Abstract
Resistance to the BCR-ABL inhibitor imatinib mesylate (IM) poses a major problem for the treatment of chronic myeloid leukemia (CML). IM resistance often results from a secondary mutation in BCR-ABL that interferes with drug binding. However, in many instances there is no mutation in BCR-ABL, and the basis of such BCR-ABL-independent IM resistance remains to be elucidated. To gain insight into BCR-ABL-independent IM resistance mechanisms, we performed a large-scale RNA interference (RNAi) screen and identified IM-sensitizing genes (IMSGs) whose knockdown renders BCR-ABL+ cells IM-resistant. In these IMSG knockdown cells, RAF/MEK/ERK signaling is sustained after IM treatment due to upregulation of PRKCH, which encodes the protein kinase C (PKC) family member PKCη, an activator of CRAF. PRKCH is also upregulated in samples from CML patients with BCR-ABL-independent IM resistance. Combined treatment with IM and trametinib, an FDA-approved MEK inhibitor, synergistically kills BCR-ABL+ IMSG knockdown cells and prolongs survival in mouse models of BCR-ABL-independent IM-resistant CML. Finally, we showed that CML stem cells contain high levels of PRKCH and this contributes to their intrinsic IM resistance. Combined treatment with IM and trametinib synergistically kills CML stem cells with negligible effect on normal hematopoietic stem cells. Collectively, our results identify a therapeutically targetable mechanism of BCR-ABL-independent IM resistance in CML and CML stem cells.
INTRODUCTION
Chronic myeloid leukemia (CML) is a hematopoietic malignancy characterized by an increase and unregulated growth of predominantly myeloid cells in the bone marrow, and their accumulation in the blood (1). A hallmark of CML is the Philadelphia chromosome, resulting from a reciprocal translocation between the long arms of chromosomes 9 and 22 (2, 3). This chromosomal translocation leads to expression of BCR-ABL, an oncogenic fusion protein with a constitutively activated ABL tyrosine kinase. BCR-ABL can transform myeloid progenitor cells and drives the development of 95% of CML cases. BCR-ABL promotes leukemogenesis by activating downstream signaling proteins that increase cell survival and proliferation (4). These pathways include, but are not limited to, the RAS/mitogen-activated protein kinase (RAF/MEK/ERK), phosphatidylinositol 3-kinase/AKT (PI3K/AKT), and JAK/STAT signaling cascades (5).
The first-line treatment for CML is imatinib mesylate (IM), which binds to the ABL kinase domain and inhibits phosphorylation of substrates (6). Although IM dramatically improves patient survival when used to treat early-stage disease, the drug is not curative. Resistance to IM can develop, especially in advanced-stage disease, leading to disease relapse and progression (7). Resistance to IM can result from multiple mechanisms that can be broadly classified as either BCR-ABL-dependent or BCR-ABL-independent (8). BCR-ABL-dependent resistance is most commonly due to the acquisition of point mutations in the ABL kinase domain that interfere with IM binding and subsequent kinase inhibition (9–11). However, in 50% or more of IM-resistant CML patients there is no mutation in BCR-ABL (12, 13), and the basis of such BCR-ABL-independent IM resistance is not understood.
CML, like several other malignancies, is propagated by a small population of stem cells, elimination of which is likely required to achieve long-term remission and cure (14, 15). An important limitation of IM treatment is that although IM inhibits BCR-ABL activity in CML stem cells, these cells do not depend on BCR-ABL activity for survival and are thus not eliminated (16, 17). These findings imply that CML stem cells use survival signals other than BCR-ABL to maintain viability in the presence of IM. Understanding the mechanism by which CML stem cells are intrinsically resistant to IM is essential for devising strategies to eradicate residual leukemia. To gain insight into how IM resistance can occur in the absence of BCR-ABL mutations, we performed an RNA interference (RNAi) screen to identify genes that regulate IM responsiveness. Our results reveal a survival pathway that promotes BCR-ABL-independent IM resistance and also contributes to the IM resistance of CML stem cells.
RESULTS
A large-scale shRNA screen identifies IM-sensitizing genes
To identify IM-sensitizing genes (IMSGs), IM-sensitive human CML K562 cells (18) were stably transduced with pools of a genome-wide human short hairpin RNA (shRNA) library (19) followed by IM treatment (Fig. 1A). Surviving cells from all pools were combined, and shRNAs corresponding to 89 genes were identified by sequence analysis. Validation experiments with individual shRNAs corresponding to those isolated from the primary screen, as well as second, unrelated shRNAs targeting the same genes, confirmed that knockdown of 25 genes conferred >2-fold increased K562 cell survival in the presence of IM relative to a control non-silencing (NS) shRNA (Fig. 1B and fig. S1 and fig. S2A). The extent of IM resistance after IMSG knockdown was roughly similar to that of the well-studied experimentally-derived IM-resistant cell line K562R and an IM-resistant patient-derived cell line, SUPB15 (fig. S2B). Quantitative real-time RT-PCR (qRT-PCR) confirmed in all cases that expression of the target gene was decreased in the corresponding K562 knockdown (KD) cell line (fig. S2, C and D).
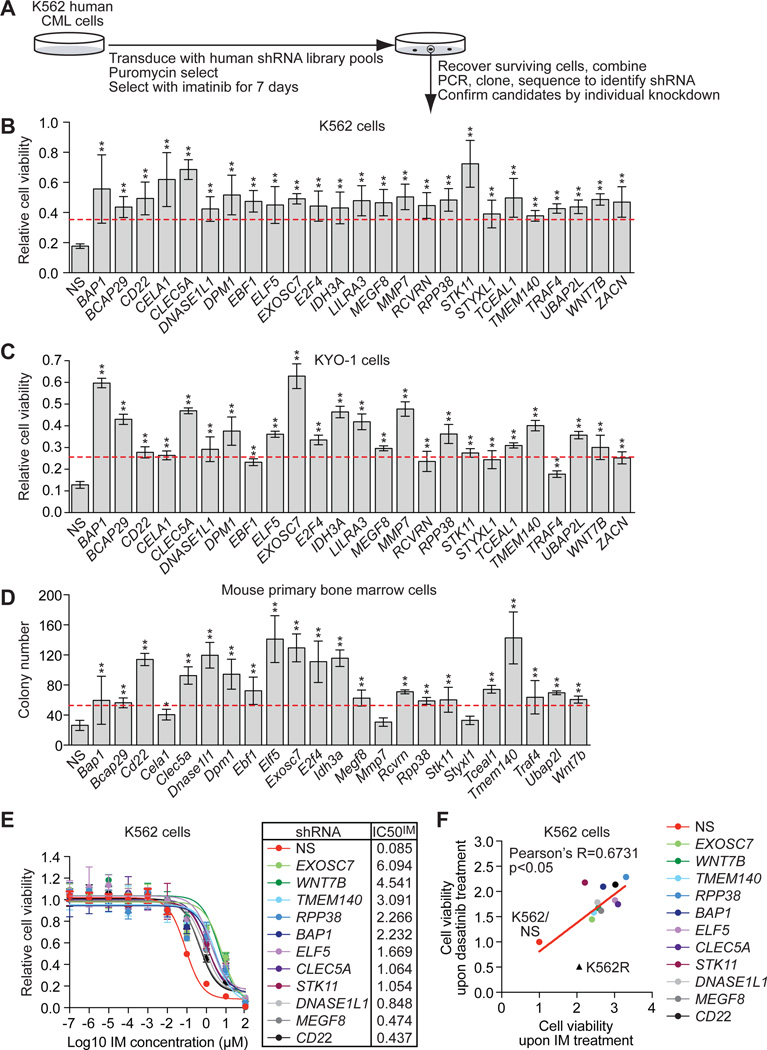
Fig. 1. A large-scale shRNA screen identifies IMSGs.
(A) Schematic summary of the screen. (B) Relative viability of IMSG KD K562 cells in the presence of IM, as measured by MTT assay (n=4). The results were normalized to that obtained with DMSO-treated cells, which was set to 1. IMSG shRNAs that conferred >2-fold increase in cell survival (indicated by the red line) relative to the NS control shRNA were considered positive. (C) Relative viability of IMSG KD KYO-1 cells in the presence of IM, as measured by MTT assay (n=4). The results were normalized and positives determined as described in (B). (D) Colony formation assay monitoring survival of BCR-ABL+ mouse primary bone marrow cells expressing an IMSG shRNA in the presence of IM (n=3). IMSG shRNAs that conferred >2-fold increase in colony number (indicated by the red line) relative to the NS control shRNA were considered positive. (E) Relative IC50IM of IMSG KD K562 cells (n=4). (F) Cell viability, as measured by MTT assay, of IMSG KD K562 cells treated with 500 nM dasatinib or 10 µM IM for 3 days (n=4). K562 cells expressing an NS shRNA (K562/NS) and IM-resistant K562 cells (K562R) were analyzed as controls. Data are represented as mean ± SD. *P≤0.05, **P≤0.01. Statistical tests and exact P values are provided in table S4.
To confirm that our results are generalizable, we analyzed the validated candidates in KYO-1 cells, another IM-sensitive human CML cell line (20). Figure 1C shows that 21 of the 25 shRNA candidates validated in KYO-1 cells. Finally, we tested whether knockdown of the validated candidates would also confer IM resistance in BCR-ABL+ mouse primary bone marrow cells. Toward this end, we induced CML-like disease in C57BL/6 mice with a BCR-ABL-expressing retrovirus (21, 22). Primary bone marrow cells were harvested, infected with a mouse candidate IMSG shRNA, and tested for their ability to form colonies in methylcellulose containing IM. We found that knockdown of 19 candidate IMSGs (fig. S3A) rendered BCR-ABL+ primary bone marrow cells IM-resistant (Fig. 1D). Equivalent results were obtained with a second, unrelated shRNA for each IMSG (fig. S3, B and C).
To quantify IM resistance, we determined the IC50 for imatinib (IC50IM) of IMSG KD K562 cells. Knockdown of 11 IMSGs increased the IC50IM greater than 5-fold (Fig. 1E and fig. S4), and we therefore focused on these IMSGs in our subsequent experiments. Notably, the IC50IMs of these 11 IMSG KD K562 cell lines are similar to those of IM-resistant cell lines derived from CML patients (23). These 11 IMSGs are involved in diverse biological processes including transcriptional regulation, signal transduction, protein metabolism and DNA/RNA metabolism (table S1).
Next, we tested whether knockdown of IMSGs would cause resistance to the second-generation tyrosine kinase inhibitor, dasatinib (24). As a control, we analyzed in parallel K562R cells, which are resistant to IM but sensitive to dasatinib due to over-expression of the Src family kinase (SFK) LYN (25). All of the IMSG shRNAs that conferred IM resistance also caused resistance to dasatinib (Fig. 1F).
Knockdown of IMSGs in BCR-ABL+ cells results in sustained RAF/MEK/ERK signaling after IM treatment
We next performed a series of experiments to identify the regulatory pathway(s) through which IMSGs promote IM sensitivity. IMSG KD K562 cell lines were cultured in the presence or absence of IM followed by immunoblotting for characteristic markers of relevant cell signaling pathways. The results in Figure 2 indicate that knockdown of IMSGs had no effect on total BCR-ABL. Moreover, in all IMSG KD K562 cell lines, IM inhibited BCR-ABL protein kinase activity, as evidenced by decreased BCR-ABL autophosphorylation and decreased phosphorylation of the BCR-ABL substrate CRKL (26). We also monitored the effect of IMSG knockdown on SFK activity, whose elevation, as mentioned above, is responsible for IM resistance in K562R cells. None of the IMSG KD K562 cell lines had elevated SFK activity or expression, consistent with their resistance to dasatinib.
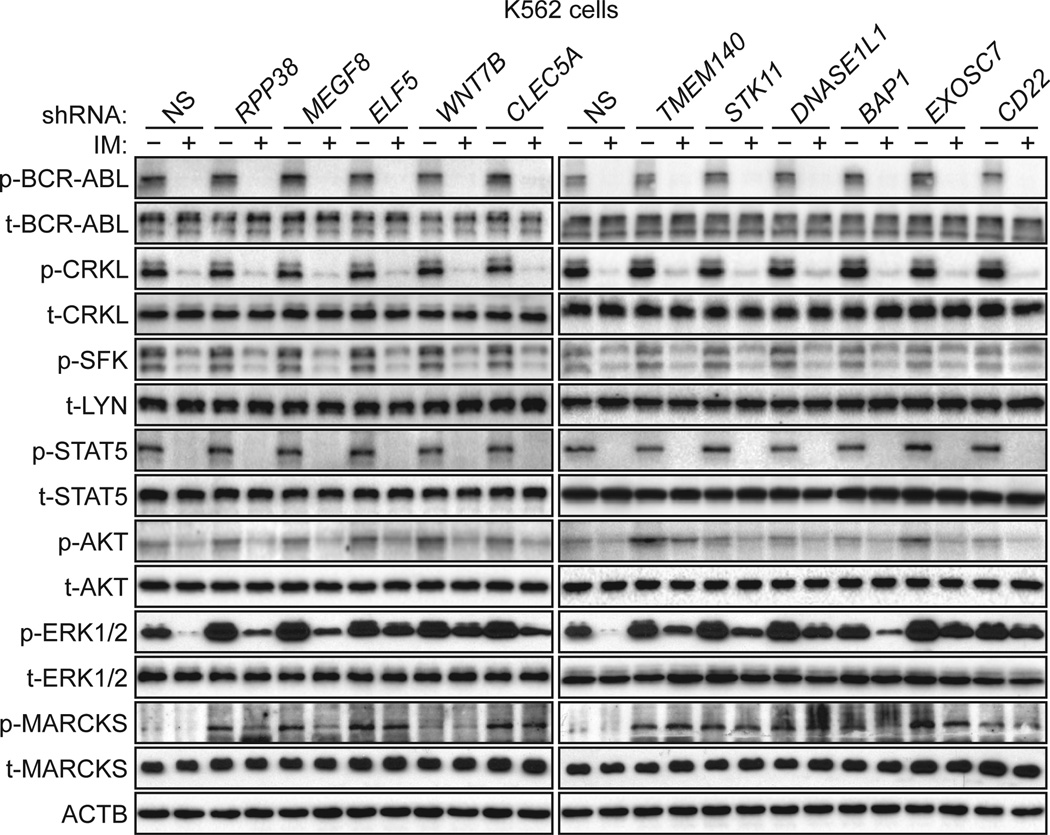
Fig. 2. Knockdown of IMSGs in BCR-ABL+ cells results in sustained RAF/MEK/ERK signaling after IM treatment.
Immunoblot analysis monitoring the activity of BCR-ABL (as measured by phosphorylated (p) and total (t) BCR-ABL and CRKL), SFKs (p-SFK and t-LYN), JAK/STAT (p- and t-STAT5), PI3K/AKT (p- and t-AKT), MEK/ERK (p- and t-ERK1/2), and PKC (p- and t-MARCKS) pathways in IMSG KD K562 cells treated in the presence or absence of IM. β-actin (ACTB) was monitored as a loading control.
We next analyzed the effect of IMSG knockdown on known downstream signaling pathways of BCR-ABL. All IMSG KD K562 cell lines had normal amounts of phosphorylated STAT5 and AKT, indicating that JAK/STAT and PI3K/AKT signaling pathways were not affected by IMSG knockdown. In contrast, most of the IMSG KD K562 cell lines had increased RAF/MEK/ERK kinase pathway activity, as evidenced by elevated phosphorylation of ERK1/2. As expected, after IM treatment of control K562 cells, there was a substantial decrease in phosphorylated ERK1/2. However, all of the IMSG KD K562 cell lines had, to varying extents, sustained phosphorylation of ERK1/2 after IM treatment. Thus, in IMSG KD K562 cell lines, there is an alternative pathway that activates RAF/MEK/ERK signaling after inhibition of BCR-ABL.
Previous studies have reported that the protein kinase C (PKC) pathway can stimulate RAF/MEK/ERK signaling (27–29). We therefore analyzed PKC pathway activity in IMSG KD K562 cells by monitoring phosphorylation of a universal PKC substrate, MARCKS (30). Phosphorylation of MARCKS was elevated in all IMSG KD K562 cell lines, indicating increased PKC activity.
PRKCH is upregulated in BCR-ABL-independent IM-resistant CML cell lines and patient samples
Next, we sought to identify the PKC family member(s) responsible for the increased PKC activity. The qRT-PCR results in Figure 3A show that PRKCH, which encodes PKCη, was upregulated in nearly all IMSG KD K562 cell lines. Similar results were obtained with a second shRNA targeting each IMSG (fig. S5A). Immunoblot analysis confirmed that PKCη protein levels were also increased in the IMSG KD K562 cell lines (fig. S5B).
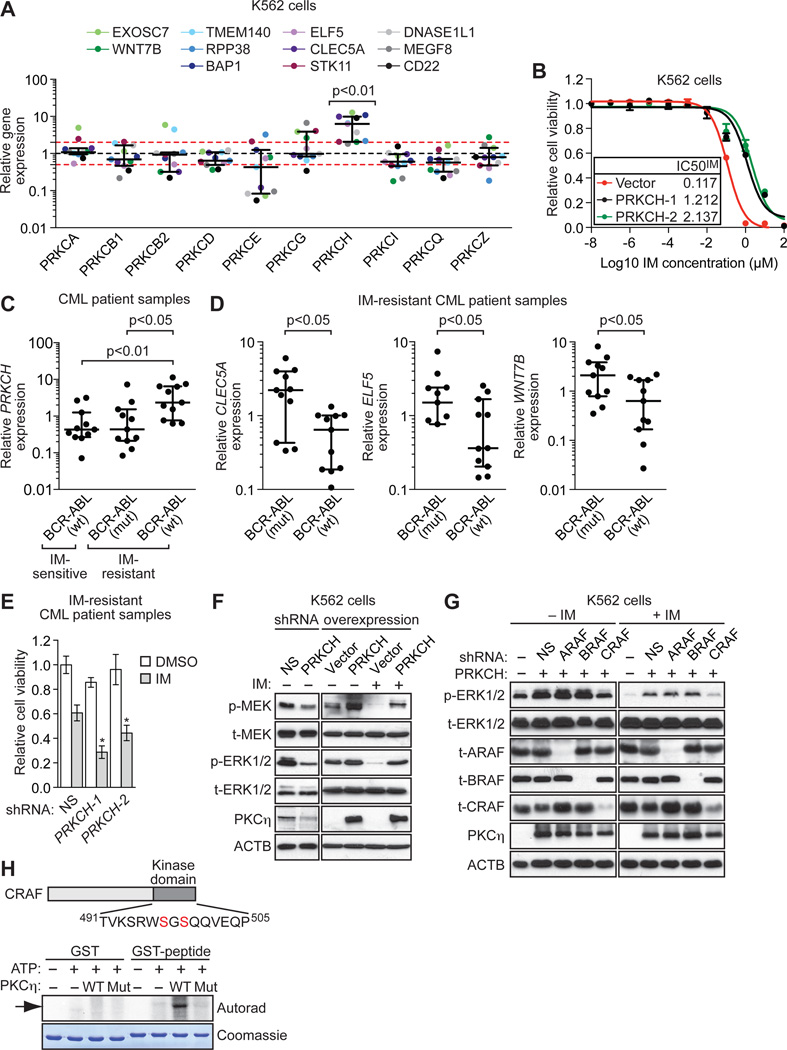
Fig. 3. IMSG knockdown increases RAF/MEK/ERK signaling through upregulation of PKCη, an activator of CRAF.
(A) qRT-PCR analysis monitoring expression of different PKC isotypes in IMSG KD K562 cells. Each colored dot represents an individual IMSG KD K562 cell line. Error bars indicate median with interquartile range. The results were normalized to that obtained with the NS control shRNA, which was set to 1. The red lines indicate >2-fold change in gene expression relative to that obtained with the NS shRNA. (B) Relative IC50IM in K562 cells expressing empty vector and in two independently derived K562 clonal cell lines ectopically expressing PRKCH (n=4). Data are represented as mean ± SD. (C) qRT-PCR analysis monitoring expression of PRKCH in BCR-ABL wild-type IM-sensitive patient samples (n=11), and BCR-ABL mutant (n=11) or BCR-ABL wild-type (n=11) IM-resistant CML patient samples. Error bars indicate median with interquartile range. (D) qRT-PCR analysis monitoring expression of three IMSGs in BCR-ABL mutant (n=11) or BCR-ABL wild-type (n=11) IM-resistant CML patient samples. For ELF5, BCR-ABL mutant (n=9). Error bars indicate median with interquartile range. (E) Relative viability, as measured by trypan blue cell counting, of primary leukemic cells from BCR-ABL independent IM-resistant CML patient samples (n=5) expressing a NS or PRKCH shRNA and treated with DMSO or IM. The results were normalized to that obtained with DMSO-treated cells expressing a NS shRNA, which was set to 1. Data are represented as mean ± SEM. *P≤0.05, **P≤0.01. Statistical tests and exact P values are provided in table S4. (F) Immunoblot analysis monitoring RAF/MEK/ERK activity (as measured by p- and t-MEK and p- and t-ERK1/2) in PRKCH KD K562 cells (left) and in K562/PRKCH-1 cells in the absence or presence of IM (right). (G) Immunoblot analysis monitoring p- and t-ERK1/2 levels in K562/PRKCH-1 cells expressing an ARAF BRAF or CRAF shRNA, treated with DMSO or IM for 1 h. (H) (Top) Schematic of CRAF showing the kinase domain bearing a potential PKC phosphorylation site at S497/S499. (Bottom) In vitro phosphorylation assay. Wild-type (WT) or kinase-dead mutant (Mut; K384R) PKCη was used in an in vitro phosphorylation reaction containing either GST or a GST-CRAF(aa491-505) fusion-protein. The phosphorylated product was visualized by autoradiography. The Coomassie-stained gel shows the abundance of each protein.
As a first step toward understanding the basis by which IMSGs regulate PRKCH expression, we further analyzed one of the IMSGs, ELF5, a known transcriptional repressor (31, 32). We used a chromatin immunoprecipitation assay and found that ELF5 was directly bound at the transcription start site of PRKCH (fig. S6A), consistent with the results of a study analyzing ELF5 occupancy genome-wide (33). Moreover, we found that expression of a PRKCH promoter-luciferase reporter construct was increased by ELF5 knockdown and, conversely, decreased by ectopic expression of ELF5 (fig. S6, B and C). Thus, ELF5 is a direct transcriptional repressor of PRKCH, explaining why decreased ELF5 levels result in increased PRKCH expression.
To verify that increased PKCη expression is responsible for the IM resistance, we derived K562 cell lines that over-expressed PRKCH (K562/PRKCH cells) to varying degrees. In several K562/PRKCH cell lines, PKCη levels were comparable to those found in IMSG KD K562 cells (fig. S7A). The elevated PRKCH expression resulted in a 10–20-fold increase in IM resistance (Fig. 3B). Conversely, knockdown of PRKCH abrogated the IM resistance of representative IMSG KD K562 cell lines (fig. S7B).
To determine the clinical relevance of these results, we analyzed PRKCH mRNA levels in IM-resistant CML patient samples harboring wild-type BCR-ABL. As a control, we also analyzed PRKCH mRNA levels in IM-resistant CML patient samples that contained a known IM-resistance mutation in BCR-ABL (table S2). The results in Figure 3C show that PRKCH mRNA levels were significantly (P<0.01) higher in IM-resistant CML patient samples containing wild-type BCR-ABL compared to those with mutant BCR-ABL. In addition, we found that the average expression of three IMSGs (CLEC5A, ELF5, and WNT7B) was significantly (P<0.01, <0.05, <0.05, respectively) lower in IM-resistant CML patient samples containing wild-type BCR-ABL compared to those with mutant BCR-ABL (Fig. 3D). Moreover, in all 11 IM-resistant CML patient samples containing wild-type BCR-ABL, at least one IMSG was down-regulated >2-fold, and in 9/11 samples at least one IMSG was down-regulated >5-fold relative to the average expression in IM-resistant mutant BCR-ABL samples (table S3). Finally, the results in Figure 3E show that knockdown of PRKCH increased IM sensitivity of leukemic cells from BCR-ABL-independent IM-resistant CML patients.
PKCη increases RAF/MEK/ERK signaling through phosphorylation and activation of CRAF
We next sought to understand in greater detail how PKCη increased RAF/MEK/ERK signaling. Figure 3F shows that even a relatively modest knockdown of PRKCH in IM-sensitive K562 cells decreased both phosphorylated MEK and ERK1/2 (see also fig. S8A) and increased IM sensitivity (fig. S7B). Conversely, K562/PRKCH cells had increased levels of both phosphorylated MEK and ERK1/2 (Fig. 3F). Most importantly, K562/PRKCH cells maintained high levels of phosphorylated MEK and ERK1/2 after IM treatment (Fig. 3F).
The finding that PKCη affected both phosphorylated MEK and ERK1/2 indicated that PKCη functioned upstream of MEK by, for example, stimulating RAF activity. There are three known RAF kinases: ARAF, BRAF, and CRAF (34). We found that in K562/PRKCH cells, knockdown of CRAF, but not ARAF or BRAF, decreased phosphorylated ERK1/2 (Fig. 3G and fig. S8B). Most importantly, in IM-treated K562/PRKCH cells, knockdown of CRAF, but not ARAF or BRAF, resulted in loss of sustained phosphorylation of ERK1/2.
To determine whether CRAF was a direct substrate of PKCη, we derived a glutathione-S-transferase (GST) fusion-protein containing a CRAF peptide bearing a potential PKC phosphorylation site at S497/S499 (35, 36). The in vitro kinase assay in Figure 3H shows that wild-type PKCη, but not a kinase-dead mutant (K384R) (37), could phosphorylate the CRAF S497/S499 site. Our results are consistent with several previous findings including phosphorylation of CRAF by PKC isoforms (35, 36, 38–40) and reduced activity of a CRAF S497A/S499A mutant (36, 38).
IM and a MEK inhibitor synergistically kill BCR-ABL-independent IM-resistant CML cells
The results presented above show that BCR-ABL-independent IM resistance can result from increased PRKCH expression, leading to sustained RAF/MEK/ERK signaling after IM treatment. An implication of this conclusion is that simultaneous inhibition of BCR-ABL and RAF/MEK/ERK signaling might efficiently kill BCR-ABL-independent IM-resistant CML cells. To investigate this possibility, we analyzed the effect of combining IM treatment with the FDA-approved MEK inhibitor trametinib (also called GSK1120212). We found that treatment with both IM and trametinib had a substantially greater effect than either drug alone in killing K562/PRKCH cells (Fig. 4A), representative IMSG KD K562 cell lines (Fig. 4B), and BCR-ABL+ mouse primary bone marrow cells over-expressing Prkch (Fig. 4C). In most instances, the effect of combined drug treatment was synergistic (table S4). The modest effect of trametinib alone on K562 cell lines likely reflects stimulation of RAF/MEK/ERK signaling by BCR-ABL. Finally, treatment with both IM and trametinib had a significantly (P<0.01) greater effect than either drug alone in killing primary leukemic cells from BCR-ABL-independent IM-resistant CML patients (Fig. 4D and fig. S9A), Moreover, these leukemic cells were killed more effectively by combined treatment with IM and trametinib than by IM and a JAK-STAT or PI3K inhibitor, and neither of these latter two drug combinations were significantly more effective than IM alone (fig. S9B).
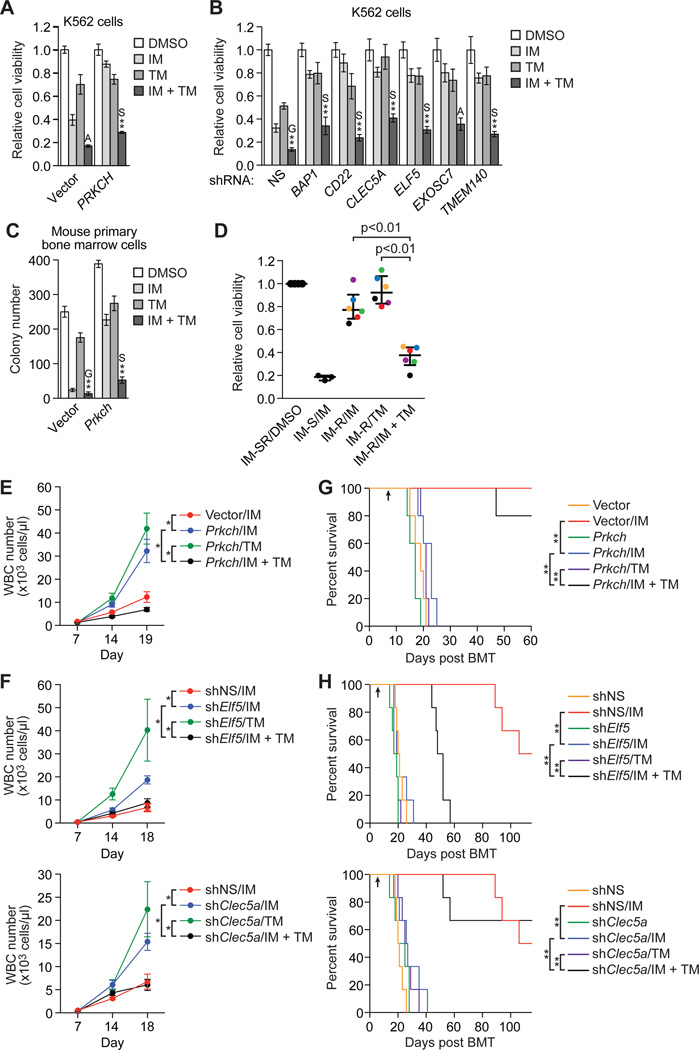
Fig. 4. Combined treatment with IM and a MEK inhibitor has beneficial effects.
(A and B) Cell viability, as measured by MTT assay, of K562/PRKCH-1 (A; n=4) or IMSG KD K562 cells (B; n=3 or 4) treated with 0.1 µM IM, 1.5 nM trametinib (TM) or a combination of the two drugs, as indicated. The results were normalized to that observed with DMSO, which was set to 1. Data are represented as mean ± SD. Asterisks indicate comparisons between the combined drug treatment and single drug treatments. Combined drug treatment was synergistic (S), additive (A) or antagonistic (G). (C) Colony formation assay monitoring survival of BCR-ABL+ mouse primary bone marrow cells ectopically expressing Prkch and treated with IM, TM or a combination, as described in (A) (n=3). Data are represented as mean ± SD. (D) Relative viability, as measured by trypan blue cell counting, of primary leukemic cells isolated from IM-sensitive (IM-S) CML patients and treated with 5 µM IM (n=3), or isolated from BCR-ABL-independent IM-resistant (IM-R) CML patients and treated with 5 µM IM, 5 µM TM or a combination (n=6). The results were normalized to those obtained by DMSO treatment of the same samples (IM-S or IM-R [IM-SR]), which was set to 1. Error bars indicate median with interquartile range. Matched samples from the same patient are indicated by dots of the same color. (E and F) White blood cell (WBC) count of leukemic mice derived by transplantation of BCR-ABL+ mouse primary bone marrow cells ectopically expressing Prkch (E) or knocked down for an IMSG (F), and treated at day 7 with either IM, TM or a combination of the two drugs as indicated (n=4 or 5 mice per group). Data are represented as mean ± SEM. The same NS control is used in the two graphs shown in (F), which were derived from a single experiment. (G and H) Kaplan-Meier survival curves of leukemic mice derived as described in (E and F). The indicated cohorts of mice (n=5 for Prkch overexpression and n=6 for IMSG knockdown) were treated with either vehicle, IM (100 mg/kg twice a day), TM (2 mg/kg once a day), or both IM and TM by oral gavage starting at day 7 (indicated by the arrow). The same NS control is used in the curves shown in (H), which were derived from a single experiment. *P≤0.05, **P≤0.01. Statistical tests and exact P values are provided in table S4.
IM and a MEK inhibitor prolong survival in mouse models of BCR-ABL-independent IM-resistant CML
Based upon the cell culture results, we analyzed the ability of this drug combination to prolong survival in mouse models of BCR-ABL-independent IM-resistant CML. Briefly, mouse primary bone marrow cells were transduced with a retrovirus co-expressing BCR-ABL and either Prkch (fig. S10A) or an shRNA targeting one of two representative IMSGs, Clec5a or Elf5 (fig. S10B–D), followed by transplantation into lethally irradiated syngeneic mice. We found that combined treatment with IM and trametinib was substantially more effective than either drug alone at suppressing leukemic progression, as evidenced by a reduced white blood cell count (Fig. 4, E and F), and prolonging survival (Fig. 4, G and H). In addition to prolonged survival, the general appearance and behavior of mice treated with IM and trametinib was normal, suggesting minimal drug toxicity.
PRKCH modulates proliferation of BCR-ABL+ cells, disease progression, and IM sensitivity
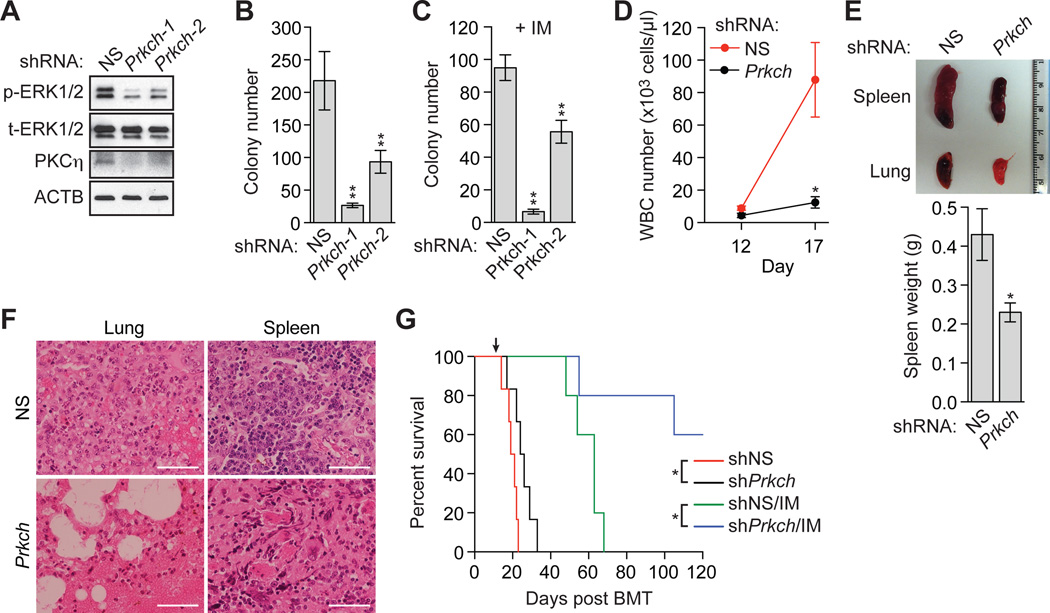
The finding that knockdown of PRKCH in K562 cells reduced phosphorylated ERK1/2 (Fig. 3F) raised the possibility that PRKCH might modulate the proliferation and survival of BCR-ABL+ cells and thus affect disease progression. To investigate this possibility, we transduced mouse primary bone marrow cells with a retrovirus co-expressing BCR-ABL and a Prkch shRNA (Fig. 5A). Prkch knockdown decreased phosphorylated ERK1/2, similar to the results in K562 cells. Figure 5B shows that knockdown of Prkch (fig. S11A) reduced the ability of untreated BCR-ABL+ mouse primary bone marrow cells to form colonies in methylcellulose (see also fig. S11B). Moreover, the colony formation assay in Figure 5C shows that knockdown of Prkch markedly increased the IM sensitivity of BCR-ABL+ mouse primary bone marrow cells.
Fig. 5. PRKCH modulates proliferation of BCR-ABL+ cells, disease progression, and IM-sensitivity.
(A) Immunoblot analysis monitoring p- and t-ERK1/2 levels in BCR-ABL+ mouse primary bone marrow cells expressing an NS shRNA or one of two Prkch shRNAs. (B) Colony formation assay after knockdown of Prkch in BCR-ABL+ mouse primary bone marrow cells (n=3). Data are represented as mean ± SD. (C) Colony formation assay monitoring survival of BCR-ABL+ mouse primary bone marrow cells expressing a NS or one of two Prkch shRNAs and treated with 0.1 µM IM. Data are represented as mean ± SD. (D) WBC count of leukemic mice derived by transplantation of Prkch KD BCR-ABL+ mouse primary bone marrow cells (n=4 or 5). Data are represented as mean ± SEM. (E) (Top) Representative spleen and lung images of leukemic mice derived as described in (D). Mice were sacrificed at day 17. (Bottom) Spleen weight of mice (n=4). Data are represented as mean ± SEM. (F) Hematoxylin and eosin (H&E) staining of spleen and lung sections from leukemic mice derived as described in (D). Scale bars, 50 µm. (G) Kaplan-Meier survival curve of untreated leukemic mice (n=6) or leukemic mice treated with IM at day 14 (indicated by the arrow) (n=5), derived as described in (D). *P≤0.05, **P≤0.01. Statistical tests and exact P values are provided in table S4.
We next transplanted the Prkch KD bone marrow cells into syngeneic mice to induce CML-like disease and analyzed the effect of Prkch knockdown on leukemic progression. We found that in untreated mice Prkch knockdown resulted in a lower white blood cell count (Fig. 5D), reduced spleen size (Fig. 5E), decreased infiltration of the lung and spleen by leukemic cells (Fig. 5F), and an increase in survival (Fig. 5G). Thus, in the absence of IM treatment, PRKCH promotes disease progression, although this effect may be relatively minor. More importantly, knockdown of Prkch markedly increased survival of IM-treated mice with CML-like disease (Fig. 5G).
IM-resistant murine and human CML stem cells contain high levels of PRKCH
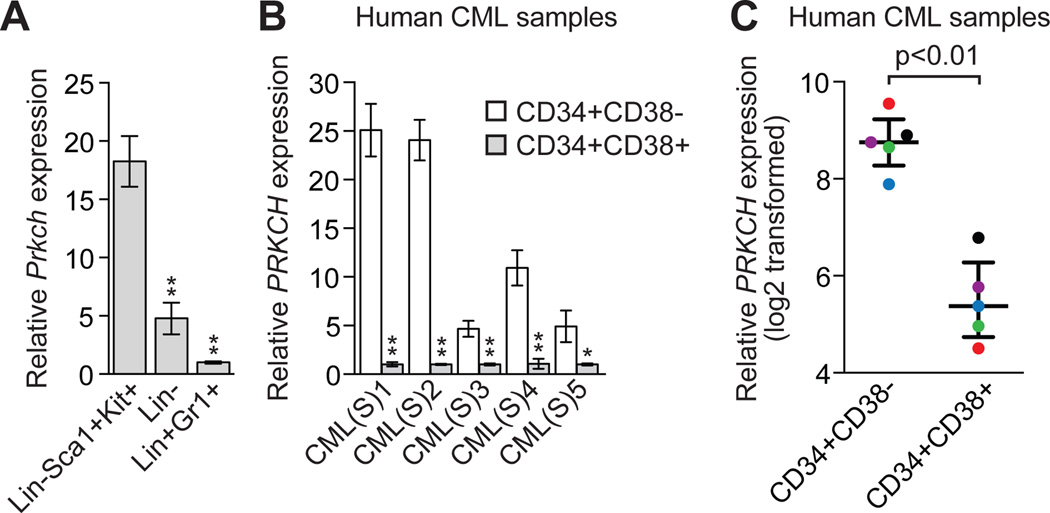
We considered the possibility that PRKCH might contribute to the intrinsic resistance of CML stem cells to IM. To investigate this idea, we induced CML-like disease in mice and isolated BCR-ABL+ murine stem cells (Lin-Sca1+Kit+), progenitor cells (Lin-) and mature cells (Lin+Gr1+) by fluorescence activated cell sorting (FACS) (17, 41). The qRT-PCR results in Figure 6A show that IM-resistant murine CML stem cells ((17, 42, 43) and see below) had substantially higher expression of Prkch compared to murine CML progenitor and mature cells, both of which are IM-sensitive ((17, 44) and fig. S12).
Fig. 6. IM-resistant murine and human CML stem cells contain high levels of PRKCH.
(A) qRT-PCR analysis monitoring Prkch expression in BCR-ABL+ murine CML stem cells (Lin-Sca1+Kit+), progenitor cells (Lin-) and mature cells (Lin+Gr1+) (n=3). Data are represented as mean ± SD. (B) qRT-PCR analysis monitoring PRKCH expression in human CML stem cells (CD34+CD38-) and progenitor cells (CD34+CD38+) isolated from CML patient samples (n=5). Data are from three technical replicates and are means ± SD. (C) PRKCH expression in CD34+CD38- and CD34+CD38+ cells, mined from a previous expression profiling study (50). Matched samples from the same patient are indicated by dots of the same color. Error bars indicate median with interquartile range. *P≤0.05, **P≤0.01. Statistical tests and exact P values are provided in table S4.
We next asked whether PRKCH expression was also high in human CML stem cells. We isolated CML stem cells (CD34+CD38−) and CML progenitor cells (CD34+CD38+) (16, 45, 46) from newly diagnosed CML patients. The qRT-PCR results in Figure 6B show that IM-resistant human CML stem cells ((15, 16, 47–49) and see below) had substantially higher expression of PRKCH compared to human CML progenitor cells, which are IM-sensitive (16, 45). Analysis of a published expression profiling study comparing highly enriched human CML stem and progenitor cell populations (50) revealed similar differences in PRKCH expression (Fig. 6C). Microarray analysis indicates that PRKCH expression is much higher in hematopoietic stem cells than in mature myeloid cells (51, 52), suggesting that high PRKCH expression may be a marker of stemness.
High Prkch expression contributes to the IM resistance of CML stem cells
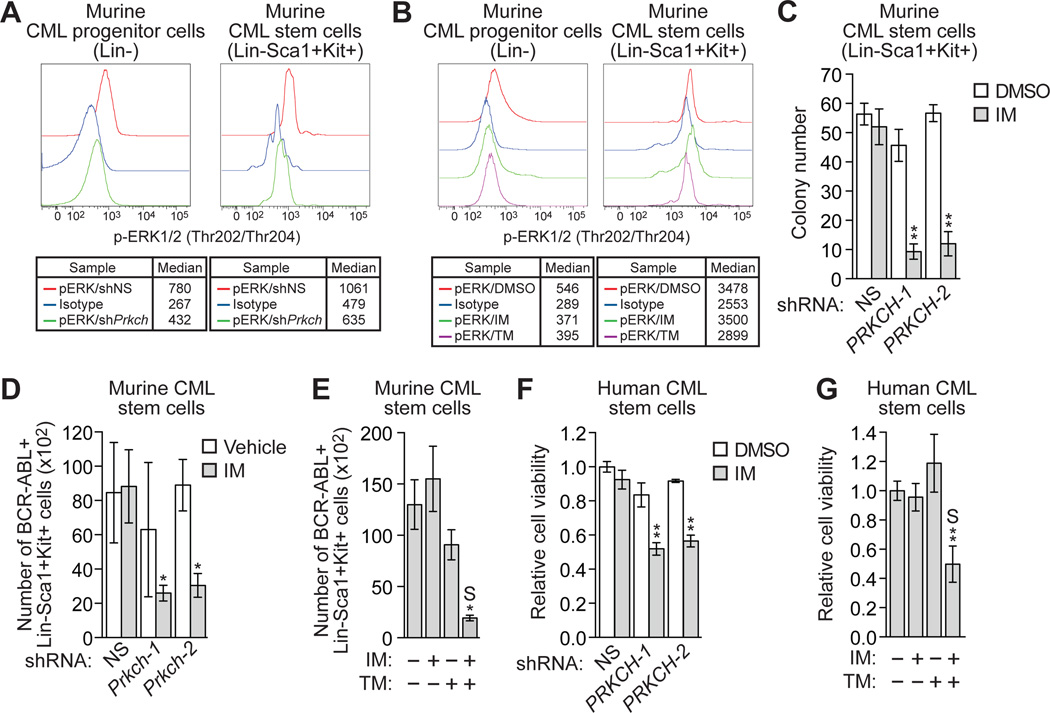
We performed several experiments to determine whether the high Prkch expression in murine CML stem cells contributes to their IM resistance. We first assessed the contribution of Prkch, and as a comparison BCR-ABL, to RAF/MEK/ERK signaling in murine CML stem cells. Prkch KD bone marrow cells were isolated from leukemic mice, permeabilized and incubated with an antibody against phosphorylated ERK1/2 or, as a negative control, IgG isotype antibody, and then analyzed by FACS to determine the phosphorylated ERK1/2 levels in CML progenitor and stem cells. Figure 7A shows that knockdown of Prkch reduced phosphorylated ERK1/2 in both CML progenitor and stem cells (see also fig. S13A).
Fig. 7. High Prkch levels contribute to the IM resistance of CML stem cells.
(A) Intracellular phosphorylated ERK1/2 levels in Lin- and Lin-Sca1+Kit+ BCR-ABL+ Prkch KD or control bone marrow cells. As a negative control, cells were incubated with a conjugated IgG isotype antibody. (B) Intracellular phosphorylated ERK1/2 levels in Lin- and Lin-Sca1+Kit+ BCR-ABL+ bone marrow cells treated with DMSO, IM or trametinib. (C) Colony formation assay monitoring survival of BCR-ABL+ murine stem cells expressing a NS or one of two Prkch shRNAs and treated with 0.1 µM IM (n=3). Data are represented as mean ± SD. (D) FACS determination of the number of BCR-ABL+ Lin-Sca1+Kit+ bone marrow cells expressing a NS or Prkch shRNA after IM treatment of mice (n=4 or 5). Data are represented as mean ± SEM. (E) FACS determination of the number of BCR-ABL+ Lin-Sca1+Kit+ bone marrow cells after treatment of mice with vehicle (n=10), IM (n=12), TM (n=10) or both IM and TM (n=12). Data are represented as mean ± SEM. Asterisks indicate comparisons between the combined drug treatment and single drug treatments. Combined drug treatment was synergistic (S). (F) Relative viability, as measured by trypan blue cell counting, of BCR-ABL+ human CML stem cells (CD34+CD38-) expressing a NS or Prkch shRNA and treated with DMSO or IM (n=3). Data are represented as mean ± SEM. (G) Relative viability of BCR-ABL+ human CML stem cells treated with DMSO, IM, TM or a combination of drugs (n=3). Data are represented as mean ± SEM. *P≤0.05, **P≤0.01. Statistical tests and exact P values are provided in table S4.
To evaluate the role of BCR-ABL, bone marrow cells were isolated from leukemic mice and treated with either IM or trametinib, and phosphorylated ERK1/2 was monitored as described above. Figure 7B shows, as expected, that trametinib reduced phosphorylated ERK1/2 in both CML progenitor and stem cells (see also fig. S13B). In contrast, IM reduced phosphorylated ERK1/2 in IM-sensitive CML progenitor cells, but not in IM-resistant CML stem cells. Collectively, these results indicate that in CML stem cells, PKCη has a more prominent role than BCR-ABL in promoting RAF/MEK/ERK signaling.
We next performed a series of experiments to determine whether Prkch affects survival of CML stem cells after IM treatment. In the first experiment, mouse primary bone marrow cells were transduced with a retrovirus co-expressing BCR-ABL and either Prkch or a control non-silencing (NS) shRNA, followed by transplantation into lethally irradiated syngeneic mice. BCR-ABL+ murine stem cells (Lin-Sca1+Kit+) were isolated from the mice and IM sensitivity determined in a colony formation assay. The results in Figure 7C show that Prkch knockdown markedly increased the IM sensitivity of CML stem cells. In the second experiment, mice with CML-like disease were treated with either vehicle or IM in parallel for two weeks, and then sacrificed at the same time followed by quantification of CML stem cells by FACS analysis. Figure 7D shows, as expected, that IM treatment had little effect on the number of CML stem cells expressing a control NS shRNA, confirming that murine CML stem cells are IM-resistant. In contrast, IM treatment markedly reduced the number of Prkch KD CML stem cells. Annexin V staining revealed that IM treatment induced a higher level of apoptosis in Prkch KD compared to control CML stem cells (fig. S14, A and B). Finally, combined treatment with IM and trametinib synergistically killed murine CML stem cells (Fig. 7E), which was due, at least in part, to the induction of apoptosis (fig. S14, C and D). By contrast, treatment with IM and trametinib had negligible effect on normal murine hematopoietic stem cells (fig. S15A).
Knockdown of PRKCH also increased the IM sensitivity of IM-resistant human CML stem cells (Fig. 7F). Moreover, treatment with both IM and trametinib had a substantially greater effect than either drug alone in killing human CML stem cells (Fig. 7G), and a negligible effect on normal human hematopoietic CD34+ cells and hematopoietic stem cells (CD34+CD38-) (fig. S15B). Collectively, these results indicate that PRKCH is expressed at relatively high levels in both mouse and human CML stem cells and this contributes to their IM resistance.
DISCUSSION
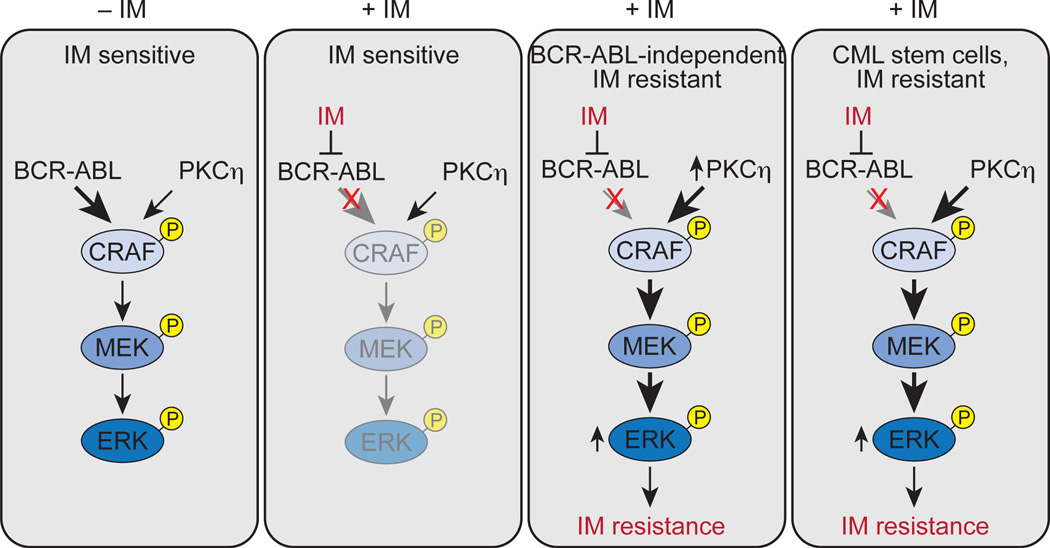
In this study, we have identified a molecular pathway whose increased activity promotes BCR-ABL-independent IM resistance and also contributes to the IM resistance of CML stem cells. Our major conclusions are summarized in the schematic model in Figure 8 and discussed below. In typical IM-sensitive CML cells, BCR-ABL is the major contributor to RAF/MEK/ERK signaling. Thus, treatment with IM substantially reduces RAF/MEK/ERK signaling, leading to inhibition of proliferation and induction of apoptosis. In BCR-ABL-independent IM-resistant CML cells, elevation of PKCη, due to decreased expression of one or more IMSGs, results in phosphorylation and activation of CRAF, thereby augmenting RAF/MEK/ERK signaling. After treatment with IM, RAF/MEK/ERK signaling is sustained, resulting in drug resistance.
Fig. 8. Elevated PKCη levels lead to IM resistance in CML and CML stem cells.
Relative contributions of BCR-ABL and PKCη to RAF/MEK/ERK signaling are indicated by arrow size and shading.
A previous study analyzing IM resistance resulting from mutations in BCR-ABL found that IM treatment "paradoxically" increased RAF/MEF/ERK signaling through a RAS-directed pathway (53). Although the IM resistance mechanism we describe, like that in (53), involves increased RAF/MEK/ERK signaling, there are several important differences. For example, in our experiments the increased RAF/MEK/ERK signaling is not dependent upon RAS but rather initiated by PKCη, is constitutive and not induced by IM, and, as discussed below, is also relevant to the intrinsic IM resistance of CML stem cells. In addition, several reports have described experimentally derived BCR-ABL-independent IM-resistant CML cell lines in which RAF/MEK/ERK signaling is increased by a mechanism that was not determined (54–56), or have provided other evidence that RAF/MEK/ERK signaling can contribute to IM resistance (57–60).
The mechanistic basis by which IMSGs regulate PRKCH expression is largely unknown. We showed that one of the IMSGs, ELF5, is directly bound at the transcription start site of PRKCH, and can decrease PRKCH expression. Thus, ELF5 is a direct transcriptional repressor of PRKCH, explaining why decreased ELF5 levels result in increased PRKCH expression. Whether other IMSGs function directly or indirectly to regulate PRKCH expression remained to be determined.
It is likely that our RNAi screen, like other large-scale RNAi screens (61), was not saturating, and thus there are probably other IMSGs and regulators of PRKCH expression that remain to be identified. Our results suggest that a variety of diverse perturbations can increase PRKCH expression. A previous expression profiling study revealed that the level of PRKCH in CML cells increased after one week of IM treatment (62), perhaps due to selection of and enrichment for cells with high PRKCH expression. This finding may also be explained by induction of PRKCH expression by IM treatment, although we found in IMSG KD K562 cell lines and CML stem cells that PRKCH is highly expressed in the absence of IM. In addition to its role in IM resistance, we found that elevated Prkch expression also accelerates disease progression in a mouse model of CML. Consistent with this idea, in a previous expression profiling study, PRKCH levels were found to increase during disease progression in CML patients. In the same study, the expression of seven of nine IMSGs analyzed decreased during disease progression (63).
The IM resistance mechanism we describe is therapeutically targetable, which we demonstrate by showing that combined treatment with IM and the FDA-approved MEK inhibitor trametinib synergistically kills BCR-ABL+ IMSG KD cells and prolongs survival in several mouse models of BCR-ABL-independent IM-resistant CML. However, analysis of patient-derived CML cell lines suggests there may be variable responsiveness to MEK inhibition (64). Our results are also relevant to another current challenge of CML treatment: the intrinsic resistance of CML stem cells to IM. We found that both human and murine CML stem cells contain high levels of PRKCH and provide evidence that this is responsible, at least in part, for their IM resistance. We further showed that the high PRKCH levels in CML stem cells promote RAF/MEK/ERK signaling, which helps explain why CML stem cells are not dependent upon BCR-ABL for survival (16, 17). Collectively, these results provide a rationale for our finding that CML stem cells, but not normal hematopoietic stem cells, are efficiently killed by combined treatment with IM and trametinib, and suggest a therapeutic strategy for their eradication.
MATERIALS AND METHODS
Study design
The overall study objective was to identify mechanisms underlying BCR-ABL-independent IM resistance in CML and CML stem cells. The study used cultured human CML cell lines, BCR-ABL+ mouse primary bone marrow cells, mouse models of BCR-ABL-independent IM-resistant CML, and bone marrow or blood samples from CML patients. Institutional Review Board (IRB) approval for the acquisition of CML patient samples was obtained from the Department of Pathology at University of Massachusetts Medical School (UMMS) (H00004484), the Druker lab at Oregon Health and Science University (OHSU) Knight Cancer Institute (4422) and the Hematology Bank at Winship Cancer Institute of Emory University (IRB00024964). All animal protocols were approved by the Institution Animal Care and Use Committee (IACUC) at UMMS (A-2300). The study consisted of a series of controlled laboratory experiments and measured multiple parameters including gene expression, cell viability, apoptosis, cell signaling pathway activity, and leukemic progression as described below. For animal experiments, mice were randomly allocated to each group for drug treatment after bone marrow transplantation, and were subsequently analyzed in a non-blinded fashion. Animal sample sizes were selected based on precedent established from previous publications and an understanding that at least n=5 is generally required to achieve statistical significance. Human CML samples were selected on the basis of sample availability and a requirement to achieve statistical significance. For mouse experiments involving shRNAs, the most efficacious shRNA of multiple shRNAs tested and validated in cell culture was used, a criterion that was established prospectively. All quantitative data were collected from experiments performed in at least triplicate. Original data for mouse and human studies are provided in table S5.
Additional experimental details are described in Supplementary Materials and Methods. Clone IDs for individual shRNAs used in this study are listed in table S6, and primer sequences used for qRT-PCR analysis are listed in table S7.
Supplementary Material
Acknowledgments
We thank N.J. Donato and T. Skorski for providing reagents, the University of Massachusetts RNAi Core for providing shRNA clones and pools and cDNA clones, and S. Deibler for editorial assistance.
Funding: R.B. is a Howard Hughes Medical Institute Medical Research Fellow. The research was supported in part by funds from the Georgia Cancer Coalition. B.J.D. is a Howard Hughes Medical Institute Investigator and receives additional research funding from NIH MERIT award 5R37 CA65823-16 and the Leukemia & Lymphoma Society. This work was supported by a grant from the National Institutes of Health (R01 CA163926) to M.R.G. M.R.G is also an investigator of the Howard Hughes Medical Institute.
Footnotes
This manuscript has been accepted for publication in Science Translational Medicine. This version has not undergone final editing. Please refer to the complete version of record at www.sciencetranslationalmedicine.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Author contributions: L.M., Z.S., S.L. and M.R.G. designed the experiments. L.M. performed the majority of the experiments, with Y.S. assisting with the CML mouse models and FACS analysis, R.B assisting with immunoblotting and qRT-PCR, and L.X. assisting with oral gavage experiments. C.A.E., L.H., J.C., B.J.D. and H.J.K. provided CML patient samples. J.O. and L.J.Z. performed statistical analyses. L.M., C.A.E., B.J.D. and H.J.K., and S.L. and M.R.G. interpreted the data. L.M. and M.R.G. wrote the manuscript, and all other authors read and commented on the manuscript.
Competing interests: OHSU has clinical trial contracts with Novartis and Bristol-Myers-Squibb to pay for patient costs, nurse and data manager salaries, and institutional overhead. B.J.D. does not derive salary, nor does his lab receive funds, from these contracts.
Data and materials availability: Requests for materials should be addressed to M.R.G. (email: michael.green@umassmed.edu).
REFERENCES
- 1.Faderl S, Talpaz M, Estrov Z, O'Brien S, Kurzrock R, Kantarjian HM. The biology of chronic myeloid leukemia. N Engl J Med. 1999;341:164–172. doi: 10.1056/NEJM199907153410306. [DOI] [PubMed] [Google Scholar]
- 2.Deininger MW, Goldman JM, Melo JV. The molecular biology of chronic myeloid leukemia. Blood. 2000;96:3343–3356. [PubMed] [Google Scholar]
- 3.Kurzrock R, Kantarjian HM, Druker BJ, Talpaz M. Philadelphia chromosome-positive leukemias: from basic mechanisms to molecular therapeutics. Ann Intern Med. 2003;138:819–830. doi: 10.7326/0003-4819-138-10-200305200-00010. [DOI] [PubMed] [Google Scholar]
- 4.Colicelli J. ABL tyrosine kinases: evolution of function, regulation, and specificity. Sci Signal. 2010;3:re6. doi: 10.1126/scisignal.3139re6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steelman LS, Pohnert SC, Shelton JG, Franklin RA, Bertrand FE, McCubrey JA. JAK/STAT, Raf/MEK/ERK PI3K/Akt and BCR-ABL in cell cycle progression and leukemogenesis. Leukemia. 2004;18:189–218. doi: 10.1038/sj.leu.2403241. [DOI] [PubMed] [Google Scholar]
- 6.An X, Tiwari AK, Sun Y, Ding PR, Ashby CR, Jr., Chen ZS. BCR-ABL tyrosine kinase inhibitors in the treatment of Philadelphia chromosome positive chronic myeloid leukemia: a review. Leuk Res. 2010;34:1255–1268. doi: 10.1016/j.leukres.2010.04.016. [DOI] [PubMed] [Google Scholar]
- 7.von Bubnoff N, Peschel C, Duyster J. Resistance of Philadelphia-chromosome positive leukemia towards the kinase inhibitor imatinib (STI571, Glivec): a targeted oncoprotein strikes back. Leukemia. 2003;17:829–838. doi: 10.1038/sj.leu.2402889. [DOI] [PubMed] [Google Scholar]
- 8.Quintas-Cardama A, Kantarjian HM, Cortes JE. Mechanisms of primary and secondary resistance to imatinib in chronic myeloid leukemia. Cancer Control. 2009;16:122–131. doi: 10.1177/107327480901600204. [DOI] [PubMed] [Google Scholar]
- 9.Jabbour E, Kantarjian H, Jones D, Talpaz M, Bekele N, O'Brien S, Zhou X, Luthra R, Garcia-Manero G, Giles F, Rios MB, Verstovsek S, Cortes J. Frequency and clinical significance of BCR-ABL mutations in patients with chronic myeloid leukemia treated with imatinib mesylate. Leukemia. 2006;20:1767–1773. doi: 10.1038/sj.leu.2404318. [DOI] [PubMed] [Google Scholar]
- 10.Shah NP, Nicoll JM, Nagar B, Gorre ME, Paquette RL, Kuriyan J, Sawyers CL. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2:117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 11.Weisberg E, Manley PW, Cowan-Jacob SW, Hochhaus A, Griffin JD. Second generation inhibitors of BCR-ABL for the treatment of imatinib-resistant chronic myeloid leukaemia. Nature reviews. Cancer. 2007;7:345–356. doi: 10.1038/nrc2126. [DOI] [PubMed] [Google Scholar]
- 12.Donato NJ, Wu JY, Stapley J, Lin H, Arlinghaus R, Aggarwal BB, Shishodia S, Albitar M, Hayes K, Kantarjian H, Talpaz M. Imatinib mesylate resistance through BCR-ABL independence in chronic myelogenous leukemia. Cancer Res. 2004;64:672–677. doi: 10.1158/0008-5472.can-03-1484. [DOI] [PubMed] [Google Scholar]
- 13.Khorashad JS, Anand M, Marin D, Saunders S, Al-Jabary T, Iqbal A, Margerison S, Melo JV, Goldman JM, Apperley JF, Kaeda J. The presence of a BCR-ABL mutant allele in CML does not always explain clinical resistance to imatinib. Leukemia. 2006;20:658–663. doi: 10.1038/sj.leu.2404137. [DOI] [PubMed] [Google Scholar]
- 14.Dean M, Fojo T, Bates S. Tumour stem cells and drug resistance. Nat Rev Cancer. 2005;5:275–284. doi: 10.1038/nrc1590. [DOI] [PubMed] [Google Scholar]
- 15.Graham SM, Jorgensen HG, Allan E, Pearson C, Alcorn MJ, Richmond L, Holyoake TL. Primitive, quiescent, Philadelphia-positive stem cells from patients with chronic myeloid leukemia are insensitive to STI571 in vitro. Blood. 2002;99:319–325. doi: 10.1182/blood.v99.1.319. [DOI] [PubMed] [Google Scholar]
- 16.Corbin AS, Agarwal A, Loriaux M, Cortes J, Deininger MW, Druker BJ. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest. 2011;121:396–409. doi: 10.1172/JCI35721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamilton A, Helgason GV, Schemionek M, Zhang B, Myssina S, Allan EK, Nicolini FE, Muller-Tidow C, Bhatia R, Brunton VG, Koschmieder S, Holyoake TL. Chronic myeloid leukemia stem cells are not dependent on Bcr-Abl kinase activity for their survival. Blood. 2012;119:1501–1510. doi: 10.1182/blood-2010-12-326843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andersson LC, Nilsson K, Gahmberg CG. K562--a human erythroleukemic cell line. International journal of cancer. Int J Cancer. 1979;23:143–147. doi: 10.1002/ijc.2910230202. [DOI] [PubMed] [Google Scholar]
- 19.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, Schlabach MR, Sheth N, Bradshaw J, Burchard J, Kulkarni A, Cavet G, Sachidanandam R, McCombie WR, Cleary MA, Elledge SJ, Hannon GJ. Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet. 2005;37:1281–1288. doi: 10.1038/ng1650. [DOI] [PubMed] [Google Scholar]
- 20.Ohkubo T, Kamamoto T, Kita K, Hiraoka A, Yoshida Y, Uchino H. A novel Ph1 chromosome positive cell line established from a patient with chronic myelogenous leukemia in blastic crisis. Leukemia Res. 1985;9:921–926. doi: 10.1016/0145-2126(85)90314-5. [DOI] [PubMed] [Google Scholar]
- 21.Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science. 1990;247:824–830. doi: 10.1126/science.2406902. [DOI] [PubMed] [Google Scholar]
- 22.Li S, Ilaria RL, Jr., Million RP, Daley GQ, Van Etten RA, The P190 P210. and P230 forms of the BCR/ABL oncogene induce a similar chronic myeloid leukemia-like syndrome in mice but have different lymphoid leukemogenic activity. J Exp Med. 1999;189:1399–1412. doi: 10.1084/jem.189.9.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Quentmeier H, Eberth S, Romani J, Zaborski M, Drexler HG. BCR-ABL1-independent PI3Kinase activation causing imatinib-resistance. J Hematol Oncol. 2011;4:6. doi: 10.1186/1756-8722-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shah NP, Tran C, Lee FY, Chen P, Norris D, Sawyers CL. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305:399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 25.Donato NJ, Wu JY, Stapley J, Gallick G, Lin H, Arlinghaus R, Talpaz M. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101:690–698. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 26.ten Hoeve J, Arlinghaus RB, Guo JQ, Heisterkamp N, Groffen J. Tyrosine phosphorylation of CRKL in Philadelphia+ leukemia. Blood. 1994;84:1731–1736. [PubMed] [Google Scholar]
- 27.Takahashi T, Ueno H, Shibuya M. VEGF activates protein kinase C-dependent, but Ras-independent Raf-MEK-MAP kinase pathway for DNA synthesis in primary endothelial cells. Oncogene. 1999;18:2221–2230. doi: 10.1038/sj.onc.1202527. [DOI] [PubMed] [Google Scholar]
- 28.Ueda Y, Hirai S, Osada S, Suzuki A, Mizuno K, Ohno S. Protein kinase C activates the MEK-ERK pathway in a manner independent of Ras and dependent on Raf. J Biol Chem. 1996;271:23512–23519. doi: 10.1074/jbc.271.38.23512. [DOI] [PubMed] [Google Scholar]
- 29.Uht RM, Amos S, Martin PM, Riggan AE, Hussaini IM. The protein kinase C-eta isoform induces proliferation in glioblastoma cell lines through an ERK/Elk-1 pathway. Oncogene. 2007;26:2885–2893. doi: 10.1038/sj.onc.1210090. [DOI] [PubMed] [Google Scholar]
- 30.Aderem A. The MARCKS brothers: a family of protein kinase C substrates. Cell. 1992;71:713–716. doi: 10.1016/0092-8674(92)90546-o. [DOI] [PubMed] [Google Scholar]
- 31.Chakrabarti R, Hwang J, Andres Blanco M, Wei Y, Lukacisin M, Romano RA, Smalley K, Liu S, Yang Q, Ibrahim T, Mercatali L, Amadori D, Haffty BG, Sinha S, Kang Y. Elf5 inhibits the epithelial-mesenchymal transition in mammary gland development and breast cancer metastasis by transcriptionally repressing Snail2. Nat Cell Biol. 2012;14:1212–1222. doi: 10.1038/ncb2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escamilla-Hernandez R, Chakrabarti R, Romano RA, Smalley K, Zhu Q, Lai W, Halfon MS, Buck MJ, Sinha S. Genome-wide search identifies Ccnd2 as a direct transcriptional target of Elf5 in mouse mammary gland. BMC Mol Biol. 2010;11:68. doi: 10.1186/1471-2199-11-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kalyuga M, Gallego-Ortega D, Lee HJ, Roden DL, Cowley MJ, Caldon CE, Stone A, Allerdice SL, Valdes-Mora F, Launchbury R, Statham AL, Armstrong N, Alles MC, Young A, Egger A, Au W, Piggin CL, Evans CJ, Ledger A, Brummer T, Oakes SR, Kaplan W, Gee JM, Nicholson RI, Sutherland RL, Swarbrick A, Naylor MJ, Clark SJ, Carroll JS, Ormandy CJ. ELF5 suppresses estrogen sensitivity and underpins the acquisition of antiestrogen resistance in luminal breast cancer. PLoS Biol. 2012;10:e1001461. doi: 10.1371/journal.pbio.1001461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wellbrock C, Karasarides M, Marais R. The RAF proteins take centre stage. Nat Rev Mol Cell Biol. 2004;5:875–885. doi: 10.1038/nrm1498. [DOI] [PubMed] [Google Scholar]
- 35.Carroll MP, May WS. Protein kinase C-mediated serine phosphorylation directly activates Raf-1 in murine hematopoietic cells. J Biol Chem. 1994;269:1249–1256. [PubMed] [Google Scholar]
- 36.Kolch W, Heidecker G, Kochs G, Hummel R, Vahidi H, Mischak H, Finkenzeller G, Marme D, Rapp UR. Protein kinase C alpha activates RAF-1 by direct phosphorylation. Nature. 1993;364:249–252. doi: 10.1038/364249a0. [DOI] [PubMed] [Google Scholar]
- 37.Suzuki T, Elias BC, Seth A, Shen L, Turner JR, Giorgianni F, Desiderio D, Guntaka R, Rao R. PKC eta regulates occludin phosphorylation and epithelial tight junction integrity. Proc Nat Acad Sci USA. 2009;106:61–66. doi: 10.1073/pnas.0802741106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai H, Smola U, Wixler V, Eisenmann-Tappe I, Diaz-Meco MT, Moscat J, Rapp U, Cooper GM. Role of diacylglycerol-regulated protein kinase C isotypes in growth factor activation of the Raf-1 protein kinase. Mol Cell Biol. 1997;17:732–741. doi: 10.1128/mcb.17.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schonwasser DC, Marais RM, Marshall CJ, Parker PJ. Activation of the mitogen-activated protein kinase/extracellular signal-regulated kinase pathway by conventional, novel, and atypical protein kinase C isotypes. Mol Cell Biol. 1998;18:790–798. doi: 10.1128/mcb.18.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sozeri O, Vollmer K, Liyanage M, Frith D, Kour G, Mark GE, 3rd, Stabel S. Activation of the c-Raf protein kinase by protein kinase C phosphorylation. Oncogene. 1992;7:2259–2262. [PubMed] [Google Scholar]
- 41.Neering SJ, Bushnell T, Sozer S, Ashton J, Rossi RM, Wang PY, Bell DR, Heinrich D, Bottaro A, Jordan CT. Leukemia stem cells in a genetically defined murine model of blast-crisis CML. Blood. 2007;110:2578–2585. doi: 10.1182/blood-2007-02-073031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang B, Strauss AC, Chu S, Li M, Ho Y, Shiang KD, Snyder DS, Huettner CS, Shultz L, Holyoake T, Bhatia R. Effective targeting of quiescent chronic myelogenous leukemia stem cells by histone deacetylase inhibitors in combination with imatinib mesylate. Cancer Cell. 2010;17:427–442. doi: 10.1016/j.ccr.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Peng C, Hu Y, Li H, Sheng Z, Chen Y, Sullivan C, Cerny J, Hutchinson L, Higgins A, Miron P, Zhang X, Brehm MA, Li D, Green MR, Li S. The Blk pathway functions as a tumor suppressor in chronic myeloid leukemia stem cells. Nat Genet. 2012;44:861–871. doi: 10.1038/ng.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li L, Wang L, Wang Z, Ho Y, McDonald T, Holyoake TL, Chen W, Bhatia R. Activation of p53 by SIRT1 inhibition enhances elimination of CML leukemia stem cells in combination with imatinib. Cancer Cell. 2012;21:266–281. doi: 10.1016/j.ccr.2011.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jiang X, Zhao Y, Smith C, Gasparetto M, Turhan A, Eaves A, Eaves C. Chronic myeloid leukemia stem cells possess multiple unique features of resistance to BCR-ABL targeted therapies. Leukemia. 2007;21:926–935. doi: 10.1038/sj.leu.2404609. [DOI] [PubMed] [Google Scholar]
- 46.Lobo NA, Shimono Y, Qian D, Clarke MF. The biology of cancer stem cells. Annu Rev Cell Dev Biol. 2007;23:675–699. doi: 10.1146/annurev.cellbio.22.010305.104154. [DOI] [PubMed] [Google Scholar]
- 47.Bhatia R, Holtz M, Niu N, Gray R, Snyder DS, Sawyers CL, Arber DA, Slovak ML, Forman SJ. Persistence of malignant hematopoietic progenitors in chronic myelogenous leukemia patients in complete cytogenetic remission following imatinib mesylate treatment. Blood. 2003;101:4701–4707. doi: 10.1182/blood-2002-09-2780. [DOI] [PubMed] [Google Scholar]
- 48.Chomel JC, Bonnet ML, Sorel N, Bertrand A, Meunier MC, Fichelson S, Melkus M, Bennaceur-Griscelli A, Guilhot F, Turhan AG. Leukemic stem cell persistence in chronic myeloid leukemia patients with sustained undetectable molecular residual disease. Blood. 2011;118:3657–3660. doi: 10.1182/blood-2011-02-335497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pellicano F, Mukherjee L, Holyoake TL. Concise review: cancer cells escape from oncogene addiction: understanding the mechanisms behind treatment failure for more effective targeting. Stem Cells. 2014;32:1373–1379. doi: 10.1002/stem.1678. [DOI] [PubMed] [Google Scholar]
- 50.Gerber JM, Gucwa JL, Esopi D, Gurel M, Haffner MC, Vala M, Nelson WG, Jones RJ, Yegnasubramanian S. Genome-wide comparison of the transcriptomes of highly enriched normal and chronic myeloid leukemia stem and progenitor cell populations. Oncotarget. 2013;4:715–728. doi: 10.18632/oncotarget.990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Konuma T, Nakamura S, Miyagi S, Negishi M, Chiba T, Oguro H, Yuan J, Mochizuki-Kashio M, Ichikawa H, Miyoshi H, Vidal M, Iwama A. Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp Hematol. 2011;39:697–709. doi: 10.1016/j.exphem.2011.03.008. e695. [DOI] [PubMed] [Google Scholar]
- 52.Bagger FO, Rapin N, Theilgaard-Monch K, Kaczkowski B, Thoren LA, Jendholm J, Winther O, Porse BT. HemaExplorer: a database of mRNA expression profiles in normal and malignant haematopoiesis. Nucleic Acids Res. 2013;41:D1034–D1039. doi: 10.1093/nar/gks1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Packer LM, Rana S, Hayward R, O'Hare T, Eide CA, Rebocho A, Heidorn S, Zabriskie MS, Niculescu-Duvaz I, Druker BJ, Springer C, Marais R. Nilotinib and MEK inhibitors induce synthetic lethality through paradoxical activation of RAF in drug-resistant chronic myeloid leukemia. Cancer Cell. 2011;20:715–727. doi: 10.1016/j.ccr.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aceves-Luquero CI, Agarwal A, Callejas-Valera JL, Arias-Gonzalez L, Esparis-Ogando A, del Peso Ovalle L, Bellon-Echeverria I, de la Cruz-Morcillo MA, Galan Moya EM, Moreno Gimeno I, Gomez JC, Deininger MW, Pandiella A, Sanchez Prieto R. ERK2, but not ERK1, mediates acquired and "de novo" resistance to imatinib mesylate: implication for CML therapy. PLoS One. 2009;4:e6124. doi: 10.1371/journal.pone.0006124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hentschel J, Rubio I, Eberhart M, Hipler C, Schiefner J, Schubert K, Loncarevic IF, Wittig U, Baniahmad A, von Eggeling F. BCR-ABL- and Ras-independent activation of Raf as a novel mechanism of Imatinib resistance in CML. Int J Oncol. 2011;39:585–591. doi: 10.3892/ijo.2011.1062. [DOI] [PubMed] [Google Scholar]
- 56.Nambu T, Araki N, Nakagawa A, Kuniyasu A, Kawaguchi T, Hamada A, Saito H. Contribution of BCR-ABL-independent activation of ERK1/2 to acquired imatinib resistance in K562 chronic myeloid leukemia cells. Cancer Sci. 2010;101:137–142. doi: 10.1111/j.1349-7006.2009.01365.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chang JS, Santhanam R, Trotta R, Neviani P, Eiring AM, Briercheck E, Ronchetti M, Roy DC, Calabretta B, Caligiuri MA, Perrotti D. High levels of the BCR/ABL oncoprotein are required for the MAPK-hnRNP-E2 dependent suppression of C/EBPalpha-driven myeloid differentiation. Blood. 2007;110:994–1003. doi: 10.1182/blood-2007-03-078303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chu S, Holtz M, Gupta M, Bhatia R. BCR/ABL kinase inhibition by imatinib mesylate enhances MAP kinase activity in chronic myelogenous leukemia CD34+ cells. Blood. 2004;103:3167–3174. doi: 10.1182/blood-2003-04-1271. [DOI] [PubMed] [Google Scholar]
- 59.Mizuchi D, Kurosu T, Kida A, Jin ZH, Jin A, Arai A, Miura O. BCR/ABL activates Rap1 and B-Raf to stimulate the MEK/Erk signaling pathway in hematopoietic cells. Biochem Biophys Res Commun. 2005;326:645–651. doi: 10.1016/j.bbrc.2004.11.086. [DOI] [PubMed] [Google Scholar]
- 60.Pellicano F, Simara P, Sinclair A, Helgason GV, Copland M, Grant S, Holyoake TL. The MEK inhibitor PD184352 enhances BMS-214662-induced apoptosis in CD34+ CML stem/progenitor cells. Leukemia. 2011;25:1159–1167. doi: 10.1038/leu.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mullenders J, Bernards R. Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene. 2009;28:4409–4420. doi: 10.1038/onc.2009.295. [DOI] [PubMed] [Google Scholar]
- 62.Bruennert D, Czibere A, Bruns I, Kronenwett R, Gattermann N, Haas R, Neumann F. Early in vivo changes of the transcriptome in Philadelphia chromosome-positive CD34+ cells from patients with chronic myelogenous leukaemia following imatinib therapy. Leukemia. 2009;23:983–985. doi: 10.1038/leu.2008.337. [DOI] [PubMed] [Google Scholar]
- 63.Radich JP, Dai H, Mao M, Oehler V, Schelter J, Druker B, Sawyers C, Shah N, Stock W, Willman CL, Friend S, Linsley PS. Gene expression changes associated with progression and response in chronic myeloid leukemia. Proc Nat Acad Sci USA. 2006;103:2794–2799. doi: 10.1073/pnas.0510423103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jing J, Greshock J, Holbrook JD, Gilmartin A, Zhang X, McNeil E, Conway T, Moy C, Laquerre S, Bachman K, Wooster R, Degenhardt Y. Comprehensive predictive biomarker analysis for MEK inhibitor GSK1120212. Mol Cancer Ther. 2012;11:720–729. doi: 10.1158/1535-7163.MCT-11-0505. [DOI] [PubMed] [Google Scholar]
- 65.Gazin C, Wajapeyee N, Gobeil S, Virbasius CM, Green MR. An elaborate pathway required for Ras-mediated epigenetic silencing. Nature. 2007;449:1073–1077. doi: 10.1038/nature06251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ihaka R, Gentleman R. R: A language for data analysis and graphics. J Comput Graph Stat. 1996;5:299–314. [Google Scholar]
- 68.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Statist Soc Ser. 1995;B 57:289–300. [Google Scholar]
- 69.Slinker BK. The statistics of synergism. J Mol Cell Cardiol. 1998;30:723–731. doi: 10.1006/jmcc.1998.0655. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.