Background: Gymnemic acids inhibit sweet taste responses in humans.
Results: Gymnemic acids and glucuronic acid inhibited human sweet receptor T1R2 + T1R3.
Conclusion: Interaction between transmembrane domains of human T1R3 and the glucuronosyl group of gymnemic acids is mainly required for the sweet-suppressing effect.
Significance: Our model may provide further insights into drug design to modify sensitivity of sweet receptor.
Keywords: Calcium Imaging, G Protein-coupled Receptor, Molecular Evolution, Molecular Modeling, Signal Transduction, Gymnemic Acids, Sweet Taste, T1R2 + T1R3
Abstract
Gymnemic acids are triterpene glycosides that selectively suppress taste responses to various sweet substances in humans but not in mice. This sweet-suppressing effect of gymnemic acids is diminished by rinsing the tongue with γ-cyclodextrin (γ-CD). However, little is known about the molecular mechanisms underlying the sweet-suppressing effect of gymnemic acids and the interaction between gymnemic acids versus sweet taste receptor and/or γ-CD. To investigate whether gymnemic acids directly interact with human (h) sweet receptor hT1R2 + hT1R3, we used the sweet receptor T1R2 + T1R3 assay in transiently transfected HEK293 cells. Similar to previous studies in humans and mice, gymnemic acids (100 μg/ml) inhibited the [Ca2+]i responses to sweet compounds in HEK293 cells heterologously expressing hT1R2 + hT1R3 but not in those expressing the mouse (m) sweet receptor mT1R2 + mT1R3. The effect of gymnemic acids rapidly disappeared after rinsing the HEK293 cells with γ-CD. Using mixed species pairings of human and mouse sweet receptor subunits and chimeras, we determined that the transmembrane domain of hT1R3 was mainly required for the sweet-suppressing effect of gymnemic acids. Directed mutagenesis in the transmembrane domain of hT1R3 revealed that the interaction site for gymnemic acids shared the amino acid residues that determined the sensitivity to another sweet antagonist, lactisole. Glucuronic acid, which is the common structure of gymnemic acids, also reduced sensitivity to sweet compounds. In our models, gymnemic acids were predicted to dock to a binding pocket within the transmembrane domain of hT1R3.
Introduction
Sweet taste mainly depends on the combination of the two receptors, taste type 1 receptor 2 (T1R2) and taste type 1 receptor 3 (T1R3) expressed in subsets of taste receptor cells on the tongue and palate (1, 2). Each subunit belongs to the class C G protein-coupled receptor family, which includes the metabotropic glutamate receptors, the calcium-sensing receptor, and other taste/olfactory receptors (1–9). On the basis of their sequence similarities to metabotropic glutamate receptors 1, 3, and 7 and rhodopsin (9–11), T1R2 + T1R3 have been proposed to consist of three main structural domains as follows: a large extracellular amino-terminal domain (ATD)2 called the Venus flytrap module containing lobes 1 and 2, which can be in an “open” or “closed” conformation; the cysteine-rich linker domain (CRD); and the heptahelical transmembrane domain (TMD) (9). The CRD connects the ATD and the TMD.
T1R2 + T1R3 are responsible for the sensitivity to structurally diverse sweet compounds such as sugars, d-amino acids, peptides, and sweet-tasting proteins. Recent studies on chimera and mutation of the sweet taste receptor revealed multiple binding sites for its ligands. For example, artificial sweeteners aspartame and neotame and other low molecular weight sweet compounds interact with the binding site formed by the bottom of lobe 1 and the top of lobe 2 of ATD in hT1R2 (12, 13). The taste-modifying protein neoculin binds the ATD of hT1R3 (14). Sensitivity to brazzein requires the CRD of hT1R3 as well as the ATD of hT1R2 (15, 16). The TMD of hT1R3 is required for sensitivity to cyclamate, neohesperidin dihydrochalcone, and the sweet inhibitor lactisole (17–19).
Gymnemic acids (GAs) are saponins of triterpene glycoside isolated from the plant Gymnema sylvestre, which is native to central and western India (Fig. 1). GAs are not a pure entity but are composed of several types of homologues (20). It is known that GAs selectively suppress taste responses to various sweet compounds without affecting responses to salty, sour, and bitter substances for 30–60 min after application of GAs in humans (21, 22). The sweet-suppressing effect of GAs is specific to humans and chimpanzees; it has no effect on sweeteners for rodents (23–27). This effect is diminished by rinsing the tongue with γ-cyclodextrin (CD) (28–30). GAs also have other physiological effects such as inhibition of intestinal glucose absorption and lowering of plasma glucose and insulin levels (31–33). Although it is thought that GAs bind to the sweet taste receptor, similar to another sweet antagonist, lactisole (18), less is known about how GAs act as a sweet inhibitor.
FIGURE 1.
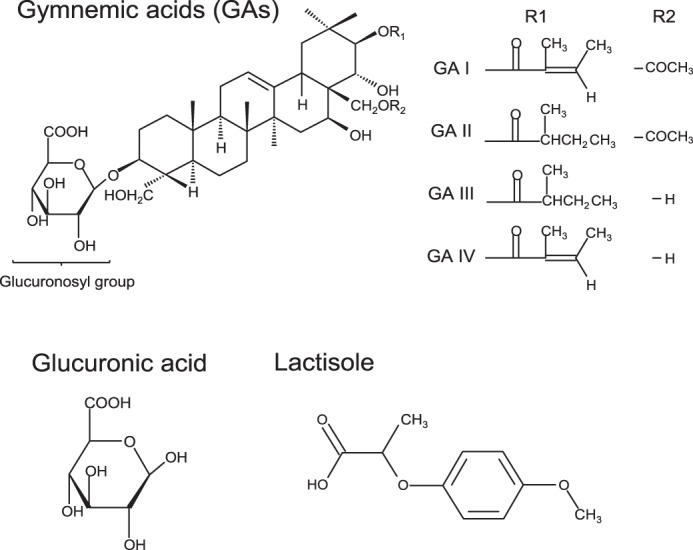
Basic molecular structures of sweet inhibitors.
In this study, we demonstrated, for the first time, the molecular mechanisms for the human-specific sweet-suppressing effect of GAs. GAs directly interact with hT1R2 + hT1R3, and this interaction is inhibited by forming an inclusion complex between GAs and γ-CD. The sensitivity to GAs depends mainly on the transmembrane domain of hT1R3 and cysteine-rich domain of hT1R2. The glucuronosyl group of GAs is the key component mediating the sweet-suppressing effect.
EXPERIMENTAL PROCEDURES
Purification of GAs
GAs are saponins with a triterpenoid structure, and more than 10 kinds of GAs and related compounds have been isolated (20, 30, 34). Because of the difficulty and tediousness of the isolation of each GA, here we used a GA preparation (herein referred to as GAs to indicate several acids are in the mixture) obtained as described below without isolation of each GAs. G. sylvestre extract was comprised of an ethanol/water extract of G. sylvestre leaves. The extract was suspended in 10 ml of methanol, and the supernatant was treated with 2.5 m H2SO4 to carry out acid precipitation at pH 2.5. The precipitate was dissolved in 30 ml of 25% methanol and applied to a Diaion HP20 column. The column was washed with 25% and then 50% methanol, and the preparation of GAs was eluted with 85% methanol. The GAs were analyzed as gymnemagenin, the aglycone of GAs. The obtained GAs were redissolved in 50% ethanol, and alkaline and acid hydrolyses were performed (31). Gymnemagenin content was determined by high performance liquid chromatography on a C18 reversed-phase column with UV detection at 213 nm using a linear gradient of acetonitrile (25–50%) in 0.1% phosphoric acid (30, 34).
Residue Numbering
The general numbering of residues in the TMD of T1R3 follows T1R3's primary sequence. Superscript residue numbers follow the generic numbering system of Ballesteros and Weinstein (35).
Preparation of Chimeras and Point Mutations
Human TAS1Rs and Gαl6-gust44 expression constructs were generated in the pEF-DEST51 Gateway vector (Invitrogen) by genomic DNA-based methods as reported previously (36). Mouse T1R2 and T1R3 were cloned as described previously (4, 37). Construction of human/mouse chimeras of T1Rs was performed by PCR using overlapping primers (15, 38). To subclone each gene into the vector, a Kozak cassette was introduced at the 5′ end before the start codon. The integrity of all DNA constructs was confirmed by DNA sequencing.
Functional Expression
HEK293 cells were cultured at 37 °C under a humidified atmosphere containing 5% CO2 in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. To obtain reproducible Ca2+ responses, cells were split every 2 days before the cells became confluent. Cells were discarded after 2 months of passages, and new cells were prepared from frozen stock. For calcium imaging experiments, cells were seeded onto a 35-mm recording chamber. After 24 h at 37 °C, confluent cells (60 and 70%) were washed in OptiMEM medium supplemented with GlutaMAX-I (Invitrogen), and plasmid DNAs were transiently cotransfected into HEK293 cells using Lipofectamine2000 reagent (Invitrogen) (2.0 μl per 1.0 μg of DNA). TAS1Rs (or their mutants) and Gαl6-gust44 were transfected using 0.3 μg of plasmids for 35-mm recording chambers. Ca2+ imaging assays were performed 24 h after transfection.
Single Cell Ca2+ Imaging
A bath perfusion system was used for determination of the kinetics of activation. Transfected cells in 35-mm recording chambers were washed in Hanks' balanced salt solution containing 10 mm HEPES, pH 7.4, and loaded with 3.0 mm Fluo-4 acetoxymethyl ester (Invitrogen) for 3 min at 37 °C. The dye-loaded cells were subjected to Ca2+ imaging. Taste solutions diluted in Hanks' balanced salt solution were applied sequentially to the cells for 30 s with a peristaltic pump at a flow rate of 1.5 ml/min, and fluorescence images were obtained using an S Fluor 620/0.75 objective lens (Nikon, Tokyo, Japan) via a cooled CCD camera (C6790, Hamamatsu Photonics K.K., Shizuoka, Japan) fitted to a TE300 microscope (Nikon). AquaCosmos software (version 1.3, Hamamatsu Photonics K.K.) was used to acquire and analyze fluorescence images. A 5-min interval was maintained between each tastant's application to ensure that the cells were not desensitized as a result of the previous application of taste solutions. Responses were measured from 11 to 43 individual responding cells. Compounds were saccharin (10 mm), SC45647 (0.3 mm), aspartame (10 mm), sodium cyclamate (30 mm), d-tryptophan (10 mm), lactisole (0.01, 0.03, 0.1, 0.3, 1, and 3 mm), and d-glucuronic acid sodium salt monohydrate (3, 10, 30, 50, 100, 200, and 300 mm). GAs (0.3, 1, 3, 10, 30, 50, and 100 μg/ml) and CD (0.01, 0.03, 0.1, 0.3, and 1 mm) were applied to the HEK293 cells for 3 and 2 min, respectively. Isoproterenol (10 μm) was used as positive control, which stimulates endogenous β-adrenergic receptors, providing that the Gα16-dependent signal transduction cascade was functional.
Data Analysis
In the analysis of single cell responses, changes in [Ca2+]i were monitored as changes in fluo-4 fluorescence. Fluorometric signals are expressed as relative fluorescence changes: ΔF/F0 = (F − F0)/F0, where F0 denotes the baseline fluorescence level. The magnitude of the calcium increases from 10 to 30 s after stimulus onset was measured and averaged. The data were expressed as the mean ± S.E. of the ΔF/F0 value and used for statistical analysis. EC50 and IC50 values were calculated from individual cumulative concentration-response data using curving-fitting routines of KaleidaGraph 4.0 (Synergy Software Inc., Essex Junction, VT). Differential inhibition of sweet compounds by GAs in wild-type receptors and in each of the chimeras was assessed in comparison with the control (before application of GAs) using a paired t test, and statistical significance was set at p < 0.05. To evaluate the effects of GAs, lactisole, and glucuronic acid on sweet responses statistically, changes in mutants compared with the wild-type control were statistically analyzed with one-way analysis of variance (ANOVA) and the post-hoc Tukey-Kramer method, with significance set at p < 0.05. A two-way ANOVA was conducted to analyze for the effect of GAs on the dose-response data in saccharin. An identical analysis was conducted to test for effects of CDs. All calculations were performed using the statistical software package IBM SPSS Statistics (IBM, New York).
Molecular Modeling
Based on the sequence alignment reported by previous studies (13, 18, 39), homology models of human/mouse T1Rs and their chimeras were constructed by residue replacement using the Modeler (40) as templates of metabotropic glutamate receptors (9, 10, 41). Structure of GA I, lactisole, and glucuronic acid were obtained from ZINC version12 (42). GA II, III, and IV were generated from Facio for the GAMESS/fragment molecular orbital method (43–45) based on the structure of GA I. GAs, lactisole, and glucuronic acid were introduced into models by using the program Autodock 4.0 (46). The models were viewed in UCSF Chimera 1.6.1 (47).
RESULTS
Sweet-suppressing Effect of GAs Requires the CRD of hT1R2 and TMD of hT1R3
In human psychophysiological studies, GAs inhibit taste sensitivity to the sweet compounds (22). To examine whether GAs directly interact with the human sweet receptor, we expressed hT1R2 + hT1R3 by transient transfection in HEK293 cells along with Gα16-gust44 and then monitored activity (calcium mobilization) to the various sweet compounds before and after application of GAs. GAs inhibited the [Ca2+]i responses of hT1R2 + hT1R3-expressing HEK293 cells to SC45647, saccharin, aspartame, cyclamate, d-tryptophan, and sucrose (Figs. 2 and 3). Calcium responses to saccharin were inhibited by GAs in a concentration-dependent manner (Fig. 11). After unit conversion from nanomolar to micrograms/ml to compare the potency, the IC50 value of GAs was smaller than that of lactisole, another inhibitor of the human sweet receptor. This indicated that GAs have high potency as compared with lactisole (Table 1). GAs did not alter the EC50 values for saccharin but reduced the magnitude of responses (Fig. 4), suggesting that GAs are noncompetitive inhibitors of saccharin. The sweet-suppressing effect of GAs (100 μg/ml) was diminished by rinsing cells with 10 mm γ-CD but not with α- and β-CD (Fig. 5). Similar to electrophysiological and behavioral studies in mice, sweet responses of HEK293 cells expressing the mouse sweet receptor (mT1R2 + mT1R3) were not suppressed by GAs (Fig. 6). To determine whether one or both hT1R subunits are needed for sensitivity to GAs, we examined the responses of human/mouse mismatched heterodimers to saccharin before and after application of GAs. One mismatched pair (hT1R2 + mT1R3) was not sensitive to GAs (Fig. 6). As described previously (15, 17, 18), the other mismatched pair (mT1R2 + hT1R3) does not produce a functional receptor. These results demonstrate that hT1R3 is required for sensitivity of GAs.
FIGURE 2.
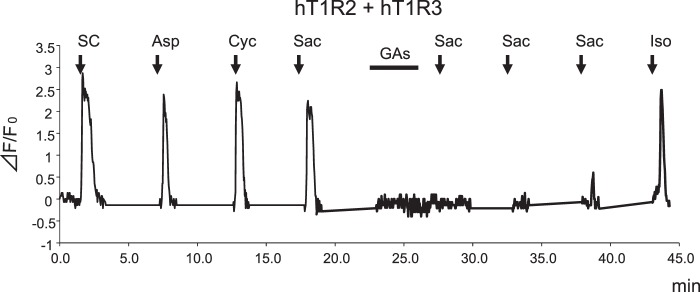
GAs inhibit the human sweet taste receptor. Typical example of single cell Ca2+ imaging is shown. HEK293 cells heterologously expressing hT1R2 + hT1R3 showed [Ca2+]i responses to SC45647 (0.3 mm SC), aspartame (10 mm Asp), cyclamate (30 mm Cyc), and saccharin (10 mm Sac). After application of GAs (100 μg/ml), responses to saccharin were inhibited. This cell showed sensitivity to isoproterenol (10 μm Iso).
FIGURE 3.
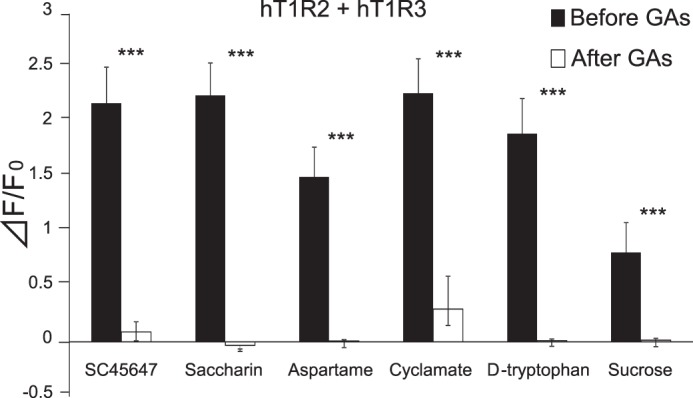
GAs blocked or reduced the activity of all sweeteners tested. The human sweet receptor (hT1R2 + hT1R3) was expressed in HEK293 cells along with a reporter G protein (G16-gust44). Calcium mobilization was measured in response to each of the following sweeteners before and after application of GAs (100 μg/ml): SC45647 (0.3 mm), saccharin (10 mm), cyclamate (30 mm), d-tryptophan (10 mm), and sucrose (100 mm). GAs blocked or reduced responses to all sweeteners. Data are expressed as means ± S.E. of 20–28 cells. Paired t test: ***, p < 0.001.
FIGURE 11.
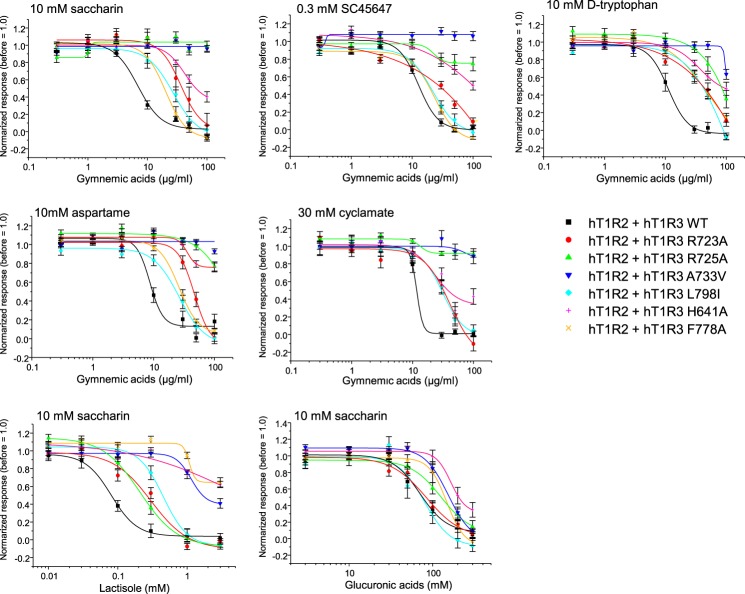
Sensitivity to GAs depends on specific residues within the TMD and EL2 of hT1R3. HEK293 cells were transiently transfected with hT1R2, hT1R3 WT or mutant (R723A, R725A, A733V, L798I, H641A, and F778A), and Gα16-gust44. The responses of the receptors to saccharin (10 mm), SC45647 (0.3 mm), d-tryptophan (10 mm), aspartame (10 mm), and cyclamate (30 mm) before and after application of GAs and the responses to saccharin (10 mm) with or without lactisole and glucuronic acid were examined. Values are means ± S.E. of 11–43 cells.
TABLE 1.
IC50 values for GAs, lactisole, and glucuronic acid obtained from WT TAS1R3 and selected mutants
Data were derived from the experiment in Fig. 11. The following abbreviations are used: Sac, saccharin; SC, SC45647; Trp, d-tryptophan; Asp, aspartame; Cyc, cyclamate.
| GAs (μg/ml) |
Lactisole (mm)/(μg/ml), Sac (10 mm) | Glucuronic acid (mm)/(μg/ml), Sac (10 mm) | |||||
|---|---|---|---|---|---|---|---|
| Sac (10 mm) | SC (0.3 mm) | Trp (10 mm) | Asp (10 mm) | Cyc (30 mm) | |||
| WT | 6.9 | 12.7 | 11.2 | 8.7 | 11.4 | 0.08/15.7 | 71/15,344.5 |
| R723ex2–51A | 36.3 | >100 | >100 | 46.5 | 41.0 | 0.30/58.9 | 84 /18,154.1 |
| R725ex2–53A | >100 | >100 | >100 | >100 | >100 | 0.21/41.2 | 126/27,231.1 |
| A7335.46V | >100 | >100 | >100 | >100 | >100 | >3/>589 | 153/33,066.4 |
| L7987.36I | 26.8 | 21.8 | 96.2 | 26.1 | 30.8 | 0.45/88.29 | 80/17,289.6 |
| H6413.33A | >100 | >100 | >100 | >100 | >100 | >3/>589 | 165/35,659.8 |
| F7786.51A | 20.1 | 22.9 | 48.9 | 26.0 | NDa | > 3/> 589 | 147/31,769.6 |
a ND means data unable to be experimentally defined.
FIGURE 4.
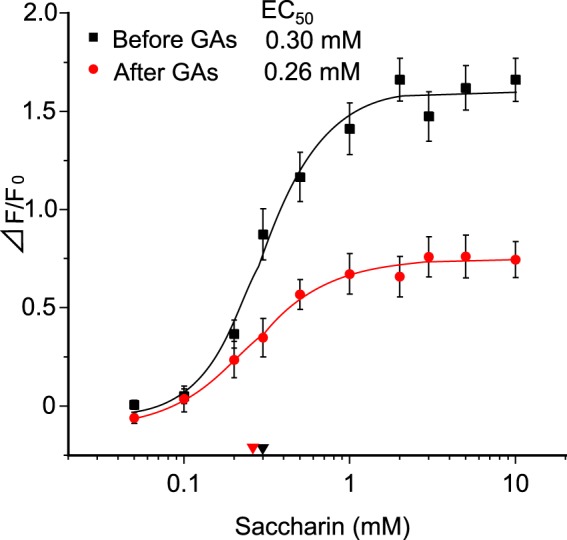
Dose-response curves for saccharin before and after application of GAs. The human sweet receptor (hT1R2 + hT1R3) was expressed in HEK293 cells along with a reporter G-protein (G16-gust44). Calcium mobilization was measured in response to a concentration series of saccharin solutions before and after application of GAs (10 μg/ml). Data are expressed as means ± S.E. of 23 cells. There was a significant effect (p < 0.0001) before and after application of GAs, as assessed by two-way analysis of variance (ANOVA, F(2, 44) = 19.47). EC50 values are shown with red (after) and black (before) arrowheads.
FIGURE 5.
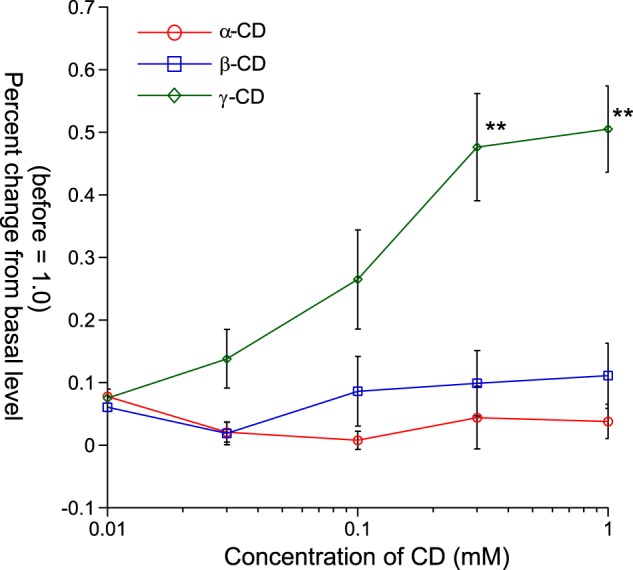
Interaction between GAs and each of CDs. Percent changes of [Ca2+]i responses to 10 mm saccharin before and after application of 100 μg/ml GAs followed by α-CD (○), β-CD (□), or γ-CD (♢) (before = 1.0). After application of GAs (100 μg/ml) followed by each of the CDs, only γ-CD showed a restorative effect in HEK293 cells expressing hT1R2 + hT1R3. Data are expressed as means ± S.E. ANOVA (F(2, 84) = 14.71) was followed by Tukey-Kramer method. **, p < 0.01.
FIGURE 6.
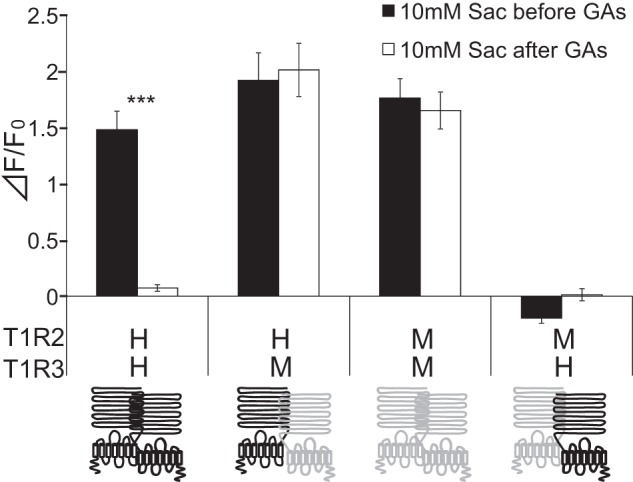
hT1R3 affects the sensitivity of human sweet receptor to GAs. Human (hT1R2 + hT1R3 as indicated by H (upper) H (lower)), mouse (mT1R2 + mT1R3 as indicated by M (upper) M (lower)), or human/mouse mismatched (hT1R2 + mT1R3 and mT1R2 + hT1R3 as indicated by HM and MH, respectively) sweet taste receptors were expressed in HEK293 cells, along with Gα16-gust44. T1R2 (left) and T1R3 (right) are indicated by schematic receptors. The human and mouse receptors are colored black and gray, respectively. The cells were assayed for their responses to saccharin (Sac) (10 mm) before and after application of GAs (100 μg/ml). Values are means ± S.E. of 20–28 cells. Paired t test: ***, p < 0.001.
To identify potential portions of hT1R2 and hT1R3 required for the sensitivity to GAs, we first examined responses of hT1R2 + human/mouse T1R3 chimeras in which varying portions of hT1R3 were fused with the complementary portion of mT1R3. Heterodimers of hT1R2 plus T1R3 chimeras containing the extracellular region of hT1R3 coupled to the TMD of mT1R3 (T1R3HHM) did not show sensitivity to GAs (Fig. 7). However, heterodimers of hT1R2 + T1R3 chimeras containing the extracellular region of mT1R3 fused with the TMD of hT1R3 (T1R3MMH) showed sensitivity to GAs. These results indicate that the sweet-suppressing effect of GAs requires the TMD of hT1R3. To determine whether hT1R2 is required for sensitivity to GAs, we examined the responses to saccharin of the mT1R3 chimera (T1R3MMH) paired with mT1R2 before and after application of GAs. This combination elicited responses to saccharin after application of GA (Fig. 7), indicating the absence of sensitivity for GAs. This suggests that hT1R2 may also be required for the sensitivity of GAs. Using combinations of full-length or chimeras of T1R2 and T1R3, we demonstrated that some combinations showed sensitivity to GAs (T1R2MMH + T1R3MMH, mT1R2 + T1R3MHH, T1R2MHM + T1R3MMH, and T1R2HHM + T1E3MMH) (Fig. 7). These results indicate that the ATD + CRD of hT1R2, the TMD of hT1R2 or the CRD of hT1R3 affect sensitivity to GAs.
FIGURE 7.
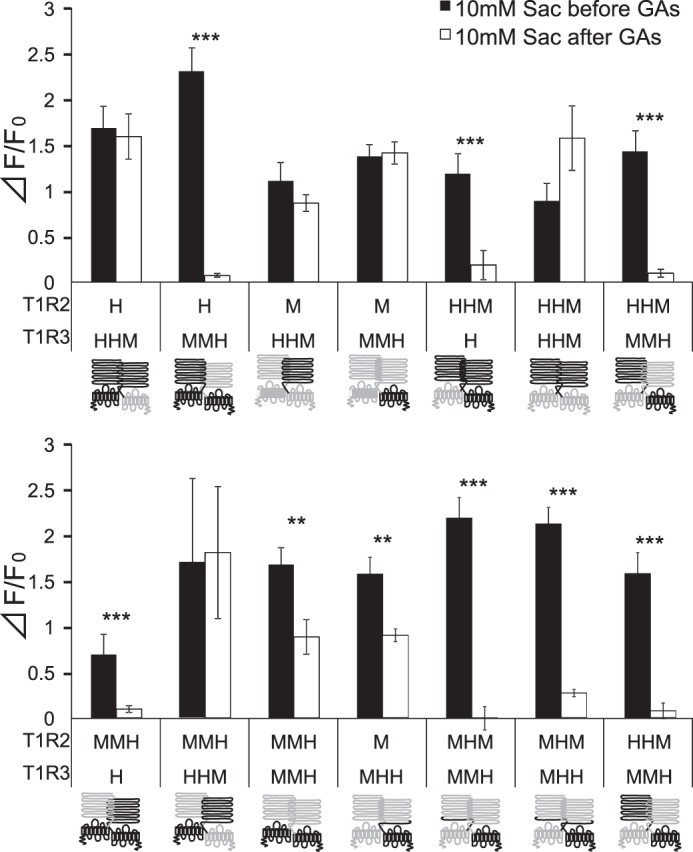
hT1R3's TMD is required for sensitivity to GAs. HEK293 cells were transiently transfected with T1R2, T1R3 (human/mouse chimeras), and Gα16-gust44. T1R2 (left) and T1R3 (right) are shown by schematic receptors. The full length of T1Rs is indicated by one letter of H (human; black) or M (mouse; gray). The chimeras of T1Rs are indicated by three letters including of H (human; black) or M (mouse; gray) (example, chimera containing ATD and CRD of human receptor coupled to the TMD of mouse receptor as for HHM). The responses of the receptors to saccharin (Sac) (10 mm) before and after application of GAs (100 μg/ml) were examined. Values are means ± S.E. of 12–34 cells. Paired t test: **, p < 0.01; ***, p < 0.001.
To identify the portion of the TMD of hT1R3 required for sensitivity to GAs, we examined the sensitivity of GAs in chimeras of T1R3 containing the ATD of hT1R3 along with varying portions of the TMD of hT1R3 fused with the complementary portion of mT1R3 in combination with hT1R2 (Fig. 8). All heterodimers of hT1R2 + these T1R3 chimeras responded to saccharin. The heterodimer of hT1R2 + T1R3 chimera containing the extracellular domain and entire TMD from hT1R3 (i.e. hT1R2h.1–604.mT1R3) showed [Ca2+]i responses to saccharin after application of GAs. Heterodimers of hT1R2 with T1R3 chimeras containing the hT1R3 ATD and hT1R3 TM helices 1–7 (i.e. hT1R2h.1–812.mT1R3) exhibited sensitivity to GAs. Heterodimers of hT1R2 with T1R3 chimeras containing the hT1R3 ATD and TM helices 1–6 (i.e. hT1R2h.1–787.mT1R3) or TM helices 1–5 (i.e. hT1R2h.1–751.mT1R3) showed weak suppression by GAs. Heterodimers of hT1R2 with T1R3 chimeras containing the hT1R3 ATD and hT1R3 TM helices 1–2 (i.e. hT1R2h.1–638.mT1R3) and TM helices 1–4 (i.e. hT1R2h.1–711.mT1R3) did not show sensitivity to GAs. These results indicate that the sweet-suppressing effect of GAs requires human-specific residues within or adjacent to human TM helices 5–7.
FIGURE 8.
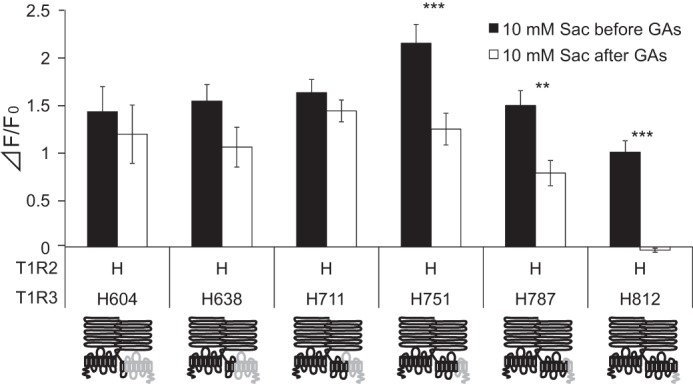
TMD5–7 from hT1R3 specifies sensitivity of GAs. HEK293 cells were transiently transfected with hT1R2, T1R3 chimeras, and Gα16-gust44. T1R2 (left) and T1R3 (right) are shown by schematic receptors. The human (H) and mouse portion of chimeras are indicated by black and gray, respectively. The responses of the receptors to saccharin (Sac) (10 mm) before and after application of GAs (100 μg/ml) were examined. Values are means ± S.E. of 10–24 cells. Paired t test: **, p < 0.01; ***, p < 0.001.
Molecular Modeling of T1Rs and GAs
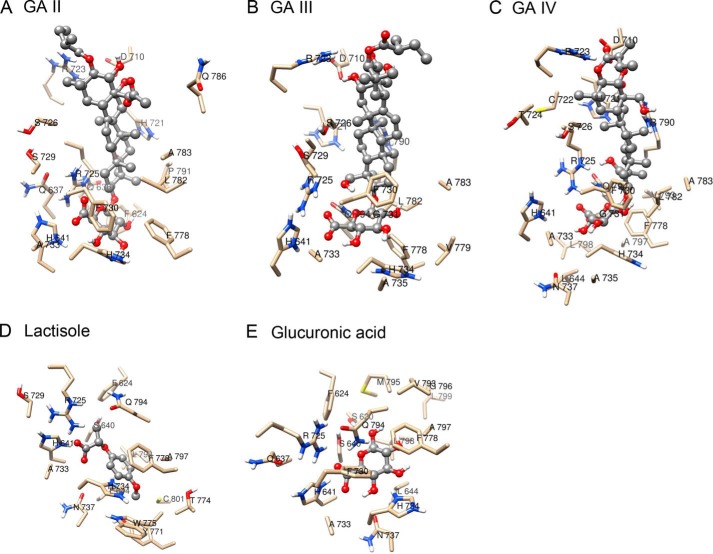
We performed molecularly modeled sweet receptors (human/mouse T1R2 and T1R3 and their chimeras) and performed docking simulations between whole parts of these receptors and GAs. GAs are composed of several types of homologues. Among derivatives, we generated the structures of GA I to IV known as sweet inhibitors (48). In our simulations, we could not observe models in which GAs interact with putative binding sites in sweet receptors (Venus flytrap modules of T1R2 and T1R3 and TMD of T1R2 and T1R3) except for TMD of hT1R3. We also performed docking simulations between CRD of T1Rs and GAs, but species-specific binding models were not observed. These results predicted that the main binding site for GAs is TMD of hT1R3. In docking experiments between TMD of hT1R3 and each of GA I–IV, each model showed similar docking features (Figs. 9 and 10). As reported previously, lactisole interacts with TMD of hT1R3 and is predicted to dock to a binding pocket within the TMD that includes key residues that, when mutated, reduced the sensitivity to lactisole (18). In our model, GAs were docked with the binding pocket within hT1R3-TMD, including key residues (Ala-7335.46 in TM5, Leu-7987.36 in TM7, His-6413.33 in TM3, and Phe-7786.51 in TM6) responsible for sensitivity to lactisole. His-6413.33 in TM helix 3 and Phe-7786.51 in TM helix 6 were close to the carboxyl group and the triterpene structure of GAs (3–5 Å) (Fig. 9). Based on our models, we examined the sensitivity to GAs with mutations that reduced sensitivity to lactisole in a previous study (18). In our control experiments, these mutations (hT1R3A7335.46V, L7987.36I, H6413.33A, and F7786.51A) in response to 10 mm saccharin significantly reduced the sensitivities to lactisole (Fig. 11 and Tables 1 and 2). The sweet-suppressing effects of GAs to various sweet compounds were also reduced in these mutants except for one residue; the mutation of F7786.51A moderately reduced or did not affect sensitivity to GAs, although this mutation abolished sensitivity to lactisole (Fig. 11 and Tables 1 and 2). Based on molecular modeling and mutation analysis, the interaction site for GAs shared the same binding pocket, including key residues that determined the sensitivity to lactisole (Figs. 9 and 10). We also found that Arg-723ex2–51 and Arg-725ex2–53 in extracellular loop (EL) 2 are close to the main structure and glucuronosyl group of GAs, respectively. Therefore, we mutated Arg-723ex2–51 and Arg-725ex2–53 to alanine to test whether the positively charged groups have a detrimental effect on the activity of GAs. Indeed, mutations of Arg-723ex2–51 and Arg-725ex2–53 modestly and greatly reduced sensitivity to GAs, respectively.
FIGURE 9.
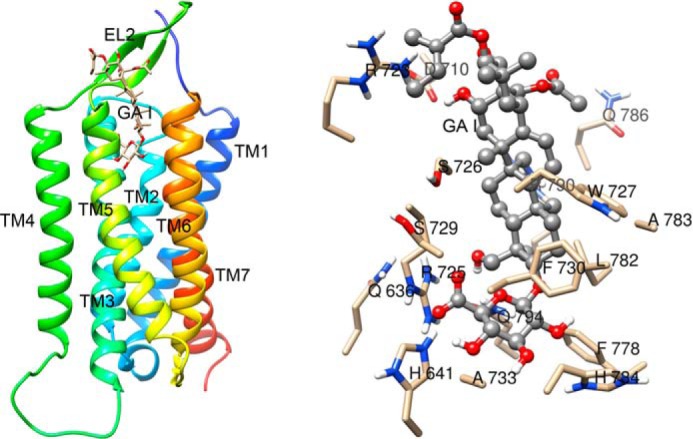
Molecular model of hT1R3 TMD binding pocket with docked GA I. An overall view (left panel) and a detailed view of the pocket (right panel) of T1R3 TMD with GA I are shown. GA I is shown in a ball-and-stick representation. Models are colored by atom type.
FIGURE 10.
Molecular models of hT1R3 TMD binding pocket with docked sweet inhibitors. Detailed views of the T1R3 TMD binding pocket with GA II (A), III (B), IV (C), lactisole (D), and GA (E) are shown. Sweet inhibitors are shown in a ball-and-stick representation. Models are colored by atom type.
TABLE 2.
Percent inhibition in response to sweet compounds before and after application of GAs and with or without lactisole and glucuronic acid obtained from WT TAS1R3 and selected mutants
All values are mean ± S.E. of 11–43 cells. Data were analyzed by one-way analysis of variance and post hoc tests, Tukey-Kramer method. *, p < 0.05; **, p < 0.01.
Data were derived from experiment in Fig. 11. The following abbreviations are used: Sac, saccharin; SC, SC45647; Trp, d-tryptophan; Asp, aspartame; Cyc, cyclamate.
| GAs (30 μg/ml) |
Lactisole (0.3 mm), Sac (10 mm) | Glucuronic acid (100 mm), Sac (10 mm) | |||||
|---|---|---|---|---|---|---|---|
| Sac (10 mm) | SC (0.3 mm) | Trp (10 mm) | Asp (10 mm) | Cyc (30 mm) | |||
| WT | 85.3 ± 2.9 | 91.7 ± 4.0 | 99.0 ± 4.1 | 79.2 ± 8.6 | 101.0 ± 3.3 | 89.9 ± 4.5 | 64.4 ± 7.5 |
| R723ex2–51A | 38.0 ± 13.6** | 49.4 ± 4.3** | 39.7 ± 6.9** | 18.8 ± 6.9** | 45.6 ± 6.6** | 47.5 ± 6.2** | 62.0 ± 6.5 |
| R725ex2–53A | −10.1 ± 4.3** | 20.7 ± 7.6** | 5.8 ± 6.2** | 3.3 ± 3.2** | 8.1 ± 2.2** | 64.1 ± 7.1* | 39.1 ± 6.7 |
| A7335.46V | −5.5 ± 3.0** | −11.5 ± 4.2** | 4.1 ± 4.1** | −4.8 ± 2.5** | −8.5 ± 9.2** | 30.2 ± 4.0** | 15.1 ± 4.6** |
| L7987.36I | 56.8 ± 8.6** | 73.5 ± 6.2 | 4.1 ± 4.0** | 60.8 ± 7.0 | 46.3 ± 7.6** | 24.5 ± 7.8** | 67.9 ± 13.8 |
| H6413.33A | 19.7 ± 2.5** | 21.7 ± 3.5** | 23.7 ± 3.1** | 0.6 ± 4.3** | 31.2 ± 6.7** | −1.1 ± 2.5** | 0.8 ± 7.9** |
| F7786.51A | 79.3 ± 8.2 | 77.1 ± 5.1 | 35.5 ± 3.7** | 53.3 ± 5.6* | NDa | −12.6 ± 3.9** | 19.4 ± 12.7* |
a ND indicates data unable to be experimentally defined.
Next, to examine which group of GAs is important for sweet antagonism, we used glucuronic acid, which is the common component of GAs and binds with the key residues that reduced sensitivity to GAs in our molecular model (Figs. 1, 9, and 10). As a result, glucuronic acid showed a suppressing effect in a concentration-dependent manner with a mixture of 10 mm saccharin, although glucuronic acid did not suppress responses to sweet compounds after washing out (Fig. 11 and Tables 1 and 2, and data not shown). In mutation analysis, A7335.46V, H6413.33A, and F7786.51A reduced sensitivity to glucuronic acid (Fig. 11 and Tables 1 and 2).
DISCUSSION
Sweet-suppressing Effect of GAs and the Recovery by γ-CD
Based on our pilot studies, GAs induced nonspecific responses in HEK293 cells in the absence of transfected receptor DNAs. To clarify the effect of GAs, we used single cell Ca2+ imaging and examined responses to sweeteners before and after application of GAs. It is known that GAs suppress perception of the sweet taste in humans but not in rodents (22, 26). In this study, we confirmed the species-specific effect of GAs showing that GAs interact with the human, but not mouse, sweet receptor. The suppressive effect of GAs in psychophysical studies is long lasting in humans (30–60 min) (21, 22), whereas the effect of lactisole was abolished immediately after washing out. In our assay, the sweet-suppressing effect of GAs lasts at least about 8 min despite the continuous washing out of the cell surface (Fig. 2), indicating a moderately long lasting effect of GAs. This lasting effect of GAs suggests that GAs might inhibit T1Rs not only extracellularly but also intracellularly. To evaluate this hypothesis, we examined potential interaction between GAs and each of the CDs. The results indicated that the sweet-suppressing effect of GAs was diminished immediately after rinsing the cells with γ-CD but not with α- and β-CD. This rapid recovery suggests that GAs may act on the receptor sites but not intracellular sites of the cells. CDs belong to a family of cyclic oligosaccharides with α-1,4 bonds. It is known that CDs form inclusion complexes with a wide variety of guest molecules, ranging from organic or inorganic compounds of neutral or ionic nature to noble gases (49). Because of the relatively high hydrophobicity and size of the cavity formed by the internal cyclic structure of CDs, chemical compounds with aromatic rings are suggested to be the best fitting guest molecules (50, 51). A previous chemical study using isothermal titration calorimetry, NMR, and dynamic light scattering demonstrated that γ-CD formed inclusion complexes with GAs (30). In our model, hydrophilic and hydrophobic groups of GAs interact with the inside and outside portion of the transmembrane domain of hT1R3, respectively. Our results suggest that γ-CD accesses the hydrophobic group of GAs from outside the receptor, and the observed reduction of the effects of GAs may be due to steric hindrance by inclusion complexes of GAs with γ-CD that may interfere with binding the GAs to human sweet taste receptors.
Interaction Site for Sweet-suppressing Effect of GAs
In this and previous heterologous expression studies, mismatched pair, chimera, and point mutation of sweet receptor and using different Gα subunits, Gα16-gust44 instead of Gnat3 may lead to different sensitivities to sweet compounds and sweet inhibitors due to conformational changes of whole receptors. Although such possibilities may affect results in this and previous studies, we demonstrated that the TMD of hT1R3 was required for sensitivity of GAs. In addition, the ATD + CRD of hT1R2, the TMD of hT1R2, or the CRD of hT1R3 were partially involved in sensitivity to GAs. As reported previously, some sweeteners interact with multiple domains of T1R2 + T1R3. For example, the activity of the protein sweetener brazzein depends on residues in ATD of hT1R2 and in CRD of hT1R3 (15, 16). Moreover, sucralose and some natural sugars have been proposed to interact with both subunits. To other artificial sweeteners, aspartame and neotame bind only to the T1R2-ATD (12, 52). In the case of GAs, the main target may be the TMD of hT1R3 because the mutation of hT1R3-TMD, which affected sensitivity to lactisole, also reduced sensitivity to GAs except for one mutation. This indicates that the interaction site for GAs shared the same binding pocket, including key residues (Ala-7335.46 in TM5, Leu-7987.36 in TM7, His-6413.33 in TM3, and Phe-7786.51 in TM6), which determined the sensitivity to lactisole. We speculate that in addition to the TMD of hT1T3, the ATD + CRD of hT1R2, TMD of hT1R2, or the CRDs of hT1R3 are partially involved in maintaining a proper conformation of hT1R3 for the interaction with GA. Although these domains may contain another direct interaction site for GA, we could not observe species-specific binding models between T1Rs and each of the GAs except for the TMD of hT1R3 in docking simulation. The reason why the heterodimer of mT1R2 and T1R3MMH does not show sensitivity to GAs is considered to be caused by the steric hindrance through the conformational change by the receptor-receptor interaction when these receptors form a heterodimer.
Although GAs are composed of several types of derivatives, all of the GAs tested showed similar features of molecular binding, indicating that these derivatives inhibit activation of sweet receptors through the common molecular mechanism. The sweet-suppressing effect of GAs is known to differ among derivatives (GA I, GA II > GA III > GA IV) (41). In our models, we could not find any particular interaction site that may cause differences in sensitivity of the sweet-suppressing effect among derivatives.
Glucuronic acid, which is the common component of GAs, also reduced sensitivities to sweet compounds, suggesting that the glucuronosyl group of GAs is important for the sweet-suppressing effect. These results are consistent with our molecular model.
How Might Mutations of His-6413.33, Ala-7335.46, Phe-7786.51, Arg-723ex2–51, and Arg-725ex2–53 Reduce Ability of GA to Inhibit Receptor Activity?
At physiological pH, His-6413.33 within TM3 is likely protonated, and its positive charge was predicted to form a salt bridge with the negatively charged carboxyl group of lactisole (18). In our models, the carboxyl group in the glucuronosyl group of GAs also binds with His-6413.33 by forming a salt bridge. Alanine substitution of His-6413.33 would eliminate this potential salt bridge, explaining the total loss of sensitivity to GAs in this mutant.
Within TM helix 5, replacement of Ala-7335.46 by valine (the mouse equivalent) greatly reduced the receptor's sensitivity to GAs as well as to lactisole. In our model, the glucuronosyl group of GAs is in close proximity with the side chain of Ala-7335.46. Larger aliphatic side chains at this position are likely to reduce sensitivity toward GAs, glucuronic acid, and lactisole by their steric effects.
Within TM helix 6, replacement of Phe-7786.51 with alanine had modest or no effect on sensitivity to GAs, although this mutant greatly reduced the receptor's sensitivity to lactisole. Our molecular models suggest that Phe-7786.51 is close to the phenoxyl ring of lactisole to form a π-π interaction similarly to a previous study (18), although GAs do not make this interaction (Figs. 9 and 10). So, our models are consistent with the results from mutant analysis. This residue would not be a common site for sweet receptor antagonists.
Replacement of Arg-723ex2–51 in EL2 with alanine reduced sensitivity to GAs as well as lactisole. In our models, Arg-723ex2–51 interacts with hydroxyl groups of triterpene structures of GAs, although lactisole does not interact with this residue. Replacement of this residue may cause conformational change of the entrance of the binding site and interfere with the access of sweet inhibitors to the binding site.
Alanine replacement of Arg-725ex2–53 in EL2 greatly reduced sensitivity to GAs, although this mutant had moderate effects on sensitivity to lactisole. In our molecular model, the negatively charged carboxyl group of GAs forms salt bridges with a positive charge of Arg-725ex2–53. We also speculate that the long lasting effect of GAs was due to high binding affinity with the human sweet receptor by this salt bridge as compared with lactisole's effect, which is diminished rapidly after washing out.
In our molecular model, GAs interact with the binding pocket formed by TM helices 3, 5, and 6 in hT1R3, which places the triterpene group of GAs in close proximity with the side chain of these residues. It could be proposed that GAs inactivate the receptor in a series of multiple step binding events per T1R2/T1R3 pair. However, less is known about such a complex inactivation scheme. Our results may not only provide insights into molecular mechanisms for the interaction of this G protein-coupled receptor with agonists and antagonists but may also be useful in the development of artificial sweeteners and sweet inhibitors.
Acknowledgments
We thank Dr. Gary K. Beauchamp (Monell Chemical Center), Dr. Ryusuke Yoshida (Kyushu University), and Dr. Atsuko Yamashita (Okayama University) for their valuable suggestions on the manuscript. We also thank Dr. Kazushige Touhara (University of Tokyo) for expert technical advice on Ca2+ imaging and Dr. Makoto Tominaga (National Institutes of Natural Sciences) for providing HEK293 cells.
This work was supported in whole or part by KAKENHI Grants 18109013, 18077004, and 23249081(to Y. N.) and 23792128 and 25861759 (to K. S.).
- ATD
- amino-terminal domain
- GA
- gymnemic acid
- CD
- cyclodextrin
- CRD
- cysteine-rich linker domain
- TMD
- heptahelical transmembrane domain
- EL
- extracellular loop
- h
- human
- m
- mouse
- TM
- transmembrane
- ANOVA
- analysis of variance.
REFERENCES
- 1. Li X., Inoue M., Reed D. R., Huque T., Puchalski R. B., Tordoff M. G., Ninomiya Y., Beauchamp G. K., Bachmanov A. A. (2001) High-resolution genetic mapping of the saccharin preference locus (Sac) and the putative sweet taste receptor (T1R1) gene (Gpr70) to mouse distal chromosome 4. Mamm. Genome 12, 13–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nelson G., Hoon M. A., Chandrashekar J., Zhang Y., Ryba N. J., Zuker C. S. (2001) Mammalian sweet taste receptors. Cell 106, 381–390 [DOI] [PubMed] [Google Scholar]
- 3. Hoon M. A., Adler E., Lindemeier J., Battey J. F., Ryba N. J., Zuker C. S. (1999) Putative mammalian taste receptors: a class of taste-specific GPCRs with distinct topographic selectivity. Cell 96, 541–551 [DOI] [PubMed] [Google Scholar]
- 4. Max M., Shanker Y. G., Huang L., Rong M., Liu Z., Campagne F., Weinstein H., Damak S., Margolskee R. F. (2001) Tas1r3, encoding a new candidate taste receptor, is allelic to the sweet responsiveness locus Sac. Nat. Genet. 28, 58–63 [DOI] [PubMed] [Google Scholar]
- 5. Sainz E., Korley J. N., Battey J. F., Sullivan S. L. (2001) Identification of a novel member of the T1R family of putative taste receptors. J. Neurochem. 77, 896–903 [DOI] [PubMed] [Google Scholar]
- 6. Montmayeur J. P., Liberles S. D., Matsunami H., Buck L. B. (2001) A candidate taste receptor gene near a sweet taste locus. Nat. Neurosci. 4, 492–498 [DOI] [PubMed] [Google Scholar]
- 7. Kitagawa M., Kusakabe Y., Miura H., Ninomiya Y., Hino A. (2001) Molecular genetic identification of a candidate receptor gene for sweet taste. Biochem. Biophys. Res. Commun. 283, 236–242 [DOI] [PubMed] [Google Scholar]
- 8. Pin J. P., Galvez T., Prézeau L. (2003) Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 98, 325–354 [DOI] [PubMed] [Google Scholar]
- 9. Kunishima N., Shimada Y., Tsuji Y., Sato T., Yamamoto M., Kumasaka T., Nakanishi S., Jingami H., Morikawa K. (2000) Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature 407, 971–977 [DOI] [PubMed] [Google Scholar]
- 10. Muto T., Tsuchiya D., Morikawa K., Jingami H. (2007) Structures of the extracellular regions of the group II/III metabotropic glutamate receptors. Proc. Natl. Acad. Sci. U.S.A. 104, 3759–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Palczewski K., Kumasaka T., Hori T., Behnke C. A., Motoshima H., Fox B. A., Le Trong I., Teller D. C., Okada T., Stenkamp R. E., Yamamoto M., Miyano M. (2000) Crystal structure of rhodopsin: A G protein-coupled receptor. Science 289, 739–745 [DOI] [PubMed] [Google Scholar]
- 12. Xu H., Staszewski L., Tang H., Adler E., Zoller M., Li X. (2004) Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl. Acad. Sci. U.S.A. 101, 14258–14263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Masuda K., Koizumi A., Nakajima K., Tanaka T., Abe K., Misaka T., Ishiguro M. (2012) Characterization of the modes of binding between human sweet taste receptor and low-molecular-weight sweet compounds. PLoS ONE 7, e35380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Koizumi A., Nakajima K., Asakura T., Morita Y., Ito K., Shmizu-Ibuka A., Misaka T., Abe K. (2007) Taste-modifying sweet protein, neoculin, is received at human T1R3 amino terminal domain. Biochem. Biophys. Res. Commun. 358, 585–589 [DOI] [PubMed] [Google Scholar]
- 15. Jiang P., Ji Q., Liu Z., Snyder L. A., Benard L. M., Margolskee R. F., Max M. (2004) The cysteine-rich region of T1R3 determines responses to intensely sweet proteins. J. Biol. Chem. 279, 45068–45075 [DOI] [PubMed] [Google Scholar]
- 16. Assadi-Porter F. M., Maillet E. L., Radek J. T., Quijada J., Markley J. L., Max M. (2010) Key amino acid residues involved in multi-point binding interactions between brazzein, a sweet protein, and the T1R2-T1R3 human sweet receptor. J. Mol. Biol. 398, 584–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang P., Cui M., Zhao B., Snyder L. A., Benard L. M., Osman R., Max M., Margolskee R. F. (2005) Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J. Biol. Chem. 280, 34296–34305 [DOI] [PubMed] [Google Scholar]
- 18. Jiang P., Cui M., Zhao B., Liu Z., Snyder L. A., Benard L. M., Osman R., Margolskee R. F., Max M. (2005) Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J. Biol. Chem. 280, 15238–15246 [DOI] [PubMed] [Google Scholar]
- 19. Winnig M., Bufe B., Kratochwil N. A., Slack J. P., Meyerhof W. (2007) The binding site for neohesperidin dihydrochalcone at the human sweet taste receptor. BMC Struct. Biol. 7, 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Suttisri R., Lee I. S., Kinghorn A. D. (1995) Plant-derived triterpenoid sweetness inhibitors. J. Ethnopharmacol. 47, 9–26 [DOI] [PubMed] [Google Scholar]
- 21. Warren R. M., Pfaffmann C. (1959) Suppression of sweet sensitivity by potassium gymnemate. J. Appl. Physiol. 14, 40–42 [DOI] [PubMed] [Google Scholar]
- 22. Kurihara Y. (1969) Antisweet activity of gymnemic acid A1 and its derivatives. Life Sci. 8, 537–543 [DOI] [PubMed] [Google Scholar]
- 23. Diamant H., Oakley B., Stroem L., Wells C., Zotterman Y. (1965) A comparison of neural and psychophysical responses to taste stimuli in man. Acta Physiol. Scand. 64, 67–74 [DOI] [PubMed] [Google Scholar]
- 24. Hellekant G., Gopal V. (1976) On the effects of gymnemic acid in the hamster and rat. Acta Physiol. Scand. 98, 136–142 [DOI] [PubMed] [Google Scholar]
- 25. Hellekant G., af Segerstad C. H., Roberts T., van der Wel H., Brouwer J. N., Glaser D., Haynes R., Eichberg J. W. (1985) Effects of gymnemic acid on the chorda tympani proper nerve responses to sweet, sour, salty and bitter taste stimuli in the chimpanzee. Acta Physiol. Scand. 124, 399–408 [DOI] [PubMed] [Google Scholar]
- 26. Hellekant G., Ninomiya Y., DuBois G. E., Danilova V., Roberts T. W. (1996) Taste in chimpanzee: I. The summated response to sweeteners and the effect of gymnemic acid. Physiol. Behav. 60, 469–479 [DOI] [PubMed] [Google Scholar]
- 27. Hellekant G., Ninomiya Y., Danilova V. (1998) Taste in chimpanzees. III: labeled-line coding in sweet taste. Physiol. Behav. 65, 191–200 [DOI] [PubMed] [Google Scholar]
- 28. Nagaoka T., Hane H., Yamashita H., Kensho I. (1990) Taste improvement of extracts from the leaves of Gymnema sylvestre. Seitou Gijutsu Kenkyukai-Shi 38, 61–70 [Google Scholar]
- 29. Imoto T. (1990) Sugar discrimination and plant-originated glycosides, gymnemic acids. Seibutsu Butsuri 30, 146–150 [Google Scholar]
- 30. Izutani Y., Kanaori K., Imoto T., Oda M. (2005) Interaction of gymnemic acid with cyclodextrins analyzed by isothermal titration calorimetry, NMR and dynamic light scattering. FEBS J. 272, 6154–6160 [DOI] [PubMed] [Google Scholar]
- 31. Nakamura Y., Tsumura Y., Tonogai Y., Shibata T. (1999) Fecal steroid excretion is increased in rats by oral administration of gymnemic acids contained in Gymnema sylvestre leaves. J. Nutr. 129, 1214–1222 [DOI] [PubMed] [Google Scholar]
- 32. Porchezhian E., Dobriyal R. M. (2003) An overview on the advances of Gymnema sylvestre: chemistry, pharmacology and patents. Pharmazie 58, 5–12 [PubMed] [Google Scholar]
- 33. Kanetkar P., Singhal R., Kamat M. (2007) Gymnema sylvestre: a memoir. J. Clin. Biochem. Nutr. 41, 77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Izutani Y., Murai T., Imoto T., Ohnishi M., Oda M., Ishijima S. (2005) Gymnemic acids inhibit rabbit glyceraldehyde-3-phosphate dehydrogenase and induce a smearing of its electrophoretic band and dephosphorylation. FEBS Lett. 579, 4333–4336 [DOI] [PubMed] [Google Scholar]
- 35. Ballesteros J. A., Weinstein H. (1995) Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G-protein-coupled receptors. Methods Neurosci. 25, 366–428 [Google Scholar]
- 36. Shigemura N., Shirosaki S., Sanematsu K., Yoshida R., Ninomiya Y. (2009) Genetic and molecular basis of individual differences in human umami taste perception. PLoS ONE 4, e6717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Li X., Staszewski L., Xu H., Durick K., Zoller M., Adler E. (2002) Human receptors for sweet and umami taste. Proc. Natl. Acad. Sci. U.S.A. 99, 4692–4696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. (1989) Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene 77, 61–68 [DOI] [PubMed] [Google Scholar]
- 39. Zhang F., Klebansky B., Fine R. M., Liu H., Xu H., Servant G., Zoller M., Tachdjian C., Li X. (2010) Molecular mechanism of the sweet taste enhancers. Proc. Natl. Acad. Sci. U.S.A. 107, 4752–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sánchez R., Sali A. (2000) Comparative protein structure modeling. Introduction and practical examples with modeller. Methods Mol. Biol. 143, 97–129 [DOI] [PubMed] [Google Scholar]
- 41. Wu H., Wang C., Gregory K. J., Han G. W., Cho H. P., Xia Y., Niswender C. M., Katritch V., Meiler J., Cherezov V., Conn P. J., Stevens R. C. (2014) Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science 344, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Irwin J. J., Shoichet B. K. (2005) ZINC–a free database of commercially available compounds for virtual screening. J. Chem. Inf. Model. 45, 177–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Suenaga M. (2005) New computational chemistry environment for PC GAMESS. J. Comput. Chem. 4, 25–32 [Google Scholar]
- 44. Suenaga M. (2008) Development of GUI for GAMESS/FMO calculation. J. Comput. Chem. 7, 33–53 [Google Scholar]
- 45. Schmidt M. W., Baldridge K. K., Boatz J. A., Elbert S. T., Gordon M. S., Jensen J. H., Koseki S., Matsunaga N., Nguyen K. A., Su S. J., Windus T. L., Dupuis M., Montgomery J. A. (1993) General atomic and molecular electronic-structure system. J. Comput. Chem. 14, 1347–1363 [Google Scholar]
- 46. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 47. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) UCSF chimera–A visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 48. Kurihara Y. (1992) Characteristics of antisweet substances, sweet proteins, and sweetness-inducing proteins. Crit. Rev. Food Sci. Nutr. 32, 231–252 [DOI] [PubMed] [Google Scholar]
- 49. Li S., Purdy W. C. (1992) Cyclodextrins and their applications in analytical chemistry. Chem. Rev. 92, 1457–1470 [Google Scholar]
- 50. Inoue Y., Miyata Y. (1981) Formation and molecular-dynamics of cycloamylose inclusion complexes with phenylalanine. Bull. Chem. Soc. 54, 809–816 [Google Scholar]
- 51. Inoue Y., Okuda T., Miyata Y. (1981) Side-chain conformational changes of some amino-acids and dipeptides having aromatic side-chains induced by complexation with cycloamyloses. J. Am. Chem. Soc. 103, 7393–7394 [Google Scholar]
- 52. Nie Y., Vigues S., Hobbs J. R., Conn G. L., Munger S. D. (2005) Distinct contributions of T1R2 and T1R3 taste receptor subunits to the detection of sweet stimuli. Curr. Biol. 15, 1948–1952 [DOI] [PubMed] [Google Scholar]