Abstract
The spliceosome, an assembly of snRNAs and proteins, catalyzes the removal of introns from premessenger RNAs. A new study identifies specific phosphates in the U2–U6 snRNA complex that position two catalytic metals. Remarkably, these correspond precisely to metal-binding phosphates in a homologous structure of Group II self-splicing introns, long proposed to be the ribozyme progenitor of spliceosome.
Widespread splicing of mRNA precursors is a signature feature of eukaryotes. The responsible enzyme is the spliceosome, consisting of five small nuclear RNAs and upwards of 100 proteins. Unlike the other two engines of eukaryotic gene expression, RNA polymerase II and the ribosome, there is no analogous apparatus in prokaryotes. However, chemical reactions like those catalyzed by the spliceosome are performed in prokaryotes by RNAs, termed Group II self-splicing introns or ribozymes. These RNAs can reverse splice into DNA where an intron-encoded reverse transcriptase converts them into DNA, effectively allowing the introns to act as mobile genetic elements. The reverse transcriptase also functions as a “maturase” that stabilizes the ribozyme to enhance splicing catalysis. Recent work from the Staley and Piccirilli groups establishes that the eukaryotic spliceosome harbors a Group II ribozyme-like active site (Fica et al., 2013).
Eukaryotic spliceosomes catalyze two reactions necessary to remove introns from the surrounding exon sequences. The first step involves nucleophilic attack by the 2′ hydroxyl of an intronic adenosine (the branchpoint) on the phosphate at the 5′ splice site with the 3′ hydroxyl of the 5′ exon serving as the leaving group (Fig. 1A). The resulting branched structure, containing the intron and 3' exon, is called the lariat-intermediate. In the second chemical step, the 3′ hydroxyl of the free 5′ exon attacks the phosphate at the 3′ splice site to form the ligated exons and release the lariat intron. Identical chemistry is used by Group II ribozymes although for some, water is used as the nucleophile for the first chemical step.
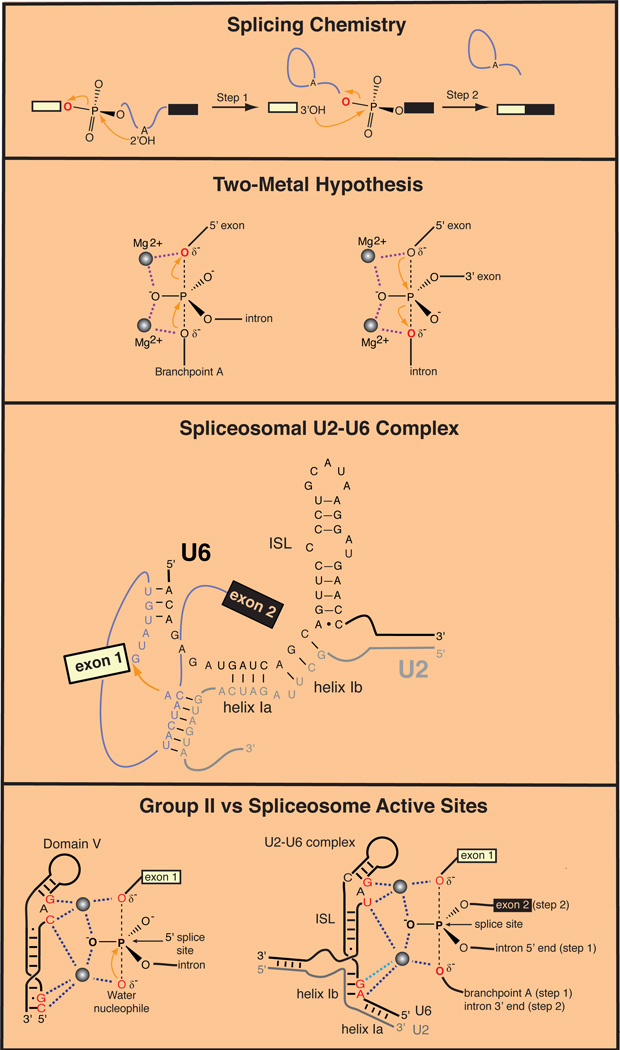
Figure 1. Catalysis of RNA Splicing.
(A) Chemical steps catalyzed by the spliceosome and Group II ribozymes.
(B) Proposed two-metal catalytic mechanism for RNA splicing (Steitz and Steitz, 1993).
(C) Structure of the spliceosomal U2–U6 complex. Attack of the branchpoint adenosine (red arrow) on the 5' exon-intron junction is shown.
(D) Comparison of active sites of a Group II ribozyme and the spliceosome. Left: Model for coordination of catalytic divalent cations in a Group II intron from Oceanobacillus iheyensis (Marcia and Pyle, 2013). Dotted lines indicate bonds with backbone phosphates inferred from high-resolution X-ray crystal structures. Right: Catalytic divalent cations positioned by the U2–U6 complex in the splceosome (Fica et al., 2013). Dark blue dotted lines indicate bonds with oxygen atoms identified by metal rescue experiments. Light blue dotted line indicates a proposed bond based on a defect in splicing produced by phosphorothioate substitution in U6.
Many protein enzymes catalyzing phosphoryl transfer reactions utilize two metal ions to stabilize the transition state through direct coordination of the oxygens of the scissile phosphate. It was proposed that such a mechanism also operates in RNA enzymes as well, including Group II introns and self-splicing Group I introns (Steitz and Steitz, 1993; Fig. 1B). A role for catalytic magnesium ions was subsequently demonstrated by structural and functional studies for these ribozyme families (Frederiksen and Piccirilli, 2009). Studies of pre-mRNA splicing have also provided strong evidence that magnesium ions function in catalysis by the spliceosome (Butcher, 2011). These mechanistic studies exploited basic chemical principles to identify positions where replacing a phosphoryl oxygen (either bridging or non-bridging) with sulfur led to improved catalysis when magnesium ions were replaced by more thiophilic ions of manganese or cadmium. However, as spliceosomal proteins had also been found to bind sequences near splice sites, many questions remained, including whether functional groups from proteins or snRNAs position the catalytic (vs. structural) magnesium ions, how many ions are involved, whether there are single or multiple active sites, and the potential relationship with Group II ribozymes.
The idea that there could be structural correspondence between spliceosomal snRNAs (U RNAs) and Group II introns emerged from the discovery that sequences around the intron branchpoint are recognized by U2 snRNA in a base-pairing interaction that bulges the adenosine nucleophile out of a helix. Domain VI of Group II introns forms an analogous bulged structure. Further evidence for a spliceosome-Group II structural connection came from studies of U6 snRNA, the most conserved snRNA in the spliceosome. U6 arrives at pre-mRNAs in a base-paired complex with U4 snRNA. This interaction is subsequently disrupted to allow formation of a base-paired structure (Helix I) between U6 and a segment of U2 snRNA just upstream of the sequence that base pairs with the intron branchpoint. Combined with an intramolecular stem-loop (ISL) in U6 snRNA, the U2–U6 complex juxtaposes highly conserved snRNA residues with the branchpoint nucleophile. The highly conserved structure produced by this RNA rearrangement displays striking similarities with the secondary structure of the most conserved domain of Group II introns, Domain V (Madhani and Guthrie, 1992; Yu et al., 1995). Further interactions between U6 and the 5′ splice site, and between the exons and U5 snRNA complete what has long been proposed to be the RNA-based active site of the spliceosome (Fig. 1C; Madhani and Guthrie, 1994). Recent work confirmed that active spliceosomes harbor the U2–U6 complex (Anokhina et al., 2013), but definitive evidence for a catalytic role for the U2–U6 complex has remained elusive.
The wait is now over. A recent study demonstrates that specific phosphates within U6 snRNA coordinate two magnesium ions that bind non-bridging and bridging oxygens of the substrate to catalyze both steps of pre-mRNA splicing (Fica et al., 2013). The authors first confirmed that cadmium is capable of rescuing the defects of mutant substrates containing sulfurs at its 5′ or 3′ splice sites, respectively. They then tested whether introducing a series of phosphorothioate substitutions in backbone residues of U6 snRNA would be capable of enhancing cadmium-mediated rescue. A positive result would imply that oxygen atoms within U6 snRNA function to position the catalytic metal ions towards the relevant phosphate of the substrate.
The experiments were performed by in vitro reconstitution of splicing in yeast extracts with synthetic snRNA and pre-mRNA. In many cases, spliceosomal fidelity mechanisms were disabled to avoid ATP-dependent discard pathways that disassemble splicing complexes containing the phosphorothioate derivatives prior to catalysis. Dose-response experiments and sophisticated metal titration experiments were used to distinguish between one- and two-metal mechanisms. An array of technical maneuvers and deductive logic together build a compelling argument that U6 snRNA positions two catalytic metals during both chemical steps of splicing. Moreover, consistent with previous thinking (Steitz and Steitz, 1993), the data support a model in which the 5′ exon of the substrate remains fixed relative to U6 snRNA at a single spliceosomal active site. For the first chemical step, the phosphate at the 5′ splice site is bound by two catalytic metal ions. This phosphate is replaced by the phosphate at the 3′ splice site for the second chemical step. Because the 5′ exon remains fixed in space during both reactions, the second chemical step is essentially the reverse reaction of the first step, such that the leaving group from the first reaction becomes the nucleophile in the second (Fig. 1D). These results are consistent with the early observations on the impact of splice site phosphorothioate substitutions on the two catalytic steps of splicing (Moore and Sharp, 1993).
The most remarkable revelation coming out of the new study is the precise correspondence between backbone phosphate oxygens in the U2–U6 complex that coordinate metal ions and those seen crystallographically to orient presumptive catalytic metals in proposed analogous positions in Domain V of Group II introns (Fig. 1D; Marcia and Pyle, 2012). Further supporting a deep phylogenetic relationship between the spliceosome and Group II ribozymes, a crystal structure of the most conserved protein component of the spliceosome, Prp8, revealed its kinship with the intron-encoded maturases of Group II introns (Galej et al., 2013). The strong implication is that the spliceosome is a ribozyme that arose via the genomic invasion of a mobile Group II intron at the birth of the eukaryotic lineage. That a single self-splicing intron and its Prp8-like reverse transcriptase/maturase evolved into one of the most complex machines in the eukaryotic cell suggests a remarkable power of mobile genetic elements to shape biological evolution.
References
- Anokhina M, Bessonov S, Miao Z, Westhof E, Hartmuth K, Luhrmann R. RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J. 2013;32:2804–2818. doi: 10.1038/emboj.2013.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher SE. The spliceosome and its metal ions. Metal ions in life sciences. 2011;9:235–251. doi: 10.1039/9781849732512-00235. [DOI] [PubMed] [Google Scholar]
- Fica SM, Tuttle N, Novak T, Li NS, Lu J, Koodathingal P, Dai Q, Staley JP, Piccirilli JA. RNA catalyses nuclear pre-mRNA splicing. Nature. 2013;503:229–234. doi: 10.1038/nature12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederiksen JK, Piccirilli JA. Identification of catalytic metal ion ligands in ribozymes. Methods. 2009;49:148–166. doi: 10.1016/j.ymeth.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galej WP, Oubridge C, Newman AJ, Nagai K. Crystal structure of Prp8 reveals active site cavity of the spliceosome. Nature. 2013;493:638–643. doi: 10.1038/nature11843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. doi: 10.1016/0092-8674(92)90556-r. [DOI] [PubMed] [Google Scholar]
- Madhani HD, Guthrie C. Dynamic RNA-RNA interactions in the spliceosome. Annual Review of Genetics. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- Marcia M, Pyle AM. Visualizing group II intron catalysis through the stages of splicing. Cell. 2012;151:497–507. doi: 10.1016/j.cell.2012.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Sharp PA. Evidence for two active sites in the spliceosome provided by stereochemistry of pre-mRNA splicing. Nature. 1993;365:364–368. doi: 10.1038/365364a0. [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:6498–6502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Maroney PA, Darzynkiwicz E, Nilsen TW. U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. RNA. 1995;1:46–54. [PMC free article] [PubMed] [Google Scholar]