Background: Tomosyn regulates vesicle fusion but its mechanism remains incompletely understood.
Results: Tomosyn uses its C-terminal domain to arrest SNARE-dependent fusion reactions, whereas its N-terminal domain is required for syntaxin interaction.
Conclusion: We have uncovered distinct roles of the N- and C-terminal domains of tomosyn in SNARE binding and fusion regulation.
Significance: Our findings shed light upon vesicle transport in the cell.
Keywords: Exocytosis, Membrane Bilayer, Membrane Fusion, Membrane Trafficking, Vesicles, Membrane, Membrane Fusion
Abstract
Tomosyn negatively regulates SNARE-dependent exocytic pathways including insulin secretion, GLUT4 exocytosis, and neurotransmitter release. The molecular mechanism of tomosyn, however, has not been fully elucidated. Here, we reconstituted SNARE-dependent fusion reactions in vitro to recapitulate the tomosyn-regulated exocytic pathways. We then expressed and purified active full-length tomosyn and examined how it regulates the reconstituted SNARE-dependent fusion reactions. Using these defined fusion assays, we demonstrated that tomosyn negatively regulates SNARE-mediated membrane fusion by inhibiting the assembly of the ternary SNARE complex. Tomosyn recognizes the t-SNARE complex and prevents its pairing with the v-SNARE, therefore arresting the fusion reaction at a pre-docking stage. The inhibitory function of tomosyn is mediated by its C-terminal domain (CTD) that contains an R-SNARE-like motif, confirming previous studies carried out using truncated tomosyn fragments. Interestingly, the N-terminal domain (NTD) of tomosyn is critical (but not sufficient) to the binding of tomosyn to the syntaxin monomer, indicating that full-length tomosyn possesses unique features not found in the widely studied CTD fragment. Finally, we showed that the inhibitory function of tomosyn is dominant over the stimulatory activity of the Sec1/Munc18 protein in fusion. We suggest that tomosyn uses its CTD to arrest SNARE-dependent fusion reactions, whereas its NTD is required for the recruitment of tomosyn to vesicle fusion sites through syntaxin interaction.
Introduction
Exocytosis, the fusion of exocytic vesicles with the plasma membrane, allows the cell to be in constant communication with the environment (1). The core engine of intracellular vesicle fusion is the soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNAREs)2 (1–4). SNAREs are membrane-associated proteins localized to both the vesicle (v-SNAREs or R-SNAREs) and the target membrane (t-SNAREs or Q-SNAREs) (5–12). Membrane fusion is initiated when the v-SNARE pairs with the t-SNAREs to form a four-helix trans-SNARE complex (or SNAREpin). N- to C-terminal assembly of the trans-SNARE complex brings the two membranes into close proximity to fuse (13–16).
Exocytosis is the basis of a wide range of fundamental biological processes including neurotransmitter release, hormone secretion, and inside-outside movement of surface transporters and receptors (1). In particular, exocytosis plays crucial roles in maintaining blood glucose homeostasis in mammals. In response to elevated blood glucose levels (e.g. after a meal), the peptide hormone insulin is released from pancreatic beta cells through exocytosis. Insulin is then transported by the bloodstream to reach its target tissues including muscles and adipocytes (17–19). Binding of insulin to its receptor on target tissues activates a cascade of signaling events that ultimately relocate GLUT4 from intracellular storage vesicles to the cell surface. The relocation is achieved by the fusion of GLUT4-containing exocytic vesicles with the plasma membrane. Once on the cell surface, GLUT4 facilitates the uptake of excess blood glucose into the cells for disposal (20, 21). Although insulin secretion and GLUT4 exocytosis are regulated by distinct trigger signals, both pathways require syntaxin-4 and SNAP-23 as the t-SNAREs, and VAMP2/synaptobrevin as the primary v-SNARE (22–26). In insulin secretion, the vesicle fusion reaction also involves another t-SNARE complex, syntaxin-1 and SNAP-25 (25–27). Another widely studied exocytic pathway is the release of neurotransmitters at chemical synapses, which serves the major form of cell-cell communication in the brain (3). The neurotransmitter release requires the t-SNAREs syntaxin-1/SNAP-25 and the v-SNARE VAMP2.
In addition to SNAREs, exocytosis also requires a group of SNARE-binding regulatory factors that control the temporal and spatial precision of the vesicle fusion reaction. One such regulatory factor is tomosyn, a 120–130-kDa protein that negatively regulates SNARE-dependent exocytosis (28–41). Originally identified as a syntaxin-binding molecule (42), tomosyn possesses two distinct domains, a large N-terminal domain containing WD40 repeats and a small C-terminal domain harboring an R-SNARE-like motif (see Fig. 6A for a schematic diagram). Mutations in tomosyn are implicated in type 2 diabetes (43).
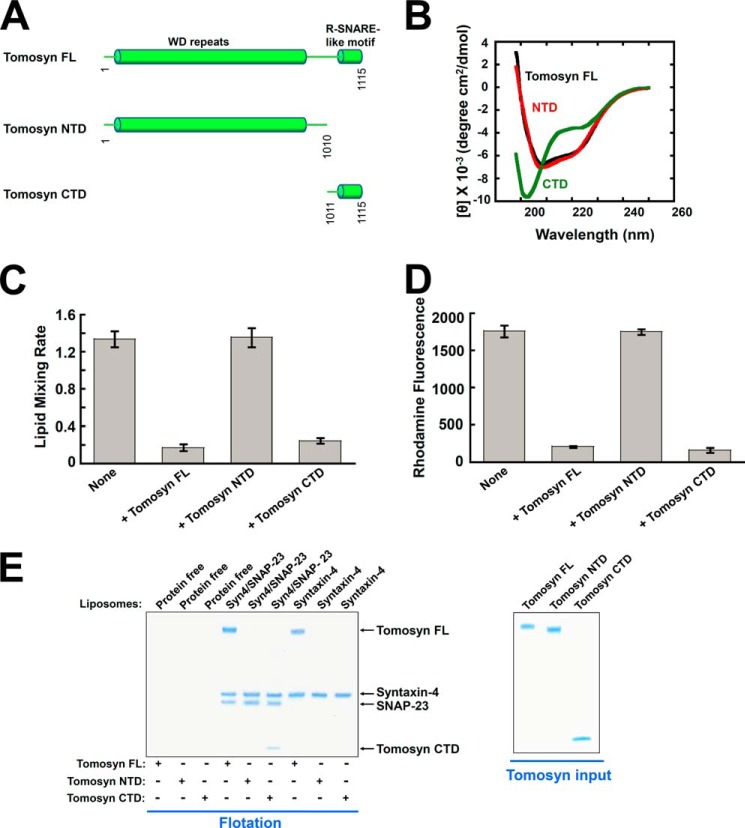
FIGURE 6.
Tomosyn uses its CTD to arrest membrane fusion whereas its NTD is necessary for binding to syntaxin monomer. A, diagrams of tomosyn FL, as well as the NTD and CTD of tomosyn. B, CD spectra of tomosyn and tomosyn fragments. C, initial lipid mixing rates of the indicated SNARE-mediated fusion reactions in the absence or presence of 5 μm tomosyn FL or tomosyn fragments. Each fusion reaction contained 5 μm t-SNAREs and 1.5 μm v-SNARE. Data are presented as percentage of fluorescence change per 10 min. Error bars indicate S.D. D, tomosyn NTD does not block SNARE-mediated liposome docking. The binding reactions were performed in the absence or presence of 5 μm tomosyn FL or tomosyn fragments. Error bars indicate S.D. E, left: Coomassie Blue-stained SDS-PAGE gel showing the binding of tomosyn FL and tomosyn fragments to the t-SNARE liposomes reconstituted with syntaxin-4/SNAP-23 or syntaxin-4 monomer. The liposomes were incubated with tomosyn FL or tomosyn fragments at 4 °C for 1 h, followed by flotation on a Nycodenz gradient. Each binding reaction contained 5 μm SNAREs. Soluble factors were added to a final concentration of 5 μm. Right, Coomassie Blue-stained SDS-PAGE gels showing recombinant tomosyn FL and tomosyn fragments.
It remains incompletely understood how tomosyn regulates SNARE-dependent exocytosis. Previous biochemical studies of tomosyn were performed mostly in static solution binding assays using truncated proteins. These truncated proteins, however, cannot recapitulate the physiological function of full-length (FL) tomosyn in exocytosis (44, 45). In this study, we expressed and purified active FL tomosyn and examined its molecular mechanism of action in reconstituted systems that recapitulate the vesicle fusion reactions of insulin secretion, GLUT4 exocytosis, and neurotransmitter release. We demonstrated that tomosyn negatively regulates SNARE-mediated membrane fusion by inhibiting the assembly of the ternary SNARE complex. Tomosyn recognizes the t-SNARE complex and prevents its pairing with v-SNARE, therefore arresting the fusion reaction at a pre-docking stage. The inhibitory function of tomosyn is dominant over the stimulatory activity of the Sec1/Munc18 (SM) protein. The inhibitory function of tomosyn is mediated by its CTD that contains an R-SNARE-liked motif, confirming previous studies carried out using truncated fragments of tomosyn. Interestingly, the NTD of tomosyn is required (although not sufficient) for the binding of tomosyn to the syntaxin monomer, indicating that FL tomosyn presents unique features not found in the widely studied CTD fragment. These findings suggest that tomosyn uses its CTD to arrest SNARE-dependent membrane fusion, whereas the NTD is required for the recruitment of tomosyn to vesicle fusion sites through syntaxin interaction.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
Recombinant t- and v-SNARE proteins were expressed in Escherichia coli and purified by affinity chromatography. The t-SNARE complexes, comprised of untagged syntaxin-4 and His6-tagged SNAP-23 or untagged syntaxin-1 and His6-tagged SNAP-25, were expressed as previously described (46–48). SNAREs were stored in a buffer containing 25 mm HEPES (pH 7.4), 400 mm KCl, 1% n-octyl-β-d-glucoside, 10% glycerol, and 0.5 mm Tris(2-carboxyethyl)phosphine.
Human tomosyn-1 gene (ATCC) was subcloned into a baculovirus transfer vector pFastBacMBP to generate a construct encoding a His6-MBP-tomosyn fusion protein separated by a tobacco etch virus protease cleavage site. The fusion protein was expressed in Sf9 cells according to the manufacturer's instruction (Bac-to-Bac Baculovirus Expression System, Invitrogen). Briefly, recombinant Bacmid DNA was obtained by transforming the pFastBacMBP-tomosyn construct into DH10Bac cells. The Bacmid DNA was transfected into Sf9 cells grown at 27 °C in Sf900-III serum-free medium (Invitrogen). Recombinant viruses were harvested 4 days after transfection and further amplified twice to obtain viral stocks with the desired titer. At the density of 2 × 106 cells/ml, Sf9 cells were infected with the recombinant virus at multiplicity of infection of 1. Cells were harvested 72 h after infection and cell pellets were stored at −80 °C. Tomosyn proteins were purified by both the nickel and amylose affinity chromatography. The His6-MBP tags were removed from tomosyn proteins by tobacco etch virus protease and the proteins were subsequently dialyzed overnight against a storage buffer (25 mm HEPES, pH 7.4, 150 mm KCl, 10% glycerol and 0.5 mm Tris(2-carboxyethyl)phosphine). The NTD of tomosyn (amino acids 1–1010) was expressed and purified in insect cells in a similar way as the FL protein. The CTD of tomosyn (amino acids 1011–1115) was expressed and purified using a procedure we previously described for Munc18-1 (46). Recombinant Munc18c protein was produced in Sf9 cells as previously described (24, 48).
Reconstitution of Proteoliposomes
All lipids were obtained from Avanti Polar Lipids Inc. For t-SNARE reconstitution, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine, and cholesterol were mixed in a molar ratio of 60:20:10:10. For v-SNARE reconstitution, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine, 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoserine, cholesterol, (N-(7-nitro-2,1,3-benzoxadiazole-4-yl)-1,2-dipalmitoyl phosphatidylethanolamine (NBD-DPPE), and N-(lissamine rhodamine B sulfonyl)-1,2-dipalmitoyl phosphatidylethanolamine (rhodamine-DPPE) were mixed at a molar ratio of 60:17:10:10:1.5:1.5. SNARE proteoliposomes were prepared by detergent dilution and isolated on a Nycodenz density gradient flotation (49–51). Complete detergent removal was achieved by overnight dialysis of the samples in Novagen dialysis tubes against the reconstitution buffer (25 mm HEPES, pH 7.4, 100 mm KCl, 10% glycerol, and 1 mm DTT). To prepare sulforhodamine-loaded liposomes, t- or v-SNARE liposomes were reconstituted in the presence of 50 mm sulforhodamine B (Sigma). Free sulforhodamine B was removed by overnight dialysis followed by liposome flotation on a Nycodenz gradient. The protein:lipid ratio was 1:200 for v-SNAREs and 1:500 for t-SNARE liposomes. To ensure the consistency in SNARE liposome preparations, we routinely monitored the sizes and morphologies of reconstituted liposomes using dynamic light scattering and electron microscopy.
FRET-based Lipid and Content Mixing Assays
A standard lipid mixing reaction contained 45 μl of unlabeled t-SNARE liposomes and 5 μl of v-SNARE liposomes labeled with NBD and rhodamine, and was conducted in a 96-well Nunc plate at 37 °C. Prior to fusion, NBD emission from the v-SNARE liposomes was quenched by neighboring rhodamine molecules through FRET. After fusion, the NBD dyes were diluted such that their emission was dequenched. Increase in NBD fluorescence at 538 nm (excitation 460 nm) was measured every 2 min in a BioTek Synergy HT microplate reader. At the end of the reaction, 10 μl of 10% CHAPSO was added to the liposomes. Fusion data were presented as the percentage of maximum fluorescence change. To assess their regulatory activities, 5 μm tomosyn was added to the fusion reaction. The maximum fusion rate within the first 10 min of the reaction was used to represent the initial rate of a fusion reaction. Full accounting of statistical significance was included for each figure based on at least three independent experiments.
For content mixing assays, unlabeled t-SNARE liposomes were directed to fuse with sulforhodamine B-loaded v-SNARE liposomes in which the sulforhodamine B fluorescence was inhibited by self-quenching. The fusion of the liposomes led to the mixing of their contents and the dequenching of sulforhodamine B fluorescence. The increase of sulforhodamine B fluorescence at 585 nm (excitation 565 nm) was measured every 2 min. Full accounting of statistical significance was included for each figure based on at least three independent experiments.
Liposome Co-flotation Assay
Association of soluble factors with liposomes was examined using a liposome co-flotation assay (46, 52, 53). A soluble factor was incubated with liposomes at 4 °C with gentle agitation. After 1 h, an equal volume of 80% Nycodenz (w/v) in reconstitution buffer was added and transferred to 5 × 41-mm centrifuge tubes. The liposomes were overlaid with 200 μl each of 35 and 30% Nycodenz, and then with 20 μl of reconstitution buffer on the top. The gradients were centrifuged for 4 h at 52,000 × g in a Beckman SW55 rotor. Samples were collected from the 0–30% Nycodenz interface (2 × 20 μl) and analyzed by SDS-PAGE.
Liposome Docking Assay
The t-SNARE liposomes were prepared in a similar way as in the liposome fusion assay except that 2% biotin-conjugated 2-dioleoyl-sn-glycero-3-phosphoethanolamine lipid was included. The biotin-labeled t-SNARE liposomes were incubated with avidin-conjugated agarose beads at room temperature for 1 h. The bead-bound t-SNARE liposomes were then used to pull down rhodamine-labeled v-SNARE liposomes. The rhodamine-labeled v-SNARE liposomes were identical to the v-SNARE liposomes used in lipid mixing assays. The pull-down reactions were performed in the liposome reconstitution buffer at 4 °C in the presence or absence of 5 μm tomosyn. After washing three times with the reconstitution buffer, 1% CHAPSO was added to solubilize the bead-bound liposomes. The avidin beads were removed by centrifugation at 4,000 × g for 2 min. Rhodamine fluorescence in the supernatant was measured in a BioTek microplate reader. In the negative control reaction, 20 μm VAMP2 CD was added to prevent the assembly of the ternary SNARE complex.
Circular Dichroism Spectroscopy
CD spectra were measured using a Jasco J-815 spectropolarimeter equipped with a 1-mm quartz cell. The readings were made at 0.1-nm intervals, and each data point represents the average of six scans at a speed of 50 nm/min over the wavelength range of 198 to 250 nm. The data were converted into mean residue weighted molar ellipticity using the following equation: [θ]MRW = 100 θ/Cnl, where C is the protein concentration (mm), θ is the measured ellipticity (millidegrees), n is the number of residues, and l is the path length (cm).
RESULTS
Tomosyn Inhibits the Assembly of the Ternary SNARE Complex, but Not the Formation of the t-SNARE Complex
The formation of the binary t-SNARE complex is a key regulatory step in exocytic vesicle fusion (3, 4). Tomosyn was initially isolated as a syntaxin-binding molecule (42), but it remains unclear how the tomosyn-syntaxin dimer regulates SNARE assembly on the membrane. There are two tomosyn genes in mammals, tomosyn-1 and tomosyn-2 (54). Because the proteins encoded by these two tomosyn genes appear to have similar domain structures and biological functions, tomosyn-1 was used in this study and hereafter referred to as tomosyn.
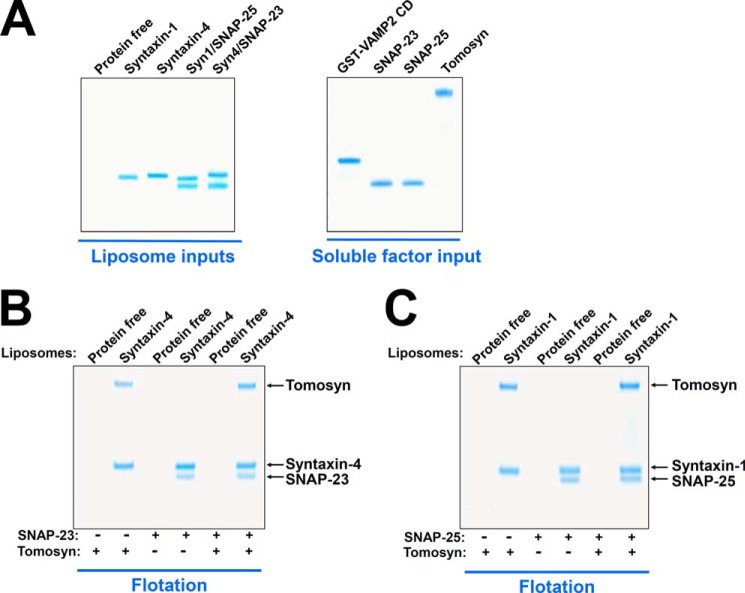
Previous biochemical and biophysical studies mostly used truncated fragments of tomosyn. We found that FL tomosyn could not be expressed in E. coli. Therefore, we expressed and purified FL tomosyn in insect cells using baculovirus. Insect cell-expressed FL tomosyn proteins were pure and highly soluble (Fig. 1A). In a liposome co-flotation assay, we observed that tomosyn bound to proteoliposomes reconstituted with either syntaxin-4 or syntaxin-1 monomer (Fig. 1, B and C). Tomosyn did not bind to protein-free liposomes (Fig. 1, B and C), indicating that the tomosyn-syntaxin interaction was specific. When added as a soluble factor, SNAP-23 or SNAP-25 readily assembled with syntaxin-4 or syntaxin-1, respectively, to form the binary t-SNARE complex (Fig. 1, B and C). Unexpectedly, we found that tomosyn prevented neither the pairing of syntaxin-4 with SNAP-23 nor the pairing of syntaxin-1 with SNAP-25 (Fig. 1, B and C). Thus, tomosyn-associated syntaxin monomers are fully competent for t-SNARE complex formation.
FIGURE 1.
Tomosyn binds to the syntaxin monomer but does not inhibit assembly of the t-SNARE complex. A, Coomassie Blue-stained SDS-PAGE gels showing the input materials of liposomes and proteins. Syn4, Syntaxin-4; Syn1, Syntaxin-1. B, Coomassie Blue-stained SDS-PAGE gel showing the binding of tomosyn and SNAP-23 to protein-free or syntaxin-4 liposomes. Syntaxin-4 liposomes were incubated with or without the recombinant tomosyn protein at 4 °C for 1 h, before SNAP-23 was added. After another hour of incubation at 4 °C, the samples were floated up on a Nycodenz gradient. C, Coomassie Blue-stained SDS-PAGE gel showing the binding of tomosyn and SNAP-25 to protein-free or syntaxin-1 liposomes. Each binding reaction contained 5 μm Syntaxin. Soluble factors were added to a final concentration of 5 μm.
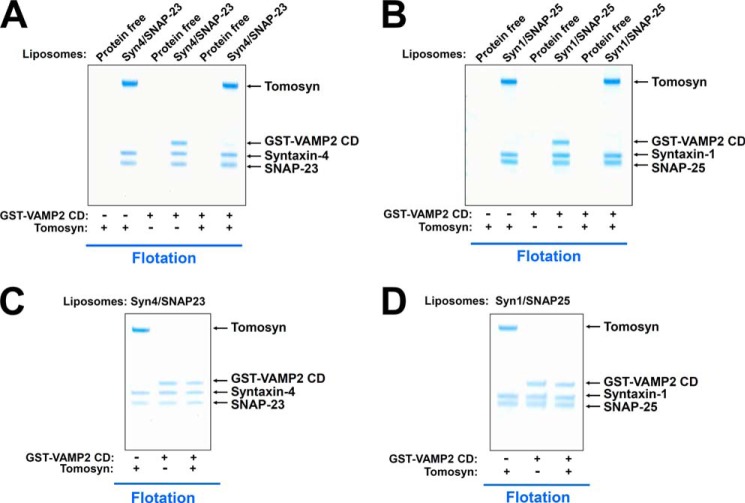
After t-SNARE complex formation, the v-SNARE assembles with the t-SNAREs to form the ternary SNARE complex that forces the two membranes into close proximity to fuse (1). Next we tested whether and how tomosyn regulates the assembly of the ternary SNARE complex. The t-SNARE complex, composed of syntaxin-4/SNAP-23 or syntaxin-1/SNAP-25, was reconstituted into proteoliposomes (Fig. 2, A and B). Addition of VAMP2 cytoplasmic domain (CD) to the t-SNARE liposomes resulted in the formation of the ternary SNARE complex on the membrane (Fig. 2, A and B). We observed that tomosyn bound to the t-SNARE complex and blocked its pairing with VAMP2 CD, therefore preventing the assembly of the ternary SNARE complex (Fig. 2, A and B). When added to pre-formed ternary SNARE complexes on the liposome membrane, tomosyn did not displace VAMP2 CD from the t-SNAREs (Fig. 2, C and D), indicating that tomosyn inhibits SNARE assembly only prior to the full formation of the ternary SNARE complex. Together, these results demonstrate that tomosyn recognizes the t-SNARE complex and inhibits the assembly of the ternary SNARE complex.
FIGURE 2.
Tomosyn binds to the binary t-SNARE complex and blocks v-SNARE entry. A, Coomassie Blue-stained SDS-PAGE gel showing the binding of tomosyn and VAMP2 CD to protein-free or t-SNARE liposomes reconstituted with syntaxin-4 and SNAP-23. To better visualize VAMP2 CD, a GST tag was included at its N terminus. The GST-tagged VAMP2 CD was fully competent for SNARE complex assembly. The t-SNARE liposomes were incubated with or without tomosyn at 4 °C for 1 h, before GST-VAMP2 CD was added. After another hour of incubation at 4 °C, the samples were floated up on a Nycodenz gradient. B, Coomassie Blue-stained SDS-PAGE gel showing binding of tomosyn and VAMP2 CD to protein-free or t-SNARE liposomes reconstituted with syntaxin-1 and SNAP-25. C, Coomassie Blue-stained SDS-PAGE gel showing the binding of tomosyn to the t-SNARE liposomes reconstituted with syntaxin-4 and SNAP-23, in the presence or absence of VAMP2 CD. The t-SNARE liposomes were incubated with or without GST-VAMP2 CD at 4 °C for 1 h before tomosyn was added. After another hour of incubation at 4 °C, the samples were floated up on a nycodenz gradient. D, Coomassie Blue-stained SDS-PAGE gel showing binding of tomosyn to t-SNARE liposomes reconstituted with syntaxin-1 and SNAP-25, in the presence or absence of VAMP2 CD. Each binding reaction contained 5 μm t-SNAREs. Soluble factors were added to a final concentration of 5 μm.
Tomosyn Negatively Regulates the SNARE-mediated Membrane Fusion Reaction
Next we sought to determine how tomosyn regulates the dynamic SNARE-dependent membrane fusion reaction. Proteoliposomes were reconstituted using SNAREs involved in insulin secretion, GLUT4 exocytosis, and neurotransmitter release, syntaxin-1/SNAP-25/VAMP2 or syntaxin-4/SNAP-23/VAMP2 (Fig. 3A). Fusion of the v- and t-SNARE liposomes was first monitored by a fluorescence/Förster resonance energy transfer (FRET)-based lipid mixing assay. We observed that the exocytic SNAREs drove efficient levels of lipid mixing (Fig. 3, B and C). When FL tomosyn was added, the SNARE-mediated lipid mixing reactions were reduced to background levels (Fig. 3, B and C). The inhibitory effects of tomosyn in membrane fusion were comparable with those of the dominant negative inhibitor VAMP2 CD (Fig. 3, B and C).
FIGURE 3.
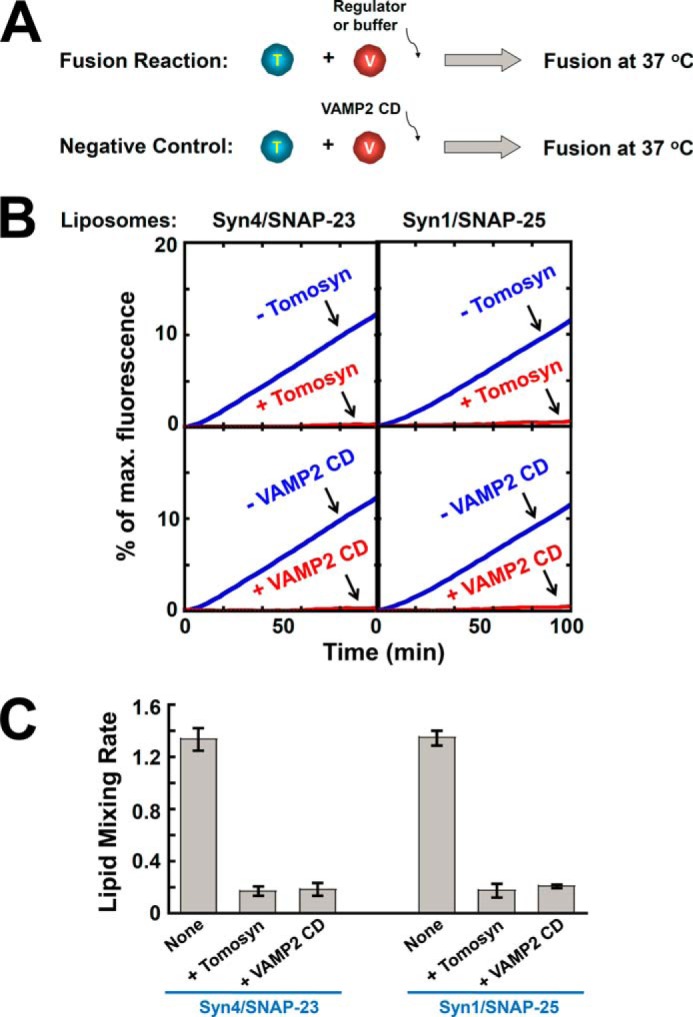
Tomosyn inhibits the SNARE-mediated lipid mixing reaction. A, illustrations of the reconstituted liposome fusion procedures. The t-SNARE liposomes were reconstituted with either syntaxin-4/SNAP-23 or syntaxin-1/SNAP-25, whereas the v-SNARE liposomes were prepared using VAMP2. B, lipid mixing of the reconstituted proteoliposomes in the absence or presence of 5 μm tomosyn. Negative controls: 20 μm VAMP2 CD was added at the beginning of the fusion reactions. Each fusion reaction contained 5 μm t-SNAREs and 1.5 μm v-SNARE. C, initial rates of the lipid mixing reactions shown in B. Data are presented as percentage of fluorescence change per 10 min. Error bars indicate S.D.
We next examined how tomosyn regulates the content mixing of SNARE liposomes. The concentrated soluble dye sulforhodamine B was encapsulated in the VAMP2 liposomes in which its fluorescence was inhibited by self-quenching. Fusion of the v-SNARE liposomes with unlabeled t-SNARE liposomes led to the dilution of sulforhodamine B and dequenching of fluorescence (48). We observed that the SNAREs drove an efficient level of content mixing, and the content mixing was strongly blocked by tomosyn. The inhibitory effects of tomosyn in the content mixing reactions were comparable with those of VAMP2 CD (Fig. 4A). In leakage controls, the sulforhodamine B dye was included in both v- and t-SNARE liposomes. The sulforhodamine B emission was not increased, indicating that content leakage did not occur in the liposome fusion reactions (Fig. 4B). Therefore, tomosyn is capable of arresting both the lipid and content mixing of SNARE liposomes. These reconstitution data are consistent with liposome co-flotation results and demonstrate that tomosyn arrests the SNARE-mediated fusion at an “off” state.
FIGURE 4.
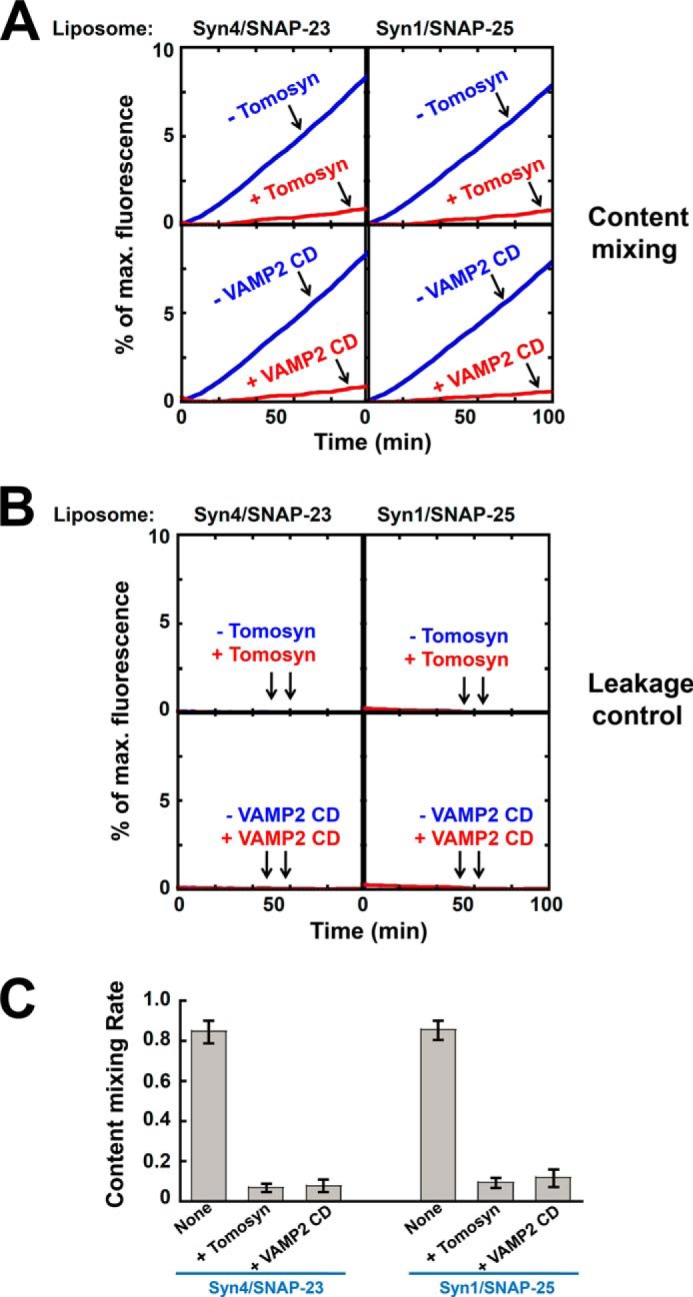
Tomosyn inhibits the content mixing of SNARE-mediated membrane fusion. A, content mixing of the reconstituted fusion reactions. The v-SNARE liposomes, in which sulforhodamine B was encapsulated, were directed to fuse with unlabeled t-SNARE liposomes in the absence or presence of 5 μm tomosyn. Each fusion reaction contained 5 μm t-SNAREs and 1.5 μm v-SNARE. Data are presented as fluorescence increase over time. In negative controls, 20 μm VAMP2 CD was added to the fusion reactions. B, the leakage controls of the content mixing reactions. Sulforhodamine B was included in both v- and t-SNARE liposomes. Increases in sulforhodamine B fluorescence were not observed, indicating that no detectable content leakage occurred during the fusion reactions. C, initial content mixing rates of the fusion reactions shown in A. Data are presented as percentage of fluorescence change per 10 min. Error bars indicate S.D.
Tomosyn Arrests the Membrane Fusion Reaction at a Pre-docking Stage
Next we sought to further dissect how tomosyn arrests SNARE-dependent membrane fusion. Since tomosyn prevents the pairing of v- and t-SNAREs, we hypothesized that it negatively regulates membrane fusion by inhibiting the initial docking of SNARE liposomes. To test this possibility, avidin-immobilized t-SNARE liposomes were used to pull down rhodamine-labeled v-SNARE liposomes (Fig. 5A). We found that the v-SNARE VAMP2 interacted with the t-SNARE complex (syntaxin-4/SNAP-23 or syntaxin-1/SNAP-25) to promote the docking of SNARE liposomes (Fig. 5B). This SNARE-dependent liposome docking was strongly inhibited by tomosyn (Fig. 5B). The ability of tomosyn to block liposome docking suggests that the v- and t-SNAREs remain unpaired in the presence of tomosyn, in agreement with liposome co-flotation findings (Fig. 1). Thus, tomosyn arrests the membrane fusion reaction at a pre-docking stage.
FIGURE 5.
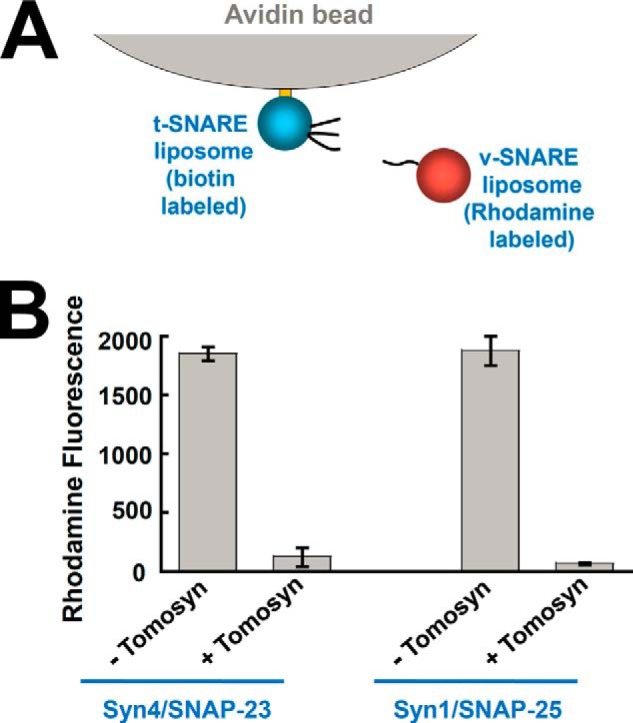
Tomosyn blocks SNARE-mediated liposome docking. A, diagram of the SNARE-liposome docking assay. B, biotin-labeled t-SNARE liposomes were anchored to avidin beads and were used to pull down rhodamine-labeled v-SNARE liposomes. The binding reactions were performed in the absence or presence of 5 μm tomosyn. The binding reaction containing 20 μm VAMP2 CD was used as a negative control to obtain the background fluorescent signal. The background fluorescence was subtracted from other binding reactions to reflect specific SNARE-dependent liposome docking. The data are presented as rhodamine fluorescence intensity. Error bars indicate S.D.
Tomosyn Uses Its CTD to Arrest SNARE-mediated Membrane Fusion Whereas Its NTD Is Required for Binding to Syntaxin Monomer
Tomosyn contains a large NTD that contains WD40 repeats, and a small R-SNARE-like motif in the CTD (Fig. 6A). Next we dissected the functional roles of the NTD and CTD in reconstituted assays. The NTD was expressed in insect cells, whereas the CTD was expressed in E. coli. Circular dichroism (CD) measurements showed that the overall folding of the NTD fragment was similar to that of the FL tomosyn protein (Fig. 6B). As expected, the CD spectrum of the CTD fragment was characteristic of a mainly unstructured polypeptide (Fig. 6B).
Here we focused on the t-SNAREs syntaxin-4/SNAP-23 and v-SNARE VAMP2, which are required for both insulin secretion and GLUT4 exocytosis. We found that the NTD of tomosyn had no effect on the SNARE-mediated lipid mixing reaction. By contrast, the CTD of tomosyn inhibited the lipid mixing with the same efficiency as FL tomosyn (Fig. 6C). These data indicate that the C-terminal R-SNARE-like motif mediates the inhibitory function of tomosyn in membrane fusion. Whereas this conclusion has been proposed in previous studies (42, 54–58), our findings using full-length tomosyn provided the direct evidence. In a liposome docking assay, the CTD, but not the NTD, of tomosyn inhibited the docking of the v- and t-SNARE liposomes (Fig. 6D).
We further examined the SNARE-tomosyn association in the liposome co-flotation assay. FL tomosyn bound to both syntaxin-4 and syntaxin-4/SNAP-23 liposomes (Figs. 1B and 6E). We observed that the CTD of tomosyn interacted with the t-SNARE liposomes but, unexpectedly, not with the syntaxin-4 monomer (Fig. 6E). Thus, the R-SNARE-like motif of tomosyn is insufficient for syntaxin association. The NTD of tomosyn, on the other hand, associated with neither the syntaxin-4 monomer nor the syntaxin-4·SNAP-23 complex in the liposome co-flotation assay (Fig. 6E). These results suggest that, whereas the CTD mediates the inhibitory function of tomosyn in SNARE-mediated fusion reaction, both the NTD and CTD are required for the binding of tomosyn to the syntaxin monomer. Thus, FL tomosyn presents unique features not found in the widely studied CTD fragment. Because syntaxin binding is an important mechanism for recruiting SNARE regulators, our data suggest that the NTD is critical to the recruitment of tomosyn to vesicle fusion sites. These findings likely explain the genetic observations that the biological function of tomosyn in exocytosis requires both the NTD and CTD (44, 45).
The Inhibitory Function of Tomosyn Is Dominant over the Stimulatory Activity of the SM Protein
SM proteins are conserved molecules required for all intracellular vesicle fusion pathways (1). We and others have shown that SM proteins promote membrane fusion through binding to their cognate SNAREs (46, 59–62). In particular, the SM protein Munc18c positively regulates insulin secretion and GLUT4 exocytosis (63–67). In reconstituted assays, Munc18c strongly accelerates the SNARE-dependent membrane fusion reaction (48). Next we examined how tomosyn and Munc18c act in concert to regulate membrane fusion. We observed that the stimulation of fusion by Munc18c was abrogated when tomosyn was added to the SNAREs (Fig. 7, A and B), indicating that tomosyn inhibits the membrane fusion reaction in the presence of Munc18c. Further analysis showed that the CTD, but not the NTD, inhibited Munc18c function. Therefore, the inhibitory function of tomosyn is dominant over the stimulatory activity of SM proteins.
FIGURE 7.
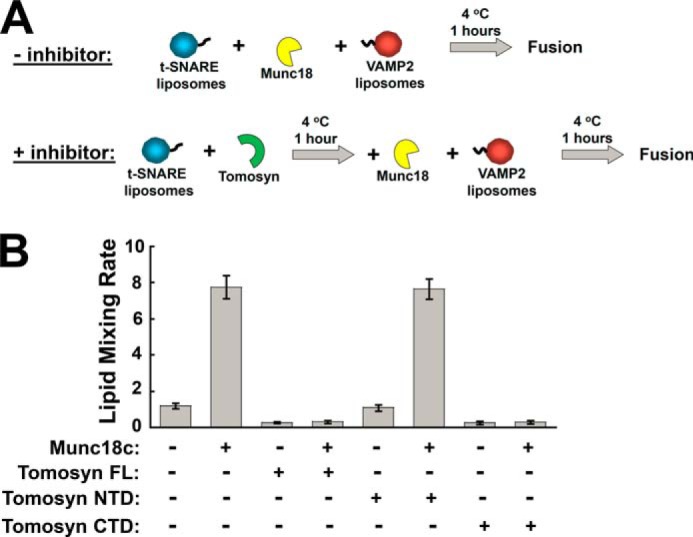
The inhibitory function of tomosyn is dominant over the stimulatory activity of Munc18c. A, diagram illustrating the experimental procedures for the reconstituted fusion reactions. B, initial lipid mixing rates of the indicated SNARE-mediated fusion reactions showing the activities of tomosyn and tomosyn fragments in the presence or absence of Munc18c. Each fusion reaction contained 5 μm t-SNAREs and 1.5 μm v-SNARE. The final concentration of each SNARE regulator was at 5 μm. Data are presented as percentage of fluorescence change per 10 min. Error bars indicate S.D.
DISCUSSION
Tomosyn negatively regulates a range of SNARE-dependent exocytic pathways and is thought to enhance the spatial and temporal precision of vesicle fusion reactions. Whereas the physiological roles of tomosyn in exocytosis are well established, its molecular mechanism remains poorly defined. In this work we have reconstituted SNARE-dependent vesicle fusion reactions using purified components to recapitulate tomosyn-regulated exocytic pathways. In these defined fusion systems, the mechanism of a SNARE regulator can be individually dissected without the complications of other molecules naturally found in the cell.
Previous biochemical studies of tomosyn mostly used truncated mutants that cannot recapitulate the function of FL tomosyn. Here, we expressed and purified active FL tomosyn protein and examined its activity in reconstituted fusion assays. We demonstrated that the SNARE regulator tomosyn arrests the SNARE-dependent fusion reaction at a pre-docking stage. Tomosyn arrests membrane fusion by inhibiting the t-SNARE complex, thereby preventing the initiation of ternary SNARE complex assembly. As a result, tomosyn inhibits the docking, lipid mixing, and content mixing of the SNARE-dependent fusion reaction. Notably, although originally isolated as a syntaxin-binding protein (42), tomosyn does not affect the formation of the t-SNARE complex on the membrane bilayer. Interestingly, tomosyn was recently found to be in complexes with both syntaxin monomers and t-SNARE complexes on the plasma membrane. It appears that the tomosyn·t-SNARE complex, rather than the tomosyn-syntaxin complex, mediates the inhibitory activity of tomosyn in exocytosis (68), in line with our reconstitution results.
Our analysis of FL tomosyn revealed novel features not found in the tomosyn fragments. Whereas not directly involved in SNARE inhibition, the NTD is required for the recruitment of tomosyn to the syntaxin monomer. We discovered that only FL tomosyn binds directly to syntaxin monomer, whereas the NTD and CTD fragments cannot. Because syntaxin binding is a key route for the recruitment of a SNARE regulator to vesicle fusion sites (69, 70), the NTD of tomosyn likely plays a key role in regulating membrane fusion. It is conceivable that, without this recruitment step, the efficiency of tomosyn in exocytosis regulation would be diminished. Indeed, the translocation of tomosyn from the cytosol to the plasma membrane appears to be accompanied by increases in tomosyn-syntaxin association (33). Our findings provide the first evidence for a biological function of the large NTD of tomosyn, and likely explain the genetic observations that both the NTD and CTD are necessary for the regulatory function of tomosyn in vivo (44, 45). It should be noted, however, certain mutations in the NTD of tomosyn appear to impair the inhibitory activity of tomosyn without altering its binding to syntaxin monomer (32). In the future, correlative in vivo and in vitro analyses will be needed to resolve this functional discrepancy.
The inhibitory mechanism of tomosyn is distinct from that of complexin in synaptic release and that of synip in GLUT4 exocytosis. Complexin recognizes the partially zippered trans-SNARE complex and appear to arrest vesicle fusion at a very late stage (71, 72). Synip, by contrast, also targets the t-SNARE complex but arrests fusion using a mechanism independent of R-SNARE-like motifs (52). Importantly, tomosyn can inhibit multiple t-SNARE complexes and therefore is capable of regulating a range of exocytic pathways.
Tomosyn may also control SNARE complex oligomerization and appears to interact with synaptotagmin-1 in neuronal exocytosis (58, 73). The synaptotagmin-binding activity, however, does not represent the core function of tomosyn because the vesicle fusion reaction of GLUT4 exocytosis does not involve synaptotagmins (74). It is thought that the NTD of tomosyn folds into a β-propeller structure similar to the fly protein lethal giant larvae and the yeast molecules Sro7 and Sro77 (75, 76). It would be interesting to determine whether lethal giant larvae and Sro7/Sro77 regulate SNAREs in a similar way as tomosyn.
Multiple molecules are involved in the regulation of SNARE-dependent membrane fusion. Our reconstituted systems provide an avenue to examine the concerted actions of multiple regulatory molecules. We observed that the inhibitory function of tomosyn is dominant over the stimulatory activity of the SM protein Munc18c. These results suggest that tomosyn arrests SNARE assembly at an upstream stage of the fusion reaction prior to membrane docking, whereas SM proteins regulate a downstream, post-docking step of the SNARE-dependent fusion reaction (46, 48, 70).
Overall, we drew two major conclusions from this study. First, we confirmed that the CTD mediates the inhibitory activity of tomosyn in the context of the FL protein. On the surface, these results seem not to extend the findings previously made using tomosyn fragments. However, we believe these data are a necessary step in tomosyn studies because FL SNARE regulators are frequently found to behave differently from truncated mutants. Second, we demonstrate that the NTD is required (although not sufficient) for the association of tomosyn with the syntaxin monomer. This novel observation likely provides a molecular explanation for the critical roles of the NTD in vivo (44, 45). It remains to be shown whether the inhibitory activity of tomosyn is influenced by cellular signaling pathways (e.g. insulin signaling). Moreover, it is also likely that tomosyn regulates vesicle fusion through a SNARE-independent mechanisms. Nevertheless, we expect that the molecular mechanisms of tomosyn in SNARE-mediated fusion reaction revealed in this work pave the path for comprehensive understanding of tomosyn activities in physiological transport processes and their associations with human disorders.
Acknowledgments
We thank Drs. David James and Edward Stuenkel for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant R01DK095367 (to J. S.).
- SNARE
- soluble N-ethylmaleimide-sensitive factor attachment protein receptor
- v-SNARE
- vesicle SNARE
- t-SNARE
- target-SNARE
- NTD
- N-terminal domain
- SM
- Sec1/Munc18
- MBP
- maltose-binding protein
- CTD
- C-terminal domain
- CHAPSO
- 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonic acid
- CD
- cytoplasmic domain.
REFERENCES
- 1. Südhof T. C., Rothman J. E. (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wickner W., Schekman R. (2008) Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rizo J., Südhof T. C. (2012) The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices-guilty as charged? Annu. Rev. Cell Dev. Biol. 28, 279–308 [DOI] [PubMed] [Google Scholar]
- 4. Jahn R., Fasshauer D. (2012) Molecular machines governing exocytosis of synaptic vesicles. Nature 490, 201–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weber T., Zemelman B. V., McNew J. A., Westermann B., Gmachl M., Parlati F., Söllner T. H., Rothman J. E. (1998) SNAREpins: minimal machinery for membrane fusion. Cell 92, 759–772 [DOI] [PubMed] [Google Scholar]
- 6. Sutton R. B., Fasshauer D., Jahn R., Brunger A. T. (1998) Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 Å resolution. Nature 395, 347–353 [DOI] [PubMed] [Google Scholar]
- 7. Schwartz M. L., Merz A. J. (2009) Capture and release of partially zipped trans-SNARE complexes on intact organelles. J. Cell Biol. 185, 535–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Burgoyne R. D., Morgan A. (2007) Membrane trafficking: three steps to fusion. Curr. Biol. 17, R255–R258 [DOI] [PubMed] [Google Scholar]
- 9. Krämer L., Ungermann C. (2011) HOPS drives vacuole fusion by binding the vacuolar SNARE complex and the Vam7 PX domain via two distinct sites. Mol. Biol. Cell 22, 2601–2611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Martens S., McMahon H. T. (2008) Mechanisms of membrane fusion: disparate players and common principles. Nat. Rev. Mol. Cell Biol. 9, 543–556 [DOI] [PubMed] [Google Scholar]
- 11. Ohya T., Miaczynska M., Coskun U., Lommer B., Runge A., Drechsel D., Kalaidzidis Y., Zerial M. (2009) Reconstitution of Rab- and SNARE-dependent membrane fusion by synthetic endosomes. Nature 459, 1091–1097 [DOI] [PubMed] [Google Scholar]
- 12. Katz L., Brennwald P. (2000) Testing the 3Q:1R “rule”: mutational analysis of the ionic “zero” layer in the yeast exocytic SNARE complex reveals no requirement for arginine. Mol. Biol. Cell 11, 3849–3858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Melia T. J., Weber T., McNew J. A., Fisher L. E., Johnston R. J., Parlati F., Mahal L. K., Sollner T. H., Rothman J. E. (2002) Regulation of membrane fusion by the membrane-proximal coil of the t-SNARE during zippering of SNAREpins. J. Cell Biol. 158, 929–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pobbati A.V., Stein A., Fasshauer D. (2006) N- to C-terminal SNARE complex assembly promotes rapid membrane fusion. Science 313, 673–676 [DOI] [PubMed] [Google Scholar]
- 15. Söllner T., Whiteheart S. W., Brunner M., Erdjument-Bromage H., Geromanos S., Tempst P., Rothman J. E. (1993) SNAP receptors implicated in vesicle targeting and fusion. Nature 362, 318–324 [DOI] [PubMed] [Google Scholar]
- 16. Gao Y., Zorman S., Gundersen G., Xi Z., Ma L., Sirinakis G., Rothman J. E., Zhang Y. (2012) Single reconstituted neuronal SNARE complexes zipper in three distinct stages. Science 337, 1340–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Eliasson L., Abdulkader F., Braun M., Galvanovskis J., Hoppa M. B., Rorsman P. (2008) Novel aspects of the molecular mechanisms controlling insulin secretion. J. Physiol. 586, 3313–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rhodes C. J. (2005) Type 2 diabetes: a matter of β-cell life and death? Science 307, 380–384 [DOI] [PubMed] [Google Scholar]
- 19. Sheu L., Pasyk E. A., Ji J., Huang X., Gao X., Varoqueaux F., Brose N., Gaisano H. Y. (2003) Regulation of insulin exocytosis by Munc13-1. J. Biol. Chem. 278, 27556–27563 [DOI] [PubMed] [Google Scholar]
- 20. Bryant N. J., Govers R., James D. E. (2002) Regulated transport of the glucose transporter GLUT4. Nat. Rev. Mol. Cell Biol., 3, 267–277 [DOI] [PubMed] [Google Scholar]
- 21. Hou J. C., Pessin J. E. (2007) Ins (endocytosis) and outs (exocytosis) of GLUT4 trafficking. Curr. Opin. Cell Biol. 19, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. D'Andrea-Merrins M., Chang L., Lam A. D., Ernst S. A., Stuenkel E. L. (2007) Munc18c interaction with syntaxin 4 monomers and SNARE complex intermediates in GLUT4 vesicle trafficking. J. Biol. Chem. 282, 16553–16566 [DOI] [PubMed] [Google Scholar]
- 23. Vicogne J., Vollenweider D., Smith J. R., Huang P., Frohman M. A., Pessin J. E. (2006) Asymmetric phospholipid distribution drives in vitro reconstituted SNARE-dependent membrane fusion. Proc. Natl. Acad. Sci. U.S.A. 103, 14761–14766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Latham C. F., Lopez J. A., Hu S. H., Gee C. L., Westbury E., Blair D. H., Armishaw C. J., Alewood P. F., Bryant N. J., James D. E., Martin J. L. (2006) Molecular dissection of the Munc18c/Syntaxin4 interaction: implications for regulation of membrane trafficking. Traffic 7, 1408–1419 [DOI] [PubMed] [Google Scholar]
- 25. Spurlin B.A., Thurmond D. C. (2006) Syntaxin 4 facilitates biphasic glucose-stimulated insulin secretion from pancreatic beta-cells. Mol. Endocrinol. 20, 183–193 [DOI] [PubMed] [Google Scholar]
- 26. Ohara-Imaizumi M., Fujiwara T., Nakamichi Y., Okamura T., Akimoto Y., Kawai J., Matsushima S., Kawakami H., Watanabe T., Akagawa K., Nagamatsu S. (2007) Imaging analysis reveals mechanistic differences between first- and second-phase insulin exocytosis. J. Cell Biol. 177, 695–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jewell J. L., Oh E., Thurmond D. C. (2010) Exocytosis mechanisms underlying insulin release and glucose uptake: conserved roles for Munc18c and syntaxin 4. Am. J. Physiol. Regul. Integr. Comp. Physiol. 298, R517–R531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Widberg C. H., Bryant N. J., Girotti M., Rea S., James D. E. (2003) Tomosyn interacts with the t-SNAREs syntaxin4 and SNAP23 and plays a role in insulin-stimulated GLUT4 translocation. J. Biol. Chem. 278, 35093–35101 [DOI] [PubMed] [Google Scholar]
- 29. Cheviet S., Bezzi P., Ivarsson R., Renström E., Viertl D., Kasas S., Catsicas S., Regazzi R. (2006) Tomosyn-1 is involved in a post-docking event required for pancreatic beta-cell exocytosis. J. Cell Sci. 119, 2912–2920 [DOI] [PubMed] [Google Scholar]
- 30. Zhang W., Lilja L., Mandic S. A., Gromada J., Smidt K., Janson J., Takai Y., Bark C., Berggren P. O., Meister B. (2006) Tomosyn is expressed in beta-cells and negatively regulates insulin exocytosis. Diabetes 55, 574–581 [DOI] [PubMed] [Google Scholar]
- 31. Yamamoto Y., Fujikura K., Sakaue M., Okimura K., Kobayashi Y., Nakamura T., Sakisaka T. (2010) The tail domain of tomosyn controls membrane fusion through tomosyn displacement by VAMP2. Biochem. Biophys. Res. Commun. 399, 24–30 [DOI] [PubMed] [Google Scholar]
- 32. Williams A. L., Bielopolski N., Meroz D., Lam A. D., Passmore D. R., Ben-Tal N., Ernst S. A., Ashery U., Stuenkel E. L. (2011) Structural and functional analysis of tomosyn identifies domains important in exocytotic regulation. J. Biol. Chem. 286, 14542–14553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gladycheva S. E., Lam A. D., Liu J., D'Andrea-Merrins M., Yizhar O., Lentz S. I., Ashery U., Ernst S. A., Stuenkel E. L. (2007) Receptor-mediated regulation of tomosyn-syntaxin 1A interactions in bovine adrenal chromaffin cells. J. Biol. Chem. 282, 22887–22899 [DOI] [PubMed] [Google Scholar]
- 34. Ashery U., Bielopolski N., Barak B., Yizhar O. (2009) Friends and foes in synaptic transmission: the role of tomosyn in vesicle priming. Trends Neurosci. 32, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen K., Richlitzki A., Featherstone D. E., Schwärzel M., Richmond J. E. (2011) Tomosyn-dependent regulation of synaptic transmission is required for a late phase of associative odor memory. Proc. Natl. Acad. Sci. U.S.A. 108, 18482–18487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gracheva E. O., Burdina A. O., Holgado A. M., Berthelot-Grosjean M., Ackley B. D., Hadwiger G., Nonet M. L., Weimer R. M., Richmond J. E. (2006) Tomosyn inhibits synaptic vesicle priming in Caenorhabditis elegans. PLoS Biol. 4, e261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gracheva E. O., Burdina A. O., Touroutine D., Berthelot-Grosjean M., Parekh H., Richmond J. E. (2007) Tomosyn negatively regulates both synaptic transmitter and neuropeptide release at the C. elegans neuromuscular junction. J. Physiol. 585, 705–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu Z., Tong X. J., Kaplan J. M. (2013) UNC-13L, UNC-13S, and Tomosyn form a protein code for fast and slow neurotransmitter release in Caenorhabditis elegans. Elife 2, e00967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gracheva E. O., Burdina A. O., Touroutine D., Berthelot-Grosjean M., Parekh H., Richmond J. E. (2007) Tomosyn negatively regulates CAPS-dependent peptide release at Caenorhabditis elegans synapses. J. Neurosci. 27, 10176–10184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dybbs M., Ngai J., Kaplan J. M. (2005) Using microarrays to facilitate positional cloning: identification of tomosyn as an inhibitor of neurosecretion. PLoS Genet. 1, 6–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gracheva E. O., Maryon E. B., Berthelot-Grosjean M., Richmond J. E. (2010) Differential regulation of synaptic vesicle tethering and docking by UNC-18 and TOM-1. Front. Synaptic Neurosci. 2, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fujita Y., Shirataki H., Sakisaka T., Asakura T., Ohya T., Kotani H., Yokoyama S., Nishioka H., Matsuura Y., Mizoguchi A., Scheller R. H., Takai Y. (1998) Tomosyn: a syntaxin-1-binding protein that forms a novel complex in the neurotransmitter release process. Neuron 20, 905–915 [DOI] [PubMed] [Google Scholar]
- 43. Bhatnagar S., Oler A. T., Rabaglia M. E., Stapleton D. S., Schueler K. L., Truchan N. A., Worzella S. L., Stoehr J. P., Clee S. M., Yandell B. S., Keller M. P., Thurmond D. C., Attie A. D. (2011) Positional cloning of a type 2 diabetes quantitative trait locus: tomosyn-2, a negative regulator of insulin secretion. PLoS Genet 7, e1002323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yizhar O., Lipstein N., Gladycheva S. E., Matti U., Ernst S. A., Rettig J., Stuenkel E. L., Ashery U. (2007) Multiple functional domains are involved in tomosyn regulation of exocytosis. J. Neurochem. 103, 604–616 [DOI] [PubMed] [Google Scholar]
- 45. Burdina A. O., Klosterman S. M., Shtessel L., Ahmed S., Richmond J. E. (2011) In vivo analysis of conserved C. elegans tomosyn domains. PLoS One 6, e26185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Shen J., Tareste D. C., Paumet F., Rothman J. E., Melia T. J. (2007) Selective activation of cognate SNAREpins by Sec1/Munc18 proteins. Cell 128, 183–195 [DOI] [PubMed] [Google Scholar]
- 47. Scott B. L., Van Komen J. S., Liu S., Weber T., Melia T. J., McNew J. A. (2003) Liposome fusion assay to monitor intracellular membrane fusion machines. Methods Enzymol. 372, 274–300 [DOI] [PubMed] [Google Scholar]
- 48. Yu H., Rathore S. S., Lopez J. A., Davis E. M., James D. E., Martin J. L., Shen J. (2013) Comparative studies of Munc18c and Munc18–1 reveal conserved and divergent mechanisms of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. U.S.A. 110, E3271–E3280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Shen J., Rathore S. S., Khandan L., Rothman J. E. (2010) SNARE bundle and syntaxin N-peptide constitute a minimal complement for Munc18–1 activation of membrane fusion. J. Cell Biol. 190, 55–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yu H., Rathore S. S., Davis E. M., Ouyang Y., Shen J. (2013) Doc2b promotes GLUT4 exocytosis by activating the SNARE-mediated fusion reaction in a calcium- and membrane bending-dependent manner. Mol. Biol. Cell 24, 1176–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Rathore S. S., Ghosh N., Ouyang Y., Shen J. (2011) Topological arrangement of the intracellular membrane fusion machinery. Mol. Biol. Cell 22, 2612–2619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yu H., Rathore S. S., Shen J. (2013) Synip arrests soluble N-ethylmaleimide-sensitive factor attachment protein receptor (SNARE)-dependent membrane fusion as a selective target membrane SNARE-binding inhibitor. J. Biol. Chem. 288, 18885–18893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Groffen A. J., Jacobsen L., Schut D., Verhage M. (2005) Two distinct genes drive expression of seven tomosyn isoforms in the mammalian brain, sharing a conserved structure with a unique variable domain. J. Neurochem. 92, 554–568 [DOI] [PubMed] [Google Scholar]
- 54. Ishiki M., Klip A. (2005) Minireview: recent developments in the regulation of glucose transporter-4 traffic: new signals, locations, and partners. Endocrinology 146, 5071–5078 [DOI] [PubMed] [Google Scholar]
- 55. Yizhar O., Matti U., Melamed R., Hagalili Y., Bruns D., Rettig J., Ashery U. (2004) Tomosyn inhibits priming of large dense-core vesicles in a calcium-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 101, 2578–2583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Masuda E. S., Huang B. C., Fisher J. M., Luo Y., Scheller R. H. (1998) Tomosyn binds t-SNARE proteins via a VAMP-like coiled coil. Neuron 21, 479–480 [DOI] [PubMed] [Google Scholar]
- 57. Yamamoto Y., Mochida S., Miyazaki N., Kawai K., Fujikura K., Kurooka T., Iwasaki K., Sakisaka T. (2010) Tomosyn inhibits synaptotagmin-1-mediated step of Ca2+-dependent neurotransmitter release through its N-terminal WD40 repeats. J. Biol. Chem. 285, 40943–40955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pobbati A. V., Razeto A., Böddener M., Becker S., Fasshauer D. (2004) Structural basis for the inhibitory role of tomosyn in exocytosis. J. Biol. Chem. 279, 47192–47200 [DOI] [PubMed] [Google Scholar]
- 59. Novick P., Schekman R. (1979) Secretion and cell-surface growth are blocked in a temperature-sensitive mutant of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 76, 1858–1862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Hata Y., Slaughter C. A., Südhof T. C. (1993) Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin. Nature 366, 347–351 [DOI] [PubMed] [Google Scholar]
- 61. Dulubova I., Khvotchev M., Liu S., Huryeva I., Südhof T. C., Rizo J. (2007) Munc18-1 binds directly to the neuronal SNARE complex. Proc. Natl. Acad. Sci. U.S.A. 104, 2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Schollmeier Y., Krause J. M., Kreye S., Malsam J., Söllner T. H. (2011) Resolving the function of distinct Munc18-1/SNARE protein interaction modes in a reconstituted membrane fusion assay. J. Biol. Chem. 286, 30582–30590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hu S. H., Latham C. F., Gee C. L., James D. E., Martin J. L. (2007) Structure of the Munc18c/Syntaxin4 N-peptide complex defines universal features of the N-peptide binding mode of Sec1/Munc18 proteins. Proc. Natl. Acad. Sci. U.S.A. 104, 8773–8778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Tellam J. T., McIntosh S., James D. E. (1995) Molecular identification of two novel Munc-18 isoforms expressed in non-neuronal tissues. J. Biol. Chem. 270, 5857–5863 [DOI] [PubMed] [Google Scholar]
- 65. Jewell J. L., Oh E., Ramalingam L., Kalwat M. A., Tagliabracci V. S., Tackett L., Elmendorf J. S., Thurmond D. C. (2011) Munc18c phosphorylation by the insulin receptor links cell signaling directly to SNARE exocytosis. J. Cell Biol. 193, 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Oh E., Spurlin B. A., Pessin J. E., Thurmond D. C. (2005) Munc18c heterozygous knockout mice display increased susceptibility for severe glucose intolerance. Diabetes 54, 638–647 [DOI] [PubMed] [Google Scholar]
- 67. Oh E., Thurmond D. C. (2009) Munc18c depletion selectively impairs the sustained phase of insulin release. Diabetes 58, 1165–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Bielopolski N., Lam A. D., Bar-On D., Sauer M., Stuenkel E. L., Ashery U. (2014) Differential interaction of Tomosyn with syntaxin and SNAP25 depends on domains in the WD40 β-propeller core and determines its inhibitory activity. J. Biol. Chem. 289, 17087–17099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Ma C., Su L., Seven A. B., Xu Y., Rizo J. (2013) Reconstitution of the vital functions of Munc18 and Munc13 in neurotransmitter release. Science 339, 421–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rathore S. S., Bend E. G., Yu H., Hammarlund M., Jorgensen E. M., Shen J. (2010) Syntaxin N-terminal peptide motif is an initiation factor for the assembly of the SNARE-Sec1/Munc18 membrane fusion complex. Proc. Natl. Acad. Sci. U.S.A. 107, 22399–22406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Giraudo C. G., Garcia-Diaz A., Eng W. S., Chen Y., Hendrickson W. A., Melia T. J., Rothman J. E. (2009) Alternative zippering as an on-off switch for SNARE-mediated fusion. Science 323, 512–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Giraudo C. G., Eng W. S., Melia T. J., Rothman J. E. (2006) A clamping mechanism involved in SNARE-dependent exocytosis. Science 313, 676–680 [DOI] [PubMed] [Google Scholar]
- 73. Sakisaka T., Yamamoto Y., Mochida S., Nakamura M., Nishikawa K., Ishizaki H., Okamoto-Tanaka M., Miyoshi J., Fujiyoshi Y., Manabe T., Takai Y. (2008) Dual inhibition of SNARE complex formation by tomosyn ensures controlled neurotransmitter release. J. Cell Biol. 183, 323–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Humphrey S. J., Yang G., Yang P., Fazakerley D. J., Stöckli J., Yang J. Y., James D. E. (2013) Dynamic adipocyte phosphoproteome reveals that Akt directly regulates mTORC2. Cell Metab. 17, 1009–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lehman K., Rossi G., Adamo J. E., Brennwald P. (1999) Yeast homologues of tomosyn and lethal giant larvae function in exocytosis and are associated with the plasma membrane SNARE, Sec9. J. Cell Biol. 146, 125–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hattendorf D. A., Andreeva A., Gangar A., Brennwald P. J., Weis W. I. (2007) Structure of the yeast polarity protein Sro7 reveals a SNARE regulatory mechanism. Nature 446, 567–571 [DOI] [PubMed] [Google Scholar]