Background: KshA is a bacterial steroid-transforming oxygenase of biocatalytic interest and a virulence determinant in Mycobacterium tuberculosis.
Results: Structures of KshA·steroid complexes were obtained for three homologs of different substrate specificity.
Conclusion: Considerable structural flexibility contributes to the respective specificities of the different KshAs.
Significance: This study provides insight into steroid catabolism and the conformational flexibility of Rieske oxygenases.
Keywords: Actinobacteria, Bacterial Metabolism, Bile Acid, Cholesterol, Metalloenzyme, Mycobacterium tuberculosis, Oxygenase
Abstract
KshA is the oxygenase component of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase involved in the bacterial degradation of steroids. Consistent with its role in bile acid catabolism, KshA1 from Rhodococcus rhodochrous DSM43269 had the highest apparent specificity (kcat/Km) for steroids with an isopropyl side chain at C17, such as 3-oxo-23,24-bisnorcholesta-1,4-diene-22-oate (1,4-BNC). By contrast, the KshA5 homolog had the highest apparent specificity for substrates with no C17 side chain (kcat/Km >105 s−1 m−1 for 4-estrendione, 5α-androstandione, and testosterone). Unexpectedly, substrates such as 4-androstene-3,17-dione (ADD) and 4-BNC displayed strong substrate inhibition (KiS ∼100 μm). By comparison, the cholesterol-degrading KshAMtb from Mycobacterium tuberculosis had the highest specificity for CoA-thioesterified substrates. These specificities are consistent with differences in the catabolism of cholesterol and bile acids, respectively, in actinobacteria. X-ray crystallographic structures of the KshAMtb·ADD, KshA1·1,4-BNC-CoA, KshA5·ADD, and KshA5·1,4-BNC-CoA complexes revealed that the enzymes have very similar steroid-binding pockets with the substrate's C17 oriented toward the active site opening. Comparisons suggest Tyr-245 and Phe-297 are determinants of KshA1 specificity. All enzymes have a flexible 16-residue “mouth loop,” which in some structures completely occluded the substrate-binding pocket from the bulk solvent. Remarkably, the catalytic iron and α-helices harboring its ligands were displaced up to 4.4 Å in the KshA5·substrate complexes as compared with substrate-free KshA, suggesting that Rieske oxygenases may have a dynamic nature similar to cytochrome P450.
Introduction
Steroids are triterpenoids containing four fused cycloalkane rings. Although they are synthesized by eukaryotes for essential roles as hormones, membrane components, and emulsifiers (1), their mineralization is largely accomplished by bacteria. The aerobic catabolism of steroids by bacteria has been studied for the biocatalytic production of high value steroids (2, 3) and for its contribution to the pathogenicity of Mycobacterium tuberculosis (4). Steroid catabolism is almost ubiquitous in mycolic acid-producing actinobacteria (5, 6) with some strains containing paralogous pathways responsible for catabolizing different steroids. For example, Rhodococcus jostii RHA1 contains four clusters of steroid catabolic genes, the first and third of which enable growth on cholesterol and cholate, respectively (5, 6). Proteobacteria, such as Comamonas testosteroni TA441 (7) and Pseudomonas sp. strain Chol1 (8–10), catabolize steroids in a similar manner, although these strains appear to be unable to degrade sterols possessing a C17 alkyl side chain such as cholesterol.
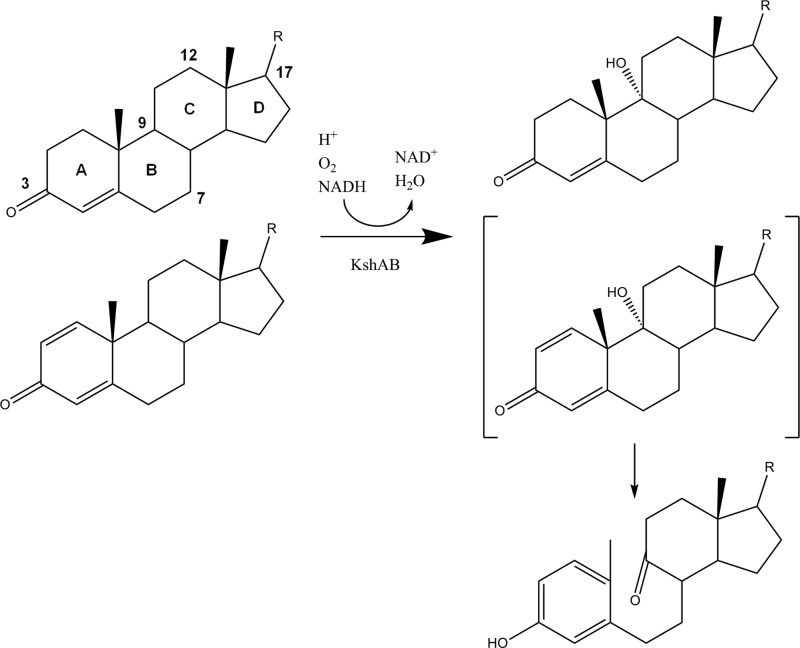
Bacterial steroid catabolism appears to be organized according to three parts of the steroid molecule (11) as follows: the side chain at C17, rings A/B, and rings C/D, respectively. The side chain is degraded by a process similar to the β-oxidation of fatty acids involving CoA thioester intermediates (12). Degradation of rings A/B involves 3-ketosteroid 9α-hydroxylase (KshAB) (13, 14), which leads to cleavage of ring B with concomitant aromatization of ring A to a phenol (Fig. 1). The latter is converted to a catechol, which is subject to extradiol cleavage (15). Rings C/D catabolism is poorly understood but appears to be encoded by the KstR2 regulon in Actinobacteria (11). This pathway shows strain- and substrate-dependent differences. For example, side chain and rings A/B degradation occur concurrently to some extent in actinobacteria based on the higher apparent specificity (kcat/Km) of KshAMtb (where Mtb is M. tuberculosis) for substrates with a CoA-thioesterified side chain (16), and the identification of a two-ringed metabolite with a partially degraded side chain and rings A/B, respectively, in the cholesterol-grown mutant of M. tuberculosis (17). Similarly, during growth on cholate, RHA1 excretes metabolites that are partially degraded in both the side chain and rings A/B before reassimilating them to complete their catabolism. By contrast, side chain degradation appears to precede rings A/B degradation in Proteobacteria (7, 10). The substrate specificities of key steroid-degrading enzymes such as KshAB should provide insight into the organization of steroid catabolic pathways.
FIGURE 1.
KshAB-catalyzed 9α-hydroxylation of 3-ketosteroids. 1,4-Diene ketosteroids (lower substrate) undergo subsequent nonenzymatic cleavage of ring B and aromatization of ring A. Carbon atoms 3, 7, 9, 12, and 17 are numbered. Cholate is hydroxylated at positions 7 and 12.
KshAB is a two-component Rieske oxygenase (RO)3 comprising an oxygenase (KshA) and a reductase (KshB) (14). ROs catalyze a variety of oxidative reactions and have considerable biocatalytic potential (18), although only a small fraction of their catalytic diversity has been explored (19). An x-ray crystallographic structure of KshAMtb solved to 2.3 Å revealed that KshA shares many features typical of ROs (13). Accordingly, KshA contains metallocenters in each of two domains as follows: the N-terminal domain harbors a Rieske [2Fe-2S] cluster coordinated by two cysteines and two histidines, and the larger C-terminal domain harbors the mononuclear iron that is coordinated by a 2-His-1-carboxylate facial triad and orchestrates the oxidative chemistry. The three KshA subunits are arranged in a head-to-tail fashion such that the [2Fe-2S] cluster and the mononuclear iron of adjoining subunits are 12 Å apart. Histidine ligands from adjacent metallocenters are bridged by a conserved aspartate that appears to mediate redox-dependent conformational changes during the RO catalytic cycle (20). Remarkably, the substrate access channel of KshA is angled at ∼90° with respect to that of other ROs characterized to date and is significantly longer (∼28 Å) as measured from the mononuclear iron to the surface of the protein (19). The channel entrance of KshA is located between the C terminus of helix α5 and the “mouth loop,” which connects strands β14 and β15. Helix α5 is the longest helix in ROs and harbors the carboxylate ligand of the mononuclear iron. By contrast, in 2-oxoquinoline 8-monooxygenase (OMO) (21), carbazole 1,9a-dioxygenase (CARDO) (22), and naphthalene dioxygenase (23), the channel entrance occurs between helix α4 (KshA numbering), containing a histidine ligand of the iron, and strand β13, the first of the central β-sheet. The different substrate access channel of KshA appears to be necessary to allow proper orientation of the large steroid substrate, including CoA thioesters, within its binding pocket (16). A better understanding of KshAs and other ROs will facilitate their engineering for biocatalytic applications.
Characterization of KshA has focused largely on the homolog from the cholesterol catabolic pathway due to its role in the virulence of M. tuberculosis (4). However, KshAs from environmental bacteria have different substrate preferences as exemplified by the five homologs of Rhodococcus rhodochrous DSM43269, KshA1 to A5 (24). KshA1 had a marked preference for substrates with a carboxylate side chain at C17, whereas KshA5 transformed a broader range of steroid substrates. KshA1 was assigned to cholate catabolism based on the enzyme's substrate preference, the up-regulation of kshA1 during growth of DSM43269 on cholate, and the ability of kshA1 to restore growth of a kshA null mutant on cholate. Moreover, KshA1 is the reciprocal best hit (72% amino acid sequence identity) with the RHA1 homolog encoded by the cholate catabolic cluster (6). By contrast, the physiological role of KshA5 is unknown (24). Expression of kshA5 was induced by a range of steroids as follows: cholesterol, cholate, progesterone, and 4-androstene-3,17-dione. Moreover, a DSM43269 mutant containing only kshA5 (i.e. disrupted in each of the kshA1 to kshA4) grew on each of these four steroids (24). It is unclear how KshA5 is able to effectively transform such a broad range of ketosteroids.
To better characterize ketosteroid hydroxylases, we investigated the substrate specificity of KshA1 and KshA5 from DSM43269 as well as substrate-bound structures of KshA1, KshA5, and KshAMtb. Steady-state kinetic parameters of KshA1 and KshA5 for each of seven steroid substrates, including 3-oxo-23,24-bisnor-cholesta-1,4-diene-22-oyl CoA (1,4-BNC-CoA), were determined, and the coupling of O2 consumption with substrate hydroxylation was investigated. X-ray crystallographic structures of substrate-free KshA5 and the following complexes were solved: KshA1·1,4-BNC-CoA, KshA5·ADD, KshA5·1,4-BNC-CoA, and KshAMtb·ADD. The findings are discussed with respect to the structural determinants of substrate specificity, the mechanism of ROs, and the different steroid catabolic pathways.
MATERIALS AND METHODS
Chemicals and Reagents
All reagents were at least analytical grade unless otherwise noted. 4-Estren-3,17-dione, testosterone, ADD, 3-oxo-23,24-bisnorchol-4-en-22-oic acid (4-BNC), and 5α-androstan-3,17-dione were purchased from Steraloids, Inc. (Newport, RI). Steroids contained <5% impurities as judged by HPLC and GC-MS. 1,4-BNC and its CoA thioester were enzymatically produced using CasI and KstD as described previously (16). Restriction enzymes and the Expand high fidelity PCR system were purchased from New England Biolabs (Ipswich, MA) and Roche Applied Science, respectively. Water for buffers was purified using a Barnstead Nanopure DiamondTM system (Dubuque, IA) to a resistance of at least 18 megohms.
DNA Manipulation
To produce proteins with N-terminal affinity tags, kshA1 (24), kshA5, and Rv3526 (13) were subcloned into pT7HP20 using standard protocols (25). The sequences of the oligonucleotides used to amplify the kshA genes were as follows: 5′-TGAGCTAGCAGCCTCGCACTTCCGAACAATC-3′ and 5′-ACTAAGCTTCTAGCCCGCGGTGGTGGACTG-3′ for kshA1; 5′-CTTGCTAGCTCCATCGACACCGCACGGTC-3′ and 5′-ACTAAGCTTCTAGGGGGTCGCGGTGGAGC-3′ for kshA5; and 5′-TAAGCTAGCAGTACCGACACGAGTGGGGTCG-3′ and 5′-ATCAAGCTTTCAGTGTTGCTCGGCGGGC-3′ for Rv3526. This minimized the extra amino acids remaining after proteolytic removal of the tag. The nucleotide sequences of kshA1, kshA5, and Rv3526 in the resulting constructs, pT7KA1, pT7KA5, and pT7KAMtb, were confirmed.
Protein Production and Purification
KshA was heterologously produced using Escherichia coli GJ1158 containing either pT7KA1, pT7KA5, or pT7KAMtb essentially as described (26). One liter of low-salt Luria broth was inoculated with 3 ml of an overnight culture and was incubated at 25 °C. When the culture reached an A600 of 0.5, kshA expression was induced by adding 50 ml of 5 m NaCl. The culture was incubated a further 18 h at 25 °C and then harvested by centrifugation. Cell pellets were washed twice with 20 mm sodium phosphate, 5% glycerol, pH 8.0, and frozen at −80 °C until use.
A cell pellet from 3 liters of culture was suspended to a final volume of 60 ml in 20 mm sodium phosphate, pH 7.4, containing 300 mm NaCl and 20 mm imidazole. A small amount of DNase I (∼0.5 mg) was added, and the cells were lysed using an Emulsiflex-05 homogenizer (Avestin, Ottawa, Canada) operated at 10,000 p.s.i. Cell debris was removed by centrifugation (10,000 × g for 45 min), and the amber-colored supernatant was transferred to a Wheaton bottle. Subsequent steps were performed anaerobically essentially as described previously for KshAMtb (13). The solution was filtered and loaded onto a nickel-Sepharose (Qiagen) column (1.8 × 4 cm). Protein was eluted using an imidazole step gradient. A brown fraction containing KshA was eluted using 20 mm sodium phosphate, pH 7.4, containing 300 mm NaCl and 300 mm imidazole. The protein was exchanged into 50 mm Tris, pH 7.5, 100 mm NaCl, and 1 mm CaCl2 and concentrated to ∼5 ml using a stirred cell concentrator equipped with a YM30 membrane (Amicon, Oakville, Ontario, Canada). For KshA1 and KshAMtb, Factor Xa (HTI, Essex Junction, VT) was added to a molar ratio of ∼1:750 Factor Xa/KshA and incubated overnight at room temperature. For KshA5, a molar ratio of 1:500 was used, and the cleavage buffer contained 0.25 m urea. The cleaved protein was loaded onto an anion-exchange resin (1 × 10 cm; SourceTM15Q (GE Healthcare)) equilibrated with 25 mm HEPES, pH 7.5, 5% glycerol, 1 mm DTT, and 0.25 mm ferrous ammonium sulfate (buffer A). The protein was eluted using a 30-ml linear gradient from 150 to 300 mm NaCl. Brown-colored fractions eluting at ∼200 mm NaCl were combined, exchanged into buffer A, concentrated to 25 mg/ml, flash frozen as beads in liquid N2, and stored at −80 °C. KshB was produced and purified as described previously (13).
Analytical Methods
Protein concentration was determined using the Micro BCATM protein assay kit (Pierce) using bovine serum albumin as a standard. UV-visible spectra were recorded using a Cary 60 spectrophotometer. The extinction coefficients of the ferrocyanide-oxidized enzymes were ϵ324 = 19.6 mm−1 cm−1 for KshA1, ϵ324 = 15.9 mm− 1cm−1 for KshA5, and ϵ324 = 24.8 mm−1 cm−1 for KshAMtb. Acid-labile sulfur and iron content of samples were determined as described previously (13). Gas chromatography-coupled mass spectrometry (GC-MS) was performed using an HP 6890 series GC system fitted with an HP-5MS 30-m × 250-μm column (Hewlett-Packard) and an HP 5973 mass-selective detector.
Steady-state Kinetic Analysis
Spectrophotometric assays were performed in 1 ml of potassium phosphate (I = 100 mm, pH 7.0) at 22 °C. Initial reaction velocities were measured by monitoring NADH oxidation at 339 nm (ϵ339 = 6.22 mm−1 cm−1). Reaction mixtures containing 0.12–0.16 μm KshA, 1.6 μm KshB, and 100 μm NADH were equilibrated for 3 min before the reaction was initiated by adding the steroid substrate. Solutions were prepared fresh daily. KshA was thawed, exchanged anaerobically into 100 mm potassium phosphate, pH 7.5, using gel filtration, and stored in a sealed vial on ice. Aliquots were withdrawn using a gas-tight syringe. The steady-state kinetic parameters for O2 were determined as described previously (13) using 4-BNC with KshA1. Steady-state kinetic parameters were evaluated by fitting either the Michaelis-Menten equation or the equivalent describing a mechanism in which substrate inhibition occurs (Equation 1) to the data using the program LEONORA (27).
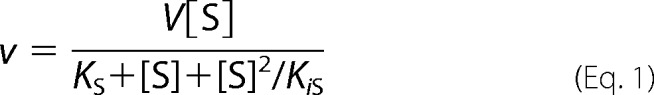 |
HPLC Analysis
Substrates and products were analyzed using a Waters 2695 Separations HPLC module equipped with a Waters 2996 photodiode array detector and a 250 × 4.60-mm C18 Aqua 5μ ODS-Prep column (Phenomenex, Torrance, CA). Reactions were quenched by adding acetic acid to a concentration of 5% and centrifuged to remove precipitated protein. Filtered reaction mixture (70 μl) was injected onto the column equilibrated with 36% MeOH in water and 0.5% phosphoric acid and eluted with a flow rate of 0.9 ml/min using a gradient of methanol from 36 to 90% over 14 min. For coupling assays, standard assay conditions were used, and NADH was monitored spectrophotometrically. Steroid substrate turnover was assessed using integrated peak areas (243 nm for 4-BNC and 244 nm for ADD) and standard curves for each substrate. l-Phenylalanine was used as an internal standard.
Crystallization
Crystals of KshA were grown aerobically at room temperature (21 °C) using the hanging drop vapor diffusion method. Drops of 1 μl contained a 1:1 ratio of 200–300 μm KshA (in 25 mm HEPES, pH 7.5, substrate (ADD or 1,4-BNC-CoA), 0.25 mm FAS, 1 mm DTT or tris(2-carboxyethyl)phosphine) and crystallization well solution.
Attempts to crystallize KshA1 alone were unsuccessful. Crystals were serendipitously obtained from a mixture of KshA1, KshA5, and 1,4-BNC-CoA. Pyramid-shaped light brown crystals grew over 2 weeks from a 1.5-μl drop containing equal volumes of KshA1 with 1,4-BNC-CoA, KshA5, and well solution containing 500 mm NaH2PO4, 125 mm K2HPO4, 4% PEG-1000, 20 mm Tris, 100 mm phosphate-citrate, pH 4.2. KshA5 was crystallized in complex with ADD using 0.8 m NaH2PO4, 0.2 m K2HPO4, 2% PEG-3000, 20 mm CHES, 0.1 m phosphate-citrate, pH 4.2. KshA5 was crystallized in complex with 1,4-BNC-CoA using 0.8 m NaH2PO4, 0.2 m K2HPO4, 0.25 m (NH4)2SO4, 20 mm CHES, 40 mm NaCl, and 0.1 m phosphate-citrate, pH 4.2. KshAMtb was crystallized in complex with ADD using 500 mm NaH2PO4, 125 mm K2HPO4, 4% PEG-1000, 20 mm Tris, 100 mm phosphate-citrate, pH 4.2. Crystals were cryo-protected with mother liquor supplemented with 20–30% glycerol and flash-frozen in liquid nitrogen prior to data collection.
X-ray Data Collection and Structure Determination
X-ray data were collected at the Canadian Light Source (Beamlines 08ID-1 and 08B1–1). Data were integrated with MOSFLM (28) and scaled and merged with AIMLESS (29), and initial resolution cutoffs were assessed according to CC1/2. Structures were solved by molecular replacement using the program PHASER (30). Models were built using BUCCANEER (31) as implemented in CCP4 (32) and refined using REFMAC (33). Ligands were built using PRODRG (34) and modeled into the density in the later stages of refinement. Models were submitted to the PDB_REDO server (35) for final refinement, and the final high resolution limit was assessed using paired refinement. Final models were validated using Molprobity (36). All structures were in the 99th percentile for Molprobity score. Electron density maps for figures were calculated using the FFT program of CCP4. The coordinates and data were deposited at the Protein Data Bank with accession codes 4QCK (KshAMtb:ADD), 4QDF (KshA5/KshA1), 4QDC (KshA5:ADD), and 4QDD (KshA5:1,4-BNC-CoA).
RESULTS
Purification of KshA
KshA1, KshA5, and KshAMtb were purified anaerobically due to their O2 lability (13, 37) to a yield of 10–40 mg/liter of culture. KshA1 preparations contained 1.7 ± 0.3 mol of sulfur and 3.9 ± 0.2 mol of iron per protomer, although those of KshA5 contained 1.7 ± 0.2 mol of sulfur and 3.8 ± 0.4 mol of iron per protomer. Interestingly, dialysis of KshA preparations against EDTA did not lower the iron content. Oxidized preparations had absorption maxima at 455 and 280 nm with a shoulder at 324 nm. The R value of KshA1 (A280/A324) was 6.1, and the specific activity of the preparation was 0.36 ± 0.02 μmol min−1mg−1 using 50 μm ADD (0.1 m potassium phosphate, pH 7.0, 22 °C). This compares with 0.75 μmol min−1 mg−1 reported for a mixed preparation of KshA1 and KshB and 200 μm ADD (50 mm Tris-HCl, pH 7.0, 22 °C) (24). The R value of KshA5 was 7.4, and the specific activity was 0.15 ± 0.02 μmol min−1 mg−1 using 50 μm ADD (0.1 m potassium phosphate, pH 7.0, 22 °C). This compares with 0.073 μmol min−1 mg−1 reported for a mixed preparation of KshA5 and KshB using 200 μm ADD (50 mm Tris-HCl, pH 7.0, 22 °C) (24). The specific activity, R value and iron and sulfur content of the KshAMtb preparation were similar to reported values (13).
Reaction Products and Coupling
KshA1 transformed ADD to a single product detected by GC-MS and HPLC. Similarly, KshA5 transformed ADD and 4-estren-3,17-dione to single products. For both enzymes, the ADD transformation product had the same GC retention time and mass spectrum as 3-hydroxy-9,10-seconandrost-1,2,5(10)-triene-9,17-dione (13). Using HPLC to measure steroid depletion and a spectrophotometer to monitor NADH oxidation, the KshA1-catalyzed hydroxylations of ADD and 4-BNC as well as the KshA5-catalyzed hydroxylations of ADD and 4-estren-3,17-dione were well coupled to NADH consumption at several substrate concentrations (Table 1). Moreover, the addition of catalase to reactions monitored using a Clark-type oxygen electrode did not detectably alter the rate of O2 consumption, indicating that H2O2 was not produced in significant amounts during steroid turnover. Finally, the reaction velocity was observed to reach zero when the amount of NADH consumed corresponded to the amount of steroid added. These data demonstrate that NADH oxidation and O2 consumption are well coupled to steroid hydroxylation in these KshAs and that the rate of steroid hydroxylation can be accurately measured by monitoring NADH or O2 consumption.
TABLE 1.
Coupling of NADH consumption to substrate turnover in KshA1 and KshA5
| Enzyme | Substrate | Conc | time | Ratio |
|---|---|---|---|---|
| μm | min | Substrate/NADH | ||
| KshA1 | ADD | 40 | 5 | 1.02 (0.07)a |
| 80 | 3 | 1.2 (0.2) | ||
| 4-BNC | 20 | 3 | 1.1 (0.1) | |
| 50 | 7 | 0.95 (0.05) | ||
| KshA5 | ADD | 20 | 3 | 1.1 (0.1) |
| 40 | 5 | 0.99 (0.01) | ||
| 4-Estren-3,17-dione | 50 | 2 | 1.0 (0.1) |
a Values in parentheses represent means ± S.E.
Steady-state Kinetic Analysis
For six steroid substrates (Fig. 2), KshA1 displayed Michaelis-Menten kinetics (Fig. 3). A seventh substrate, 5α-androstan-3,17-dione, was not detectably transformed. KshA1 had the highest apparent specificity (kcat/Km) for steroids possessing an isopropionyl side chain at C17 (Table 2), consistent with the enzyme's previously reported substrate preference. For example, the apparent kcat/Km values for 4-BNC was 3 orders of magnitude higher than for 4-estren-3,17-dione. KshA1's low specificity for 1,4-BNC-CoA is interesting inasmuch as such CoA thioesters are produced during steroid side chain degradation. Indeed, the apparent kcat/Km value of KshAMtb for 1,4-BNC-CoA was 45 times higher than for 1,4-BNC (16). In contrast to the variation in the apparent kcat/Km values, the apparent kcat values of KshA1 varied by only 5-fold.
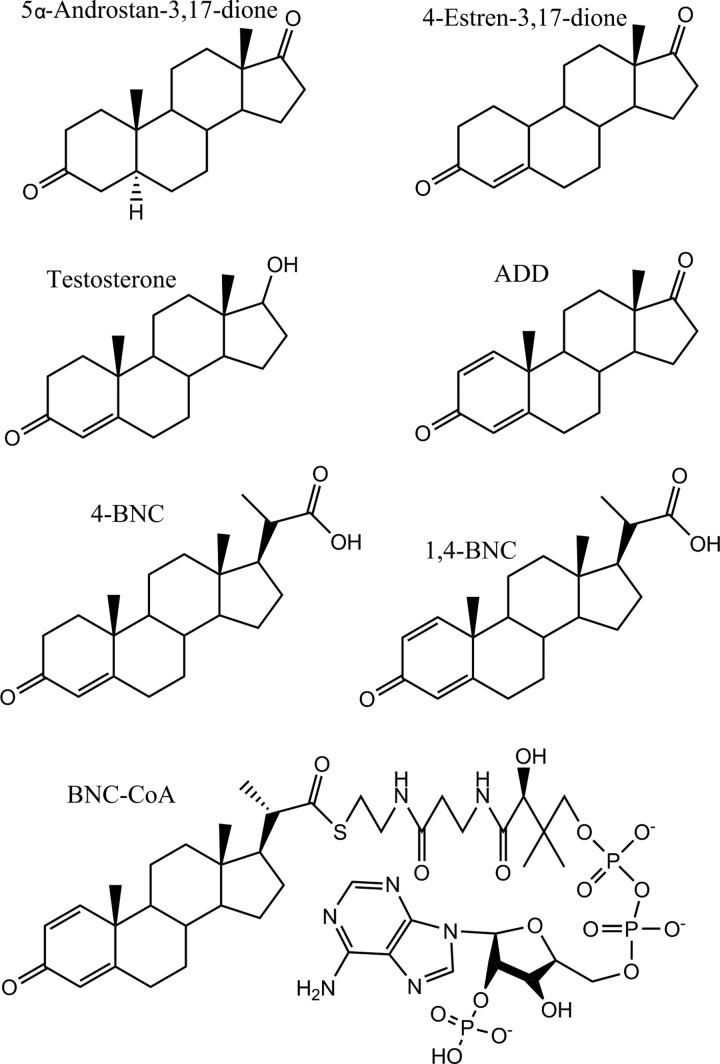
FIGURE 2.
Compounds investigated as KshA substrates. 5α-Androstan-3,17-dione; 4-estren-3,17-dione; testosterone; 1,4-androstadiene-3,17-dione (ADD); 3-oxo-23,24-bisnorchol-4-en-22-oate (4-BNC); 3-oxo-23,24-bisnorchola-1,4-dien-22-oate (1,4-BNC); and 1,4-BNC-CoA.
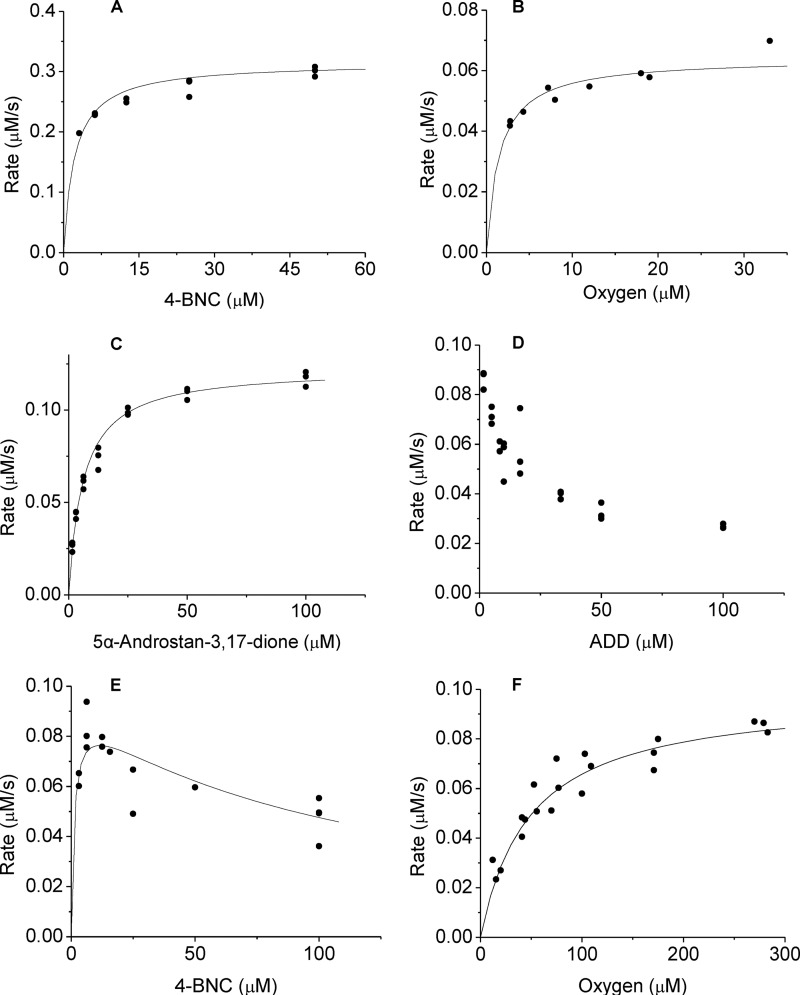
FIGURE 3.
Steady-state kinetic curves of KshA1 and KshA5. Shown is the dependence of the initial velocity of NADH consumption by KshA1 on 4-BNC (A) and O2 (B), and by KshA5 on 5α-androstan-3,17-dione (C), ADD (D), 4-BNC (E), and O2 (F). Assays were performed using 100 μm NADH and potassium phosphate (I = 100 mm, pH 7.0) at 22 °C. B, steroid substrate was 50 μm 4-BNC; F, steroid substrate was 5 μm 4-estren-3,17-dione. Curves represent the best fit of the Michaelis-Menten equation using LEONORA.
TABLE 2.
Apparent steady-state kinetic parameters of KshA1 and KshA5
Assays were performed using potassium phosphate (I = 0.1 m, pH 7.0) at 22 °C containing 100 μm NADH (∼260 μm O2). Errors in parentheses were calculated from the weighted mean of the error in the enzyme concentration, and the standard errors in the parameters were estimated using LEONORA.
| Substrate | KshA1 |
KshA5 |
|||||
|---|---|---|---|---|---|---|---|
| kcat | Km | kcat/Km | kcat | Kma | kcat/Km | KiS | |
| s−1 | μm | s−1 mm−1 | s−1 | μm | s−1 mm−1 | μm | |
| 5α-Androstan-3,17-dione | 0.8 (0.1) | 6.1 (0.3) | 140 (20) | ||||
| 4-Estren-3,17-dione | 0.5 (0.1) | 400 (200) | 1.2 (0.3) | 0.8 (0.1) | 0.8 (0.1) | 1000 (200) | |
| Testosterone | 1.4 (0.3) | 110 (20) | 13 (2) | 0.8 (0.1) | 0.5 (0.1) | 1500 (300) | 130 (30) |
| ADD | 1.3 (0.2) | 50 (10) | 27 (4) | 0.7 (0.1)b | |||
| 4-BNC | 2.6 (0.5) | 2.2 (0.2) | 1200 (20) | 0.6 (0.1) | 1.2 (0.5) | 500 (200) | 110 (20) |
| 1,4-BNC | 0.9 (0.2) | 1.0 (0.2) | 900 (200) | 0.5 (0.1) | 0.5 (0.3) | 1000 (500) | 40 (20) |
| 1,4-BNC-CoA | 1.1 (0.3) | 2 (1) | 600 (400) | 1.7 (0.6) | 500 (200) | 3.7 (0.8) | |
| O2c | 1.7 (0.3) | 1.5 (0.2) | 1100 (200) | 0.8 (0.1) | 49 (6) | 17 (3) | |
a For parameters calculated using the substrate inhibition equation, the Km values represent Ks values.
b Data were calculated by averaging the maximal rates from Fig. 3.
c Data were determined using 50 μm 4-BNC and 5 μm 4-estren-3,17-dione for KshA1 and KshA5, respectively.
KshA5 displayed Michaelis-Menten kinetics with three 3-ketosteroid substrates and substrate inhibition with four (Fig. 3 and Table 2). For ADD, the inhibition was too strong to permit determination of an apparent Km value (i.e. Ks). Consistent with reversible substrate inhibition, ADD did not inactivate KshA5 when incubated with the enzyme, and at low concentrations of ADD, an increase in the steady-state reaction velocity was observed as ADD was consumed (Fig. 4). Despite the different behavior of KshA5 with the ketosteroids, the apparent kcat values for the different substrates were remarkably similar. KshA5 had similar apparent specificity for three of the ketosteroids (kcat/Km ∼106 m−1 s−1) as follows: 4-estren-3,17-dione, testosterone, and 1,4-BNC. This value was about 300-fold higher than the enzyme's apparent specificity for 1,4-BNC-CoA, the poorest of the tested substrates.
FIGURE 4.
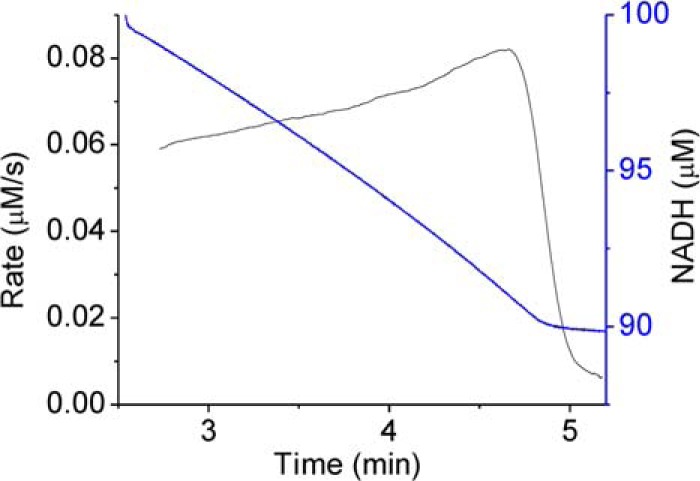
Progress curve of KshA5 with ADD. The rate of NADH consumption (calculated over 10-s intervals) is displayed in black, and the amount of NADH is shown in blue. Assay was performed using 8.3 μm ADD and 100 μm NADH in potassium phosphate (I = 0.1 m, pH 7.0), 22 °C.
The ability of KshA1 and KshA5 to utilize O2 was investigated in the presence of saturating amounts of 4-BNC and 4-estren-3,17-dione, respectively (Fig. 3 and Table 2). The latter was used because inhibition was not observed in the presence of this steroid (Fig. 3 and Table 2). The apparent Km, O2 value of KshA1 was over an order of magnitude lower than the values for KshA5 and KshAMtb (16). Similarly, the apparent kcat/Km, O2 value was 30–60 times that of KshAMtb and KshA5.
Structural Determination
To investigate the structural basis of catalysis and specificity in KshA, we determined structures of the enzymes in complex with substrates. Crystals of KshAMtb·ADD belonged to space group P32 and diffracted x-rays to ∼2.5 Å. The structure was solved by molecular replacement using substrate-free KshAMtb as a search model (PDB 2ZYL). Crystals grown in a mixture of KshA1, KshA5, and 1,4-BNC-CoA diffracted to ∼2.5 Å had space group F23 and two protomers in the asymmetric unit. The electron density clearly resembled that of the sequences of KshA1 for one protomer and KshA5 for the second. Both were modeled using KshAMtb for molecular replacement. Side chains were removed before further refinement and rebuilt into the density using the amino acid sequence of each homolog. Inspection of the crystallographic symmetry revealed that each homolog is arranged in the α3 structure typical of ROs to provide independent KshA1 and KshA5 trimers, and in turn, four trimers form a higher order structure described by the crystal symmetry (Fig. 5). The sole point of contact between the respective KshA1 and KshA5 protomers is at the C-terminal helix with 3 Å between Gln-372 of KshA5 and Thr-359 of KshA1. Otherwise, the protomers do not approach within 3.5 Å. Superposition of KshAMtb with either protomer revealed similar positioning of the backbones and many residues at the interface between KshA1 and KshA5, indicating that the crystal packing does not significantly impact the structures. KshA5 exhibited no substrate in its active site and a closed active site mouth loop (discussed below). Crystals of KshA5·ADD belonged to space group P63 and diffracted to ∼1.9 Å. The structure was again solved by molecular replacement using KshAMtb. Finally, crystals of KshA5·1,4-BNC-CoA belonged to space group P63 and diffracted to 2.6 Å. The structure was solved by molecular replacement using KshA5·ADD as a search model. Data collection and refinement statistics are listed in Table 3.
FIGURE 5.
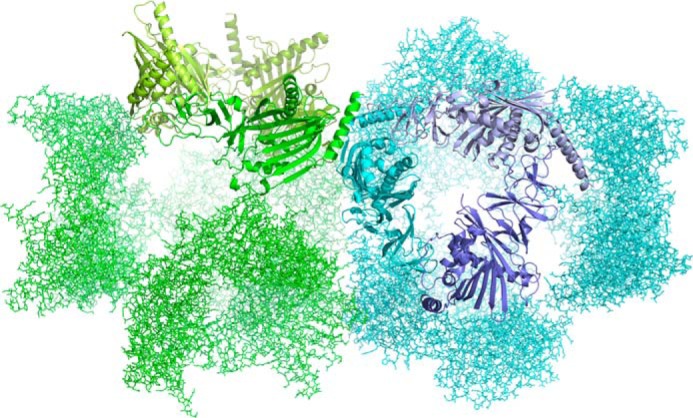
Crystal arrangement of KshA1·1,4-BNC-CoA and KshA5. The closest contact between the respective trimers in the mixed crystal is 3 Å. This occurs between Gln-372 of KshA5 (green) and Thr-359 of KshA1 (blue), both of which are located in the proteins' C-terminal helices. Figure was generated using PyMOL (50).
TABLE 3.
Data collection and refinement statistics
Values in parentheses are for highest resolution shell.
| KshAMtb·ADD | A5·ADD | A5·1,4-BNCCoA | A5/A1·1,4-BNC-CoA | |
|---|---|---|---|---|
| Data collection | ||||
| Wavelength (Å) | 0.92 | 0.92 | 0.98 | 0.98 |
| Space group | P32 | P63 | P63 | F23 |
| Cell dimensions | ||||
| a, b, c (Å) | 115.8, 115.8, 80.81 | 162.2, 162.2, 46.97 | 163.18, 163.18, 47.09 | 273.6, 273.6, 273.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 47.07–2.46(2.52–2.46) | 46.82–1.92(1.97–1.92) | 53.41–2.6(2.74–2.6) | 68.41–2.45(2.52–2.45) |
| Rsyma | 0.161(1.15) | 0.124(1.318) | 0.244(1.278) | 0.176(1.524) |
| Rpimb | 0.071(0.582) | 0.043(0.494) | 0.101(0.551) | 0.067(0.586) |
| CC1/2 | 0.997(0.412) | 0.999(0.453) | 0.983(0.197) | 0.998(0.536) |
| I /σI | 13.77(1.61) | 18(2.0) | 7.1(1.5) | 12.6(2.0) |
| Completeness (%) | 99.95(99.87) | 88.1(88.1) | 100(100) | 100(100) |
| Redundancy | 9.7(9.4) | 9.3(7.9) | 6.7(6.2) | 14.7(14.3) |
| Refinement | ||||
| Resolution (Å) | 2.46 | 1.92 | 2.6 | 2.45 |
| No. of reflections (unique) | 23062(1502) | 49275(1620) | 22485(3248) | 64010(4518) |
| Rworkc/Rfreed | 0.177/0.235 | 0.187/0.217 | 0.225/0.256 | 0.185/0.214 |
| No. of atoms | ||||
| Protein | 2900 | 2986 | 2959 | 5828 |
| Ligand/ion | 31 | 50 | 37 | 60 |
| Water | 128 | 276 | 23 | 201 |
| B-factors | ||||
| Protein | 47.4 | 29.7 | 51.5 | 49.8 |
| Ligand/ion | 40.2 | 37.3 | 57.4 | 71.1 |
| Water | 41.0 | 35.9 | 39.5 | 43.0 |
| r.m.s.d. | ||||
| Bond lengths (Å) | 0.007 | 0.141 | 0.007 | 0.008 |
| Bond angles (°) | 1.266 | 1.680 | 0.966 | 1.256 |
| Ramachandran plot | ||||
| Outliers | 0.00% | 0.54% | 0.83% | 0.14% |
| Allowed | 1.99% | 1.63% | 5.80% | 2.81% |
| Favored | 98.01% | 97.83% | 93.37% | 97.05% |
a Rsym = ΣhΣl|Ihl − 〈Ih〉|/ΣhΣl〈Ih〉.
b Rpim = Σh/(nh − 1)Σl|Ihl − 〈Ih〉|/ΣhΣl〈Ih〉.
c Rwork = Σ‖Fo| − |Fc‖/Σ|Fo|, where Fo and Fc represent the observed and calculated structure factors, respectively.
d Rfree is the Rwork value for 5% of the reflections excluded from the refinement.
KshAMtb·ADD Complex
The final model of the complex contains residues 14–375 except for residues 216–222 of the mouth loop. The model also contains the Rieske cluster, the mononuclear iron, and 201 waters. All major structural features are very similar to those of KshAMtb (13) as follows: the superimposed models had an r.m.s.d. of 0.20 Å over 319 Cα atoms, with residues of the Rieske center and active site pocket positioned nearly identically in the two structures. Indeed, the maximum distance between Cα was 0.2 Å, and most active site side chain atoms were within 0.5 Å. Electron density was insufficient for placement of residues 216–222, whereas residues 283–284 exhibited higher B-factors in the KshAMtb·ADD structure and included the only significant differences in the backbone as compared with the substrate-free model as summarized below. Similarly, the only side chain that could be reliably positioned differently was Asp-178 at the subunit interface as discussed below.
Electron density in the active site was modeled well with ADD at 100% occupancy with little residual density and B-factors similar to those of the surrounding protein atoms (Fig. 6A). The substrate is positioned such that C9 of the steroid is closest to the mononuclear iron at a distance of 3.9 Å, and the C17 oxo is oriented toward the solvent. The substrate is cradled by the β-sheet composed of strands β12 to β19, with most of the first shell residues provided by strands β15 and β16 (Fig. 7). Above this sheet, the pocket is bound on one side by helix α5, which runs transverse to the β-strands and harbors iron ligand Asp-304 at its C terminus, and helix α4, which harbors the iron ligands His-181 and His-186. The substrate's C3 oxo is hydrogen bonded to a water at 2.9 Å, which is similarly positioned in the substrate-free structure and is in turn hydrogen bonded to the hydroxyl of Tyr-232 (2.5 Å) and the backbone carbonyl of Ile-172 (2.7 Å). The 15-residue loop between strands β14 and β15, which partly seals off the pocket in the substrate-free structure, is disordered in the ADD-bound structure. It is possible that this loop has multiple conformations in the presence of ADD. More importantly, the pocket of KshAMtb·ADD would easily accommodate the CoA thioester as modeled in KshAMtb (13).
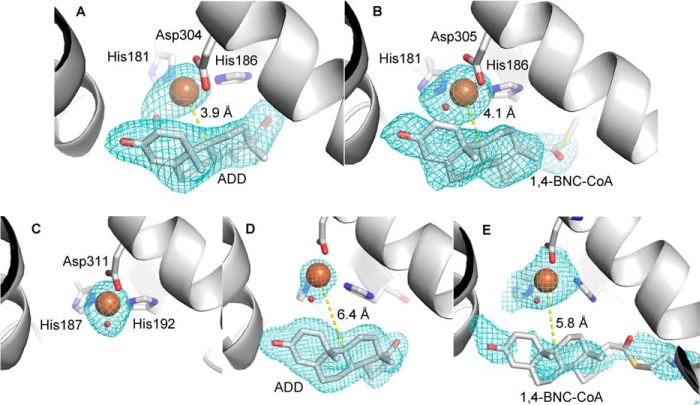
FIGURE 6.
Electron density in the active sites of KshAMtb·ADD (A), KshA1·1,4-BNC-CoA (B), KshA5 (C), KshA5·ADD (D), and KshA5·1,4-BNC-CoA (E). Fo − Fc mesh calculated in absence of iron, ligands, and substrate is contoured at 3σ in A, 2σ in B, 5σ in C, 5σ in D, and 2σ in E. Iron and water are orange and red spheres, respectively. Carbon, nitrogen, oxygen, and sulfur atoms are shown in gray, blue, red, and yellow, respectively. Figure was generated using PyMOL (50).
FIGURE 7.
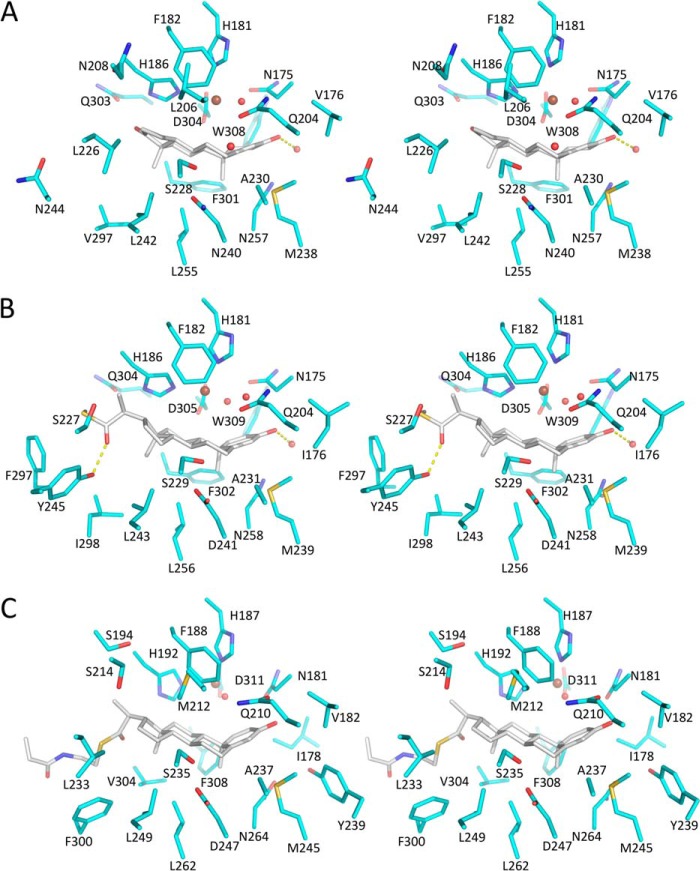
Stereo substrate-binding pockets of KshAMtb·ADD (A), KshA1·1,4-BNC-CoA (B), and KshA5·1,4-BNC-CoA (C). The carbon atoms of the enzyme and steroid are shown in cyan and gray, respectively. Nitrogen, oxygen, and sulfur are dark blue, red and yellow, respectively. The mononuclear iron is a brown sphere and waters are red spheres. H-bonds are shown as yellow dashed lines. Figure was generated using PyMOL (50).
The catalytic iron had a B-factor of 70 Å2 when modeled at full occupancy, approximately twice those of the adjacent protein atoms. The significance of this is unclear and may indicate some flexibility in the iron's positioning. Density adjacent to the iron was clearly that of a single atom and modeled well as a water at 2.6 Å. The mononuclear iron is coordinated by His-181, His-186, Asp-304 (bidentate), and an exogenous ligand in a five-coordinate distorted tetragonal geometry. Although this is similar to the coordination sphere of the substrate-free KshAMtb (13), the exogenous ligand is shifted ∼1.5 Å away from the presumed O2-binding site in KshAMtb·ADD, and the metal-ligand bond length is slightly longer. The presumed coordination site for O2 is trans to His-181. Beyond the coordination sphere, Asp-178, which bridges His-181 and FeS cluster ligand His-89, is rotated 135° around its Cα–Cβ bond (Fig. 8).
FIGURE 8.
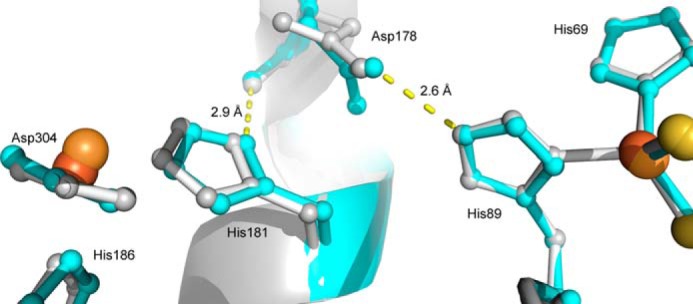
Superposition of the subunit interface in KshAMtb and KshAMtb·ADD. The mononuclear iron, its ligands, and the bridging Asp-178, at left, are in one subunit. The Rieske center and its ligands, at right, are in the neighboring subunit. KshAMtb and KshAMtb:ADD are in gray and cyan, respectively. The iron ions of KshAMtb·ADD and KshAMtb are brown and orange spheres, respectively. Sulfur atoms are represented by yellow spheres. Figure was generated using PyMOL (50) and PDB code 2ZYL (13).
KshA1·1,4-BNC-CoA Complex
The refined model of the complex includes residues 20–384 except for mouth loop residues 216–223. The model also contains the Rieske cluster, the mononuclear iron, and 128 waters. KshA1 has all the major structural features of KshAMtb, including the nine α-helices and 19 β-strands of the fold and the metal coordination spheres. As in KshAMtb, electron density for loop residues 216–223 was insufficient to model. Superpositioning KshA1·1,4-BNC-CoA and KshAMtb·ADD yielded an r.m.s.d. of 0.47 Å over 294 Cα atoms.
Electron density resembling a steroid nucleus with a C17 extension of several atoms was observed in the active site. This was modeled using 1,4-BNC and the first six atoms of the CoA (Fig. 6B). The occupancy of the substrate was lowered to 75% to match the B-factors of the surrounding protein atoms (30–50 Å2). The lack of obvious electron density for the remaining portion of the CoA may be due to its flexibility, although it is also possible that some degradation occurred during crystallization. The mononuclear iron was refined at 100% occupancy, yielding a B-factor of 55 Å2. Density adjacent to the iron corresponded to that of a single atom, and modeled well as a fully occupied water at 1.8 Å.
The active site of KshA1·1,4-BNC-CoA is remarkably similar to that of KshAMtb·ADD. The similarity of the five-coordinate mononuclear iron is highlighted in Table 4. The similarity extends to the conserved solvent species and its hydrogen bonding network at the C3 oxo, the residues that contact the 4-ringed nucleus, as well as the positioning of the steroid substrate, with C17 oriented toward the solvent (Fig. 7). The latter are conserved in the two enzymes except for Asp-241 (Asn-240 in KshAMtb) and Ile-176 (Val-176 in KshAMtb). Nevertheless, steroid rings C and D lie closer to the β-sheet in the KshA1·1,4-BNC-CoA structure, such that C17 is 1.6 Å closer to helix α5 in KshAMtb·ADD. This is consistent with the main structural differences in the complexes occurring at the opening of the substrate-binding pocket, where Asn-208, Leu-226, Asn-244, and Gly-296 of KshAMtb are Gly-208, Ser-227, Tyr-245, and Phe-297, respectively, in KshA1. Of the latter, the hydroxyl of Tyr-245 is within 2.4 Å of the 1,4-BNC-CoA C-22 carbonyl and could interact with the carboxylates of 4-BNC and 1,4-BNC, the preferred substrate of KshA1.
TABLE 4.
Metal-ligand and metal-substrate distances for mononuclear iron
Distances are provided in Å.
| Enzyme | Substrate | His-181a Nϵ2 | His-186 Nϵ2 | Asp-304 Oδ1 | Asp-304 Oδ2 | S1b | C9 |
|---|---|---|---|---|---|---|---|
| KshAMtbc | 2.2 | 2.1 | 2.5 | 2.1 | 2.1 | ||
| KshAMtb | ADD | 2.4 | 2.6 | 2.4 | 2.4 | 2.6 | 3.9 |
| KshA1 | 1,4-BNC-CoA | 2.2 | 2.3 | 2.5 | 2.3 | 1.8 | 4.1 |
| KshA5 | 1.9 | 2 | 2.4 | 2.2 | 1.6 | ||
| KshA5 | ADD | 2.2 | 2.4 | 2.8 | 2.3 | 2.4 | 6.4 |
| KshA5 | 1,4-BNC-CoA | 2.3 | 2.2 | 2.5 | 2.7 | 1.9 | 5.8 |
a Residues are numbered for KshAMtb. The corresponding residues in KshA1 are His-181, His-186, and Asp-305. In KshA5, they are His-187, His-192, and Asp-311.
b These are solvent species.
c Data are from Ref. 19.
Strikingly, the substrate binding pocket of KshAMtb is longer and more solvent-accessible than that of KshA1 despite the smaller substrate in the former's active site. This is due in part to Phe-297, located on helix α5, which occludes the pocket in KshA1. The corresponding residue in KshAMtb, Gly-296, opens the pocket along the length of the helix, presumably to accommodate the CoA moiety of the preferred substrate.
KshA5 Overall Structure
The KshA5 structure was obtained from the KshA1·KshA5 co-crystal and is described as substrate-free because no significant electron density was observed in the active site. The refined model includes residues 25–384, the [2Fe-2S] cluster, the mononuclear iron, and 201 waters. All major features of KshAMtb and KshA1 are conserved, including the nine α-helices and 19 β-strands. Indeed, the substrate-binding residues within the active site pocket are positioned nearly identically. The active site iron is fully occupied in KshA5, as evidenced by similar B-factors in surrounding atoms. Moreover, the metal ion's coordination sphere is similar to that observed in KshAMtb with distorted octahedral geometry by His-187, His-192, Asp-311 (bidentate), and a solvent species (Fig. 6C). Additional positive density within the substrate pocket was modeled as water molecules.
Structural alignment of KshA5 to KshAMtb revealed that nearly all of the backbone is positioned similarly between the two enzymes (r.m.s.d. 0.44 Å) with the exception of the mouth loop positioned at the entrance of the substrate-binding pocket. This loop is one residue longer in KshA5 (residues 216–231, inclusive) than in KshAMtb, and the electron density is better defined (Cα B-factors of 50–70 Å2 versus >100 Å2). In KshA5, the loop occludes the substrate-binding pocket from the solvent (Fig. 9A), in part due to the positioning of Tyr-226, although in KshAMtb it is positioned away from the active site opening.
FIGURE 9.
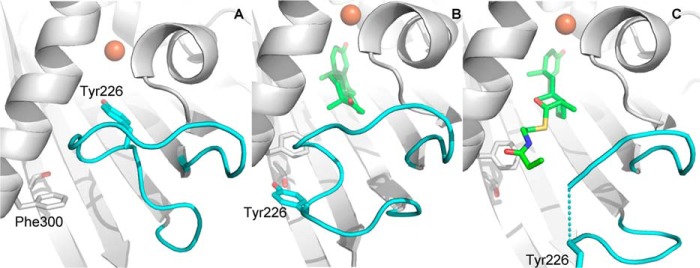
Mouth loop arrangements of KshA5. Comparison of the mouth loop in KshA5 (A), KshA5·ADD (B), and KshA5·1,4-BNC-CoA (C). Mouth loop residues are colored cyan, and substrate is colored green. Substrate (C atoms in green), Tyr-226 (cyan), and Thr-260 and Phe-300 (white) are represented as sticks, and the iron ion is an orange sphere. Figure was generated using PyMOL (50).
KshA5·ADD Complex
The refined model of the complex includes residues 15–383. ADD fits well to the electron density in the active site with B-factors of ∼20 Å2 (Fig. 6D). The overall KshA5·ADD structure is similar to that in the absence of substrate (r.m.s.d. 0.88 Å), as is the coordination sphere of the mononuclear iron. However, KshA5·ADD differs strikingly from the substrate-free KshA5 structure in a few well defined regions (Fig. 10). First, to accommodate the bound substrate, the mouth loop adopts a significantly different conformation with some residues shifted up to 11 Å in KshA5·ADD, although the loop still closes over the substrate channel (B-values of 30–40 Å2 for Cα). Notably, Tyr-226, displaced by ADD from the substrate-binding site, shifts more than 8 Å, packing against Ile-258 and hydrogen bonding with Thr-260. Interestingly, comparison with the substrate-free structure reveals that, in the presence of ADD, the aromatic side chain of Tyr-226 occupies the same position as the side chain of Phe-300 in the absence of substrate (Fig. 9B). Furthermore, Tyr-226 is able to insert into the pocket previously occupied by Phe-300 due to the displacement of helix α5 (∼4.5 Å shift (Fig. 11)), with Phe-300 now projecting toward the substrate-binding pocket and forming a direct contact with ADD. Strikingly, helix α5 also harbors Asp-311, one of the mononuclear iron ligands, and this helix displacement results in the iron being shifted 3.3 Å away from the substrate-binding pocket. The remaining iron ligands (His-187 and His-192) on helix α4 also move to maintain the metal coordination, and the side chain of Asn-181 is rotated 50° around Cβ to remain within 4 Å of the mononuclear iron. Because the helices are displaced orthogonally to the [2Fe-2S] cluster (i.e. helix α5 is displaced in the direction of its C terminus and slightly toward the Rieske domain), the distance between the metallocenters is not significantly different in the presence of ADD (∼12.5 Å). Finally, although the mouth loop changes and the helix displacement appear to be concerted, in neither the KshA5 nor the KshA5·ADD structures does the conformation of the mouth loop allow entry or egress of the substrate.
FIGURE 10.
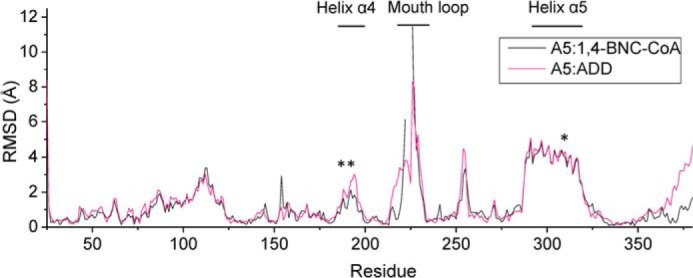
Per residue r.m.s.d. between the substrate-bound and substrate-free structures of KshA5. After a structural alignment of the three KshA5 structures, the distances between Cα of the substrate-bound and substrate-free structures was measured. The ligands of the mononuclear iron (His-187, His-192, and Asp-311) are indicated with an asterisk.
FIGURE 11.
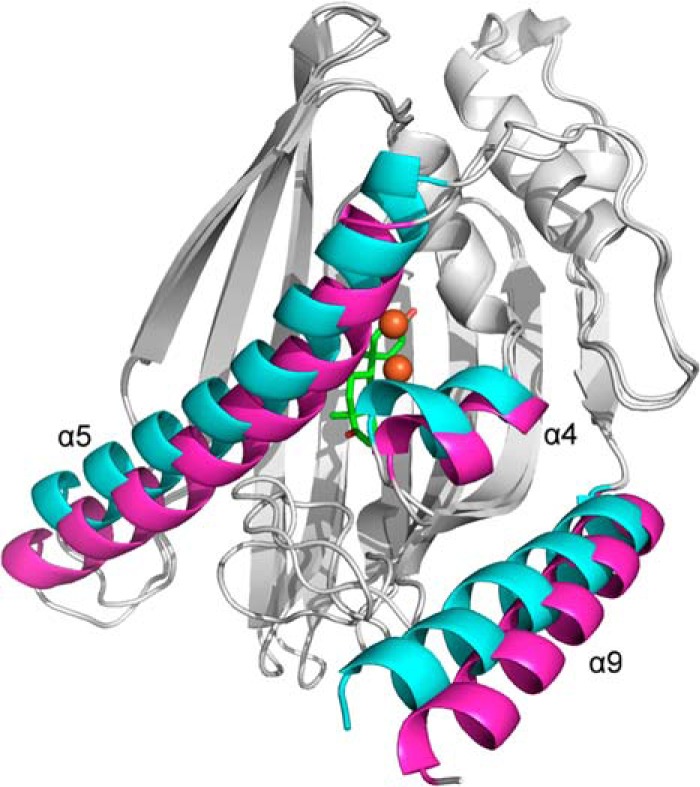
Superposition of the C-terminal domains of KshA5·ADD and KshA5. Helices α4, α5, and α9 are in magenta for KshA5 and cyan for KshA5·ADD. ADD and the mononuclear iron are colored green and orange, respectively. Figure was generated using PyMOL (50).
Despite these dramatic shifts, ADD is bound in the same manner as the steroid substrates in the KshA1·1,4-BNC-CoA and KshAMtb·ADD complexes (Fig. 7). Thus, the C17 oxo is oriented toward the solvent, and all residues that interact with the steroid nucleus are conserved. These include Val-182, Gln-210, Met-245, Phe-308, and Trp-315 as well as Tyr-239, which interacts with the 3-oxo via a water. One difference is Asp-247, which is Asn in KshA1 and KshAMtb. The above-described shift in the mononuclear iron in KshA5·ADD leaves the site of hydroxylation of ADD, C9, 6.4 Å from the mononuclear iron with an intervening water molecule. Indeed, three ADD carbon atoms are closer to the metal ion as follows: C2, C1, and C11 are 5.3, 5.4, and 6.2 Å away, respectively (Fig. 6D). In contrast, C9 is the steroid carbon atom closest to the iron in the KshA1·1,4-BNC-CoA and KshAMtb·ADD complexes (∼4 Å; Fig. 6). Finally, the configuration of Asp-184, which bridges the metallocenters, is similar to that seen in KshA1·1,4-BNC-CoA and KshAMtb·ADD.
KshA5·1,4-BNC-CoA Complex
The refined model of the complex contains residues 15–383 and 23 waters but is missing three residues of the mouth loop (223–225). Additional electron density in the active site was fit with the steroid nucleus of 1,4-BNC-CoA and the first eight atoms of the CoA moiety (B-factors ∼60 Å2; Fig. 6E). Superpositioning reveals that the model is more similar to KshA5·ADD (r.m.s.d. 0.47) than KshA5 (r.m.s.d. 0.78). This reflects the similar positioning of the displaced α-helices and the catalytic metallocenter in the structures of the enzyme·substrate complexes (Fig. 10). In KshA5·1,4-BNC-CoA, C9 of the substrate is 5.8 Å from the mononuclear iron. Similar to KshA5·ADD, C1, C2, C11, and C12 are closer to the iron. The mouth loop is less well defined in KshA5·1,4-BNC-CoA, with no defined electron density for residues 223–225 and B-factors of 100 Å2 for the Cα of residues 222 and 227. Nevertheless, it is clear that the loop does not close over the substrate-binding pocket (Fig. 9C) as in the other KshA5 structures and is displaced farther from the active site than in KshA5·ADD. This conformation is similar to that observed in KshAMtb·ADD and appears to be due in part to the CoA moiety. Thus, the first eight atoms of the CoA moiety have clear electron density that exits the substrate pocket beside residue Phe-300 of helix α5.
Sequences of Cholesterol- and Cholate-degrading KshAs
An alignment of KshA sequences to a template based on the crystallographic structures of KshA1, KshA5, and KshAMtb indicates that Tyr-245 and Phe-297 are conserved in predicted actinobacterial cholate-degrading orthologs, such as KshA3 from R. jostii RHA1 (6) and a KshA from Thermomonospora curvata (Tcur_3509). In Nocardia farcinica Nfa33560, the Tyr is also conserved, and Phe is substituted with Trp. Genome analyses revealed that the genes encoding these predicted cholate-degrading enzymes cluster with known bile acid catabolic genes such as casG and casI (11). By contrast, the known cholesterol-degrading orthologs, such as KshA from R. jostii RHA1 (5), KshA from Gordonia sp. KTR9 (38), and KshA3 from DSM43269, have smaller residues in place of Tyr-245 and Phe-297. Finally, these residues are not conserved in KshAs from bile acid-degrading Proteobacteria (7, 8).
DISCUSSION
This study provide insight into the structural basis of the substrate specificity of ketosteroid-transforming ROs. The steady-state kinetic studies corroborate and extend previous studies of KshA1 that has been implicated in bile acid catabolism based on substrate preference (24), gene deletion studies, and phylogenetic analysis (39). These studies further establish KshA5's broad substrate specificity for short side chain steroids and, unexpectedly, its strong substrate inhibition. This specificity is borne out by the structural studies showing that the three enzymes bind the steroid very similarly but that KshAMtb has a larger pocket that can accommodate a substrate with a longer side chain. Finally, the remarkable conformational flexibility observed in KshA5 may contribute to the enzyme's broad specificity and has important implications for the catalytic cycle of ROs.
Implication of KshA Specificity for Steroid Catabolism
Importantly, KshA1 does not possess the increased apparent specificity (kcat/Km) for CoA thioesters such as KshAMtb despite the involvement of both enzymes in catabolizing steroids with C17 side chains. The different apparent specificities are intriguing considering the following: (a) cholesterol catabolism in M. tuberculosis (16) and RHA1 and cholate catabolism in RHA1 (40) all involve concurrent side chain and rings A/B degradation; and (b) side chain degradation involves CoA thioesters. The specificities of KshA1 for 4-BNC and 1,4-BNC are consistent with concurrent side chain and rings A/B degradation in cholate catabolism by DSM43269, i.e. the low specificity of KshA1 for steroids lacking a C17 side chain such as ADD indicates that ring B is likely opened before complete catabolism of the side chain. This specificity further suggests that not all intracellular side chain degradation catabolites are CoA thioesters and thus that cholate and cholesterol catabolism are organized differently. This is consistent with the occurrence of metabolites such as 3,7(R),12(S)-trihydroxy-9-oxo-9,10-seco-23,24-bisnorchola-1,3,5(10)-trien-22-oate in the culture supernatant of R. jostii RHA1 growing on cholate (40) and may reflect the toxicity of cholate metabolites.
The greater specificity (kcat/Km) of KshA5 for substrates with shorter C17 side chains and the strong substrate inhibition by ADD apparently contradict the conclusion that KshA5 is involved in the catabolism of a range of steroids (24). However, KshA5 turned over steroids at similar maximal rates, consistent with the reported activities determined at single substrate concentrations (24). Indeed, the steady-state kinetic parameters only appear to rule out 1,4-BNC-CoA as a physiological substrate for KshA5 despite the strong substrate inhibition for other steroids. Even ADD, for which a kcat/Km value could not be determined, is turned over efficiently at very low concentrations. More particularly, the apparent kcat/Km values of KshA5 for various steroids are within those of ROs for their physiological substrate, which range from 0.036 × 106 m−1 s−1 (aminopyrrolnitrin oxygenase) to 2.4 × 106 m−1 s−1 (BPDOLB400) (41, 42). Overall, the steady-state kinetic data are consistent with the proposal that KshA5 is involved in degrading a range of steroids.
The ability of KshA5 to support the growth of a rhodococcal strain on cholesterol (24) is unexpected given the enzyme's very low kcat/Km value for 1,4-BNC-CoA. In the cholesterol catabolic pathways examined to date, ring A/B and side chain degradation occur concurrently, with KshA acting on a CoA thioester substrate (16, 17). It is possible that such concurrent degradation is more efficient but not obligate and that KshA5 acts on a side chain-degraded cholesterol catabolite. Further studies are required to determine the precise role of KshA5 in steroid catabolism.
The significance of KshA1's low apparent Km, O2 value is not clear. This value is over an order of magnitude lower than that of KshAMtb (90 μm in the presence of 1,4-BNC-CoA (16)) and KshA5. Moreover, it is one of the lowest Km, O2 values reported for an RO, comparable with that of toluate dioxygenase measured in the presence of p-toluate (8.4 μm (43)). The low apparent Km, O2 would allow KshA1 to turnover substrate at significant rates at low O2 concentrations, as might occur in the natural soil environment of rhodococci. Nevertheless, the different ability of the paralogs to utilize O2 is striking and may be related to the different physiological roles of these enzymes.
Specificity Determinants
The structural and sequence analyses identify Tyr-245 and Phe-297 as determinants of substrate specificity in actinobacterial cholate-degrading KshAs. The residues that interact with the steroid nucleus are remarkably conserved in cholesterol- and cholate-degrading enzymes despite the 7- and 12-hydroxyl groups of cholate (Fig. 1). These hydroxyls interact with Gln-204, Ser-229, and His-186 in docking experiments (results not shown), residues that are conserved in KshAMtb and KshA5. This may reflect the role of KshA1 and its homologs in degrading bile acids that are not hydroxylated at these positions, such as lithocholate. Finally, it is possible that residues in the loop also contribute to specificity. However, this is not possible to evaluate in the current structural models due to the absence of the loop residues. It is possible that the cholesterol-degrading KshAs are unique in their preference for CoA thioesters.
Comparison of the KshA structures further identifies the entrance of the substrate-binding pocket, including the mouth loop as a major determinant of substrate specificity. The role of the mouth loop had been described in studies of chimeras of KshA1 and KshA5 (39). Thus, the broad substrate preference of KshA5 was eliminated by substituting its mouth loop with that of KshA1. However, the chimeric enzyme did not possess KshA1's strong preference for 4-BNC. Moreover, reverse chimeras of KshA1 with the KshA5 loop or strands β14–17, including the loop, did not possess KshA5's broader substrate preference, although both had decreased activity for 4-BNC. This indicates that the β-sheet and mouth loop are necessary but not sufficient for KshA5's broad substrate preference. This is consistent with other residues at the mouth entrance contributing to substrate specificity such as Tyr-245 and Phe-297 of KshA1. Interestingly, the corresponding residues are smaller in both KshA5 (Ser-251 and Gly-303) and KshAMtb (Asn-244 and Gly-296), enzymes that prefer substrates with shorter and longer C17 side chains, respectively, suggesting that the role of these residues in determining substrate specificity depends on their context.
Metallocenters
The structures of the mononuclear iron and Reiske cluster in each of the KshA structures is consistent with what has been reported in ROs in which the Rieske center is oxidized, such as OMOox in the absence and presence of its substrate, oxoquinoline (21). Of particular note is the configuration of the bridging acidic residue; in ROs in which the Rieske cluster is reduced, such as OMOred (21) and stachydrine demethylase (Stc2) (44), the carboxylate bridges the ligands of the metallocenters. Nevertheless, it is unclear why the KshA Rieske clusters would be oxidized because the x-ray beam is expected to reduce the clusters (45).
The position of the steroid C9 atom relative to the catalytic iron in both KshA1 and KshAMtb substrate complexes is similar to that of the corresponding substrate atom, C8, in the OMOox·oxoquinoline complex (21), although the atom to be hydroxylated is positioned 0.6–0.8 Å closer to the iron in the two KshA complexes. The mononuclear iron shifts up to 0.8 Å away from the substrate-binding site in response to Rieske cluster reduction in OMO (21) and following substrate binding in CARDO (22). It has been proposed that these shifts ensure that O2 reactivity at the mononuclear iron site is gated by both substrate binding and FeS cluster reduction (21, 46, 47). Importantly, the open site in the mononuclear iron's coordination sphere in the KshA1 and KshAMtb complexes is appropriately positioned to enable the binding and activation of O2 relative to C9 as has been proposed in other ROs (21, 23).
Although the shifts in the mononuclear iron and its ligands are much greater in the KshA5·substrate complexes, they are similar in nature to those described above. Specifically, the shift in the mononuclear iron is ∼4.4 Å in KshA5·ADD and KshA5·1,4-BNC-CoA, and the distance between the substrate carbon to be hydroxylated and the iron is ∼2 Å greater than what has been observed in other ROs. This distance is likely too long for efficient hydroxylation at C9. It is possible that these structures represent a substrate-inhibited form of the enzyme. Nevertheless, the efficient coupling of O2 activation with ADD and 4-estren-3,17-dione hydroxylation suggests that the C9 is positioned close to the activated oxygen intermediate. It therefore seems likely that further adjustment to the substrate and/or mononuclear iron position occurs subsequent to O2 binding.
Conformational Flexibility
The structures of KshA5 in three states (Fig. 10) provide clear evidence that the mouth loop of KshA can exist in multiple conformations to sequester the active site from bulk solvent and to contribute to the specificity of the enzyme. The mouth loops in the KshA1 and KshAMtb structures are disordered and are most similar to the open state observed in the KshA5·1,4-BNC-CoA complex. Moreover, their disorder is likely due to their ability to adopt multiple conformations, as captured in the KshA5 structures. Nevertheless, the conformations adopted by the loop likely reflect the substrate specificity of the parent KshA. For example, the mouth of the substrate pocket in KshAMtb is very large but must accommodate a CoA thioester that extends into the solvent. In this respect, it is possible that KshA5 has unusual conformational flexibility, contributing not only to the enzyme's substrate specificity but also to its strong substrate inhibition, i.e. the conformational flexibility of the mouth loop might allow the binding of a second inhibitory substrate molecule to KshA5 at the entrance of the substrate-binding pocket.
It is unclear whether the entrance of the substrate-binding pocket of other ROs has the same conformational flexibility as the KshA mouth loop. As noted above, the substrate-binding pocket of KshA is “straight-in,” in contrast to the pocket in other ROs, which has an elbow bend. Although the straight-in nature of the KshA pocket appears to be necessary to accommodate the 4-ringed substrate, the entrance of the substrate-binding pocket of the other ROs occurs on a different surface of the enzyme with no homologous mouth loop. Structural studies of CARDO revealed that the substrate-binding pocket is closed in the presence of substrate due to shifts in residues of up to 3.3 Å (22, 48), much less than the shifts in KshA. It is possible that the mouth loop provides a KshA-specific device to sequester the substrate from the solvent.
The shifts in the active site of KshA5 are a manifestation of the significant displacement of helices α4 and α5 (i.e. >4 Å), which harbor the iron ligands (Fig. 11). By contrast, the β-sheet bearing many of the conserved substrate-binding residues shifts minimally. We cannot rule out that the displacement is a crystal packing artifact because helix α5 forms crystal contacts at its N and C termini. However, the concerted movement of the mouth loop and helices α5/α4 suggest the observed conformational changes are biologically relevant. Movement of this type and magnitude has not been reported in any of the other 12 ROs structurally characterized to date. Moreover, displacement of these helices was not observed in KshA1 or KshAMtb, and it remains to be seen whether this conformational change is unique to KshA5. Nevertheless, the high functionality of the KshA1/KshA5 chimeras (39) suggests that KshAs undergo similar conformational changes. More generally, our data suggest that conformational changes in the RO active site during the catalytic cycle may be larger than has been recognized.
The conformational space sampled by KshA5 is reminiscent of what has been reported in cytochrome P450cam, a heme-dependent monooxygenase (49). In P450cam, an open conformation observed in the absence of substrate differs from the closed, substrate-bound state by a 5–6 Å displacement of two helices, F and G, and a 9 Å retraction of the FG loop connecting them at the opening of the substrate channel. The displacement of the helices in KshA5 and P450cam are not entirely analogous. First, the KshA5 structure does not represent an open state that allows entry of the substrate to the active site. Second, the displaced helices of P450cam do not harbor key active site residues, such as Thr-252 or the heme ligand Cys-357. As such, displacement of helices F and G in P450cam does not implicate dramatic shifts in the catalytic machinery. Nevertheless, the observation of similar conformational mobility in two different classes of oxygenases suggests that it may be a general property of these types of enzymes.
By identifying the substrate-binding residues of KshAs and identifying determinants of specificity in KshA, this study provides a framework for engineering an important class of monooxygenases for biocatalytic applications (2, 39). Further studies should also establish the occurrence and significance of helix displacement in ROs.
Acknowledgments
Drs. Robert van der Geize and Mirjam Petrusma provided kshA1 and kshA5 constructs. X-ray synchrotron data were collected at the Canadian Light Source, which is supported by Natural Sciences and Engineering Council of Canada, Canadian Institutes for Health Research, the National Research Council Canada, the Province of Saskatchewan, Western Economic Diversification Canada, and the University of Saskatchewan.
This work was supported by operating grants from the Canadian Institutes for Health Research and Natural Sciences and Engineering Council of Canada (to L. D. E.).
The atomic coordinates and structure factors (codes 4QCK, 4QDF, 4QDC, and 4QDD) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- RO
- Rieske oxygenase
- 1,4-BNC
- 3-oxo-23,24-bisnorcholesta-1,4-diene-22-oate
- ADD
- 4-androstene-3,17-dione
- OMO
- 2-oxoquinoline 8-monooxygenase
- CARDO
- carbazole 1,9a-dioxygenase
- r.m.s.d.
- root mean square deviation
- Ches
- 2-(cyclohexylamino)ethanesulfonic acid
- PDB
- Protein Data Bank.
REFERENCES
- 1. Wollam J., Antebi A. (2011) Sterol regulation of metabolism, homeostasis, and development. Annu. Rev. Biochem. 80, 885–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fernandes P., Cruz A., Angelova B., Pinheiro H. M., Cabral J. M. (2003) Microbial conversion of steroid compounds: recent developments. Enzyme Microb. Technol. 32, 688–705 [Google Scholar]
- 3. van der Geize R., Dijkhuizen L. (2004) Harnessing the catabolic diversity of rhodococci for environmental and biotechnological applications. Curr. Opin. Microbiol. 7, 255–261 [DOI] [PubMed] [Google Scholar]
- 4. Hu Y., van der Geize R., Besra G. S., Gurcha S. S., Liu A., Rohde M., Singh M., Coates A. (2010) 3-Ketosteroid 9α-hydroxylase is an essential factor in the pathogenesis of Mycobacterium tuberculosis. Mol. Microbiol. 75, 107–121 [DOI] [PubMed] [Google Scholar]
- 5. Van der Geize R., Yam K., Heuser T., Wilbrink M. H., Hara H., Anderton M. C., Sim E., Dijkhuizen L., Davies J. E., Mohn W. W., Eltis L. D. (2007) A gene cluster encoding cholesterol catabolism in a soil actinomycete provides insight into Mycobacterium tuberculosis survival in macrophages. Proc. Natl. Acad. Sci. U.S.A. 104, 1947–1952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mohn W. W., Wilbrink M. H., Casabon I., Stewart G. R., Liu J., van der Geize R., Eltis L. D. (2012) Gene cluster encoding cholate catabolism in Rhodococcus spp. J. Bacteriol. 194, 6712–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Horinouchi M., Hayashi T., Kudo T. (2012) Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 129, 4–14 [DOI] [PubMed] [Google Scholar]
- 8. Birkenmaier A., Holert J., Erdbrink H., Moeller H. M., Friemel A., Schoenenberger R., Suter M. J., Klebensberger J., Philipp B. (2007) Biochemical and genetic investigation of initial reactions in aerobic degradation of the bile acid cholate in Pseudomonas sp. strain Chol1. J. Bacteriol. 189, 7165–7173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Birkenmaier A., Möller H. M., Philipp B. (2011) Identification of a thiolase gene essential for β-oxidation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1. FEMS Microbiol. Lett. 318, 123–130 [DOI] [PubMed] [Google Scholar]
- 10. Holert J., Kulić Ž., Yücel O., Suvekbala V., Suter M. J., Möller H. M., Philipp B. (2013) Degradation of the acyl side chain of the steroid compound cholate in Pseudomonas sp. strain Chol1 proceeds via an aldehyde intermediate. J. Bacteriol. 195, 585–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Casabon I., Zhu S. H., Otani H., Liu J., Mohn W. W., Eltis L. D. (2013) Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol. Microbiol. 89, 1201–1212 [DOI] [PubMed] [Google Scholar]
- 12. Nesbitt N. M., Yang X., Fontán P., Kolesnikova I., Smith I., Sampson N. S., Dubnau E. (2010) A thiolase of Mycobacterium tuberculosis is required for virulence and production of androstenedione and androstadienedione from cholesterol. Infect. Immun. 78, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Capyk J. K., D'Angelo I., Strynadka N. C., Eltis L. D. (2009) Characterization of 3-ketosteroid 9α-hydroxylase, a Rieske oxygenase in the cholesterol degradation pathway of Mycobacterium tuberculosis. J. Biol. Chem. 284, 9937–9946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. van der Geize R., Hessels G. I., van Gerwen R., van der Meijden P., Dijkhuizen L. (2002) Molecular and functional characterization of kshA and kshB, encoding two components of 3-ketosteroid 9α-hydroxylase, a class IA monooxygenase, in Rhodococcus erythropolis strain SQ1. Mol. Microbiol. 45, 1007–1018 [DOI] [PubMed] [Google Scholar]
- 15. Gibson D. T., Wang K. C., Sih C. J., Whitlock H., Jr. (1966) Mechanisms of steroid oxidation by microorganisms. IX. On the mechanism of ring A cleavage in the degradation of 9,10-seco steroids by microorganisms. J. Biol. Chem. 241, 551–559 [PubMed] [Google Scholar]
- 16. Capyk J. K., Casabon I., Gruninger R., Strynadka N. C., Eltis L. D. (2011) Activity of 3-ketosteroid 9α-hydroxylase (KshAB) indicates cholesterol side chain and ring degradation occur simultaneously in Mycobacterium tuberculosis. J. Biol. Chem. 286, 40717–40724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Thomas S. T., VanderVen B. C., Sherman D. R., Russell D. G., Sampson N. S. (2011) Pathway profiling in Mycobacterium tuberculosis: elucidation of cholesterol-derived catabolite and enzymes that catalyze its metabolism. J. Biol. Chem. 286, 43668–43678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ferraro D. J., Gakhar L., Ramaswamy S. (2005) Rieske business: structure-function of Rieske non-heme oxygenases. Biochem. Biophys. Res. Commun. 338, 175–190 [DOI] [PubMed] [Google Scholar]
- 19. Capyk J. K., Eltis L. D. (2012) Phylogenetic analysis reveals the surprising diversity of an oxygenase class. J. Biol. Inorg. Chem. 17, 425–436 [DOI] [PubMed] [Google Scholar]
- 20. Tarasev M., Pinto A., Kim D., Elliott S. J., Ballou D. P. (2006) The “bridging” aspartate 178 in phthalate dioxygenase facilitates interactions between the Rieske center and the iron(II)-mononuclear center. Biochemistry 45, 10208–10216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martins B. M., Svetlitchnaia T., Dobbek H. (2005) 2-Oxoquinoline 8-monooxygenase oxygenase component: active site modulation by Rieske-[2Fe-2S] center oxidation/reduction. Structure 13, 817–824 [DOI] [PubMed] [Google Scholar]
- 22. Ashikawa Y., Fujimoto Z., Usami Y., Inoue K., Noguchi H., Yamane H., Nojiri H. (2012) Structural insight into the substrate- and dioxygen-binding manner in the catalytic cycle of Rieske nonheme iron oxygenase system, carbazole 1,9α-dioxygenase. BMC Struct. Biol. 12, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Karlsson A., Parales J. V., Parales R. E., Gibson D. T., Eklund H., Ramaswamy S. (2003) Crystal structure of naphthalene dioxygenase: Side-on binding of dioxygen to iron. Science 299, 1039–1042 [DOI] [PubMed] [Google Scholar]
- 24. Petrusma M., Hessels G., Dijkhuizen L., van der Geize R. (2011) Multiplicity of 3-ketosteroid-9α-hydroxylase enzymes in Rhodococcus rhodochrous DSM43269 for specific degradation of different classes of steroids. J. Bacteriol. 193, 3931–3940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, 2nd Ed., Cold Spring Harbor, NY [Google Scholar]
- 26. Vaillancourt F. H., Han S., Fortin P. D., Bolin J. T., Eltis L. D. (1998) Molecular basis for the stabilization and inhibition of 2, 3-dihydroxybiphenyl 1,2-dioxygenase by t-butanol. J. Biol. Chem. 273, 34887–34895 [DOI] [PubMed] [Google Scholar]
- 27. Cornish-Bowden A. (1995) Analysis of Enzyme Kinetic Data, Oxford University Press, New York [Google Scholar]
- 28. Leslie A. G. W., Powell H. R. (2007) Processing diffraction data with Mosflm. in Evolving Methods of Macromolecular Crystallography, Vol. 245 (Read R. J., Sussman J. L, eds) pp. 41–51, Springer Netherlands, Amsterdam [Google Scholar]
- 29. Evans P. R., Murshudov G. N. (2013) How good are my data and what is the resolution? Acta Crystallogr. D Biol. Crystallogr. 69, 1204–1214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McCoy A. J. (2007) Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. D Biol. Crystallogr. 63, 32–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 32. Winn M. D., Ballard C. C., Cowtan K. D., Dodson E. J., Emsley P., Evans P. R., Keegan R. M., Krissinel E. B., Leslie A. G., McCoy A., McNicholas S. J., Murshudov G. N., Pannu N. S., Potterton E. A., Powell H. R., Read R. J., Vagin A., Wilson K. S. (2011) Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 67, 235–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 34. Schüttelkopf A. W., van Aalten D. M. (2004) PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr. D Biol. Crystallogr. 60, 1355–1363 [DOI] [PubMed] [Google Scholar]
- 35. Joosten R. P., Joosten K., Murshudov G. N., Perrakis A. (2012) PDB-REDO: Constructive validation, more than just looking for errors. Acta Crystallogr. D Biol. Crystallogr. 68, 484–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen V. B., Arendall W. B., 3rd., Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Petrusma M., Dijkhuizen L., van der Geize R. (2009) Rhodococcus rhodochrous DSM 43269 3-ketosteroid 9α-hydroxylase, a two-component iron-sulfur-containing monooxygenase with subtle steroid substrate specificity. Appl. Environ. Microbiol. 75, 5300–5307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen H. P., Zhu S. H., Casabon I., Hallam S. J., Crocker F. H., Mohn W. W., Indest K. J., Eltis L. D. (2012) Genomic and transcriptomic studies of an RDX (hexahydro-1,3,5-trinitro-1,3,5-triazine)-degrading actinobacterium. Appl. Environ. Microbiol. 78, 7798–7800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petrusma M., Dijkhuizen L., van der Geize R. (2012) Structural features in the KshA terminal oxygenase protein that determine substrate preference of 3-ketosteroid 9α-hydroxylase enzymes. J. Bacteriol. 194, 115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Swain K., Casabon I., Eltis L. D., Mohn W. W. (2012) Two transporters essential for the reassimilation of novel cholate metabolites by Rhodococcus jostii RHA1. J. Bacteriol. 194, 6720–6727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lee J., Simurdiak M., Zhao H. (2005) Reconstitution and characterization of aminopyrrolnitrin oxygenase, a Rieske N-oxygenase that catalyzes unusual arylamine oxidation. J. Biol. Chem. 280, 36719–36727 [DOI] [PubMed] [Google Scholar]
- 42. Gómez-Gil L., Kumar P., Barriault D., Bolin J. T., Sylvestre M., Eltis L. D. (2007) Characterization of biphenyl dioxygenase of Pandoraea pnomenusa B-356 as a potent polychlorinated biphenyl-degrading enzyme. J. Bacteriol. 189, 5705–5715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ge Y., Eltis L. D. (2003) Characterization of hybrid toluate and benzoate dioxygenases. J. Bacteriol. 185, 5333–5341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Daughtry K. D., Xiao Y., Stoner-Ma D., Cho E., Orville A. M., Liu P., Allen K. N. (2012) Quaternary ammonium oxidative demethylation: x-ray crystallographic, resonance Raman, and UV-visible spectroscopic analysis of a Rieske-type demethylase. J. Am. Chem. Soc. 134, 2823–2834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Karlsson A., Parales J. V., Parales R. E., Gibson D. T., Eklund H., Ramaswamy S. (2000) The reduction of the Rieske iron-sulfur cluster in naphthalene dioxygenase by x-rays. J. Inorg Biochem. 78, 83–87 [DOI] [PubMed] [Google Scholar]
- 46. Yang T. C., Wolfe M. D., Neibergall M. B., Mekmouche Y., Lipscomb J. D., Hoffman B. M. (2003) Substrate binding to NO-ferro-naphthalene 1,2-dioxygenase studied by high-resolution Q-band pulsed 2H-ENDOR spectroscopy. J. Am. Chem. Soc. 125, 7056–7066 [DOI] [PubMed] [Google Scholar]
- 47. Wolfe M. D., Parales J. V., Gibson D. T., Lipscomb J. D. (2001) Single turnover chemistry and regulation of O2 activation by the oxygenase component of naphthalene 1,2-dioxygenase. J. Biol. Chem. 276, 1945–1953 [DOI] [PubMed] [Google Scholar]
- 48. Inoue K., Usami Y., Ashikawa Y., Noguchi H., Umeda T., Yamagami-Ashikawa A., Horisaki T., Uchimura H., Terada T., Nakamura S., Shimizu K., Habe H., Yamane H., Fujimoto Z., Nojiri H. (2014) Structural basis of the divergent oxygenation reactions catalyzed by the rieske nonheme iron oxygenase carbazole 1,9a-dioxygenase. Appl. Environ. Microbiol. 80, 2821–2832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lee Y.-T., Wilson R. F., Rupniewski I., Goodin D. B. (2010) P450cam visits an open conformation in the absence of substrate. Biochemistry 49, 3412–3419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. DeLano W. (2002) The PyMOL Molecular Graphics System, Version 1.5.0.4, Schrödinger, LLC [Google Scholar]