Abstract
GammaPNA oligomers having one or two repeats of the sequence AATCCC were designed to hybridize to DNA having one or more repeats of the complementary TTAGGG sequence found in the human telomere. UV melting curves and surface plasmon resonance experiments demonstrate high affinity and cooperativity for hybridization of these miniprobes to DNA having multiple complementary repeats. Fluorescence spectroscopy for Cy3-labeled miniprobes demonstrate increases in fluorescence intensity for assembling multiple short probes on a DNA target compared with fewer longer probes. The fluorescent γPNA miniprobes were then used to stain telomeres in metaphase chromosomes derived from U2OS cells possessing heterogeneous long telomeres and Jurkat cells harboring homogenous short telomeres. The miniprobes yielded comparable fluorescence intensity to a commercially available PNA 18mer probe in U2OS cells, but significantly brighter fluorescence was observed for telomeres in Jurkat cells. These results suggest that γPNA miniprobes can be effective telomere-staining reagents with applications toward analysis of critically short telomeres, which have been implicated in a range of human diseases.
Hybridization probes are central to many biotechnological applications involving nucleic acids, including microarrays, PCR amplification and diagnostics, fluorescence in situ hybridization (FISH), affinity purification and antisense regulation of gene expression. Modified backbones such as phosphorothioate, morpholino and locked nucleic acids (LNA) have established footholds in various applications due to superior performance relative to unmodified DNA oligonucleotides. For example, the nuclease stability of morpholino oligomers has led to their common use as antisense agents to block translation of specific mRNAs in microinjected zebrafish embryos1, 2, while LNA is gaining increasing use due in large part to its exceptionally high affinity for complementary DNA and RNA3.
Peptide nucleic acid (PNA, Chart 1) is another synthetic nucleic acid analogue that has found applications, primarily in FISH analyses. Considerable interest in PNA has arisen from its high affinity for complementary DNA and RNA in spite of its radically altered backbone structure relative to DNA.4 (The enhanced affinity is due, at least in part, to the lack of a negative charge on the PNA backbone.5) A fluorescent PNA 18mer complementary to three repeats of the human telomere sequence is widely used for telomere staining and quantitative length determination.6 PNA FISH probes targeted to ribosomal RNA are also commonly used in clinical pathogen identification assays.7
Chart 1.
Chemical structures of PNA, γPNA and the guanidino G-clamp nucleobase.
An important advance in the development of PNA came with the reports of γPNA, which exhibits further enhancement of affinity,8 comparable to LNA,9 but also improved water solubility with appropriate substituents at the gamma carbon10 (Chart 1). The high affinity of γPNA has been attributed to its right-handed helical structure, which pre-organizes γPNA for hybridization to complementary DNA and RNA.8, 11 The added affinity extends the strand invasion capability first reported for unmodified PNA12, 13 to virtually any sequence of double-stranded DNA.14, 15
The high affinity of γPNA also provides an opportunity to address an ongoing concern with FISH probes, namely brightness. In order to sensitively detect genomic DNA or low abundance RNA targets in situ without signal amplification, bright probes are needed. cDNA probes prepared by enzymatic methods can contain multiple fluorescent dyes by virtue of using labeled dNTPs, although such long, polyanionic probes typically require lengthy incubation steps due to slow hybridization kinetics. Alternatively, relatively short (20mer), synthetic 2'-O-methoxy probes, each bearing a fluorescent dye and targeted to a different subsequence within a transcript, have been used to stain mRNA.16 Multimer probes consisting of up to four copies of a biotinylated probe assembled via a streptavidin core have also been used to achieve single molecule imaging of endogenous mRNAs.17
In the context of nucleic acid hybridization, higher affinity can be translated into shorter probe length to realize comparable equilibrium constant and melting temperature as lower affinity probes. These “miniprobes” offer a new opportunity to increase the brightness of a labeled DNA or RNA target: more probes and, therefore, more dyes can be delivered to a target sequence of a given length. This strategy can be particularly effective for staining repeating sequence targets, such as telomeric DNA. Herein, we describe the DNA hybridization properties of γPNA miniprobes and demonstrate effective staining of telomeres in two human cell lines.
RESULTS AND DISCUSSION
Design
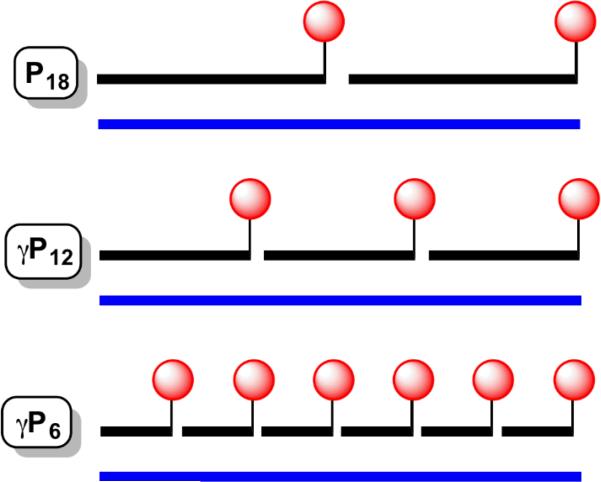
γPNA miniprobes were designed to be complementary to the repeating hexamer sequence found in human telomeric DNA: 5’-TTAGGG-3’. Three oligomers were synthesized (Table 1): a 12mer complementary to two telomere repeats, a 6mer complementary to a single repeat, and a 6mer in which two of the cytosine bases were replaced by guanidino G-clamp, a synthetic nucleobase that forms more stable base pairs with G than does C, by virtue of additional hydrogen bonding and pi stacking ability.18, 19 Ly and coworkers reported that incorporation of guanidino G-clamp residues into γPNA significantly enhances strand invasion into duplex DNA.20 As shown in Scheme 1, hybridization of the 12mer or 6mer γPNA miniprobes to a complementary DNA of a given length will result in 1.5 or 3 times as many dyes being delivered to the target nucleic acid as the conventional PNA probe.
Table 1.
Sequences of PNA, γPNA and DNA oligomers used in this study.
| Name | Sequence† |
|---|---|
| P18 | H2N-Lys-(AATCCC)3-Cy3 |
| γP12 | H2N-Lys-(AATCCC)2-Cy3 |
| γP6 | H2N-Lys-AATCCC-Cy3 |
| γP6-clamp§ | H2N- Lys-AATXCX-Cy3 |
| Telo-n (DNA) | 5’-(TTAGGG)n-3’ |
PNA and γPNA sequences are written in the C-to-N direction.
X = G-clamp nucleobases
Scheme 1.
γPNAs were synthesized by standard solid-phase methods,21, 22 end-labeled with Cy3 prior to cleavage from the polystyrene support, purified by reversed phase HPLC and characterized by MALDI-ToF mass spectrometry (supporting information).
UV Melting Analysis
The DNA hybridization properties of the γPNAs were assessed using UV melting curve analysis. We first compared formation of 1:1 duplexes between each of the γPNAs and a complementary DNA oligonucleotide consisting of the corresponding number of telomeric repeats. For example γP12 was hybridized to Telo-2, consisting of two telomeric repeats, while γP6 and γP6-clamp were hybridized to Telo-1. For comparison, P18 was hybridized to Telo-3.
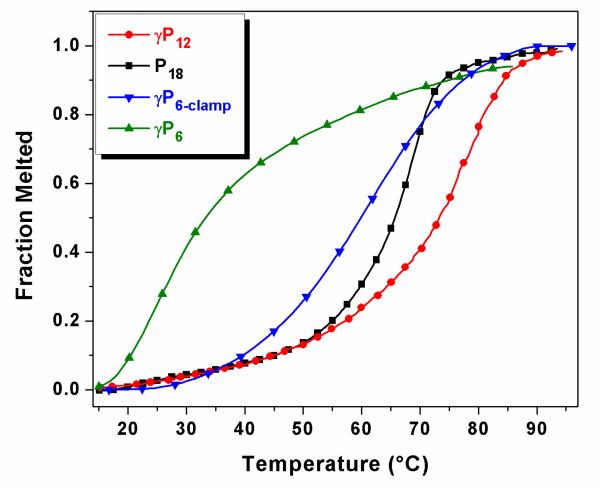
Figure 1 shows UV melting curves recorded at 260 nm for samples containing PNA or γPNA hybridized to the complementary DNA in a Tris buffer containing 100 mM KCl. Under these conditions, the traditional PNA probe P18 exhibits a melting temperature Tm = 66 °C with the complementary Telo-3 probe. The 12mer γPNA miniprobe γP12 forms a more stable duplex with its complementary DNA Telo-2 (Tm = 77 °C) in spite of having 6 fewer base pairs, illustrating the exceptionally high affinity of the gamma-modified backbone. In contrast, the γP6/Telo-1 duplex has a much lower stability (Tm = 24 °C), although substituting two G-clamp bases into the γPNA significantly increases the stability to the point where the 6mer γPNA-DNA duplex is almost as stable as the 18mer PNA-DNA duplex (Tm = 62 °C versus 66 °C).
Fig. 1.
UV melting curves recorded for 1:1 duplexes between PNA or γPNA and the complementary DNA strands Telo-1, Telo-2 or Telo-3. Samples contained 4 μM each of PNA/γPNA and DNA repeat sequences, as explained in the Experimental section.
We next characterized the ability of multiple γPNA miniprobes to hybridize to a single DNA oligonucleotide having multiple repeating complementary target sites. The experiments included hybridization of γP12 to either Telo-2 (1:1 duplex) or Telo-4 (2:1 duplex), and of γP6 or γP6-clamp to Telo-1/Telo-4 (1:1 up to 4:1 duplexes). The experiments were designed to maintain the same total base pair concentration (24 μM) by adjusting the concentration of the DNA complement. For example, γP6 and Telo-1 were mixed in 4 μM concentration, whereas Telo-4 was used at 1 μM concentration.
Melting curves are shown in Supplementary Figures 1 and 2 and melting temperatures are reported in Table 2. It is evident that the stabilities of the duplexes formed by γP12 and γP6-clamp increase with the length of the DNA complement, in spite of the entropic penalty inherent to formation of 2:1, 3:1 or 4:1 duplexes compared with 1:1 duplexes. These results indicate considerable cooperativity in hybridization of γPNA miniprobes to adjacent sites on the DNA complement, with end-to-end stacking of the adjacent probes likely responsible for the added stability for the longer DNA complements. A similar phenomenon was proposed to account for the higher thermal stability of a PNA2-DNA triplex formed on a polymeric deoxyadenosine template relative to a dA8 single strand.23 Cooperative hybridization has also been proposed as a possible explanation for the ability of antisense oligonucleotides to inhibit translation of huntingtin mRNA bearing greater numbers of triplet CAG repeats relative to alleles with fewer repeats24. It is also worth noting that Telo-4 folds into a G-quadruplex structure at room temperature, but this is far less stable (Tm = 40 °C, Supplementary Figure 3) than the corresponding duplexes formed with γP12 and γP6-clamp. (γP6 initially shows a substantial increase in Tm for Telo-2 versus Telo-1, but this is reduced at longer DNA lengths, perhaps due in part to competitive G-quadruplex formation, at least for Telo-4.) Taken together, the UV melting experiments reveal that the γPNA miniprobes can hybridize to complementary DNA oligonucleotides to form heteroduplexes stabilized by cooperative end-stacking of adjacent miniprobes.
Table 2.
UV melting temperature (Tm, °C) values recorded for PNA- and γPNA-DNA duplexes.
| P18 | γP12 | γP6 | γP6-clamp | |
|---|---|---|---|---|
| Telo-1 | nd | nd | 24.3 ± 1.7 | 62.3 ± 1.1 |
| Telo-2 | nd | 77.3 ± 3.2 | 42.8 ± 1.9 | 72.4 ± 0.4 |
| Telo-3 | 66.0 | nd | 38.4 ± 0.7 | 78.4 ± 0.3 |
| Telo-4 | nd | 82.5 ± 0.3 | 38.2 ± 1.3 | 81.3 ± 0.7 |
nd = not determined
Fluorescence Spectroscopy
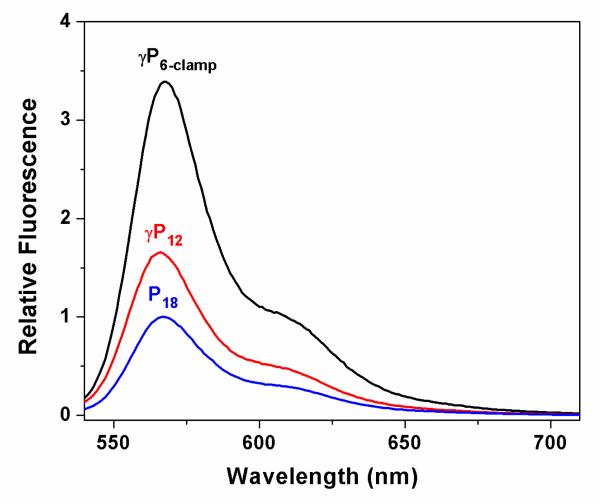
The advantage of miniprobes for fluorescent labeling is the ability to deliver more dyes per target nucleic acid (Scheme 1). To demonstrate this effect, we compared fluorescence spectra recorded for P18, γP12 and γP6-clamp after hybridization to a 36mer DNA bearing 6 repeats of the AATCCC complementary sequence. Thus, the three probes should hybridize in 2:1, 3:1 and 6:1 stoichiometries, respectively (Scheme 1). Figure 2 shows the spectra, normalized to P18. The fluorescence intensity increases by factors of 1.7 and 3.4 for the 12mer and 6mer γPNA miniprobes, respectively, relative to the PNA 18mer, slightly exceeding the theoretical 1.5- and 3-fold increases based on the stoichiometries. These results demonstrate that fluorescently labeled γPNA miniprobes as short as 6mers can be hybridized directly adjacent to one another along a DNA target with no apparent loss of quantum yield for the fluorophore.
Fig. 2.
Fluorescence emission spectra recorded for samples containing 20 nM Telo-6 DNA and 40 nM P18. 60 nM γP12 or 120 nM γP6-clamp.
SPR Analysis
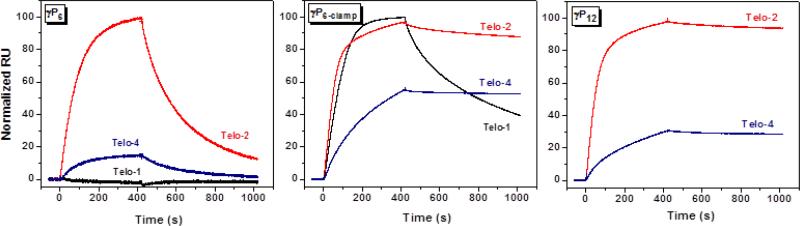
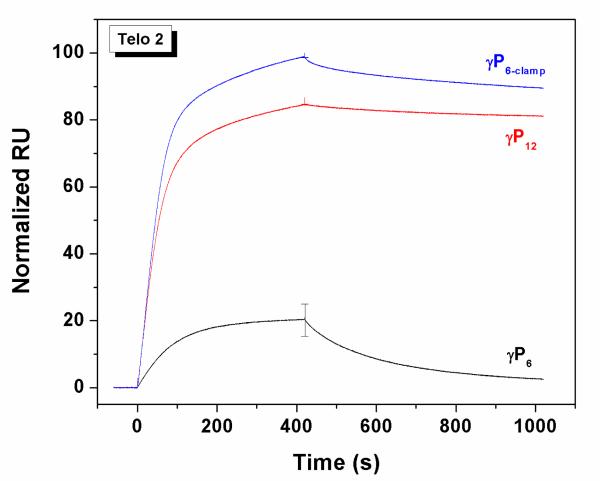
We also used surface plasmon resonance experiments to compare the kinetics of hybridization and dissociation for the various PNA and γPNA oligomers to immobilized DNA targets. For this experiment, approximately the same mass of biotinylated Telo-1, Telo-2 and Telo-4 was immobilized on separate flow cells of a streptavidin-coated SPR chip. Since the molecular weight of the three DNAs approximately doubles with each increase in length, this insures that the same number of binding sites is present on each flow cell, although the surface density of the DNA varies. Each of the γPNA miniprobes was flowed over the surface of the chip at a concentration of 50 nM for a period of 420 sec, then the flow solution was changed to γPNA-free buffer to promote dissociation of the bound γPNA. Sensorgrams representing the average of three trials are shown in Figures 3 and 4 and initial on- and off-rates are shown in Tables 3 and 4.
Fig. 3.
SPR sensorgrams recorded for association and dissociation of γP6, γP6-clamp or γP12 to immobilized DNA targets. 10 nM γPNA was flowed over the SPR chip during the association phase (400 s) before switching to γPNA-free buffer to promote dissociation.
Fig. 4.
SPR sensorgrams for association and dissociation of 50 nM γP6, γP6-clamp and γP12 to immobilized. Telo-2
Table 3.
Initial on-rates and off-rates (RU/s) of duplexes formed in 150 mM KCl. (Values in parentheses are normalized to the γP12 data for each column.)
| Telo-1 | Telo-2 | Telo-4 | ||||
|---|---|---|---|---|---|---|
| On-rate | Off-rate | On-rate | Off-rate | On-rate | Off-rate | |
| γP6 | 0 | 0 | 0.768 (0.8) | 0.427 (33) | 0.117 (0.7) | 0.68 (34) |
| γP6-clamp | 0.789 | 0.250 | 1.016 (1.0) | 0.036 (2.7) | 0.276 (1.8) | 0.011 (5.5) |
| γP12 | 1.015 (1.0) | 0.013 (1.0) | 0.155 (1.0) | 0.002 (1.0) | ||
Figure 3 (left) shows SPR sensorgrams for the hexamer miniprobe γP6. No binding is observed with Telo-1, consistent with the low melting temperature recorded for this duplex, even at much higher concentration (Tm = 24 °C, Table 2). Extending the DNA target to include two complementary sites (Telo-2) leads to observable binding of γP6, although the γPNA also dissociates relatively quickly from the immobilized DNA. Binding of the γPNA probe to Telo-4 is significantly slower than to Telo-2 under these conditions, presumably due to folding of Telo-4 into a G-quadruplex structure on the chip, as we observed previously for a related DNA oligonucleotide.25 This hypothesis was supported by the observation that replacing KCl with LiCl, where the quadruplex would be destabilized,26 resulted in a large increase in γPNA hybridization (Supplementary Figure 4).
The same experiment was performed using the higher affinity miniprobe γP6-clamp (Figure 3, center). In contrast to γP6, γP6-clamp binds rapidly to Telo-1, followed by slower dissociation. Binding to Telo-2 appears to occur in two phases, a faster phase that slightly exceeds that of Telo-1, followed by a slower phase. Since there are two binding sites for the γPNA on Telo-2, one possible explanation for these observations is that the kinetics reflect differences in rates for binding to the two sites. Within this model, the fast rate could correspond to binding to the distal site, i.e. the site furthest from the chip surface, while the slower rate could be due to lower accessibility of the site proximal to the streptavidin-coated surface. Regardless of the order of hybridization to the two sites, once bound, the γPNA molecules strongly resist dissociation. Although the slower off-rate from Telo-2 relative to Telo-1 could reflect the presence of two binding sites per DNA strand (which would promote re-capture of a dissociated γPNA), another contribution likely comes from the stabilizing effect of end-stacking between adjacent γPNA molecules hybridized to the same strand. Notably, while association of γP6-clamp is slower for Telo-4, those γPNA molecules that bind to the immobilized DNA are even slower to dissociate than for Telo-2. In addition, comparing the data for γP6-clamp and γP6 with Telo-2, these results indicate that the guanidine G-clamp nucleobase modifications exert their affinity-enhancing effect primarily through a slower dissociation rate.
We next performed SPR analysis of the 12mer miniprobe γP12. Figure 3 (right) compares binding and dissociation from Telo-2 and Telo-4. Even the 1:1 complex between γP12 and Telo-2 shows very slow dissociation, as we reported previously for γPNA-DNA duplexes.10 Interestingly, four-fold less γP12 binds to the quadruplex target Telo-4 versus the unfolded Telo-2, whereas this difference for γP6-clamp is only two-fold (compare the central and right panels of Figure 3), perhaps indicating a kinetic advantage of shorter probes for invading structured targets.
The data in Figure 3 were normalized to the highest binding signal within each set of data. To compare different γPNA miniprobes for binding to a single target, we renormalized the sensorgrams for binding to Telo-2 (Figure 4). These data show the slightly faster binding of the γP6-clamp versus γP12 but slower dissociation of the longer probe, where twice as many base pairs need to be disrupted in order to allow the probe to dissociate.
Table 3 shows the initial on- and off-rates determined from the SPR sensorgrams. Several conclusions can be drawn from the data, which reinforce the qualitative observations made from the sensorgrams. First, the data for Telo-2 illustrate similar on-rates for the three γPNAs but off-rates that vary by a factor of 33 from the fastest (γP6) to the slowest (γP12). These results illustrate the helix-stabilizing effects of the guanidine G-clamp nucleobase for γP6-clamp and the longer probe length for γP12. Next, comparing data across a row, e.g. γP6-clamp with Telo-1, -2 and -4, The increasing kinetic stability with target length is evident as the off-rates get progressively slower as more probes can hybridize adjacent to one another. In particular, comparing Telo-1 with Telo-2, the on-rate is only 29% greater for the longer target, whereas the off-rate is nearly 7-fold slower, connecting the cooperativity of hybridization to greater resistance to dissociation. In addition, the much slower on-rates due to the inherent quadruplex structure for Telo-4 are also evident in comparing the absolute values with those of Telo-2.
Telomere Analysis
Since the initial report by Lansdorp and coworkers6, the 18mer Cy3-labeled PNA probe P18 has become widely used for telomere imaging and length analysis. However, growing epidemiological evidence indicating that critically short telomeres are implicated in a wide range of aging-related diseases27, 28 motivates the development of brighter telomere probes, since shorter telomeres will bind fewer probes and therefore give lower fluorescence intensity relative to longer telomeres. Our hypothesis was that the higher affinity of γPNA would permit 12mer or perhaps even 6mer versions of the original 18mer PNA probe to be used for telomere analysis since this would allow a greater number of probes to bind to a telomere of a given length (Scheme 1).
To test the ability of the fluorescent γPNA miniprobes to stain telomeres using quantitative fluorescence in situ hybridization (qFISH), two human cell lines, U2OS osteosarcoma and Jurkat T-lymphocytes, were stained using standard telomere-FISH (teloFISH) protocols.29 These cell lines were chosen for their divergent telomere maintenance mechanisms. Approximately 85% of human cancers maintain stable telomere lengths through the activation of the holoenzyme telomerase.30 Conversely, approximately 15% of cancers maintain their telomeres in the absence of telomerase through a recombination-based pathway known as alternative lengthening of telomeres (ALT).30 This includes U2OS cells, which have relatively long mean telomere lengths (~21kb) compared to normal human cell lines.31-33 ALT-active cells exhibit highly variable telomere lengths, leading to a notable number of signal free ends (SFEs) in U2OS cells despite their long mean length. In contrast, Jurkat cells are telomerase positive and have shorter average telomere lengths than U2OS cells.31-33
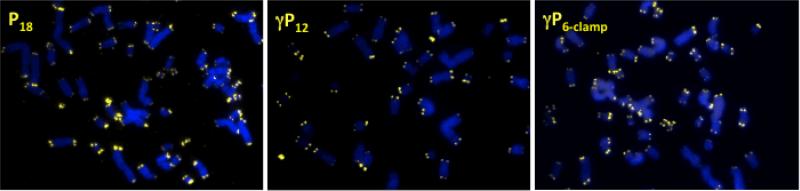
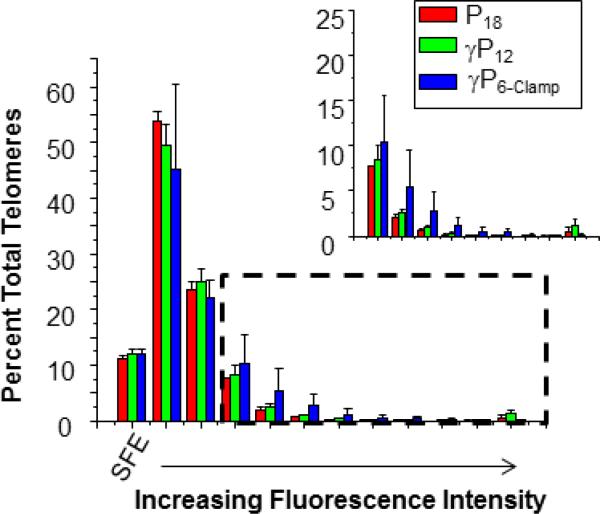
Cells were stalled in metaphase by the addition of colcemid and metaphase chromosome spreads were prepared and fixed as previously described.34 Chromosomes were hybridized for two hours with P18, γP12 or γP6-clamp and imaged for telomere staining. The fluorescence intensities of the Cy3 spots were determined using TFL-Telo quantitation software. Results for U2OS cells are shown in Figures 5 and 6. The yellow fluorescent spots observed at the ends of the chromosomes (counterstained with DAPI and colored blue) indicate successful telomere staining by all three probes (Figure 5). Quantitative analysis (Figure 6) revealed similar performance for the three probes in the low intensity region of the histogram, including equal number of unstained termini (SFEs), which result from loss of a telomere or telomeres that are too short to bind sufficient probe for detection. However, at higher fluorescence intensities, slightly higher percentages of telomeres were stained by the γPNA probes, particularly γP6-clamp (Figure 6, inset).
Fig. 5.
Telomere staining of U2OS metaphase chromosome spreads using Cy3-labeled PNA and γPNA probes. Staining by fluorescent in situ hybridization under denaturing conditions was achieved using P18 (left), γP12 (center), and γP6-clamp (right).
Fig. 6.
Quantitation of FISH results for U2OS cells. Percent values represent the average of three independent experiments. Error bars represent standard error of the mean. Bars represent fluorescence values quantified by bins of 30,000 arbitrary fluorescence units (dashed box indicates inset).
The similarity in SFEs detected using all three probes (18, 12 and 6-mer), despite the improved fluorescence intensity of the γ-modified probes, implies that the shortest telomeres in U2OS cells may be below the threshold for detection even for the shorter probes. Alternatively, these SFEs may represent chromatid ends that lack telomeric DNA or contain degenerative repeats such as those in subtelomeric DNA, since telomere lengthening is not templated by telomerase in these cells (due to the absence of telomerase).35
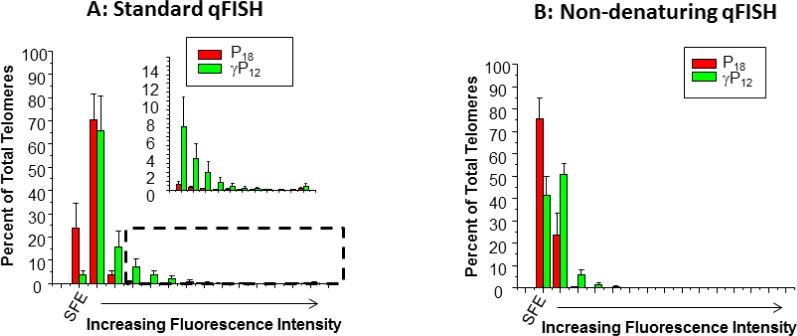
Similar experiments were performed with Jurkat cells (Figure 7A and Supplementary Figure 5), which rely on telomerase for telomere maintenance and exhibit stable lengths that average 6 kb.32. Considerable non-telomeric staining of chromosomes was observed for γP6-clamp (data not shown) so the analysis is restricted to P18 and γP12. In contrast to U2OS cells, staining Jurkat cells with γP12 resulted in approximately 5-fold fewer SFEs than staining with P18, consistent with improved staining of critically short telomeres. The γP12 staining also revealed a significantly greater percentage of telomeres with brighter signals compared to P18. The increase in the number of signal free ends and brighter overall fluorescence in this case is likely due to hybridization of more γP12 probes relative to P18 and suggests that analyzing the shortest telomeres in Jurkat cells is improved by using a more sensitive probe.
Fig. 7.
Quantitative analysis of telomere staining in Jurkat cells by fluorescent PNA and γPNA probes under (A) denaturing and (B) non-denaturing conditions. SFE = signal-free ends. Percent values represent the average of three independent experiments. Error bars represent standard error of the mean. Bars represent fluorescence values quantified by bins of 10,000 arbitrary fluorescence units (dashed box indicates inset).
The standard qFISH protocol requires denaturation of the chromosomal DNA prior to hybridization of the probe to the telomere, which consists of a relatively short single-stranded region extending into a much longer double-stranded region, both of which contain TTAGGG repeats complementary to the probe. However, previous studies have shown that γPNA affinity for complementary DNA is sufficiently high to allow direct invasion of double-stranded DNA in order to access target sites.15 Therefore, we hypothesized that the denaturing step could be omitted when using the γP12 probe to stain the metaphase chromosome preparations. To test this, we compared unmodified P18 and γP12 under non-denaturing hybridization conditions, in which formamide was omitted from the hybridization buffer and heating and RNAse A treatment steps were deleted from the protocol. Under these conditions, ca. 75% of the telomeres were unstained (i.e. SFEs) by the unmodified P18 probe whereas this number was reduced to 40% for the higher affinity γP12 (Figure 7B and Supplementary Figure 6). For those telomeres that were stained, γP12 gave considerably brighter signals than P18. Thus, while overall staining efficiency was reduced for both probes under non-denaturing conditions, the γP12 probe yielded a greater percentage of telomeres with brighter signals compared to the unmodified P18 probe. Overall, these results demonstrate that, in metaphase chromosome spreads prepared from Jurkat cells, the shorter γP12 probe exhibited significantly improved telomere staining under both denaturing and non-denaturing conditions. This finding opens potential new applications of qFISH using nondenaturing conditions, such as flow cytometry coupled with FISH. This technique is useful for examining telomere lengths in specific cell subpopulations in human blood including granulocytes, T cells and or B cells, which are identified with specific antibodies against antigens that would not withstand harsh denaturing conditions needed for hybridization of lower affinity probes.36
Another potential application of γPNA miniprobes is in studies of telomere dysfunction, which rely on observation of co-localization of DNA damage response (DDR) proteins with telomeres, leading to inappropriate recognition as a DNA double strand break due to uncapping. These DDR markers include 53BP1 protein and the phosphorylation of histone H2AX at the telomeres which are detected with antibodies or GFP-tagging of exogenous protein.37, 38 Current protocols require that cells or chromosome spreads are immuno-stained with DDR-specific antibodies and then fixed prior to subjecting the samples to denaturing FISH conditions for PNA staining of the telomeric DNA. γPNA hybridization under milder non-denaturing conditions should allow more reliable detection of fluorescent protein tags or antibody-conjugated fluorophores or substrates.
Conclusion
The results described above demonstrate that γPNA miniprobes as short as 6 nucleobases assemble cooperatively on repeating sequence DNA targets to form stable γPNADNA heteroduplexes. The miniprobes successfully stained telomeric DNA in metaphase chromosome spreads. FISH results for Jurkat cells stained with the 12mer probe γP12 are particularly promising, with the observation of significantly fewer signal free ends (SFEs) and a greater number of telomeres exhibiting brighter signals, compared with a conventional 18mer PNA probe. Our findings support the hypothesis that shorter probes can produce more intense fluorescence signal and allow better sensitivity and detection of short telomeres by allowing more probes, and thus more fluorophores, to bind to the same number of DNA bases. The imaging and quantitation data show that this is a viable application of fluorescent miniprobes, although the extent of improvement depends on the experimental conditions and cell line. Future work will involve testing the miniprobes in additional cell lines to see if the performance in qFISH assays depends on the telomere maintenance mechanism (ALT versus telomerase).
Any hybridization probe requires optimization in order to maximally capture or detect its target. Short probes have the benefit of being more sensitive to single mismatches (i.e. a mismatch has a greater destabilizing effect than in a long probe), but are more likely to find perfect-match sequences at unintended sites. This is a possible reason for the less optimal performance of the 6mer miniprobes: without the G-clamp modification, the probe did not have sufficient affinity to stain telomeres under our washing conditions, whereas with two G-clamp bases, the affinity was so high that staining of other sites in the genome is likely to occur, particularly under denaturing conditions. It is also worthwhile to recognize the considerably higher amount of effort required to synthesize the G-clamp γPNA monomer for a miniprobe that would be, at best, only twofold brighter than the 12mer version which does not need any modified bases. On the other hand, long probes are less likely to find multiple perfect-match sites but are more likely to hybridize to single-mismatch sites. Ongoing work is directed toward optimizing the length and affinity of γPNA miniprobes to obtain maximum telomere brightness while minimizing background staining of nontelomeric genomic regions. (It is also possible that, in the absence of an RNase treatment, telomeric RNA hybridizes to PNA and γPNA probes.)
Finally, we note that γPNA miniprobes should be useful for chromosome or mRNA “painting”, in which sets of fluorescent probes targeted to different regions of the same chromosome39 or mRNA16 are applied in combination, resulting in brighter and/or multicolor fluorescence. The short length and lack of a negative charge on the γPNA backbone should permit faster hybridization relative to oligonucleotide or cDNA FISH probes.
METHODS
Materials
DNA oligonucleotides were purchased from Integrated DNA Technologies and used after standard desalting by gel filtration chromatography. P18 was obtained from Panagene. DNAs contained different numbers of telomeric repeats Telo-n (5’-TTAGGG-3’)n, where n = 1-4 or 6. γ-minipeg-t-Boc-protected PNA monomers and oligomers were synthesized according to published procedures,10, 21 purified using reverse-phase high performance liquid chromatography (HPLC), and characterized by matrix-assisted laser desorption/ionization time of flight (MALDI-ToF) mass spectrometry (Applied Biosystems, Voyager DE sSTR) using α-cyano-4-hydroxycinnamic acid as the matrix. Cy3 was a generous gift from B. Schmidt of the Molecular Biosensor and Imaging Center at Carnegie Mellon University. For Cy3 labeling, a carboxylic acid derivative of Cy3 was coupled to the γPNA prior to cleavage using similar procedures to all of the other monomers. Samples were run with linear detection and positive ionization. γPNA characterization data are shown in Figures S7-S9.
All PNA, γPNA and DNA concentrations were determined by measuring the absorbance at 260 nm at 90°C on a Cary 3 Bio spectrophotometer. At high temperature, all the bases are assumed to be unstacked, and the extinction coefficient can be calculated as the sum of the individual bases. Extinction coefficients for PNA monomers were obtained from Applied Biosystems whereas DNA extinction coefficients were used as reported in the literature. Calculated extinction coefficients (ε260, M−1cm−1) are P18 = 167,400; γP6 = 55,800; Quantitation of FISH results = 65,400 and γP12 = 111,600.
UV Melting Experiments
UV-vis measurements were conducted on a Varian Cary 300 Bio UV-vis spectrophotometer equipped with thermoelectrically controlled multicell holders. Samples were prepared in a buffer containing 10mM Tris-HCl (pH 7) and 0.1 mM Na2EDTA. For LiCl experiments, a stock solution of 10 mM Li2EDTA was used. Thermal experiments were conducted in the presence of either 100 mM KCl or 100 mM LiCl. The solutions were heated to 95 °C followed by cooling to 15 °C at a rate of 1 °C min−1. The samples were allowed to equilibrate for 5 minutes at 15 °C and then heated back to 95 °C at the same rate. The absorbance at 260 nm for complementary binding or 295nm for G-quadruplex formation was recorded at an interval of 0.5 °C. All samples contained concentrations of 4 μM PNA and DNA repeat sequences, for example 2 μM γP12 (2 repeat sequences) + 2 μM Telo-2 (2 repeat sequences) or 1 μM Telo-4 (4 repeat sequence); 4 μM γP6 or Quantitation of FISH results (1 repeat sequence) + 4 μM Telo-1, 2 μM Telo-2, 1.3 μM Telo-3 or 1 μM Telo-4. All experiments were run in triplicate and the averaged data were plotted as the fraction of melted PNA-DNA duplex as a function of temperature. Melting transition temperature (Tm), the temperature at which PNA-DNA hybrid was half-melted, was determined as described by Marky and Breslauer.40
Fluorescence Spectroscopy
Fluorescence emission spectra were collected on Cary Eclipse Fluorescence Spectrophotometer at 25°C. Spectra for samples containing a final concentration of 4 μM (PNA + DNA repeats) buffered in 10 mM Tris-HCl (pH=7), 0.1 mM EDTA and 100 mM KCl were recorded with excitation at 530 nm after annealing of the samples from 95-5°C.
Surface Plasmon Resonance Experiments
SPR experiments were performed on a Biacore T100 system with a four-channel sensor chip coated with carboxymethylated dextran on the gold surface. Approximately 10000 response units (RU) of streptavidin was immobilized on the chip’s surface using standard N-hydroxysuccinimide/1-ethyl-3-[3-(dimethylamino)propyl]-carbodiimide hydrochloride (NHS/EDC) coupling. Then 120 RU of 5’-biotinylated DNAs (25 nM) were attached to separate flow cells via non-covalent capture to streptavidin-coated surface. Before sample injection, sensor chips were conditioned with five consecutive 1 min-injections of 1M NaCl, 50 mM NaOH followed by multiple washing flow with LiCl/HBS-EP buffer (0.01 M HEPES, 0.15 M LiCl, 3mM EDTA, and 0.005% Surfactant P-20, pH 7.4). Biotinylated DNA in LiCl/HBS-EP buffer was then injected at a flow rate of 2 μL/min to achieve long contact times with the surface until a sufficient amount of DNA was immobilized. Synthesized PNAs were dissolved in water and then diluted to a working stock solution of 5 μM. Samples of varying PNA concentration were then prepared in filtered and degassed running buffers by serial dilutions (10-50 nM in 10 nM increments) from stock solutions. Samples were run in triplicate at a flow rate of 50 L/min with either LiCl/HBS-EP or KCl/HBS-EP running buffers. The sensor surface was then regenerated with two injections of 10 mM NaOH/1 M NaCl. Buffer injections were conducted to remove remaining regeneration solution before the next sample was injected. Prior to data analysis, we corrected the raw sensorgrams by performing “double referencing”. Briefly, we subtracted control flow cell 1 from all sample flow cells to remove changes in RU contributed by bulk shift. Within the same flow cell, we subtracted an average of the two buffer injections from PNA injections to account for any nonspecific binding or baseline shifts due to experimental artifacts.
Cell Culture
Jurkat cells and U2OS cells were cultured in standard cell culture conditions. Cells were grown in a temperature controlled incubator at 37°C and 5% CO2 in Dulbecco's Modified Eagle's Medium (DMEM) with 10% Fetal Bovine Serum (FBS), 50U Penicillin/mL and 50 μg Streptomycin/mL.
Quantitative Fluorescence In-situ Hybridization (qFISH)
For staining and imaging of telomeres in metaphase chromosome spreads the cells were treated with 0.05μg/mL colcemid for 10 hours. 1×106 cells (1×105 cell/mL) were subsequently harvested and treated with 75 mM KCl hypotonic buffer for 12 min at 37°C and then fixed and stored at −20 °C in methanol/acetic acid fixative (3:1). Lysed cells were dropped onto water-coated glass microscope slides and dried at 42 °C for 3 minutes to spread the chromosomes and were then incubated overnight at room temperature. The prepared microscope slides were fixed in 4% formaldehyde and then treated with 0.1% Pepsin in 0.01N HCl for 10 minutes at 37 °C. Fixation and washing were repeated. Slides were then dehydrated in successive ethanol solutions of 70%, 90%, and 100% for 5 minutes each. Slides were then treated with 500μg/mL of RNAse A for 10 minutes at 37 °C followed by an additional dehydration series. Samples were denatured for 3 min at 80 °C, in a hybridization mixture containing 70% deionized formamide, unless otherwise indicated, 10% NEN blocking reagent [Roche], 0.1 M Tris-HCl [pH=7.4], MgCl2 buffer [82 mM NaH2PO4, 9 mM citric acid, 20 mM MgCl2], and 0.5 μg/ml of the indicated PNA or γPNA. Non-denaturing conditions were identical except formamide was omitted and samples were incubated at room temperature rather than 80°C. After 2h hybridization at room temperature, the samples were washed twice with wash solution (70% deionized formamide, 0.1% BSA, and 10 mM Tris-HCl [pH=7.4]), and three times with a second was solution (100 mM Tris-HCl [pH=7.4], 150 mM NaCl, 0.1% Tween 20). Samples were finally counterstained with DAPI and mounted with coverslips.
Image Analysis
Stained metaphase spreads were imaged using a Nikon Ti90 epifluorescence microscope (Nikon Inc., NY) equipped with PlanApo 60×/1.40 oil immersion objective and run by Nikon Elements. A series of nine Z-stack 0.25 μm cross sections were captured with 80ms exposure for DAPI and a 300ms exposure time for Cy3 unless otherwise indicated. Images were converted to 8-bit .TIFF files using Image J, and optimal Z-stacks were analyzed for telomere brightness by employing TFL-Telo software.41 The telomere threshold was set to three for U2OS cells and two for Jurkat cells. The chromosome threshold was set to 9. Telomere spots and chromosomes were inspected and adjusted for accuracy as needed. Telomere fluorescence was binned by brightness, and calculated as a ratio of telomere objects per bin to total telomere objects in the metaphase using Excel (Microsoft). Error was calculated as standard error of the means from three repeats of each experiment (two repeats for nondenaturing conditions). Column graphs were generated using Origin software.
Supplementary Material
Acknowledgment
The authors gratefully acknowledge generous support for this research from the U.S. National Institutes of Health (R43GM108187 to BAA and PLO), the National Science Foundation (CHE 1012467 to DHL) and from the DSF Charitable Foundation. SPR instrumentation was purchased with support from NSF Major Research Instrumentation Award CHE 0821296.
Footnotes
Supporting Information. UV melting curves, SPR sensorgrams, microscope images and MALDI-TOF spectra and HPLC chromatograms for γPNA oligomers. This material is available free-of-charge via the Internet at http://pubs.rsc.org.
References
- 1.Bedell VM, Westcot SE, Ekker SC. Briefings Func. Genom. 2011;10:181–188. doi: 10.1093/bfgp/elr021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang P, Zhu Z, Lin S, Zhang B. J. Genet. Genom. 2012;39:421–433. doi: 10.1016/j.jgg.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 3.Lundin KE, Hoejland T, Hansen BR, Persson R, Bramsen JB, Kjems J, Koch T, Wengel J, Smith CIE. Adv. Genet. 2013;82:47–107. doi: 10.1016/B978-0-12-407676-1.00002-0. [DOI] [PubMed] [Google Scholar]
- 4.Egholm M, Buchardt O, Christensen L, Behrens C, Freier SM, Driver DA, Berg RH, Kim SK, Nordén B, Nielsen PE. Nature. 1993;365:566–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 5.Tomac S, Sarkar M, Ratilainen T, Wittung P, Nielsen PE, Nordén B, Gräslund A. J. Am. Chem. Soc. 1996;118:5544–5552. [Google Scholar]
- 6.Lansdorp PM, Verwoerd NP, van de Rijke FM, Dragowska V, Little M-T, Dirks RW, Raap AK, Tanke HJ. Hum. Mol. Gen. 1996;5:685–691. doi: 10.1093/hmg/5.5.685. [DOI] [PubMed] [Google Scholar]
- 7.Stender H, Fiandaca M, Hyldig-Nielsen JJ, Coull J. J. Microbiol. Methods. 2002;48:1–17. doi: 10.1016/s0167-7012(01)00340-2. [DOI] [PubMed] [Google Scholar]
- 8.Dragulescu-Andrasi A, Rapireddy S, Frezza BM, Gayathri C, Gil RR, Ly DH. J. Am. Chem. Soc. 2006;128:10258–10267. doi: 10.1021/ja0625576. [DOI] [PubMed] [Google Scholar]
- 9.Wengel J. Acc. Chem. Res. 1999;32:301–310. [Google Scholar]
- 10.Sahu B, Sacui I, Rapireddy S, Zanotti KJ, Bahal R, Armitage BA, Ly DH. J. Org. Chem. 2011;76:5614–5627. doi: 10.1021/jo200482d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yeh JI, Shivachev B, Rapireddy S, Crawford MJ, Gil RR, Du S, Madrid M, Ly DH. J. Am. Chem. Soc. 2010;132:10717–10727. doi: 10.1021/ja907225d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen PE, Egholm M, Berg RH, Buchardt O. Science. 1991;254:1498–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 13.Lohse J, Dahl O, Nielsen PE. Proc. Natl. Acad. Sci. USA. 1999;96:11804–11808. doi: 10.1073/pnas.96.21.11804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He G, Rapireddy S, Bahal R, Sahu B, Ly DH. J. Am. Chem. Soc. 2009;131:12088–12090. doi: 10.1021/ja900228j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bahal R, Sahu B, Rapireddy S, Lee CM, Ly DH. ChemBioChem. 2012;13:56–60. doi: 10.1002/cbic.201100646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Nature Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lifland AW, Zurla C, Santangelo PJ. Bioconjugate Chem. 2010;21:483–488. doi: 10.1021/bc9003876. [DOI] [PubMed] [Google Scholar]
- 18.Wilds CJ, Maier MA, Tereshko V, Manoharan M, Egli M. Angew. Chem. Int. Ed. 2002;41:115–117. doi: 10.1002/1521-3773(20020104)41:1<115::aid-anie115>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 19.Ortega J-A, Blas JR, Orozco M, Grandas A, Pedroso E, Robles J. Org. Lett. 2007;9:4503–4506. doi: 10.1021/ol701826x. [DOI] [PubMed] [Google Scholar]
- 20.Rapireddy S, Bahal R, Ly DH. Biochemistry. 2011;50:3913–3918. doi: 10.1021/bi2002554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christensen L, Fitzpatrick R, Gildea B, Petersen KH, Hansen HF, Koch T, Egholm M, Buchardt O, Nielsen PE, Coull J, Berg RH. J. Pept. Sci. 1995;3:175–183. doi: 10.1002/psc.310010304. [DOI] [PubMed] [Google Scholar]
- 22.Koch T. Nielsen PE, editor. Peptide Nucleic Acids: Protocols and Applications. Horizon Bioscience, Norfolk. (2nd edn.) 2004:37–60. [Google Scholar]
- 23.Kim SK, Nielsen PE, Egholm M, Buchardt O, Berg RH, Nordén B. J. Am. Chem. Soc. 1993;115:6477–6481. [Google Scholar]
- 24.Gagnon KT, Watts JK, Pendergraff HM, Montmaillier C, Thai D, Potier P, Corey DR. J. Am. Chem. Soc. 2011;133:8404–8407. doi: 10.1021/ja200312y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roy S, Zanotti KJ, Murphy CT, Tanious FA, Wilson WD, Ly DH, Armitage BA. Chem. Commun. 2011;47:8524–8526. doi: 10.1039/c1cc12805a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Williamson JR, Raghuraman MK, Cech TR. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 27.Sanders JL, Newman AB. Epidemiol. Rev. 2013;35:112–131. doi: 10.1093/epirev/mxs008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aviv A. J. Gerontol. A Biol. Sci. Med. Sci. 2008;63:979–983. doi: 10.1093/gerona/63.9.979. [DOI] [PubMed] [Google Scholar]
- 29.Poon SS, Lansdorp PM. Curr. Protoc. Cell Biol. 2001 doi: 10.1002/0471143030.cb1804s12. Chapter 18, Unit 18 14. [DOI] [PubMed] [Google Scholar]
- 30.Cesare AJ, Reddel RR. Nat. Rev. Genet. 2010;11:319–330. doi: 10.1038/nrg2763. [DOI] [PubMed] [Google Scholar]
- 31.Bryan TM, Englezou A, Dalla-Pozza L, Dunham MA, Reddel RR. Nature Med. 1997;3:1271–1274. doi: 10.1038/nm1197-1271. [DOI] [PubMed] [Google Scholar]
- 32.Sarzotti-Kelsoe M, Daniell XG, Whitesides JF, Buckley RH. Immunol. Res. 2011;49:44–48. doi: 10.1007/s12026-010-8192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Pan X, Kalmbach K, Seth-Smith ML, Ye X, Antumes DMF, Yin Y, Liu L, Keefe DL, Weissman SM. Proc. Natl. Acad. Sci. USA. 2013;110:E1906–E1912. doi: 10.1073/pnas.1306639110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu F-J, Barchowsky A, Opresko PL. PLoS One. 2010:5. [Google Scholar]
- 35.Coleman J, Baird DM, Royle NJ. Hum. Mol. Gen. 1999;8:1637–1646. doi: 10.1093/hmg/8.9.1637. [DOI] [PubMed] [Google Scholar]
- 36.Baerlocher GM, Vulto I, de Jong G, Lansdorp PM. Nature Protocols. 2006;1:2365–2376. doi: 10.1038/nprot.2006.263. [DOI] [PubMed] [Google Scholar]
- 37.Cesare AJ, Kaul Z, Cohen SB, Napier CE, Pickett HA, Neumann AA, Reddel RR. Nature Struct. Mol. Biol. 2009;16:1244–1251. doi: 10.1038/nsmb.1725. [DOI] [PubMed] [Google Scholar]
- 38.Takai H, Smogorzewska A, de Lange T. Curr. Biol. 2003;13:1549–1556. doi: 10.1016/s0960-9822(03)00542-6. [DOI] [PubMed] [Google Scholar]
- 39.Ried T, Schröck E, Ning Y, Wienberg J. Hum. Mol. Gen. 1998;7:1619–1626. doi: 10.1093/hmg/7.10.1619. [DOI] [PubMed] [Google Scholar]
- 40.Marky LA, Breslauer KJ. Biopolymers. 1987;26:1601–1620. doi: 10.1002/bip.360260911. [DOI] [PubMed] [Google Scholar]
- 41.Poon SS, Lansdorp PM. In: Methods in Cell Biology: Flow Cytometry. 64 edn. Darzynkiewicz Z, Crissman HA, Robinson JP, editors. Vol. 64. Academic Press; San Diego, CA: 2001. pp. 69–96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.