by primepreptm Genomic DNA isolation Kit (GeNet Bio, Korea) according to its protocol. DNA was treated by sodium bisulfate using epitect bisulfate kit (Qiagen, Germany), according to its instruction. The PCR reaction were carried out in thermal cycler EP-gradien PCR (Ependorf, Germany): 94oC for 5 min followed by 40 cycles of 94oC for 30s, 60oC for 30s and 72oC for 30s and the final extension 72oC for 5 min. PCR products were transferred to electrophoresis on a 2% agarose gel and visualized with staining by DNA green viewer. Methylated and unmethylated RASSF1A primers were
M1 (forward) 5’-GTGTTAACGCGTTGCGTATC-3’; M (reverse) 5’AACCCCGCGAACTAAAAACGA-3’; U1 (forward) 5’-TTTGGTTGGAGTGTGTTAATGTG-3’; U2 (reverse) 5’-CAAACCCCACAAACTAAAAACAA- 3’.[22]
RNA extraction and Real-time PCR
Total RNA was isolated by RNeasy mini kit (Qiagen, USA). RNA samples were treated by RNase free DNase (Qiagen, USA) to eliminate the genomic DNA. The RNA concentration was measured using a Biophotometer (Eppendorf Germany). 5 μl of total RNA was reverse-transcribed to cDNA. The cDNA construction was performed by RevertAidTM Kit (Fermentas EU) according to the protocols. Real-time RT-PCR was carried out by the Maxima SYBR Green Rox qPCR master mix kit (Fermentase EU). The Real-Time primers were designed using AlleleID software (Primer Biosoft) which generated the following sequences:
RASSF1A (Forward -TCATCTGGGGCGTCGTG, reverse- CGTTCGTGTCCCGCTCC),
P21 (forward- GACCAGCATGACAGATTTCTACCA, reverse-AACTGAGACTAAGGCAGAAGATG), ACTB (forward- GTTGTCGACGACGAGCG, reverse- GCACAGAGCCTCGCCTT).
Real-time PCR reactions were performed using the Steponeplus quantitative real-time PCR detection system (Applied Biosystem USA). The PCR amplification conditions consisted of 10 min at 95oC followed by 40 cycles of denaturation step at 95oC for 15 sec and annealing and extension for 1 min at 60oC. The expression level of each target gene was calculated as 2-ΔΔCt, as previously described. Briefly the epression mRNA levels of RASSF1-A, p21 and Bax were calculated by determining a ratio between the amount of them and endogenous control (ACT B). Melting curve analysis (60oC → 95oC increment at 0.3oC/S) was used to determine melting temperature of specific amplification products and primer. These experiments were carried out in triplicate and independently repeated at least three times.[21,22]
Statistical analysis
All the quantitative data were presented as the mean ± standard deviations. Kolmogorov-Simonov test was used for assessing normal distribution of variables. One-way-analysis of variance (ANOVA) with LSD post hoc test was performed to determine statistical significance among different groups by using SPSS software package16.0. Significance was accepted at (P < 0.05).
RESULT
5-Aza-CdR
In this study, different concentration of 5-Aza-CdR were tested during first, second and third days but no death result was shown by MTT. At the fourth days of treatment, it demonstrated around 50% cell death at the concentration of 5 μM (55%) and 10 μM (45%). It was accordance to Stresemann 2006 [Figures 1a and 2b].
Figure 1.
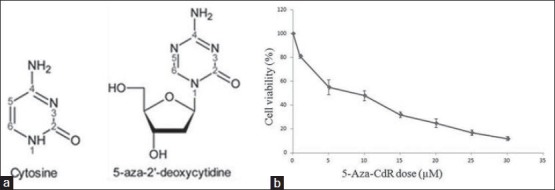
(a) The structures of cytosine and 5-aza-2’-deoxycytine derived from cytosine. (b) Effect of 5-Aza-CdR in PANC-1 cell lines as individual dose-response curves; cell lines with monophasic curve
Figure 2.
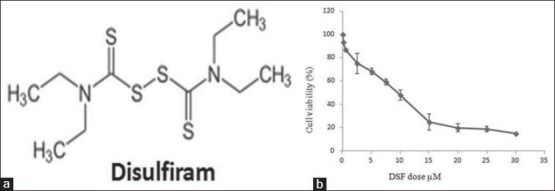
(a) Structure of disulfiram. (b) Effect of disulfiram in PANC-1 cell lines as individual dose-response curves; cell lines with monophasic curve
DSF
As cell death was observed in the first day of treatment IC50 value in PANC-1 was determined after 24 hours of treatment by DSF. The reduction of cell living was dependent on DSF concentration as shown by the IC50 index (half-maximal inhibitory concentration). The IC50 values for the DSF were established and the results showed that the essential DSF concentration to achieve the IC50 in PANC-1 cells at 24 hours was 13 μM [Figures 2a and b].
MS-PCR
The result of MS-PCR showed that in the control (untreated) sample methylated DNA band was amplified by using methylated RASSF1A primers and no DNA band was detected. Treatment by 5-Aza-CdR in 10 μM for 4 days showed partially methylated compared to nontreated PANC-1. (Partially methylated showed both methylated and un-methylated bands because some of the RASSF1A promoters are converted into demethylation and become re-expressed). Treatment by 13 μM DSF for 24 hours showed no unmethylated band in MS-PCR [Figures 3a and b].
Figure 3.
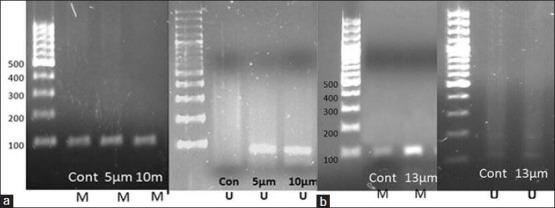
(a) Detection of methylation and unmethylation bands in promoter region of RASSF1A by MS-PCR in 5-Aza-CdR treated PANC-1. (b) Detection of methylation band in promoter region of RASSF1A by MS-PCR in DSF treated PANC-1. *M: Product by using methylated primers, *U: Product by using unmethylated primers. The M and U primer sets generated 94-bp and 108-bp products, respectively
Real-time PCR
Real-time PCR was used to evaluate RASSF1A re-expression induced by 5-Aza-CdR and DSF in mentioned concentration, and following p21 up-regulation afterward. The result demonstrated that 10 μM 5-Aza-CdR treatment (for 4 days) induced RASSF1A re-expression significantly (P < 0.01) but not in 5 μM concentration (P > 0.05). Subsequently, P21 expression was significantly up regulated in the 10 μM 5-Aza-CdR treated PANC-1 compared to control (P < 0.01), but there wasn’t significant up regulation with 5 μM treated one (P > 0.05).
On one hand DSF treatment for 24 hours didn’t show any significant alteration of RASSF1A re-expression in 13 μM (P > 0.05) but on the other hand the significant increase of p21 expression was demonstrated in 13 μM (P < 0.01) [Figures 4a and b].
Figure 4.
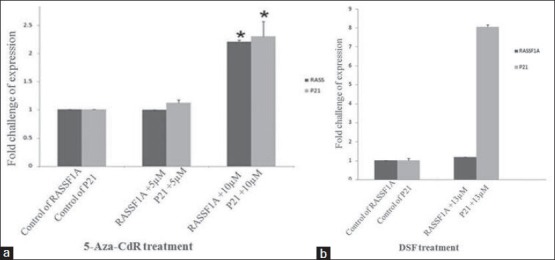
(a) 5-Aza-CdR treatment on PANC-1. 10 μM 5-Aza-CdR made re-expression of RASSF1A and up-regulation of p21 (P < 0.01). (b) 13 μM DSF treated PANC-1 for 24 h showed no re-expression of RASSF1A (P > 0.05) but made the up-regulation of p21 (P < 0.01)
DISCUSSION
We have studied the potency and functional mechanism of 5-Aza-CdR and DSF on re-expression of RASSF1A at mentioned concentrations on PANC-1. Our result showed that 5-Aza-CdR as a nucleoside DNMTi can demethylate the RASSF1A promoter hypermethylation while there was no epigenetic effect by DSF.
RASSF1A is one of the tumor suppressor genes and its inactivation induces tumor development. Many studies demonstrated RASSF1A expression in all normal tissues but silencing in different cancers.[23,24] Some studies have showed that RASSF1A performs its duties without any clear enzymatic activity but may serve as a scaffold for activation of other signaling complexes.[17,25,26]
In this study, PANC-1 was chosen as it contains RASSF1A promoter hypermethylation and P53 mutation. Stresemann demonstrated 3-day incubation is sufficient for epigenetic reversion.[20] After treatment by 5-Aza-CdR the MSP, the result demonstrated partially unmethylated band which was confirmed by RASSF1A re-expression by Real time (P < 0.01). Present data were verified by different studies such as Harada in 2002 and Shen in 2008 who displayed RASSF1A promoter hypermethylation is critical point of cancer pathogenesis.[27,28] Consequently, reversing epigenetic changes has been considered as a valuable therapeutic approach. 5-Aza-CdR has been used in different fields such as mammalian development, cell differentiation, cancer therapy, and reverse of cancer malignant phenotype as a nucleoside DNMT inhibitor.[8,29,30,31] Similar approaches were carried out by Shivakumar in 2002, Agathangglou in 2005, and Zuo in 2007 in neuroblastoma, rhabdomyosarcoma, and retinoblastoma cell lines.[16,32,33] Furthermore, 5-azaC and 5-aza-dC have been approved by the FDA for cancers and other solid malignancies treatment.[34]
Since 5-Aza-CdR has been used for a long term and incorporated to DNA during S-phase, so it is toxic and carcinogenesis. It can inhibit DNMT by permanent trapping of DNA and DNA mutations.[35] Clinical trials proved that nucleoside analogues have poor clinical activity in late stage of solid tumor malignancies. Hence, small-molecule and non-nucleoside DNMTs inhibitors are safer for prolonged epigenetic therapy.[18] DSF has strong thiol-reactive functional groups; it has a long history of alcohol abuse treatment but its anti-cancer effect is novel. DSF is an oral agent so is more suitable than the existing nucleoside analogous.[21]
In the present study, RASSF1A MSP result showed poor epigenetic reversion and there was no un-methylated band after treatment and real time result demonstrated no RASSF1A re-expression (P > 0.05) while cell death was observed.
As it is known, cancer development is closely related to the deregulation of G1/S checkpoint of cell cycle and one of the important roles of RASSF1A, as an important tumor suppressor gene is cell cycle arrest; moreover, P21 is a key protein of CKI and is expressed after DNA damage and p53 induction.[18,36] So this paper investigated p21 up-regulation after 5-Aza-CdR treatment. The results showed significant correlation between P21 and RASSF1A re-expression (P < 0.01). Present data were confirmed by Thalar in 2009. They found that RASSF1A induces cell death up regulation of p21 in vitro and in vivo due to age. Additionally, they found that re-expression of RASSF1A enhances nuclear accumulation of p21.[19]
Although DSF demethylating effect is weaker than 5-Aza-CdR to detect the unmethylation band by MS-PCR, it makes p21 up-regulation in other non-epigenetic ways. This result has controversy to Lin in 2010 who demonstrated DSF epigenetic therapy in prostate cancer cell line but confirmed by previous studies that pronounced non-nucleoside DNMTi has lower potentially of DNA demethylation effect.[37,38] It was also in accordance with Lloyd, Roper and Sewing who demonstrated that arrested fibroblasts and keratinocytes had high expression of p21.[39,40,41] Also, Latha Shivakumar et al. showed that in H1299 cells, p21 can contribute to negative regulation of CDK2 so it helps to control cell cycle in H1299 cells.[16] Furthermore, present results were accordance with Wangin in 2003, Chen in 2006, Wickstrom in 2007, and Cho in 2007 who showed that DSF eliminates various cancer cell lines such as melanoma, leukemia, small lung cell cancer, osteosarcoma, cervical adenocarcinoma, and colorectal adenocarcinoma cell lines through non-epigenetic mechanism.[42,43,44,45]
In conclusion, we can say that although DSF did not show the potentially of DNA demethylation effect, its multi-potential antineoplastic effects induce tumor-specific toxicity and favorable safety which is worth to its prolonged use compared with the other group in epigenetic cancer therapy.
ACKNOWLEDGMENT
The authers appreciate Dr. Ali Valiani, Dr. Mohammad Reza Salahshoor, Dr. Fatemeh sadat Mostafavi and Mrs. Maryam Aliakbari for their sincere help. Without their contributions, this study could not be performed.
Footnotes
Source of Support: Nil
Conflict of Interest: None declared.
REFERENCES
- 1.Hariharan D, Saied A, Kocher HM. Analysis of mortality rates for pancreatic cancer across the world. HPB (Oxford) 2008;10:58–62. doi: 10.1080/13651820701883148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li C, Heidt DG, Dalerba P, Burant CF, Zhang L, Adsay V, et al. Identification of pancreatic cancer stem cells. Cancer Res. 2007;67:1030–7. doi: 10.1158/0008-5472.CAN-06-2030. [DOI] [PubMed] [Google Scholar]
- 3.Hruban RH, Brune K, Fukushima N, Maitra A. Pancreatic intraepithelial neoplasia. Pancreatic Cancer. 2008;378:41–51. [Google Scholar]
- 4.Dammann R, Schagdarsurengin U, Liu L, Otto N, Gimm O, Dralle H, et al. Frequent RASSF1A promoter hypermethylation and K-ras mutations in pancreatic carcinoma. Oncogene. 2003;22:3806–12. doi: 10.1038/sj.onc.1206582. [DOI] [PubMed] [Google Scholar]
- 5.Arlt A, Müerköster SS, Schäfer H. Targeting apoptosis pathways in pancreatic cancer. Cancer Lett. 2013;332:346–58. doi: 10.1016/j.canlet.2010.10.015. [DOI] [PubMed] [Google Scholar]
- 6.Cohen Y, Singer G, Lavie O, Dong SM, Beller U, Sidransky D. The RASSF1A tumor suppressor gene is commonly inactivated in adenocarcinoma of the uterine cervix. Clin Cancer Res. 2003;9:2981–4. [PubMed] [Google Scholar]
- 7.Abouzeid HE, Kassem AM, Abdel Wahab AH, El-mezayen HA, Sharad H, Abdel Rahman S. Promoter hypermethylation of RASSF1A, MGMT, and HIC-1 genes in benign and malignant colorectal tumors. Tumour Biol. 2011;32:845–52. doi: 10.1007/s13277-011-0156-7. [DOI] [PubMed] [Google Scholar]
- 8.Fandy TE. Development of DNA methyltransferase inhibitors for the treatment of neoplastic diseases. Curr Med Chem. 2009;16:2075–85. doi: 10.2174/092986709788612738. [DOI] [PubMed] [Google Scholar]
- 9.Cortez CC, Jones PA. Chromatin, cancer and drug therapies. Mutat Res. 2008;647:44–51. doi: 10.1016/j.mrfmmm.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 11.Momparler RL. Cancer epigenetics. Oncogene. 2003;22:6479–83. doi: 10.1038/sj.onc.1206774. [DOI] [PubMed] [Google Scholar]
- 12.Billam M, Sobolewski MD, Davidson NE. Effects of a novel DNA methyltransferase inhibitor zebularine on human breast cancer cells. Breast Cancer Res Treat. 2010;120:581–92. doi: 10.1007/s10549-009-0420-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barth KS, Malcolm RJ. Disulfiram: An old therapeutic with new applications. CNS Neurol Disord Drug Targets. 2010;9:5–12. doi: 10.2174/187152710790966678. [DOI] [PubMed] [Google Scholar]
- 14.Iljin K, Ketola K, Vainio P, Halonen P, Kohonen P, Fey V, et al. High-throughput cell-based screening of 4910 known drugs and drug-like small molecules identifies disulfiram as an inhibitor of prostate cancer cell growth. Clin Cancer Res. 2009;15:6070–8. doi: 10.1158/1078-0432.CCR-09-1035. [DOI] [PubMed] [Google Scholar]
- 15.Donninger H, Vos MD, Clark GJ. The RASSF1A tumor suppressor. J Cell Sci. 2007;120:3163–72. doi: 10.1242/jcs.010389. [DOI] [PubMed] [Google Scholar]
- 16.Shivakumar L, Minna J, Sakamaki T, Pestell R, White MA. The RASSF1A tumor suppressor blocks cell cycle progression and inhibits cyclin D1 accumulation. Mol Cell Biol. 2002;22:4309–18. doi: 10.1128/MCB.22.12.4309-4318.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sherwood V, Recino A, Jeffries A, Ward A, Chalmers AD. The N-terminal RASSF family: A new group of Ras-association-domain-containing proteins, with emerging links to cancer formation. Biochem J. 2009;425:303–11. doi: 10.1042/BJ20091318. [DOI] [PubMed] [Google Scholar]
- 18.Kierszenbaum AL. E-Book. 2nd ed 2007. Histology and Cell Biology: An Introduction to Pathology. [Google Scholar]
- 19.Thaler S, Hähnel PS, Schad A, Dammann R, Schuler M. RASSF1A mediates p21Cip1/Waf1-dependent cell cycle arrest and senescence through modulation of the Raf-MEK-ERK pathway and inhibition of Akt. Cancer Res. 2009;69:1748–57. doi: 10.1158/0008-5472.CAN-08-1377. [DOI] [PubMed] [Google Scholar]
- 20.Stresemann C, Brueckner B, Musch T, Stopper H, Lyko F. Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res. 2006;66:2794–800. doi: 10.1158/0008-5472.CAN-05-2821. [DOI] [PubMed] [Google Scholar]
- 21.Lin J, Haffner MC, Zhang Y, Lee BH, Brennen WN, Britton J, et al. Disulfiram is a DNA demethylating agent and inhibits prostate cancer cell growth. Prostate. 2011;71:333–43. doi: 10.1002/pros.21247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avramouli A, Tsochas S, Mandala E, Katodritou E, Ioannou M, Ritis K, et al. Methylation status of RASSF1A in patients with chronic myeloid leukemia. Leuk Res. 2009;33:1130–2. doi: 10.1016/j.leukres.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Yang B, House MG, Guo M, Herman JG, Clark DP. Promoter methylation profiles of tumor suppressor genes in intrahepatic and extrahepatic cholangiocarcinoma. Mod Pathol. 2005;18:412–20. doi: 10.1038/modpathol.3800287. [DOI] [PubMed] [Google Scholar]
- 24.Schagdarsurengin U, Wilkens L, Steinemann D, Flemming P, Kreipe HH, Pfeifer GP, et al. Frequent epigenetic inactivation of the RASSF1A gene in hepatocellular carcinoma. Oncogene. 2003;22:1866–71. doi: 10.1038/sj.onc.1206338. [DOI] [PubMed] [Google Scholar]
- 25.Vos MD, Ellis CA, Bell A, Birrer MJ, Clark GJ. Ras uses the novel tumor suppressor RASSF1 as an effector to mediate apoptosis. Science's STKE. 2000;275:35669. doi: 10.1074/jbc.C000463200. [DOI] [PubMed] [Google Scholar]
- 26.Lund AH, van Lohuizen M. Epigenetics and cancer. Genes Dev. 2004;18:2315–35. doi: 10.1101/gad.1232504. [DOI] [PubMed] [Google Scholar]
- 27.Shen WJ, Dai DQ, Teng Y, Liu HB. Regulation of demethylation and re-expression of RASSF1A gene in gastric cancer cell lines by combined treatment of 5-Aza-CdR and NaB. World J Gastroenterol. 2008;14:595–600. doi: 10.3748/wjg.14.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada K, Toyooka S, Maitra A, Maruyama R, Toyooka KO, Timmons CF, et al. Aberrant promoter methylation and silencing of the RASSF1A gene in pediatric tumors and cell lines. Oncogene. 2002;21:4345–9. doi: 10.1038/sj.onc.1205446. [DOI] [PubMed] [Google Scholar]
- 29.Brueckner B, Kuck D, Lyko F. DNA methyltransferase inhibitors for cancer therapy. Cancer J. 2007;13:17–22. doi: 10.1097/PPO.0b013e31803c7245. [DOI] [PubMed] [Google Scholar]
- 30.Qiu H, Yashiro M, Shinto O, Matsuzaki T, Hirakawa K. DNA methyltransferase inhibitor 5-aza-CdR enhances the radiosensitivity of gastric cancer cells. Cancer Sci. 2009;100:181–8. doi: 10.1111/j.1349-7006.2008.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chiam K, Centenera MM, Butler LM, Tilley WD, Bianco-Miotto T. GSTP1 DNA methylation and expression status is indicative of 5-aza-2’-deoxycytidine efficacy in human prostate cancer cells. PLoS One. 2011;6:e25634. doi: 10.1371/journal.pone.0025634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Agathanggelou A, Cooper WN, Latif F. Role of the Ras-association domain family 1 tumor suppressor gene in human cancers. Cancer Res. 2005;65:3497–508. doi: 10.1158/0008-5472.CAN-04-4088. [DOI] [PubMed] [Google Scholar]
- 33.Zuo S, Chen Y, Xu L, Tang Q, Zou S. Re-expression of RASSF1A by 5-Aza-CdR induced demethylation of the promoter region in human biliary tract carcinoma cells. J Huazhong Univ Sci Technolog Med Sci. 2007;27:281–4. doi: 10.1007/s11596-007-0316-6. [DOI] [PubMed] [Google Scholar]
- 34.Yu JN, Xue CY, Wang XG, Lin F, Liu CY, Lu FZ, et al. 5-AZA-2’-deoxycytidine (5-AZA-CdR) leads to down-regulation of Dnmt1o and gene expression in preimplantation mouse embryos. Zygote. 2009;17:137–45. doi: 10.1017/S0967199408005169. [DOI] [PubMed] [Google Scholar]
- 35.Christman JK. 5-Azacytidine and 5-aza-2’-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–95. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 36.Rui L, Xue WJ, Li P, Wang ZW, Wang P, Li HX. [Inhibitory effect of wild-type RASSF1A gene expression on proliferation of hepatocellular carcinoma QGY-7703 cells] Ai Zheng. 2008;27:924–8. [PubMed] [Google Scholar]
- 37.Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005;97:1498–506. doi: 10.1093/jnci/dji311. [DOI] [PubMed] [Google Scholar]
- 38.Lin X, Asgari K, Putzi MJ, Gage WR, Yu X, Cornblatt BS, et al. Reversal of GSTP1 CpG island hypermethylation and reactivation of pi-class glutathione S-transferase (GSTP1) expression in human prostate cancer cells by treatment with procainamide. Cancer Res. 2001;61:8611–6. [PubMed] [Google Scholar]
- 39.Lloyd AC, Obermüller F, Staddon S, Barth CF, McMahon M, Land H. Cooperating oncogenes converge to regulate cyclin/cdk complexes. Genes Dev. 1997;11:663–77. doi: 10.1101/gad.11.5.663. [DOI] [PubMed] [Google Scholar]
- 40.Roper E, Weinberg W, Watt FM, Land H. p19ARF-independent induction of p53 and cell cycle arrest by Raf in murine keratinocytes. EMBO Rep. 2001;2:145–50. doi: 10.1093/embo-reports/kve020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sewing A, Wiseman B, Lloyd AC, Land H. High-intensity Raf signal causes cell cycle arrest mediated by p21Cip1. Mol Cell Biol. 1997;17:5588–97. doi: 10.1128/mcb.17.9.5588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wickström M, Danielsson K, Rickardson L, Gullbo J, Nygren P, Isaksson A, et al. Pharmacological profiling of disulfiram using human tumor cell lines and human tumor cells from patients. Biochem Pharmacol. 2007;73:25–33. doi: 10.1016/j.bcp.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 43.Chen D, Cui QC, Yang H, Dou QP. Disulfiram, a clinically used anti-alcoholism drug and copper-binding agent, induces apoptotic cell death in breast cancer cultures and xenografts via inhibition of the proteasome activity. Cancer Res. 2006;66:10425–33. doi: 10.1158/0008-5472.CAN-06-2126. [DOI] [PubMed] [Google Scholar]
- 44.Wang W, McLeod HL, Cassidy J. Disulfiram-mediated inhibition of NF-kappaB activity enhances cytotoxicity of 5-fluorouracil in human colorectal cancer cell lines. Int J Cancer. 2003;104:504–11. doi: 10.1002/ijc.10972. [DOI] [PubMed] [Google Scholar]
- 45.Cho HJ, Lee TS, Park JB, Park KK, Choe JY, Sin DI, et al. Disulfiram suppresses invasive ability of osteosarcoma cells via the inhibition of MMP-2 and MMP-9 expression. J Biochem Mol Biol. 2007;40:1069–76. doi: 10.5483/bmbrep.2007.40.6.1069. [DOI] [PubMed] [Google Scholar]


