Background: SARS-CoV nsp10 binds and stimulates both the nsp14 and nsp16 activities.
Results: We mapped the nsp10 surface interacting with nsp14 and demonstrated that this surface plays a critical role in SARS-CoV replication.
Conclusion: The core interaction domain of nsp10 is essential for SARS-CoV replication.
Significance: This nsp10 core represents an attractive target for antiviral drug development against various pathogenic coronaviruses.
Keywords: Genetics, Protein-Protein Interaction, RNA Methyltransferase, RNA Virus, Viral Replication, Viral Transcription
Abstract
The RNA-synthesizing machinery of the severe acute respiratory syndrome Coronavirus (SARS-CoV) is composed of 16 non-structural proteins (nsp1–16) encoded by ORF1a/1b. The 148-amino acid nsp10 subunit contains two zinc fingers and is known to interact with both nsp14 and nsp16, stimulating their respective 3′-5′ exoribonuclease and 2′-O-methyltransferase activities. Using alanine-scanning mutagenesis, in cellulo bioluminescence resonance energy transfer experiments, and in vitro pulldown assays, we have now identified the key residues on the nsp10 surface that interact with nsp14. The functional consequences of mutations introduced at these positions were first evaluated biochemically by monitoring nsp14 exoribonuclease activity. Disruption of the nsp10-nsp14 interaction abrogated the nsp10-driven activation of the nsp14 exoribonuclease. We further showed that the nsp10 surface interacting with nsp14 overlaps with the surface involved in the nsp10-mediated activation of nsp16 2′-O-methyltransferase activity, suggesting that nsp10 is a major regulator of SARS-CoV replicase function. In line with this notion, reverse genetics experiments supported an essential role of the nsp10 surface that interacts with nsp14 in SARS-CoV replication, as several mutations that abolished the interaction in vitro yielded a replication-negative viral phenotype. In contrast, mutants in which the nsp10-nsp16 interaction was disturbed proved to be crippled but viable. These experiments imply that the nsp10 surface that interacts with nsp14 and nsp16 and possibly other subunits of the viral replication complex may be a target for the development of antiviral compounds against pathogenic coronaviruses.
Introduction
Viruses in the order Nidovirales, which includes the Coronaviridae, Arteriviridae, Roniviridae, and (proposed) Mesoniviridae families, possess the largest RNA genomes known to date (1, 2). Coronaviruses (CoVs)9 are respiratory and enteric pathogens of humans and domesticated animals and also appear to be omnipresent in wildlife, in particular in bats and rodents (3–5). The capability to cross species barriers appears to be a widespread CoV feature, and all endemic human CoVs are thought to have been transmitted from animal hosts at some point in the past. CoVs can cause life-threatening zoonotic infections, and the emergence in humans, less than a decade apart, of CoVs causing severe acute respiratory syndrome (SARS) (6) and Middle East respiratory syndrome (MERS) (7, 8) emphasizes the significant pandemic potential of members of this virus family. SARS-CoV presumably emerged from bats in China in 2002, spread around the globe in a few months, and resulted in >8000 registered cases with a fatality rate of ∼10% (3, 9, 10). MERS-CoV emerged in the Middle East in 2012 and has thus far resulted in >800 laboratory-confirmed cases with a fatality rate around 35% (7, 8, 11, 12).
The SARS-CoV genome is a single-stranded, positive-sense RNA of ∼29.7 kb, which is 5′-capped and 3′-polyadenylated. After virus entry, genome translation initiates a complex gene expression program (13, 14) that is regulated at the translational, post-translational, and transcriptional levels. Open reading frames (ORFs) 1a and 1b, which occupy the 5′-proximal two-thirds of the genome, are translated directly from the genome to yield two large polyprotein (pp) precursors called pp1a and pp1ab. The latter is derived from extension of pp1a with the ORF1b-encoded sequence after a −1 ribosomal frameshift near the 3′ end of ORF1a, which gives rise to a fixed ratio of pp1a over pp1ab synthesis (15–30% ribosomal frameshift efficiency reported; Refs 15–17). The two replicase polyproteins are cleaved into a total of 16 functional proteins (see Fig. 1) called nonstructural protein 1 (nsp1) to nsp16. Viral RNA synthesis entails genome replication, which proceeds via a full-length minus-strand RNA, and the synthesis of an extensive nested set of subgenomic mRNAs, each from their own subgenome-length minus-stranded template (13, 18–20). Subgenomic mRNAs are used to express structural and accessory proteins encoded in the 3′-proximal third of the genome (see Fig. 1).
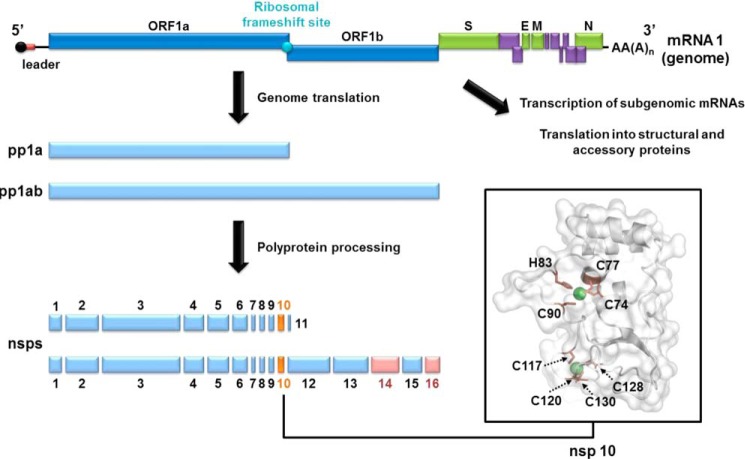
FIGURE 1.
Schematic representation of SARS-CoV genome and nsp10 structure. Nsp10 (orange) and nsp14/16 (pink) are highlighted. The structural (green) and accessory (purple) protein genes are expressed from a nested set of subgenomic mRNAs. The schematic of the nsp10 structure (PDB code 2FYG) was generated using PyMOL software. Zinc ions are shown as green spheres, and residues forming the two nsp10 zinc fingers are labeled and depicted as brown sticks.
After replicase polyprotein cleavage, the SARS-CoV nsps assemble into a multienzyme replication-transcription complex that is associated with membrane structures derived from the modification of the host cell endoplasmic reticulum (21, 22). Although the functions of some CoV nsps have remained elusive thus far (e.g. nsp2 and nsp9), enzymatic activities have been attributed to many others. Some of these are commonly found in the RNA virus world (e.g. protease, helicase, and RNA polymerase functions), whereas others are rare or even unique, including e.g. an endoribonuclease of unknown function in nsp15 and the nsp14 exoribonuclease, which has been implicated in a primitive form of proofreading that enhances the fidelity of CoV RNA synthesis (for more details please refer to reviews in Refs. 14 and 23).
The CoV genome also encodes proteins required for the formation and modification of the cap structure present at the 5′ end of all coronaviral mRNAs. In eukaryotic cells most mRNAs of invading viruses and the host itself are modified by the addition of such a cap, which protects from degradation by 5′-3′ exoribonucleases, ensures efficient translation, and in the case of viral mRNAs helps to prevent recognition by the host innate immune system (24–27). In the eukaryotic cell, the cap is added co-transcriptionally in the nucleus by the sequential activity of four enzymes: (i) an RNA triphosphatase, (ii) a guanylyltransferase, (iii) an N7-methyltransferase (MTase), and (iv) a 2′-O-MTase yielding a so-called cap-1 structure (7MeGpppN2′-O-Me) (24, 28). The biological importance of the latter structure is underlined by the fact that numerous viruses have evolved strategies to secure the presence of cap-1 at the 5′ terminus of their mRNAs (for a review, see Ref. 29).
Positive-stranded RNA viruses, like SARS-CoV, replicate in the cytoplasm and are, therefore, presumed not to have access to the host cell capping machinery. Nevertheless, the 5′ end of CoV mRNAs is thought to carry a type-1 cap (30–32), which is produced by the concerted action of several viral enzymes (33, 34). As in many other positive-strand RNA viruses, an RNA triphosphatase activity is embedded in the RNA helicase carried by nsp13 and is likely to mediate the first step of viral cap synthesis (35). The guanylyltransferase activity involved in CoV cap synthesis has remained elusive thus far. In contrast, two CoV methyltransferase activities were identified and assigned to the C-terminal domain of nsp14 ((N7-guanine)-methyltransferase) (33, 34) and nsp16 (2′-O-MTase) (36). We recently demonstrated that the in vitro activity of the SARS-CoV nsp16 2′-O-MTase depends on an interaction with nsp10. The nsp10-nsp16 complex exhibits a potent MTase activity specifically converting cap-0 (7MeGpppN) into cap-1 structures (34). Nsp10 is 148 residues long, contains two zinc fingers, and is overexpressed relative to nsp14 and nsp16 as it is encoded in ORF1a. Although its structure was solved almost 10 years ago (Fig. 1), no enzymatic function was identified or proposed for the protein (37, 38). Together with other replicase subunits, nsp10, nsp14, and nsp16 were implicated in the formation of the CoV replication-transcription complex (39), a process that is likely based on a large repertoire of specific interactions of nsps with other nsps and with viral RNA sequences. The nsp10-nsp16 interaction was characterized biochemically and structurally to decipher the molecular basis of nsp10 function in activating the nsp16-mediated 2′-O-MTase activity. Several key residues for the nsp10-nsp16 interaction were identified, and these findings were subsequently supported by the crystal structure of the complex (40, 41). The recent observation that 2′-O-methylation of the RNA cap may be mandatory to avoid or delay viral RNA recognition by innate immune sensors highlights the importance of the nsp10-nsp16 2′-O-MTase complex in CoV-host interplay (42–44).
Using bioinformatics and biochemistry, a 3′-5′ exoribonuclease (ExoN) activity has been identified in CoV nsp14, with its catalytic residues (DEDD) mapping to the N-terminal part of the protein (6, 45). Subsequently, an (N7-guanine)-methyltransferase activity involved in viral mRNA capping was identified in the C-terminal domain of the same protein (33, 46). The nsp14 (N7-guanine)-methyltransferase domain does not seem to be functionally separable from the ExoN domain (33, 47). A predicted interaction between nsp10 and nsp14 (48, 49) was recently confirmed in vitro by Bouvet et al. (34, 47) in a study demonstrating a second regulatory role for SARS-CoV nsp10. Upon interacting with nsp10, the ExoN activity of nsp14 increased by >35-fold. Using diverse RNA substrates, the nsp10-nsp14 ExoN activity was shown to specifically degrade double-stranded (ds) RNAs with a preference for substrates carrying a single non-hybridized nucleotide at their 3′ end. This study together with the 15–20-fold increased mutation rate observed for reverse engineered ExoN-deficient nsp14 mutants (50, 51) strongly suggested the involvement of ExoN in a unique RNA proofreading mechanism allowing CoVs to safeguard the integrity of their unusually large RNA genome. Interestingly, nsp10 does not show any effect on nsp14 (N7-guanine)-methyltransferase activity in vitro (34). Considering the importance of the nsp10 protein in the regulation of viral enzymatic activities and replication-transcription complex assembly, it constitutes an attractive target for drug design programs.
In this context we now combined several approaches to map the nsp10-nsp14 interaction at the molecular level. Using an extended mutagenesis approach and by coupling in cellulo bioluminescence resonance energy transfer (BRET) studies to in vitro functional assays, we identified a continuous and specific nsp10 surface involved in the interaction with nsp14. Key surface residues involved in this interaction were found to control SARS-CoV nsp14 ExoN activity and also appear to be part of the nsp10 domain that interacts with nsp16. Thus, nsp10 possesses overlapping interaction surfaces for the activation of two important viral enzymes. Reverse genetics experiments targeting key residues of this domain revealed that they are indeed critical for SARS-CoV replication, thus validating this surface as a potential target for antiviral drug development.
EXPERIMENTAL PROCEDURES
Reagents
All radioactive reagents were purchased from PerkinElmer Life Sciences. S-Adenosyl-l-methionine was purchased from New England Biolabs.
Plasmids
All cloning experiments were performed using Gateway® technology (Invitrogen). For BRET experiments, plasmid constructions are described in Lugari et al. (41). The SARS-CoV nsp10 and nsp14 expression plasmids (pDest14/His6-nsp10 and pTXB1-nsp14) were described previously (34). Nsp10 and nsp14 genes were also cloned into a dual-promoter expression plasmid as described previously (52). In this backbone SARS-CoV nsp10 is expressed in fusion with an N-terminal Strep-TagII (named Strep-nsp10), whereas nsp14 is fused to an N-terminal hexahistidine tag (named nsp14HN). Single point mutant plasmids were generated by PCR using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions.
Cell Culture
HEK 293T cells were grown in accordance with ATCC recommendations in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2 mm l-glutamine, 50 units/ml penicillin, and 50 μg/ml streptomycin. Vero-E6 cells (ATCC: CRL-1586) were cultured in Eagle's minimal essential medium (EMEM; Lonza) with 8% fetal calf serum (PAA) and antibiotics. BHK-Tet-SARS-N cells (53) were cultured in Eagle's minimum essential medium with 8% fetal calf serum (PAA), antibiotics, and 100 μg/ml of G418.
BRET Assays
Cells in 6-well plates were transfected using FuGENE® 6 transfection reagent (Roche Applied Science) according to the manufacturer's protocol. Cells were plated with 300,000 cells per well, 8 h before transfection. Transfections were made with 300 ng of pNRLuc-nsp14 vector and various amounts of pEYFP-nsp10 vectors (50–100-300–600-900 ng) complemented with pUC19 vector for a total of 1.5 μg of DNA transfected per well. Cells were then incubated at 37 °C, 5% CO2 for 48 h prior to the BRET assays. BRET assays were performed in living cells according to Lugari et al. (41). In each experiment transfections of pNRLuc-nsp14 alone or plus pEYFP were performed as controls. Coelenterazine H (Tebu-Bio) was added on cells at a 5 μm final concentration, and cells were incubated for 20 min at room temperature before reading. BRET measurements were performed at 25 °C by sequentially integrating luminescence signals at 480 and 530 nm for 1 s. The BRET ratio is defined as: [(emission at 530 nm) − (emission at 485 nm) × Cf]/(emission at 485 nm), where Cf corresponds to (emission at 530 nm)/(emission at 485 nm) for the Renilla luciferase (Rluc) fusion protein expressed alone under the same experimental conditions. All experiments were performed at least three times, and BRET signals were normalized according to the fluorescence signals of nsp10-EYFP mutants compared with wild type.
Antibodies
Anti-GFP antibodies (a mix of clones 7.1 and 13.1 was used) were purchased from Roche Applied Science. Anti-Renilla luciferase antibodies (mAb4400 and mAb4410) were purchased from Chemicon. Secondary antibodies coupled to horseradish peroxidase were purchased from Dako. Alexa-conjugated antibodies for immunostaining are from Jackson ImmunoResearch laboratories.
Expression and Purification of SARS-CoV nsp10 and nsp14 Proteins
Expression and purification of His6-nsp10 from Escherichia coli transformed with pDest14/His6-nsp10 and untagged nsp14 from E. coli carrying pTXB1-nsp14 were performed as previously described (34, 47). SARS-CoV nsp10-nsp14 co-expression was performed as described for nsp10-nsp16 in Debarnot et al. (52).
RNA Synthesis and Purification
Synthetic RNA H4 was purchased from Jena Bioscience (HPLC grade). H4 RNA was radiolabeled using polynucleotide kinase (New England Biolabs) and [γ-32P]ATP according to the manufacturer's instructions.
Exonuclease Assay
The reactions were performed as described by Bouvet et al. (47). Briefly, nsp10 and nsp14 were incubated in a 4:1 molecular ratio with radiolabeled RNA (oligo H4, 5′-UGACGGCCCGGAAAACCGGGCC-3′) (47). After 30 min the reaction was stopped. The reaction products were separated on denaturing polyacrylamide gels and visualized using phosphorimaging (Fluorescent Image Analyzer FLA3000; Fuji). The relative ExoN activity was quantified using Image Gauge software.
Pulldown Assay
For pulldown assays, SARS-CoV nsp10 and nsp14 were co-expressed in E. coli using a dual promotor approach. In this system SARS-CoV nsp10 expression is under control of a tet promoter and produces a protein in fusion with an N-terminal Strep-TagII, whereas nsp14 is expressed from a T7 promoter and carries an N-terminal His6 tag. E. coli C41 (DE3) cells (Avidis SA, France) harboring the pLysS plasmid (Novagen) were transformed with the various expression vectors and grown in 2YT medium containing antibiotics. Protein expression was induced by adding 50 μm isopropyl 1-thio-β-d-galactopyranoside and 200 μg/liter anhydrotetracycline, then cells were incubated for 16 h at 24 °C. Bacterial cell pellets were frozen and resuspended in lysis buffer (50 mm HEPES, pH 7.5, 500 mm NaCl, 5 mm MgSO4) supplemented with 1 mm PMSF, 10 μg/ml DNase I, and 0.5% Triton X-100. After sonication and clarification, proteins were purified by chromatography with Strep-Tactin-Sepharose (IBA GmbH, Göttingen, Germany). After 3 washes in high salt buffer (1 m NaCl) and 3 washes in low salt buffer (500 mm NaCl), bound proteins were eluted with 2.5 mm d-desthiobiotin in binding buffer. The purified proteins were analyzed and quantified by capillary electrophoresis (Caliper LabChip, PerkinElmer Life Sciences).
SARS-CoV Reverse Genetics
Using “en passant recombineering” (recombineering by mutagenesis) (54), mutations in the nsp10-, nsp14-, and nsp16-coding regions of SARS-CoV isolate Frankfurt-1 were engineered in prSCV, a pBeloBac11 derivative containing a full-length cDNA copy of the viral genome (55). The DNA of such BAC clones was linearized with NotI, extracted with phenol-chloroform, and transcribed with the mMessage-mMachine® T7 (Ambion) using 2 μg of DNA template in a 20-μl reaction. Full-length viral RNA was precipitated with LiCl according to the manufacturer's protocol, and 6 μg was electroporated into 5 × 106 BHK-Tet-SARS-N cells, which express the SARS-CoV N protein after >4 h of induction with 2 μm doxycycline (53). Electroporation was done using the Amaxa Nucleofector (Lonza), Nucleofector Kit T, and program T-020 according to the manufacturer's instructions. Cells were mixed in a 1:1 ratio with Vero-E6 cells and seeded on coverslips for immunofluorescence microscopy and for analysis of virus production. Each mutant was launched twice from independently generated BAC clones. All work with live SARS-CoV was performed inside biosafety cabinets in a biosafety level 3 facility at Leiden University Medical Center.
RT-PCR Analysis of the nsp10, nsp14, and nsp16 Coding Region
Fresh Vero-E6 cells were infected with harvests taken at 42 h post transfection, and cells were incubated overnight. Intracellular RNA was isolated from the infected cells by using TriPure Isolation Reagent (Roche Applied Science) as described by the manufacturer's instructions and amplified by RT-PCR using random hexamers to prime the RT reaction, and primers 5′-TTGCCTACTATAACAATTCG-3′ and 5′-GTTATAAATAGTCTCTTCATGTTGG-3′ for PCR amplification of the nsp10-coding region. The nsp14 region was amplified by using primers 5′-GGTTCTGAATATGACTATGTCATATTC-3′ and 5′-CCTGTCCTTCCACTCTACCATC-3′; the primers for amplification of the nsp16-coding region were 5′-CTATGCTGAAATTTCATTCATGC-3′ and 5′-TGGTGCACCGGTCAAGGTCACTACC-3′. Amplicons were sequenced to verify the presence of the original mutations and/or putative (second site) reversions.
In both experiments with mutant nsp10-M44A, no sign of virus replication was observed at early time points (18–48 h post transfection) using either immunofluorescence microscopy or titration of viral progeny. In one experiment, however, all cells died from viral infection in a dish used to obtain a 72 h post-transfection harvest. This material was used to infect fresh Vero-E6 cells for RNA isolation and amplification of the nsp10-coding region by RT-PCR. Sequencing of this product revealed that the original M44A mutation had been converted into M44V.
Immunofluorescence Microscopy
To monitor the progression of SARS-CoV infection, transfected cells on coverslips were fixed at various time points post transfection. Immunofluorescence assays were done after a previously described protocol (56) using a rabbit antiserum against nsp4 (21) and mAb against N (kindly provided by Ying Fang, South Dakota State University; Ref. 57). As the expression level of N in BHK-Tet-SARS-N cells is much lower than the level reached in infected cells, it is possible to discriminate between SARS-CoV-positive and -negative cells.
Titration of Virus Progeny and Plaque Reduction Assays
For plaque assays, Vero-E6 cells seeded in 6-well clusters were infected with serial 10-fold dilutions (in PBS containing 0.005% DEAE and 2% FCS) of supernatants from transfected cells. The inoculum was removed after 1 h at 37 °C and replaced with an overlay of 1.2% Avicel (FMC BioPolymer) in DMEM supplemented with 50 mm HEPES, 2% FCS. Cells were incubated at 37 °C, and after 3 days cell layers were fixed with 7.4% formaldehyde in PBS and stained with crystal violet to visualize plaques. Titers are expressed in plaque forming units per ml. Plaque reduction assays were essentially performed as plaque assays with the following modifications; each well of a 6-well cluster was infected with the same amount of virus (∼15–30 plaque forming units per well). Subsequently, the overlays that were applied contained increasing concentrations of 5-fluorouracil (Sigma), ranging from 0 to 250 μm.
RESULTS
Mapping of nsp10 Surface Residues Involved in the Interaction with nsp14
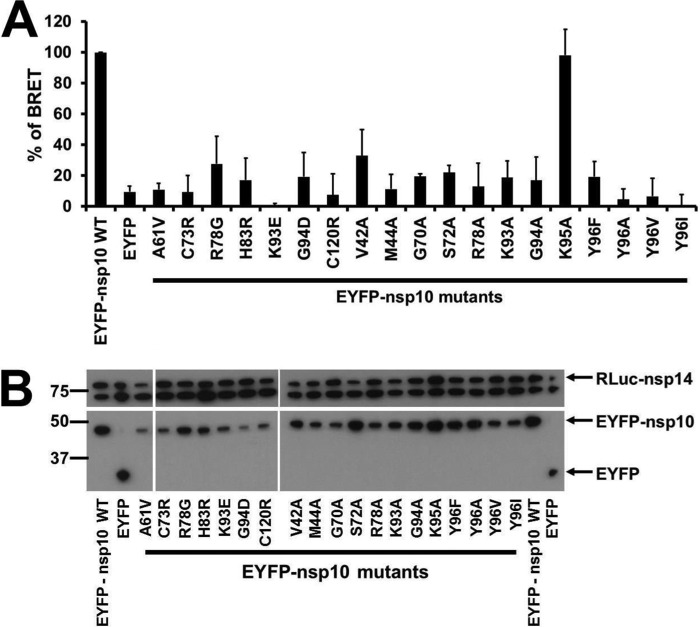
We recently showed that the interaction of nsp10 with nsp14 induces a >35-fold stimulation of nsp14 ExoN activity (47). This qualitative study pinpointed nsp10 residues involved in this interaction that are also important for the nsp10-nsp16 interaction regulating SARS-CoV 2′-O-MTase activity (40, 41, 47). To assess the overlap between these interaction surfaces, we first tested the impact of nsp10 mutations on the nsp10-nsp14 interaction using in cellulo BRET assays (for an overview of BRET strategy and set-up, see Ref. 58). Nsp10 mutants were fused to the C terminus of enhanced yellow fluorescent protein (EYFP), and nsp14 was fused to the C terminus of RLuc, all placed under the control of the cytomegalovirus promoter. BRET signal was measured in human HEK 293T cells at 48 h after plasmid transfection (Fig. 2A). As the expression level of EYFP-nsp10 mutants was quite variable, the amount of transfected plasmid DNA for each mutant was adjusted to achieve comparable protein expression levels (Fig. 2B). The nsp10 mutations selected to be tested in BRET assays mainly targeted surface residues described in Lugari et al. (41), and most of them were known to affect or prevent the nsp10-nsp16 interaction.
FIGURE 2.
BRET characterization of the interaction of wild-type and mutant nsp10 with nsp14 in mammalian cells. A, BRET interaction assays were performed in HEK 293T cells after co-transfection of plasmids expressing EYFP-nsp10 mutants with an RLuc-nsp14 expressing plasmids. The experiments were performed three times, and the relative interaction of each mutant is calculated compared with the interaction of wild-type nsp10 with nsp14 (which was taken to be 100%). The BRET signals were further normalized according to the fluorescence signal measured for nsp10-EYFP mutants compared with wild-type control. B, Western blot analysis confirming similar levels of protein expression for the two interaction partners. Levels of RLuc-nsp14 and EYFP-nsp10 were determined using anti-luciferase and anti-GFP antibodies, respectively. The anti-luciferase antibody also recognized an ∼70-kDa host cell protein, which could conveniently serve as a loading control for the cell lysates.
Almost all nsp10 mutants tested with BRET were impaired in their interaction with nsp14 (relative BRET values below 50%), suggesting their involvement in the nsp10-nsp14 interaction (Fig. 2). The K95A mutant, in which the mutation resides outside the interaction surface, yielded a BRET value comparable with that of wild-type nsp10 and could thus be used as a negative control for the BRET assay. Because most of the mutations tested concerned surface residues, it is assumed that they do not alter the three-dimensional structure of the protein as supported by the 1H,15N heteronuclear single quantum correlation NMR experiments of Lugari et al. (41). Three mutants, however, C73R, H83R, and C120R, target residues close to or involved in the formation of the two zinc fingers of nsp10 (Fig. 1). Replacement of those residues abrogated the interaction with nsp14 (BRET values between 10 and 20%), probably due to misfolding of nsp10. We, therefore, discarded those mutants for further studies. Finally, the Y96F mutant showed a 80% reduction of the nsp10-nsp14 interaction (BRET value of 21 ± 7.9%) as opposed to its stimulation of the nsp10-nsp16 interaction (41).
Reduced nsp10-nsp14 Interaction Correlates with Loss of nsp14 ExoN Activity
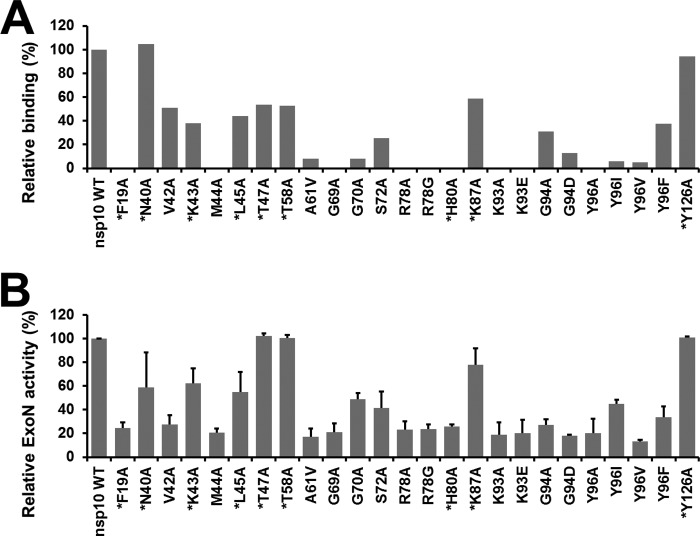
We next performed pulldown assays to measure the binding efficiency of each nsp10 mutant to nsp14 in vitro (Fig. 3A). Strep-tagged nsp10 and His6-tagged nsp14 were co-expressed in E. coli. Nsp10 was purified using Strep-Tactin beads, whereas nsp14 was co-purified in a complex that was subsequently quantitated by capillary electrophoresis. In the set of residues targeted by mutagenesis, we now included alanine mutants of residues Phe-19, Asn-40, Lys-43, Leu-45, Thr-47, Thr-58, Gly-69, His-80, and Lys-87, which surround the nsp10 surface as defined by BRET assay, and as a negative control residue Tyr-126, which is positioned on the opposite side of the nsp10 structure. For most of the nsp10 mutants tested (20 of 25), nsp14 binding was reduced by at least 50%, with approximately half of them displaying a complete loss of nsp14 co-purification. Mutations V42A, L45A, T47A, T58A, and K87A reduced the relative binding efficiency to ∼50% compared with wild-type nsp10. Finally, for two of the mutants, N40A and the negative control Y126A, the interaction with nsp14 was not or only slightly altered in vitro (Fig. 3A).
FIGURE 3.
Effect of nsp10 mutations on nsp10-nsp14 complex formation and on nsp14-ExoN activity in vitro. A, bar graph showing the relative nsp14 binding to each nsp10 mutant, as measured by in vitro pulldown assays. Nsp10 was purified by affinity chromatography and analyzed using capillary electrophoresis. The amount of nsp14 interacting with nsp10 was then quantified and normalized using nsp10. The binding activities were compared with the interaction of wild-type nsp10 with nsp14, which was arbitrarily set to 100%. B, relative nsp14 ExoN activities in the presence of a panel of nsp10 mutants. The ExoN activity obtained in the presence of wild-type nsp10 was arbitrarily set to 100%. Each experiment was repeated two times independently. Residues surrounding the nsp10 surface (as defined by BRET assay) that were newly included at this stage of the study are marked with an asterisk.
We next studied the functional importance of the residues described above for stimulation of the nsp14 ExoN activity. The capability of mutant nsp10 proteins to modulate the nsp14 ExoN activity was assayed by measuring the hydrolysis of radiolabeled RNA oligonucleotide H4 by (mutant) nsp10 and nsp14. As described previously, this RNA substrate forms a double-stranded RNA structure that can be readily degraded by the nsp10/nsp14 complex (47). Most nsp10 mutants displaying a reduced affinity for nsp14, as judged by BRET analysis and the in vitro binding assay, showed reduced stimulation of the nsp14 ExoN activity compared with wild-type nsp10 (Fig. 3B).
The 15 BRET-identified mutations hampering the nsp10-nsp14 interaction caused a reduction in both in vitro nsp14 binding and ExoN activity (≤50% in both assays). The mutagenesis to alanine of residues Thr-47, Thr-58, and Lys-87, which surround the nsp10 surface as defined by the BRET study, resulted in a loss of at least 40% of the binding capacity but did not significantly affect nsp14 ExoN activation. Several mutants abrogated the nsp10-nsp14 interaction (F19A, M44A, G69A, R78A, R78G, H80A, K93A, K93E, and Y96A) and resulted in a relative nsp14 ExoN activity below 30%, suggesting an important role for these residues in nsp14 recognition. Two other mutants, K43A and L45A, showing a relative nsp14 binding of ∼40%, affected nsp14 ExoN activity less dramatically (ExoN activity of 62 ± 13 and 55 ± 17% respectively), suggesting a smaller contribution to the nsp10-nsp14 interaction. Finally, nsp10 mutant Y126A, used as negative control, does not seem to be involved in the nsp10-nsp14 interaction, as nsp14 ExoN activity and in vitro binding values were close to 100%. We also confirmed that inactivation of the nsp14 ExoN active site (nsp14 catalytic residues Asp-90 and Glu-92 mutated to alanine), used as negative control, resulted in a complete loss of exoribonuclease activity in this assay (47).
Nsp14 and nsp16 Bind to the Same Core Interaction Surface on nsp10
To interpret our results, we mapped the residues found to be important by BRET assay by in vitro interaction analysis and by ExoN activity assays on the three-dimensional structure of nsp10 (PDB code 2FYG (37)) (Fig. 4, A–C, respectively). The three methods clearly define a common nsp10 surface for the interaction with nsp14. Interestingly, it has previously been shown that (part of) this same nsp10 surface is also involved in the interaction with nsp16 and that this surface is well conserved among coronaviruses (41).
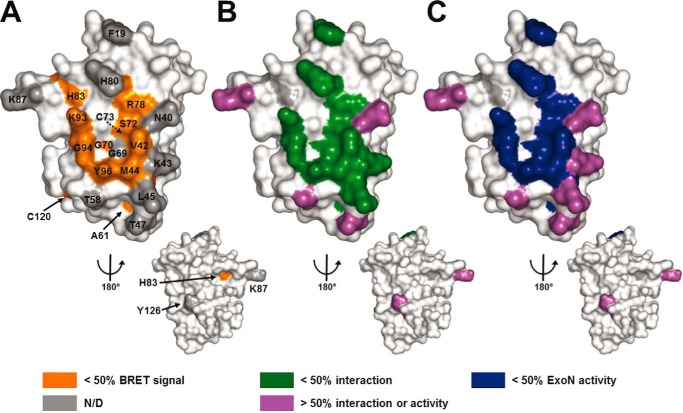
FIGURE 4.
Three-dimensional structure of nsp10 highlighting residues involved in the interaction with nsp14. SARS-CoV nsp10 (PDB code 2FYG (37)) is depicted in white as a surface representation. A, residues that were found to be involved in interaction between nsp10 and nsp14 by the in vivo BRET assay (>50% effect) are colored in orange. B, residues that were found to be involved in the nsp10-nsp14 interaction according to the in vitro binding capacity (pulldown) assays (>50% effect) are colored in green. C, residues that were found to be involved in the nsp10 and nsp14 interaction on the basis of ExoN activity (>50% reduction) are colored in dark blue. Residues that could be mutated without significantly altering the nsp10-nsp14 interaction or the ExoN activity (<50% effect) are depicted in purple. Residues not tested in BRET experiments compared with other assays are displayed in gray (N/D). All figures were generated using PyMOL.
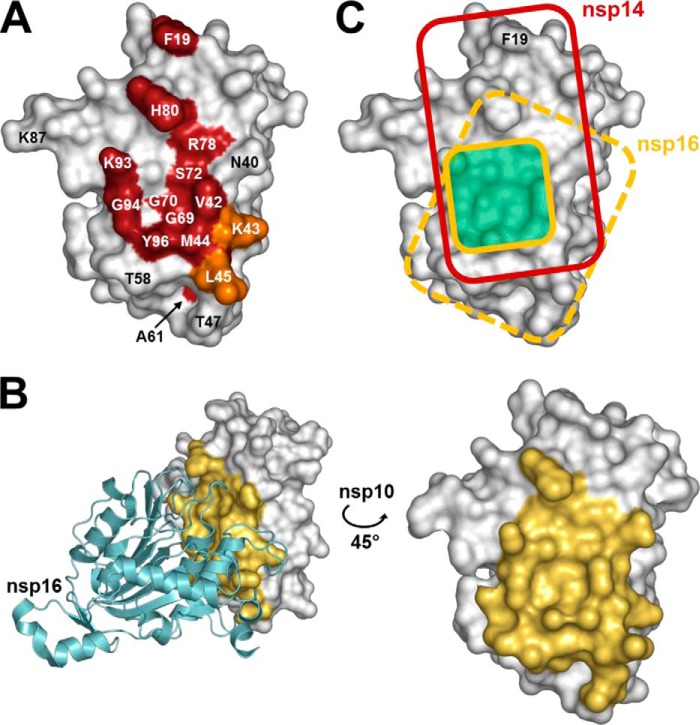
To assess the degree of overlap between nsp10 interaction surface with nsp14 and nsp16, we first mapped the residues involved in the nsp10-nsp14 interaction identified above to the three-dimensional structure of nsp10 (Fig. 5A). Using the structure of the nsp10-nsp16 complex (PDB code 2XYQ (40)), we also mapped the nsp10 residues involved in the interaction with nsp16. For this purpose, we highlighted all nsp10 residues within a 5 Å radius of nsp16 (Fig. 5B). The areas corresponding to the nsp14 and nsp16 binding surfaces were then marked on the same nsp10 structure model (Fig. 5C). This figure distinctly highlights the fact that nsp10 interaction surfaces have a substantial overlap. The surface that interacts with nsp14 seems to be more extended than the one involved in the interaction with nsp16, as residue Phe-19, that is not involved in the nsp10-nsp16 interaction, plays a critical role in the nsp10-nsp14 interaction. In the nsp10 structure, as depicted in Fig. 5C, the nsp14 interaction surface covers most of the nsp16 interaction surface and extends toward the upper part of nsp10. Table 1 summarizes the data obtained for the nsp10-nsp14 interaction compared with the data from the previously described nsp10-nsp16 interaction analysis (41).
FIGURE 5.
Comparison of the nsp14 and nsp16 interaction domains on the nsp10 surface. A, schematic representation of nsp10 residues that are engaged in the nsp10-nsp14 interaction. SARS-CoV nsp10 (PDB code 2FYG (37)) is shown as a white surface representation. Residues that significantly affect the nsp10-nsp14 interaction when mutated are colored in red (>50% decrease of BRET values, binding affinity, and ExoN activity). Residues Lys-43 and Leu-45, which impair the nsp10-nsp14 interaction with a smaller effect when mutated (both binding affinity and ExoN activity ∼50% of wild type), are shown in orange. B, representation of nsp10 residues involved in the nsp10-nsp16 interaction in the structure of the complex (PDB code 2XYQ (40)). Nsp16 is shown as a schematic representation, colored in cyan. Nsp10 residues that are present within a 5 Å radius of nsp16 are depicted in yellow. C, schematic representation of nsp10 functional interacting surface with nsp14 (red rectangle) and nsp16 (dark yellow square, based on Lugari et al. (41)). The structural nsp10-nsp16 interaction surface is depicted as a dashed yellow rectangle on nsp10. The overlapping functional interaction surface is depicted in pale green. All figures were generated using PyMOL.
TABLE 1.
Effects of a variety of nsp10 surface mutations on the nsp10-nsp14 and nsp10-nsp16 interactions and on ExoN (nsp14) and 2′-O-MTase (nsp16) enzyme activities
Mutations tested in reverse genetics experiments are shown as bold underlined characters. ND, not defined.
| nsp10 mutants | nsp14 |
nsp16a |
||||
|---|---|---|---|---|---|---|
| % BRET | % Interaction | % Activity ExoN | % BRET | % Interaction | % Activity 2′-O-MTase | |
| WT | 100 | 100 | 100 | 100 | 100 | 100 |
| F19Ab | ND | 0 | 24 ± 4.9 | ND | 108 | 92 ± 5.3 |
| N40A | ND | 105 | 59 ± 29.4 | ND | 64 | ND |
| V42A | 33 ± 17 | 50 | 28 ± 8 | 12 ± 2.3 | 1 | 1 ± 0.2 |
| K43A | ND | 38 | 62 ± 12.6 | ND | 98 | 84 ± 13.6 |
| M44A | 11 ± 9.8 | 0 | 21 ± 3.7 | 8 ± 5.1 | 0 | 0 ± 0.1 |
| L45A | ND | 44 | 55 ± 17 | ND | 6 | ND |
| T47A | ND | 53 | 102 ± 2,1 | ND | 66 | ND |
| T58A | ND | 52 | 100 ± 2,5 | ND | 93 | ND |
| A61V | 11 ± 4.2 | 8 | 17 ± 7 | 3 ± 5 | ND | ND |
| G69A | ND | 0 | 21 ± 7.1 | ND | 92 | ND |
| G70A | 19 ± 1.9 | 8 | 49 ± 5 | 31 ± 7.4 | 50 | 32 ± 0.8 |
| S72A | 22 ± 4.6 | 26 | 42 ± 13.6 | 60 ± 8.6 | 6 | 22 ± 1.1 |
| C73R | 9 ± 10.6 | ND | ND | 2 ± 0.6 | ND | ND |
| R78A | 13 ± 15.2 | 0 | 23 ± 6.8 | 9 ± 4.5 | 10 | 2 ± 0.1 |
| R78G | 28 ± 18 | 0 | 23 ± 4.1 | 35 ± 1.8 | 6 | 9 ± 0.3 |
| H80Ab | ND | 0 | 26 ± 1.8 | ND | 78 | 60 ± 3.1 |
| H83R | 17 ± 14.3 | ND | ND | 13 ± 11.3 | ND | ND |
| K87A | ND | 59 | 78 ± 14.1 | ND | 98 | ND |
| K93A | 19 ± 10.6 | 0 | 19 ± 10.4 | 35 ± 2.2 | 54 | 9 ± 1.2 |
| K93E | 0 ± 1.8 | 0 | 20 ± 11 | 7 ± 5.8 | 16 | 0 ± 0 |
| G94A | 17 ± 15.1 | 31 | 27 ± 4.7 | 59 ± 2.4 | 80 | 85 ± 1.2 |
| G94D | 19 ± 15.8 | 13 | 18 ± 0.8 | 16 ± 4.9 | 7 | 0 ± 0.1 |
| K95A | 98 ± 16.8 | ND | ND | 80 ± 3.9 | 83 | 71 ± 2.6 |
| Y96Ab | 5 ± 6.8 | 0 | 20 ± 12 | 30 ± 6.2 | 6 | 15 ± 0.6 |
| Y96I | 1 ± 7.9 | 6 | 45 ± 3.6 | 12 ± 2.3 | 3 | 4 ± 0.4 |
| Y96V | 7 ± 11.9 | 5 | 13 ± 1.6 | 20 ± 3.4 | 0 | 5 ± 0.1 |
| Y96F | 19 ± 10.6 | 38 | 33 ± 9.4 | 123 ± 18 | 124 | 163 ± 10 |
| C120R | 8 ± 13.5 | ND | ND | 49 ± 2.7 | ND | ND |
| Y126A | ND | 94 | 101 ± 0.9 | ND | 99 | ND |
| EYFP | 9 ± 3.8 | 14 ± 2.2 | ||||
a Taken from Lugari et al. (41) except for residues Phe-19, Lys-43, and His-80, which were added in this study.
b Mutations prohibiting virus replication.
Nsp10 Residues Involved in the Interaction with nsp14 Are Essential for SARS-CoV Replication
A selection of nsp10 mutations described above was reverse-engineered into the SARS-CoV genome using a BAC-based full-length cDNA clone from which recombinant virus can be launched (55). We chose to include mutations affecting the nsp10-nsp14 interaction only (F19A, K43A, and H80A) or targeting both the nsp10-nsp14 and nsp10-nsp16 interactions (M44A and Y96A). The Y96F mutant was also included, because it was shown to have a stronger in vitro interaction with nsp16 compared with wild-type nsp10. As additional phenotypic controls, we constructed mutants with inactivated nsp14 ExoN (D90A/E92A) (50) or nsp16 2′-O-MTase (D130A) (34) activities. In addition, an nsp10-nsp16 interaction knock-out mutant was made in which nsp16 residue Met-247, which was previously shown to interact with nsp10 in the crystal structure of the nsp10-nsp16 complex (40), was substituted by alanine.
Mutant viruses were launched by electroporation of in vitro transcribed full-length RNA into BHK-Tet-SARS-N cells (53), which express the viral nucleocapsid protein. These cells can be transfected efficiently and produce progeny virions but do not support further viral spread because they lack the ACE2 receptor used by the virus. For this reason transfected cells were mixed with (SARS-CoV-susceptible) Vero-E6 cells, and the replication of mutant viruses was studied at different time points using immunofluorescence microscopy and progeny virus titration by plaque assays. For each virus mutant, two independent mutant full-length cDNA clones were generated and found to yield identical results. The reverse genetics data are summarized in Table 2, and plaque phenotypes of replication-competent mutants are shown in Fig. 6.
TABLE 2.
Reverse genetics phenotypes of SARS-CoV mutants
| Nsp | Mutant | IFAa | Virus titerb | Plaque phenotype | Summary |
|---|---|---|---|---|---|
| WT | + | 8 × 107 | Wild type | Wild-type control | |
| nsp10 | F19A | − | Non-viable | ||
| K43A | + | 5 × 106 | Intermediate | Crippled | |
| M44A | − | Non-replicating in one experiment; late pseudo-reversion to M44V in one experiment | |||
| H80A | − | Non-viable | |||
| Y96A | − | Non-viable | |||
| Y96F | + | 3.108 | Wild type | Similar to wild-type control | |
| nsp14 | D90A/E92A | + | 3 × 104 | Small | ExoN knockout mutant. Crippled |
| nsp16 | D130A | + | 2 × 106 | Small | 2′-O-MTase knockout mutant. Crippled |
| M247A | + | 2 × 105 | Small | No interaction with nsp10. Crippled |
a Immunofluorescence assay, t = 10–72 hpt.
b t = 24 h post transfection, plaque forming units/ml.
FIGURE 6.
Plaque morphology of viable SARS-CoV nsp10, nsp14, and nsp16 mutants. Small-plaque phenotypes were observed for mutants nsp14-D90A/E92A, nsp16-D130A, and nsp16-M247A, an intermediate plaque size for mutant nsp10-K43A, whereas the plaque size of mutant nsp10-Y96F was similar to that of the wild-type control.
Our in vivo data fully support the notion that residues in nsp10 that are involved in the interaction with nsp14 are important for SARS-CoV replication in cell culture: three of the nsp10 mutations that abolished this interaction in vitro (F19A, H80A, and Y96A) resulted in a non-viable virus phenotype, whereas a fourth mutation of this type (M44A) pseudo-reverted at a late time point in one of two experiments (the original M44A mutation had been changed to valine (M44V) by 72 h post transfection). The K43A mutation, which decreased but did not block the nsp10-nsp14 interaction and nsp14 ExoN activity in vitro, yielded a crippled virus with ∼15-fold reduced titers compared with the wild-type control 24 h post transfection. Plaques produced by this virus were slightly smaller than wild-type plaques and more heterogeneous in size compared with the wild-type virus or other mutants (Fig. 6).
The non-viable and late-reverting phenotypes of several of the nsp10 mutants cannot be attributed solely to a reduction or lack of ExoN activity as measured in vitro (Table 1), as the ExoN active site mutant nsp14-D90A/E92A, despite being crippled, was clearly replication competent (Fig. 6; and also previously reported by Eckerle et al. (50)). The interaction between nsp10 and nsp16 apparently plays a less critical role in basic CoV replication, as also illustrated by the replication-competent phenotype of mutant nsp16-M247A, which displayed a small-plaque phenotype and gave an ∼2-log reduced virus titer at 24 h post transfection. Its plaque phenotype is similar to that of the 2′-O-MTase active site mutant nsp16-D130A, although the virus titers of the latter mutant were only ∼1 log lower. The replication-competent but crippled phenotype of the 2′-O-MTase active site mutant is comparable with that of the corresponding mutants previously described for SARS-CoV, MHV, and HCoV-229E (44, 59). It has been postulated that the conversion of cap 0 to cap 1 on viral mRNAs plays an important role in escape from innate immune recognition but is not essential for viral replication per se. Mutation nsp10-Y96F, which decreased the interaction of nsp10 with nsp14 and increased its interaction with nsp16 in vitro, gave rise to virus titers and plaque sizes similar to those of the wild-type control. These results strongly suggest that the replication defect of the non-viable nsp10 mutants is not caused by a reduction of the nsp10-based stimulation of either the nsp14 ExoN activity or the nsp16 2′O MTase function. Apparently, the nsp10 core domain plays a pleiotropic role during virus replication, as also previously deduced from data obtained in the MHV model (63).
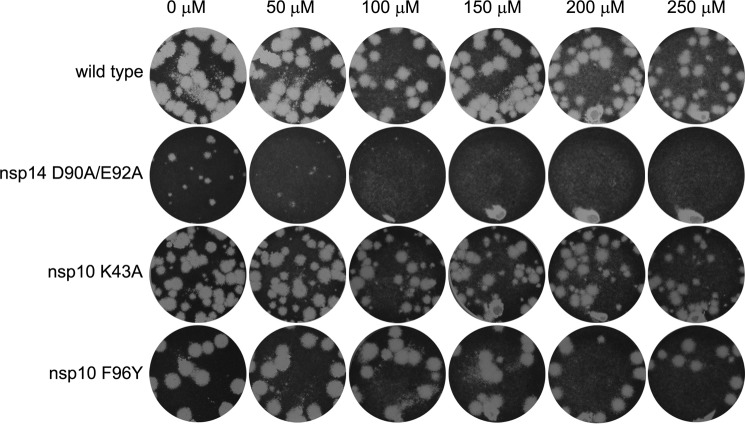
Given the fact that they compromised both the nsp10-nsp14 interaction and the nsp10-mediated stimulation of the nsp14-ExoN activity in vitro, we investigated the possibility that the nsp10 mutations K43A and Y96F increased the SARS-CoV mutation frequency (“mutator phenotype”) in a manner similar to what has been reported for the nsp14-D90A/E92A ExoN knock-out mutant (50). To test whether the K43A and Y96F mutants also display such a phenotype, plaque reduction assays were performed in the presence of increasing concentrations of the mutagen 5-fluorouracil (5FU) (60). As previously reported (60), increased 5FU sensitivity was readily observed, even at the lowest dose tested (50 μm), for the nsp14-D90A/E92A ExoN knock-out mutant, which was included as a positive control in this experiment (Fig. 7). However, nsp10 mutants K43A and Y96F were found to be equally insensitive to the drug as the wild-type virus. Only at the highest 5FU dose tested (250 μm), a slight reduction in plaque size and numbers was observed for both wild-type virus and nsp10 mutants (Fig. 7). This observation is inconsistent with the hypothesis that these nsp10 mutants would have a mutator phenotype, which should have markedly increased their sensitivity to a mutagen-like 5FU.
FIGURE 7.
Lack of 5′-fluorouracil sensitivity of SARS-CoV mutants nsp10-K43A and nsp10-Y96F suggests they do not exhibit a mutator phenotype. Plaque reduction assays were performed in the presence of increasing concentrations of 5-fluorouracil. Each well was infected with the same amount of virus, after which cell layers were overlaid with a semi-solid medium containing the indicated increasing concentrations of 5FU. Cell layers were incubated for 3 days, fixed, and stained to reveal plaque formation. Like the wild-type virus, nsp10 mutants K43A and Y96F were insensitive to 5FU up to a dose of 250 μm. On the other hand, plaque size and number for mutant nsp14-D90A/E92A (the ExoN knock-out mutant known to exhibit a mutator phenotype) were strongly reduced even at the lowest 5FU concentration tested.
DISCUSSION
SARS-CoV nsp14 ExoN activity was recently shown to be strongly stimulated by the presence of its interaction partner nsp10 (47). This feature is likely to be highly relevant for the unique RNA proofreading mechanism that is thought to allow coronaviruses to reliably replicate their long RNA genome (60). Accordingly, the replacement of ExoN catalytic residues creates a mutator phenotype that may in the long run threaten CoV genetic stability and survival (51, 61).
In this study we identified nsp10 residues involved in the interaction with nsp14. BRET methodology, considered as one of the most versatile techniques to study the dynamics of protein-protein interactions in living cells (62) as well as in vitro pulldown and enzymatic assays allowed the mapping of key residues involved in the formation of the nsp10-nsp14 complex and the nsp10-driven stimulation of the ExoN activity (Figs. 2 and 3). Interestingly, a large majority of the mutants for which the nsp10-nsp14 interaction was disrupted lost their nsp14 ExoN-activating potential, revealing the critical nature of this interaction for ExoN activity.
The nsp10 surface deduced to be involved in the nsp10-nsp14 interaction is conserved across all four genera of the Coronaviridae subfamily (Fig. 8) (41) and overlaps with the nsp10-nsp16 interaction surface previously described (Fig. 5; Refs. 40 and 41). Nsp10 molecules may thus act as a platform that recruits nsp14 or nsp16 to the replication-transcription complex to either boost nsp14 ExoN activity or to switch on nsp16 2′-O-MTase activity. Although the overlapping interaction surfaces suggest an exclusive interaction of nsp10 with either nsp14 or nsp16, the protein is produced at a much higher rate due to the fact that, unlike nsp14 and nsp16, it is encoded in ORF1a, just upstream of the ORF1a/1b ribosomal frameshift site (6, 19, 63). Even though little is known about the relative turnover of CoV nsps in general and although nsp10 may have additional binding partners that remain to be identified, it is fair to assume that nsp10 levels in infected cells are substantially higher than those of nsp14 and nsp16, implying that the latter two proteins would not necessarily be competing to bind nsp10 (16). Thus, nsp10-nsp14 and nsp10-nsp16 complexes may coexist, possibly as part of larger nsp assemblies engaged in different steps of viral RNA synthesis.
FIGURE 8.
Conservation of the nsp10 sequence across the Coronaviridae subfamily. Alignment of nsp10 sequences from a representative set of Coronavirinae subfamily viruses including members of each of the four genera (Alpha-, Beta-, Gamma-, and Deltacoronavirus). Residues that are conserved in all sequences and whose replacement is lethal to SARS-CoV and MHV (Phe-19, Met-44, Gly-69, and Ser-72) are boxed in blue and indicated by triangles. Residues that are conserved in >80% of the sequences and whose replacement is lethal to SARS-CoV (His-80 and Tyr-96) are labeled in blue and indicated by asterisks. Sequences were aligned using the ESPript program (67). National Center for Biotechnology Information (NCBI) accession numbers for replicase polyprotein sequences including nsp10 are: SARS-CoV, AY345988; MERS-CoV, JX869059; HCoV-HKU1, AY884001; MHV, AY700211; BtCoV-HKU5, bat Coronavirus HKU5–1 (EF065509); HCoV-229E, NC_002645; HCoV-NL63, DQ445911; FcoV, feline Coronavirus (DQ010921); IBV, avian infectious bronchitis virus (NC_001451); BWCoV-SW1, beluga whale Coronavirus SW1 (EU111742); ACoV-HKU11, bulbul Coronavirus HKU11-796 (FJ376620); ACoV-HKU13, munia Coronavirus HKU13-3514 (NC_011550); ACoV-HKU12, thrush Coronavirus HKU12-600 (NC_011549).
One nsp10 residue, Tyr-96, is of particular interest. The aromatic nature of Tyr-96 plays a crucial role in the nsp10-nsp16 interaction and in the activation of the nsp16 2′-O-MTase activity (41). This residue is specific for SARS-CoV nsp10 and is a phenylalanine in most other Coronavirus homologues (Fig. 7). As the nsp10 Y96F mutation resulted in an enhanced binding to nsp16 correlating with an increased 2′-O-MTase activity, residue Tyr-96 was hypothesized to fine-tune the strength of the interaction between nsp10 and nsp16 (41). However, the underlying mechanism could not be clearly defined at the molecular level, not even using the structure of the nsp10-nsp16 complex (40). In this study we show that Tyr-96 is involved in the interaction with nsp14 and that the nsp10 Y96F mutation interferes with nsp14 binding and decreases ExoN activity in vitro. This may explain why a tyrosine residue has been selected in the course of SARS-CoV evolution, whereas a phenylalanine has been preferred at this position in other CoVs to balance the relative affinities of nsp10 for nsp14 and nsp16. Nevertheless, in reverse genetics experiments the phenotype of the nsp10 Y96F mutant virus was found to be similar to that of the wild-type virus, indicating that the presence of a phenylalanine residue at this position does not strongly affect viral replication in the type of short term cell culture experiments included in this study.
Because complete inactivation of the CoV nsp14 ExoN function yields a crippled but replicating virus (Refs. 50 and 51 and confirmed in this study), it was highly surprising that several nsp10 mutations that disrupt the nsp10-nsp14 interaction proved to be lethal. Clearly, the non-viable phenotype of these mutants cannot be explained by the loss of nsp14 ExoN activity alone, which raises important questions about the functionality of nsp10 or the nsp10-nsp14 complex in CoV replication. Nsp14 is a bifunctional enzyme, and its interaction with nsp10 may also influence its critical (N7-guanine)-methyltransferase activity, although no positive or negative influence of nsp10 on this activity was observed in in vitro assays (34). Alternatively, nsp10 or the nsp10-nsp14 complex may have additional yet unknown functions that are vital for CoV replication. Previously, based on an MHV study, an indirect role for nsp10 in proteolytic processing of replicase polyproteins and viral RNA synthesis was proposed (64, 65). It is conceivable that the non-viable phenotype of several SARS-CoV nsp10 mutants in the present study could be attributed to critically influencing one or more of these as yet poorly defined additional nsp10 functions.
Many of the SARS-CoV nsp10 residues found to affect the nsp10-nsp14 interaction and viral replication (Gly-69, Gly-70, Ser-72, His-80, and Tyr-96) correspond to residues that localize within the core of nsp10, as previously defined by Donaldson et al. (64) for MHV nsp10. This core does not appear to tolerate mutations (64), and the non-viable phenotype of some of the MHV mutants tested by Donaldson et al. (64) again seems to reflect the importance of the nsp10-nsp14 interaction surface in coronaviruses. For instance, conserved residues Gly-69, Gly-70, and Ser-72 were targeted for MHV and were all found to be critical for virus replication, in line with the data from the in vitro interaction studies presented here for SARS-CoV nsp10. Still, the non-viable phenotypes could also be explained by alternative scenarios, like the nsp10 surface being involved in another function that is critical for coronaviruses replication. The available data lead us to propose that specific “hot spot” residues (Phe-19, Met-44, Gly-69, Ser-72, His-80, and Tyr-96) within and around the nsp10 core can be targeted to disrupt nsp10-nsp14 interactions and inhibit nsp14 ExoN activity and/or other nsp10 functions, potentially interfering with CoV replication. Strikingly, those residues are highly conserved across coronaviruses, including the recently emerged and potentially life-threatening MERS-CoV (Fig. 8). Considering its importance for virus replication, highlighted by this and other studies, nsp10 represents an attractive target for anti-Coronavirus drug discovery. Specific molecules or peptides targeting the described nsp10 surface and inhibiting the interaction with nsp14 and nsp16 could be developed to reduce or prevent Coronavirus replication (66). Because nsp10 is well conserved among coronaviruses, such compounds could then be used for treatment or in a prophylactic approach to prevent MERS-CoV infection, possible outbursts of SARS-CoV, or any other newly emerging lethal Coronavirus.
Acknowledgments
We thank Barbara Selisko and Bruno Coutard for helpful discussions, Françoise Debart for RNA synthesis, and Ying Fang for kindly providing a monoclonal antibody recognizing the SARS-CoV N protein.
This work was supported by French National Research Agency ANR-08-MIEN-032, ANR 12 BSV3 07-1, -2, and -3, Fondation pour la Recherche Médicale (Programme Équipe FRM), by the European Union Seventh Framework Programme FP7/2007-2013 (Project SILVER (small inhibitor leads against emerging RNA viruses) Grant 260644), Infectiopole Sud, and by the Netherlands Organization for Scientific Research (NWO; TOP-GO Grant 700.10.352).
- CoV
- coronaviruses
- SARS
- severe acute respiratory syndrome
- nsp
- non-structural proteins
- MERS
- Middle East respiratory syndrome
- HCoV
- human CoV
- 2′-O-MTase
- (nucleoside-2′-O-)-methyltransferase
- ExoN
- exoribonuclease
- EYFP
- enhanced EFP
- RLuc
- Renilla luciferase
- MHV
- mouse hepatitis virus
- BRET
- bioluminescence resonance energy transfer
- 5FU
- 5-fluorouracil.
REFERENCES
- 1. Gorbalenya A. E., Enjuanes L., Ziebuhr J., Snijder E. J. (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res. 117, 17–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nga P. T., Parquet Mdel C., Lauber C., Parida M., Nabeshima T., Yu F., Thuy N. T., Inoue S., Ito T., Okamoto K., Ichinose A., Snijder E. J., Morita K., Gorbalenya A. E. (2011) Discovery of the first insect Nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog. 7, e1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Peiris J. S., Guan Y., Yuen K. Y. (2004) Severe acute respiratory syndrome. Nat. Med. 10, S88–S97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Smith I., Wang L. F. (2013) Bats and their virome: an important source of emerging viruses capable of infecting humans. Curr. Opin. Virol. 3, 84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Corman V. M., Kallies R., Philipps H., Göpner G., Müller M. A., Eckerle I., Brünink S., Drosten C., Drexler J. F. (2014) Characterization of a novel Betacoronavirus related to middle East respiratory syndrome Coronavirus in European hedgehogs. J. Virol. 88, 717–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Snijder E. J., Bredenbeek P. J., Dobbe J. C., Thiel V., Ziebuhr J., Poon L. L., Guan Y., Rozanov M., Spaan W. J., Gorbalenya A. E. (2003) Unique and conserved features of genome and proteome of SARS-Coronavirus, an early split-off from the Coronavirus group 2 lineage. J. Mol. Biol. 331, 991–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Boheemen S., de Graaf M., Lauber C., Bestebroer T. M., Raj V. S., Zaki A. M., Osterhaus A. D., Haagmans B. L., Gorbalenya A. E., Snijder E. J., Fouchier R. A. (2012) Genomic characterization of a newly discovered Coronavirus associated with acute respiratory distress syndrome in humans. MBio 3, e00473–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zaki A. M., van Boheemen S., Bestebroer T. M., Osterhaus A. D., Fouchier R. A. (2012) Isolation of a novel Coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 367, 1814–1820 [DOI] [PubMed] [Google Scholar]
- 9. Stadler K., Masignani V., Eickmann M., Becker S., Abrignani S., Klenk H. D., Rappuoli R. (2003) SARS: beginning to understand a new virus. Nat. Rev. Microbiol. 1, 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rota P. A., Oberste M. S., Monroe S. S., Nix W. A., Campagnoli R., Icenogle J. P., Peñaranda S., Bankamp B., Maher K., Chen M. H., Tong S., Tamin A., Lowe L., Frace M., DeRisi J. L., Chen Q., Wang D., Erdman D. D., Peret T. C., Burns C., Ksiazek T. G., Rollin P. E., Sanchez A., Liffick S., Holloway B., Limor J., McCaustland K., Olsen-Rasmussen M., Fouchier R., Günther S., Osterhaus A. D., Drosten C., Pallansch M. A., Anderson L. J., Bellini W. J. (2003) Characterization of a novel Coronavirus associated with severe acute respiratory syndrome. Science 300, 1394–1399 [DOI] [PubMed] [Google Scholar]
- 11. de Groot R. J., Baker S. C., Baric R. S., Brown C. S., Drosten C., Enjuanes L., Fouchier R. A., Galiano M., Gorbalenya A. E., Memish Z. A., Perlman S., Poon L. L., Snijder E. J., Stephens G. M., Woo P. C., Zaki A. M., Zambon M., Ziebuhr J. (2013) Middle East respiratory syndrome Coronavirus (MERS-CoV): announcement of the Coronavirus Study Group. J. Virol. 87, 7790–7792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bermingham A., Chand M. A., Brown C. S., Aarons E., Tong C., Langrish C., Hoschler K., Brown K., Galiano M., Myers R., Pebody R. G., Green H. K., Boddington N. L., Gopal R., Price N., Newsholme W., Drosten C., Fouchier R. A., Zambon M. (2012) Severe respiratory illness caused by a novel Coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro. Surveill. 17, 20290. [PubMed] [Google Scholar]
- 13. Masters P. S. (2006) The molecular biology of coronaviruses. Adv. Virus. Res. 66, 193–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Denison M. R., Graham R. L., Donaldson E. F., Eckerle L. D., Baric R. S. (2011) Coronaviruses: an RNA proofreading machine regulates replication fidelity and diversity. RNA Biol. 8, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brierley I., Boursnell M. E., Binns M. M., Bilimoria B., Blok V. C., Brown T. D., Inglis S. C. (1987) An efficient ribosomal frame-shifting signal in the polymerase-encoding region of the Coronavirus IBV. EMBO J. 6, 3779–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Brierley I., Digard P., Inglis S. C. (1989) Characterization of an efficient Coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57, 537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Plant E. P., Sims A. C., Baric R. S., Dinman J. D., Taylor D. R. (2013) Altering SARS Coronavirus frameshift efficiency affects genomic and subgenomic RNA production. Viruses 5, 279–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pasternak A. O., Spaan W. J., Snijder E. J. (2006) Nidovirus transcription: how to make sense? J. Gen. Virol. 87, 1403–1421 [DOI] [PubMed] [Google Scholar]
- 19. Sawicki S. G., Sawicki D. L., Siddell S. G. (2007) A contemporary view of Coronavirus transcription. J. Virol. 81, 20–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hussain S., Pan J., Chen Y., Yang Y., Xu J., Peng Y., Wu Y., Li Z., Zhu Y., Tien P., Guo D. (2005) Identification of novel subgenomic RNAs and noncanonical transcription initiation signals of severe acute respiratory syndrome Coronavirus. J. Virol. 79, 5288–5295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. van Hemert M. J., van den Worm S. H., Knoops K., Mommaas A. M., Gorbalenya A. E., Snijder E. J. (2008) SARS-Coronavirus replication/transcription complexes are membrane-protected and need a host factor for activity in vitro. PLoS Pathog. 4, e1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Knoops K., Kikkert M., Worm S. H., Zevenhoven-Dobbe J. C., van der Meer Y., Koster A. J., Mommaas A. M., Snijder E. J. (2008) SARS-Coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 6, e226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Subissi L., Imbert I., Ferron F., Collet A., Coutard B., Decroly E., Canard B. (2014) SARS-CoV ORF1b-encoded nonstructural proteins 12–16: replicative enzymes as antiviral targets. Antiviral Res. 101, 122–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gu M., Lima C. D. (2005) Processing the message: structural insights into capping and decapping mRNA. Curr. Opin Struct. Biol. 15, 99–106 [DOI] [PubMed] [Google Scholar]
- 25. Shuman S. (2001) Structure, mechanism, and evolution of the mRNA capping apparatus. Prog. Nucleic Acid Res. Mol. Biol. 66, 1–40 [DOI] [PubMed] [Google Scholar]
- 26. Shuman S. (2002) What messenger RNA capping tells us about eukaryotic evolution. Nat. Rev. Mol. Cell Biol. 3, 619–625 [DOI] [PubMed] [Google Scholar]
- 27. Yoneyama M., Fujita T. (2010) Recognition of viral nucleic acids in innate immunity. Rev. Med. Virol 20, 4–22 [DOI] [PubMed] [Google Scholar]
- 28. Shuman S. (2001) The mRNA capping apparatus as drug target and guide to eukaryotic phylogeny. Cold Spring Harb. Symp. Quant. Biol. 66, 301–312 [DOI] [PubMed] [Google Scholar]
- 29. Decroly E., Ferron F., Lescar J., Canard B. (2012) Conventional and unconventional mechanisms for capping viral mRNA. Nat. Rev. Microbiol. 10, 51–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lai M. M., Patton C. D., Stohlman S. A. (1982) Further characterization of mRNA's of mouse hepatitis virus: presence of common 5′-end nucleotides. J. Virol. 41, 557–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lai M. M., Stohlman S. A. (1981) Comparative analysis of RNA genomes of mouse hepatitis viruses. J. Virol. 38, 661–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. van Vliet A. L., Smits S. L., Rottier P. J., de Groot R. J. (2002) Discontinuous and non-discontinuous subgenomic RNA transcription in a Nidovirus. EMBO J. 21, 6571–6580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Y., Cai H., Pan J., Xiang N., Tien P., Ahola T., Guo D. (2009) Functional screen reveals SARS Coronavirus nonstructural protein nsp14 as a novel cap N7 methyltransferase. Proc. Natl. Acad. Sci. U.S.A. 106, 3484–3489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bouvet M., Debarnot C., Imbert I., Selisko B., Snijder E. J., Canard B., Decroly E. (2010) In vitro reconstitution of SARS-Coronavirus mRNA cap methylation. PLoS Pathog 6, e1000863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ivanov K. A., Ziebuhr J. (2004) Human Coronavirus 229E nonstructural protein 13: characterization of duplex-unwinding, nucleoside triphosphatase, and RNA 5′-triphosphatase activities. J. Virol. 78, 7833–7838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. von Grotthuss M., Wyrwicz L. S., Rychlewski L. (2003) mRNA cap-1 methyltransferase in the SARS genome. Cell 113, 701–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Joseph J. S., Saikatendu K. S., Subramanian V., Neuman B. W., Brooun A., Griffith M., Moy K., Yadav M. K., Velasquez J., Buchmeier M. J., Stevens R. C., Kuhn P. (2006) Crystal structure of nonstructural protein 10 from the severe acute respiratory syndrome Coronavirus reveals a novel fold with two zinc binding motifs. J. Virol. 80, 7894–7901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Su D., Lou Z., Sun F., Zhai Y., Yang H., Zhang R., Joachimiak A., Zhang X. C., Bartlam M., Rao Z. (2006) Dodecamer structure of severe acute respiratory syndrome Coronavirus nonstructural protein nsp10. J. Virol. 80, 7902–7908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sawicki S. G., Sawicki D. L., Younker D., Meyer Y., Thiel V., Stokes H., Siddell S. G. (2005) Functional and genetic analysis of Coronavirus replicase-transcriptase proteins. PLoS Pathog. 1, e39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. (2011) Crystal structure and functional analysis of the SARS-Coronavirus RNA cap 2′-O-methyltransferase nsp10/nsp16 complex. PLoS Pathog. 7, e1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lugari A., Betzi S., Decroly E., Bonnaud E., Hermant A., Guillemot J. C., Debarnot C., Borg J. P., Bouvet M., Canard B., Morelli X., Lécine P. (2010) Molecular mapping of the RNA cap 2′-O-methyltransferase activation interface between severe acute respiratory syndrome Coronavirus nsp10 and nsp16. J. Biol. Chem. 285, 33230–33241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li S. H., Dong H., Li X. F., Xie X., Zhao H., Deng Y. Q., Wang X. Y., Ye Q., Zhu S. Y., Wang H. J., Zhang B., Leng Q. B., Zuest R., Qin E. D., Qin C. F., Shi P. Y. (2013) Rational design of a Flavivirus vaccine through abolishing viral RNA 2′-O-methylation. J. Virol. 87, 5812–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Daffis S., Szretter K. J., Schriewer J., Li J., Youn S., Errett J., Lin T. Y., Schneller S., Zust R., Dong H., Thiel V., Sen G. C., Fensterl V., Klimstra W. B., Pierson T. C., Buller R. M., Gale M., Jr., Shi P. Y., Diamond M. S. (2010) 2′-O methylation of the viral mRNA cap evades host restriction by IFIT family members. Nature 468, 452–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Züst R., Cervantes-Barragan L., Habjan M., Maier R., Neuman B. W., Ziebuhr J., Szretter K. J., Baker S. C., Barchet W., Diamond M. S., Siddell S. G., Ludewig B., Thiel V. (2011) Ribose 2′-O-methylation provides a molecular signature for the distinction of self and non-self mRNA dependent on the RNA sensor Mda5. Nat. Immunol. 12, 137–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Minskaia E., Hertzig T., Gorbalenya A. E., Campanacci V., Cambillau C., Canard B., Ziebuhr J. (2006) Discovery of an RNA virus 3′ → 5′ exoribonuclease that is critically involved in Coronavirus RNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 5108–5113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y., Tao J., Sun Y., Wu A., Su C., Gao G., Cai H., Qiu S., Wu Y., Ahola T., Guo D. (2013) Structure-function analysis of SARS Coronavirus RNA cap guanine-N7 methyltransferase. J. Virol. 87, 6296–6305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bouvet M., Imbert I., Subissi L., Gluais L., Canard B., Decroly E. (2012) RNA 3′-end mismatch excision by the severe acute respiratory syndrome Coronavirus nonstructural protein nsp10/nsp14 exoribonuclease complex. Proc. Natl. Acad. Sci. U.S.A. 109, 9372–9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Imbert I., Snijder E. J., Dimitrova M., Guillemot J. C., Lécine P., Canard B. (2008) The SARS-Coronavirus PLnc domain of nsp3 as a replication/transcription scaffolding protein. Virus Res. 133, 136–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pan J., Peng X., Gao Y., Li Z., Lu X., Chen Y., Ishaq M., Liu D., Dediego M. L., Enjuanes L., Guo D. (2008) Genome-wide analysis of protein-protein interactions and involvement of viral proteins in SARS-CoV replication. PLoS ONE 3, e3299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Eckerle L. D., Becker M. M., Halpin R. A., Li K., Venter E., Lu X., Scherbakova S., Graham R. L., Baric R. S., Stockwell T. B., Spiro D. J., Denison M. R. (2010) Infidelity of SARS-CoV Nsp14-exonuclease mutant virus replication is revealed by complete genome sequencing. PLoS Pathog. 6, e1000896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Eckerle L. D., Lu X., Sperry S. M., Choi L., Denison M. R. (2007) High fidelity of murine hepatitis virus replication is decreased in nsp14 exoribonuclease mutants. J. Virol. 81, 12135–12144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Debarnot C., Imbert I., Ferron F., Gluais L., Varlet I., Papageorgiou N., Bouvet M., Lescar J., Decroly E., Canard B. (2011) Crystallization and diffraction analysis of the SARS Coronavirus nsp10-nsp16 complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 67, 404–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang G. H., Dividson A., Lin L., Wilson M., Siddell S. G., Zhu Q. Y. (2010) Establishment of the eukaryotic cell lines for inducible control of SARS-CoV nucleocapsid gene expression. Virol. Sin. 25, 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tischer B. K., Smith G. A., Osterrieder N. (2010) En passant mutagenesis: a two step markerless red recombination system. Methods Mol. Biol. 634, 421–430 [DOI] [PubMed] [Google Scholar]
- 55. Pfefferle S., Krähling V., Ditt V., Grywna K., Mühlberger E., Drosten C. (2009) Reverse genetic characterization of the natural genomic deletion in SARS-Coronavirus strain Frankfurt-1 open reading frame 7b reveals an attenuating function of the 7b protein in vitro and in vivo. Virol J. 6, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. van der Meer Y., Snijder E. J., Dobbe J. C., Schleich S., Denison M. R., Spaan W. J., Locker J. K. (1999) Localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J. Virol. 73, 7641–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Fang Y., Pekosz A., Haynes L., Nelson E. A., Rowland R. R. (2006) Production and characterization of monoclonal antibodies against the nucleocapsid protein of SARS-CoV. Adv. Exp. Med. Biol. 581, 153–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Xu Y., Piston D. W., Johnson C. H. (1999) A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc. Natl. Acad. Sci. U.S.A. 96, 151–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Menachery V. D., Yount B. L., Jr., Josset L., Gralinski L. E., Scobey T., Agnihothram S., Katze M. G., Baric R. S. (2014) Attenuation and restoration of SARS-CoV mutant lacking 2′ O methyltransferase activity. J. Virol. 88, 4251–4264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Smith E. C., Blanc H., Vignuzzi M., Denison M. R. (2013) Coronaviruses lacking exoribonuclease activity are susceptible to lethal mutagenesis: evidence for proofreading and potential therapeutics. PLoS Pathog. 9, e1003565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Graham R. L., Becker M. M., Eckerle L. D., Bolles M., Denison M. R., Baric R. S. (2012) A live, impaired-fidelity Coronavirus vaccine protects in an aged, immunocompromised mouse model of lethal disease. Nat. Med. 18, 1820–1826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Bacart J., Corbel C., Jockers R., Bach S., Couturier C. (2008) The BRET technology and its application to screening assays. Biotechnol. J. 3, 311–324 [DOI] [PubMed] [Google Scholar]
- 63. Snijder E. J., van der Meer Y., Zevenhoven-Dobbe J., Onderwater J. J., van der Meulen J., Koerten H. K., Mommaas A. M. (2006) Ultrastructure and origin of membrane vesicles associated with the severe acute respiratory syndrome Coronavirus replication complex. J. Virol. 80, 5927–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Donaldson E. F., Sims A. C., Graham R. L., Denison M. R., Baric R. S. (2007) Murine hepatitis virus replicase protein nsp10 is a critical regulator of viral RNA synthesis. J. Virol. 81, 6356–6368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Donaldson E. F., Graham R. L., Sims A. C., Denison M. R., Baric R. S. (2007) Analysis of murine hepatitis virus strain A59 temperature-sensitive mutant TS-LA6 suggests that nsp10 plays a critical role in polyprotein processing. J. Virol. 81, 7086–7098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ke M., Chen Y., Wu A., Sun Y., Su C., Wu H., Jin X., Tao J., Wang Y., Ma X., Pan J. A., Guo D. (2012) Short peptides derived from the interaction domain of SARS Coronavirus nonstructural protein nsp10 can suppress the 2′-O-methyltransferase activity of nsp10/nsp16 complex. Virus Res. 167, 322–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]