Summary
Chromatin remodelling is the ATP-dependent change in nucleosome organisation driven by Snf2 family ATPases. The biochemistry of this process depends on the behaviours of ATP dependent motor proteins and their dynamic nucleosome substrates, which brings significant technical and conceptual challenges. Steady progress has been made in characterizing the polypeptides that these enzymes are comprised of. Divergence in the sequences of different subfamilies of Snf2 related proteins suggests that the motors are adapted for different functions. Recently structural insights have suggested that the Snf2 ATPase acts as a context-sensitive DNA translocase. This may have arisen as a means to enable efficient access to DNA in the high density of the eukaryotic nucleus. How the enzymes engage nucleosomes and how the network of non-covalent interactions within the nucleosome respond to the force applied remains unclear, and it remains prudent to recognise the potential for both DNA distortions and dynamics within the underlying histone octamer structure.
Introduction
Chromatin remodelling is the directed alteration of genome packaging in the eukaryotic cell nucleus. The term is usually used to describe ATP-dependent changes in nucleosome organisation driven by Snf2 family ATPases, although it predates the discovery of those factors and is sometimes also applied to changes such as large scale nuclear reconfiguration or non-ATP driven nucleosome rearrangements.
The chromatin remodelling activity of Snf2 family ATPases was uncovered independently in Saccharomyces cerevisiae screens for factors contributing to expression of the HO nuclease required for mating type switching [1, 2], and sucrose fermentation by invertase encoded at SUC2 [3]. Both the switching and sucrose non-fermenting (SWI and SNF) screens revealed the involvement of a gene at locus YOR290C encoding a large ATPase, since named SNF2 [4]. Suppressors of snf2 mutants in turn revealed SWI/SNF independent Sin− mutants including point mutants of histone genes and other chromatin components [5-7]. Together with the observed changes to chromatin structure in snf2 mutants at target loci [8], this suggested Snf2p affected chromatin structure [9]. The hypothesis was confirmed by mutagenesis in vivo and in vitro observation of the activity of complexes containing Snf2 family ATPases. Biochemical purifications and bioinformatic sequence comparisons have since revealed that Snf2 family members are ubiquitous and numerous across eukaryotes [10].
Although early identifications concentrated on Snf2 family members as general transcription factors, the SNF2 locus had previously been identified as contributing to protection against DNA damage [11]. Subsequent investigations have shown that Snf2 family ATPases are involved in a wide variety of genomic processes including transcription, replication, repair and recombination. This suggests that ATP-dependent chromatin remodelling is a fundamental functional requirement in the nucleosome-packaged genomes of eukaryotes. However, the occurrence of Snf2 family ATPases in bacteria and archaea illustrates that the DNA-dependent ATP-driven translocase activity of the Snf2 family does not necessarily act on nucleosomes alone.
Biochemistry of chromatin remodelling
The biochemistry of ATP-dependent remodelling has been an active area of investigation for almost two decades. The challenges for this field arise because the enzymatic activities are provided by large protein complexes whose function often appears redundant and whose members are ubiquitous, and because the chromatin substrate is dynamic and its properties are incompletely understood. Although much of the published work has focused on the mechanism of remodelling on nucleosomes, several genetic screens have implicated Snf2 family members in a diverse range of functions.
Composition and structures of remodelling complexes
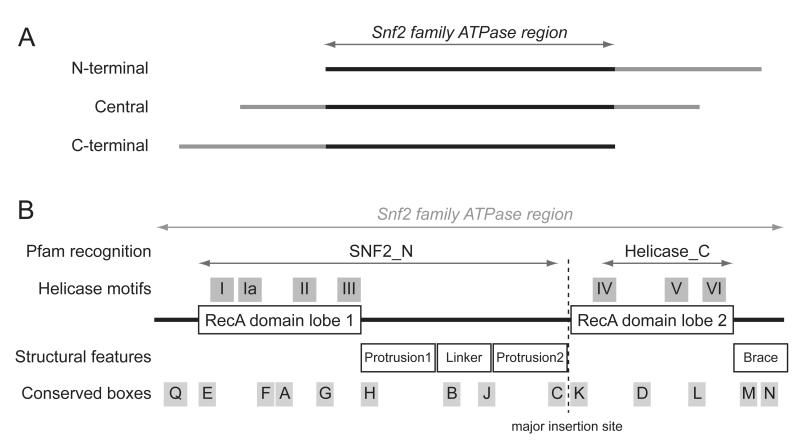
All recognised ATP-dependent chromatin remodelling complexes contain a large core polypeptide which includes a region of homology to the helicase-related Snf2 family ATPase (Fig. 1A). Having such a central molecular motor provides scope for a large number of potential mechanisms to drive the remodelling process. Therefore, a clear understanding of the composition and structure of remodelling complexes is crucial for constraining the many possible mechanistic models that can be imagined for remodelling.
Fig 1.
Sequence and structural features of the Snf2 family ATPase region. (A) Snf2 ATPase region is embedded in full length polypeptide, and can be central, N-terminal or C-terminal. (B) Motifs, conserved boxes and structural elements within the Anf2 ATPase region as defined by Flaus et al [26] are shown relative to paired RecA domain lobes. Not to scale.
The direct purification of remodelling complexes is technically challenging because many are large multi-protein associations comprising many small subunits (Table 1). A further complication is that some core Snf2 family polypeptides such as S. cerevisiae Sth1 and Isw1 participate in multiple variant complexes differing by only one or a few subunits [12, 13].
Table 1.
S. cerevisiae Snf2 family abundance and complex involvement.
| Snf2 family member | Grouping | Molecules/cell | Nucleosome/remodeller | Complex | Subunits/complex | Refs |
|---|---|---|---|---|---|---|
| Snf2 | Snf2-like | 217 | 337 | SWI/SNF | 11 | [21, 94, 95] |
| Sth1 | Snf2-like | 1990 | 37 | RSC | 15 | [12, 96, 97] |
| Isw1 | Snf2-like | 1500 | 49 | Isw1a/Isw1b | 2/3 | [13] |
| Isw2 | Snf2-like | 1520 | 48 | Isw2 | 4 | [80, 98] |
| Chd1 | Snf2-like | 1620 | 45 | Chd1 | 1 | [99, 100] |
| Irc5 | Snf2-like | - | - | Lsh | ? | |
| Swr1 | Swr1-like | 656 | 112 | Swr1 | ~12 | [101-104] |
| Fun30 | Swr1-like | 6800 | 11 | Fun30 | 1 | [69, 105] |
| Ino80 | Swr1-like | 6280 | 12 | Ino80 | ~12 | [106, 107] |
| Rad54 | Rad54-like | - | - | Rad54 | 1-2 | [108, 109] |
| Rdh54 | Rad54-like | 1270 | 58 | Rdh54 | ? | [109-111] |
| Rad5 | Rad5/16-like | 1520 | 48 | Rad5 | 3 | [112, 113] |
| Rad16 | Rad5/16-like | 358 | 204 | Rad16 | 2-3 | [114, 115] |
| Irc20 | Rad5/16-like | 143 | 512 | ? | ? | [116] |
| Ris1 | Rad5/16-like | - | - | Ris1 | ? | [109, 117] |
| Rad26 | Sso1653-like | - | - | Rad26 | ? | [118, 119] |
| Mot1 | Sso1653-like | 6260 | 12 | Mot1 | 2 | [120-122] |
Characterising purified complexes is also challenging because of the difficulty of creating specific in vitro activity assays using typical methods such as changes restriction enzyme or nuclease accessibility, or in native gel mobility, which have low resolution and lack standardised kinetic parameters. Lack of clarity about the relevant biological function of complexes means defining an appropriate substrate can be confusing, and assembling substrates with specific histone post-translational modifications or defined nucleosomal arrays is technically difficult. In fact, most detailed in vitro assays are performed on nucleosomes comprising the “601” DNA sequence artificially selected for unusual stability [14] and core histones with sequence from the frog Xenopus laevis that lack any post-translational modifications because they are prepared in Escherichia coli [15].
Low resolution EM structural envelopes have been determined for the large RSC [16-19], PBAF [20] and Swi/Snf [21] remodelling complexes. These all reveal a large bowl-like shape with a central depression of appropriate size to hold a nucleosome [22]. Evidence for density consistent with the nucleosome is seen in the RSC structure [19]. A dimeric complex of Iswi subfamily member ACF on a nucleosome has also been observed [23].
Sequence classification of the Snf2 family ATPases
Due to the large number of genetic or functional investigations and the difficulty of biochemically characterising them, the core Snf2 family polypeptide is typically used as an identifier for chromatin remodeller complexes. Their sequences carry a characteristic conserved Snf2 family ATPase region, which can be located anywhere within the polypeptide (Fig. 1A). Consistent with this, exchange of the Snf2 family ATPase region has been shown to carry with it the properties of the remodelling complex [24].
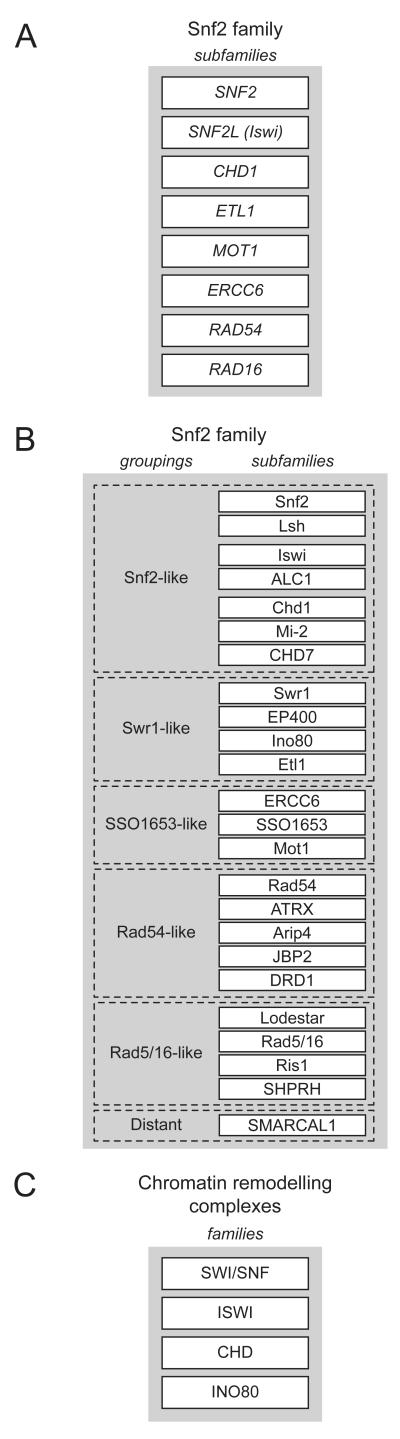
An early analysis of the Snf2 family ATPase region based on only 30 sequences proposed 8 distinct subfamilies (Fig. 2A) [25]. A subsequent comprehensive classification was carried out using over 1300 family members that became available through the extraordinary recent progress in genome sequencing [26]. This confirmed the principles identified by Eisen et al [25], and revealed an expanded phylogeny of 23 subfamilies with the same effective topology as the original study (Fig. 2B). The equivalence based on this helicase region and the origins of nomenclature for the 17 proteins identified in S. cerevisiae and the 32 in the human genomes is shown in table 2. The subfamilies fall into 5 groupings, two of which contain the subfamilies most related to the archetypal S. cerevisiae Snf2p. These two ‘Snf2-like’ and ‘Swr1-like’ groupings were based on some 400 sequences and encompass 7 and 4 subfamilies respectively.
Fig 2.
Snf2 family classification schemes. (A) Original subfamilies defined by sequence comparison of Eisen et al using ATPase and flanking sequences [25]. (B) Schematic of subfamilies defined by expanded phylogenomic comparison of Flaus et al using Snf2 family ATPase region [26]. Subfamilies take name and nomenclature from first biochemically identified archetype. (C) Example of an empirical classification of chromatin remodelling enzymes into separate families [10].
Table 2.
Nomenclature of Snf2 family members in S. cerevisiae and Human
| Snf2 subfamily | Grouping | S. cerevisiae genes | S. cerevisiae nomenclature | Human genes | Human nomenclature |
|---|---|---|---|---|---|
| Snf2 | Snf2-like | SNF2, STH1 | Sucrose Non Fermenting, Snf Two Homolog | SMARCA4/BR G1, SMARCA2/hB RM | Brm-Related Gene, human BRahMa-like |
| Iswi | Snf2-like | ISW1, ISW2 | Imitation SWitch | SMARCA1/SN F2L, SMARCA5/hS NF2H | SNF2-Like, human SNF2 Homologue |
| Lsh | Snf2-like | IRC5 | Increased Recombination Centres | HELLS/SMAR CA6 | HELicase, Lymphoid-Specific |
| ALC1 | Snf2-like | - | ALC1 | Amplified in Liver Cancer | |
| Chd1 | Snf2-like | Chd1 | Chromo Domain containing | CHD1L | CHD1-Like |
| Mi-2 | Snf2-like | - | CHD3, CHD4, CHD5 | CHD1-like | |
| CHD7 | Snf2-like | - | CHD6, CHD7, CHD8, CHD9 | Chd1-like | |
| Swr1 | Swr1-like | SWR1 | ? | SRCAP | Snf2-related CREBBP activator protein |
| EP400 | Swr1-like | - | EP400 | E1A binding protein p400 | |
| Ino80 | Swr1-like | INO80 | INOsitol biosynthesis | INO80 | INO80 homolog |
| Etl1 | Swr1-like | FUN30 | Function UNknown | SMARCAD1 | SMARCA containing DEAD/H box |
| Rad54 | Rad54-like | RAD54 | RADiation sensitive | RAD54L, RAD54B | RAD54-Like, RAD54 homologue B |
| ATRX | Rad54-like | - | ATRX | Alpha Thalassemia/mental Retardation syndrome X-linked | |
| Arip4 | Rad54-like | - | RAD54L2 | RAD54-Like 2 | |
| Rad5/16 | Rad5/16-like | Rad5, Rad16 | RADiation sensitive | HLTF/SMARC A3 | Helicase-Like Transcription Factor |
| Ris1 | Rad5/16-like | ULS1 | Ubiquitin Ligase for SUMO conjugates | - | |
| Lodestar | Rad5/16-like | - | TTF2 | Transcription Termination Factor, RNA polymerase II | |
| SHPRH | Rad5/16-like | IRC20 | Increased Recombination Centres | SHPRH | SNF2 Histone linker PHD RING Helicase |
| Mot1 | Sso1653-like | MOT1 | Modifier Of Transcription | BTAF1 | B-TFIID Transcription Associated Factor |
| ERCC6 | Sso1653-like | RAD26 | RADiation sensitive | ERCC6, ERCC6L, C9orf102 | Excision Repair Cross-Complementing rodent repair deficiency, complementation group 6, chromosome 9 open reading frame 102 |
| SMARCAL1 | Smarcal1-like | - | SMARCAL1, ZRANB3 | SMARCA Like, Zinc finger, RAN-Binding domain containing 3 |
Nomenclature of Snf2-related proteins using subfamily and grouping classifications from [26] based on alignments of the helicase-like region. Subfamilies are named from the first reported archetype in any organism. S. cerevisiae nomenclature is from Saccharomyces Genome Database and human nomenclature is from ENSEMBL using official Human Genome Naming Commission symbols. Some members are known by a SMARCA acronym standing for for “SWI/SNF-related, Matrix-associated, Actin-dependent Regulator Chromatin group A”. Where common alternatives are used alongside SMARCA nomenclature, the official HGNC name is shown first.
More recently, an alternative classification has been adopted grouping remodelling complexes into a four nominal but separate families, typically SWI/SNF, ISWI, CHD and INO80 (Fig. 2C) [10, 27]. This classification based on empirical assignments of the ATPase and flanking regions is a simplification, obscuring diversity within the CHD grouping and making it is difficult to categorise enzymes such as Alc1 [28], Lsh [29] and ATRX [30] which are likely to have chromatin related functions. It also ignores relationships with the broader range of Snf2-related ATPases. Flanking domains are presently poorly categorised and standard domain finding tools leave large areas unassigned in many sequences, making this a difficult basis for classification.
The availability of a large number of sequences enables the explicit definition of the Snf2 family by multiple alignment. Early studies had identified the ATPase was a member of helicase-like superfamily 2 (SF2) through the presence of seven helicase motifs (Fig. 1B) [31], and this formed the basis for biochemical investigations demonstrating the underlying DNA translocase activity [32]. The Snf2 family ATPases are distinguished from other SF2 members by an extended span of sequence between the two RecA domains [33]. Together with historical quirks in seed alignments, this has led to the Snf2 ATPase being recognised by common motif-finding algorithms as bipartite “Snf2_N” and “HelicaseC” regions. Some Snf2 family members such as those in the Swr1-like grouping (Fig. 2B) contain very large sequence regions at a specific major insertion site that generate an expansion of such scale that it confound simple alignment algorithms [26] and they have been termed ‘split ATPases’ [34].
Sequence-structure relationships in Snf2 family ATPase region
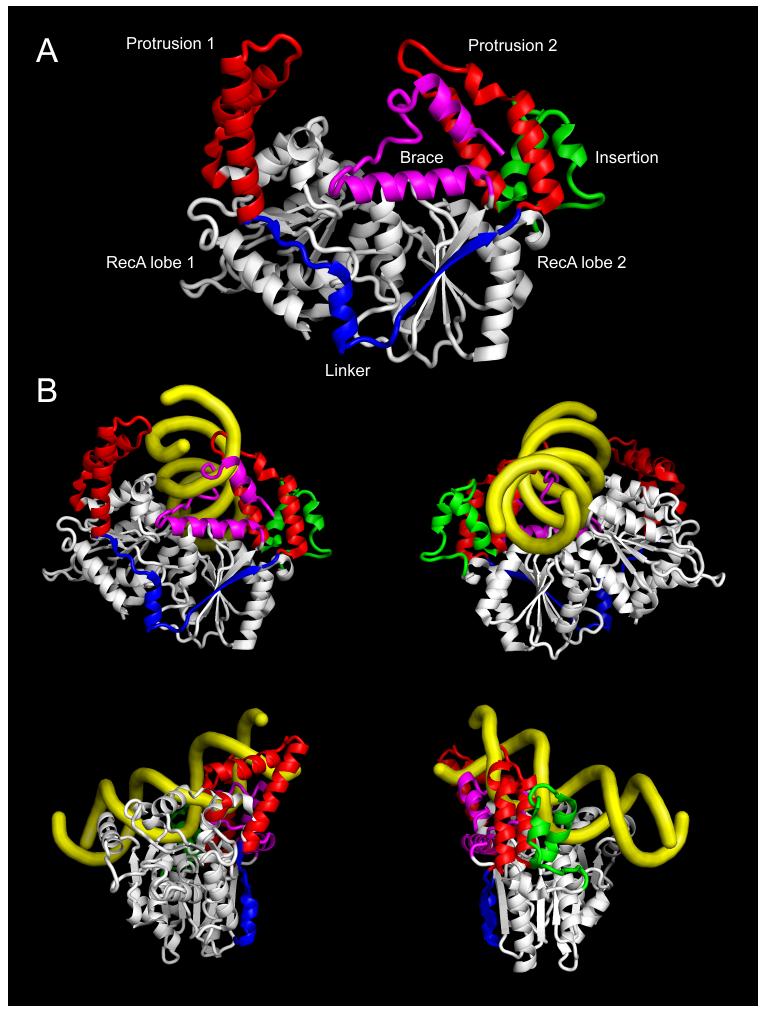
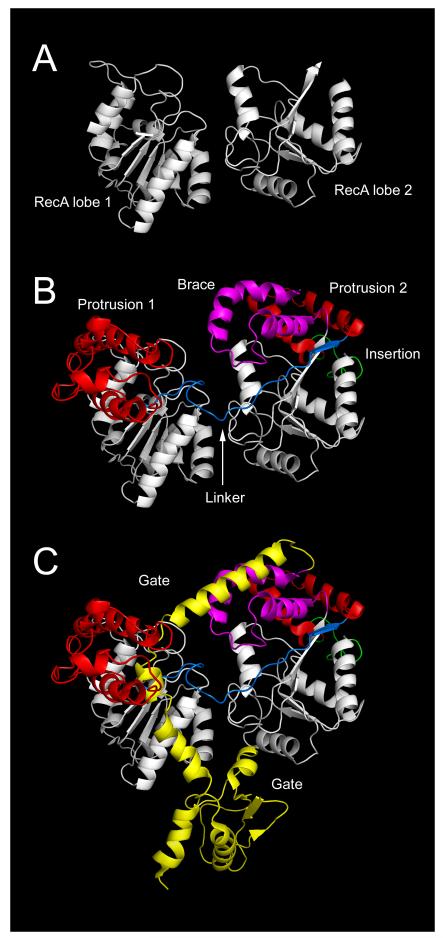
The characteristic sequence features within Snf2 ATPases (Fig. 1B) can be interpreted through the Zebrafish Rad54 structure which remains the most relevant atomic resolution model of the Snf2 family (table 3) [35]. At its core the structure is composed of the same pair of RecA domain lobes as all helicase superfamily members (Fig. 3A). The extended span of sequence between the RecA domains contributes to two alpha helical ‘protrusions’ from the spherical RecA domains. The sequence of these protrusions is not itself conserved, but amino acid residues around their bases which stabilise or ‘glue’ the protrusions to the RecA domains comprise conserved boxes H, C, J and K (Fig. 1B). Box B within the flexible ‘linker’ passing across the groove between the RecA domains contains a pair of absolutely conserved arginine residues which are essential for function [36]. The major insertion region which accommodates such variation in length is situated behind the second RecA domain, but would be adjacent to DNA towards the back of the ATPase (Fig. 3). An additional region of alpha helical structure containing conserved boxes extends from the C-terminus of the second RecA domain, forming a ‘brace’ that stretches towards the modelled DNA and includes highly conserved charged residues in boxes M and N (Fig. 3).
Table 3.
Structures related to Snf2 family ATPase region.
| Protein | Organism | PDB | Resolution | Snf2 family homology | Identity/Similarity to Snf2p | Ref |
|---|---|---|---|---|---|---|
| Chd1 | S. cerevisiae Budding yeast | 3mwy | 3.7Å | e−144 | 45%/65% | [39] |
| Rad54 | D. rerio Zebrafish | 1z3i | 3.0Å | e−128 | 33%/54% | [35] |
| Sso1653 | S. sulfotaricus Archaea | 1z63 | 3.0Å | e−125 | 26%/44% | [37] |
| RapA | E. coli Bacteria | 3dmq | 3.2Å | e−36 | 27%/46% | [49] |
| Hef | P. furiosus Archaea | 1wp9 | 2.9Å | e−16 | 28%/49% | [51] |
| XPB | A. fulgidus Archaea | 2fwr | 2.6Å | e−16 | 15%/34% | [124] |
| Vasa | D. melanogaster Fruit fly | 2db3 | 2.2Å | e−13 | 19%/38% | [125] |
Helicase-like superfamily 2 (SF2) structures in Protein Data Bank (PDB) related to Snf2 family by homology of ATPase region. Similarity shown as expectation value for hmmsearch hit with Snf2 family model [26] to PDB database. Identity and similarity for global alignment to Snf2 residues 767-1222 using EMBOSS stretcher. PDB code is for most relevant structure where multiple related accessions deposited.
Fig 3.
Structural components of Zebrafish Rad54 Snf2 family ATPase region. (A) Snf2 ATPase region showing RecA domain lobes (white), protrusions (red), linker (blue), brace (magenta) and insertion (green). From PDB code 1Z3I. (B) Modelling of DNA path on Zebrafish Rad54 by alignment of RecA lobe 1 of S. solfataricus SSO1653 structure. Structure PDB codes 1Z3I and 1Z63 [35, 37].
A structure of a Snf2 family ATPase enzyme SSO1653 from the archaeal Sulfolobus sulfotaricus has determined in complex with double-stranded DNA (table 3) [37]. The first RecA lobe appears to be engaged in a relevant position based on other helicase complex structures, although the second RecA lobe is probably in an non-functional orientation [38]. Using the DNA and first SSO1653 RecA lobe enables the Rad54 structure to be oriented so that DNA can be modeled on it, and confirms that Snf2 family enzymes are likely to function by the same enzymatic mechanism as other SF2 helicase-like DNA translocases (Fig. 3B).
Snf2 family proteins as allosterically regulated ATPases
Surprisingly, modeling the DNA-bound structure did not reveal an obvious mechanistic role for the characteristic protrusions, brace or linker region of the Snf2 family. Clues are provided by the recent structural investigation of the S. cerevisiae Chd1 protein which can remodel nucleosomes without additional subunits (Fig. 4A) [39]. Although the Chd1 diffraction was at sub-atomic resolution, the Rad54 coordinates could be used to model the high resolution orientation. This and parallel biochemical experiments show that the chromo domains flanking the ATPase region contact the protrusions and are likely to block activity by occluding DNA from its binding site on the ATPase (Figs. 4A, 4B). This is consistent with earlier observations showing genetic linkages between adjacent domains and protrusion residues in Sth1 [40]. Hauk et al propose that this represents ‘modular allostery’ [41] whereby DNA binding and ATPase activity is inhibited by structurally independent domains encoded either in sequences flanking the Snf2 ATPase or in independent subunits of the chromatin remodeling complex. The domains act as a switch or ‘gate’ for translocase activity.
Fig 4.
Modular allostery by inhibitory gate domain to prevent DNA binding to Snf2 ATPase. (A) S. cerevisiae Chd1 ATPase region with equivalent orientation and colouring to Rad54 in fig 3A. Chromowedge domain (yellow) at rear. (B) View from A rotated 120° to illustrate Chromowedge domain association with protrusion 2 and RecA lobe 2. (C) Schematic of modulation by switching of ‘gate’ domain (yellow) on Snf2 ATPase through introduction of an alternative binding site (light grey). (D) Modulation by competition with ‘gate’ domain (yellow) through introduction of a competitor for Snf2 ATPase binding (dark grey). Structure PDB code 3MWY [39].
Since all Snf2 family polypeptides contain at least one domain-size region adjacent to the ATPase [26], this suggests a fundamental property of the Snf2 family could be as DNA-dependent ATPases whose activity can be modulated by adjacent inhibitory ‘gate’ domains (Figs. 4C, 4D). The highly conserved alpha helical organisation but not sequences of protrusions observed in the sequence analysis [26] could provide distinctive surfaces for interactions with the inhibitory domains to set up the gating.
The high local ‘concentration’ of an adjacent inhibitory domain from the same polypeptide or complex means only weak interfaces are required to stabilise inhibitory binding of the gate domain to the ATPase. This allows dynamic switching of the gate across a low energy barrier when an alternative interaction for the inhibitory domain is brought into proximity (Fig. 4C). Such a switch in gating could be driven by a higher affinity epitope for the inhibitory gate domain such as methylated histones for the Chromo domain in Chd1, or the ARID domain of BAF250 for the HSA domain in Snf2 [42]. Alternatively, the switch could occur when a feature brought into proximity competes with the inhibitory binding of the gate domain and displaces it (Fig. 4D), as preferred by Hauk et al to explain their biochemical observations for S. cerevisiae Chd1 [39].
Snf2 family as context-sensitive DNA-dependent ATPases
Tight regulation of DNA-dependent ATPases makes intuitive sense in biochemical terms. The expansion in genome size during the early evolution of eukaryotes required chromatin organisation to package DNA at high density into a membrane-enclosed nucleus [43]. Adaptation of existing archael histones as nucleosomes [44] for this task would in turn require a remodelling machinery which could be provided in part by the Snf2 family of ATP-driven, DNA-stimulated DNA translocases that also existed in archaea and bacteria [25, 26]. Their distinctive modular allosteric regulation would reduce wasteful turnover of ATP in the high concentration of DNA substrate by providing a context-dependent switch via the inhibitory gate domain [39]. Subsequent specialisation in this context-dependence would explain the diversity of Snf2 family members in eukaryotes that targets chromatin remodelling for highly specific functions.
The need to remodel the chromatin substrate for genomic access provides a large number of possible roles for Snf2 family proteins and is understandably the focus of most functional investigations. However, there are a number of other potential uses for DNA translocases regulated by context in the eukaryotic nucleus. Examples include the Rad54 subfamily role in establishment and progress of homologous recombination repair [45], the Mot1 subfamily role in TBP cycling at promoters [46], and the ERCC6 subfamily roles in RNA polymerase passage through DNA lesions [47]. Interestingly, Mot1 and ERCC6 subfamily members are the most similar to non-eukaryotic Snf2 family proteins (Fig. 2B) [26].
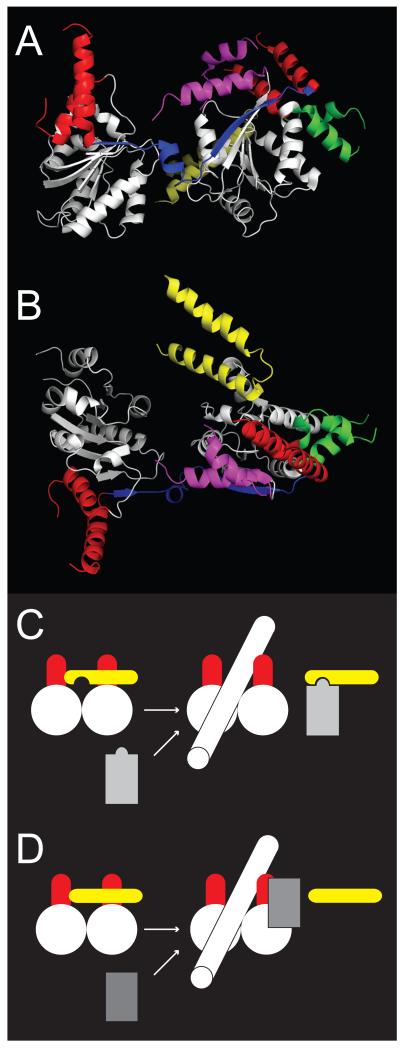
The biological function of S. sulfotaricus SSO1653 is unknown and the type strain has an inactivating transposon inserted in this gene. More distantly related bacterial RapA proteins are found at the edge of the Snf2 family and their E. coli archetype has been shown to have a role in RNA polymerase recycling at promoters [48]. The structure of RapA has been solved (table 3) [49] and its core retains a Snf2 family-like organisation with a protrusion and brace on RecA lobe 2, although the protrusion on lobe 1 is less similar to other structures (Fig. 5). The RapA structure contains additional domains that could act as allosteric gates (Fig. 5C), and biochemical observations are consistent with gating [50]. The yet more distant archael Pyrococcus furiosus Hef protein related to Mph1 in S. cerevisiae and human FANCM is a true DNA helicase and also has a protrusion and potential brace on RecA lobe 2 [51] (see supplementary data in [26]). Two distantly related non-Snf2 family helicases, archaeal Archaeoglobus fulgidis XPB and Drosophila melanogaster Vasa, mainly retain similarity to helicase motifs in the core RecA domains of the Snf2 family and lack any additional motifs in common with the Snf2 family (table 3).
Fig 5.
Similarity of Rad54 and RapA structures. (A) Conserved RecA lobes in equivalent orientation to fig 3. (B) Characteristic Snf2 family structures including protrusions (red), linker (blue), insertion region (green) and brace (magenta). (C) Additional structure (yellow) potentially acting as modulatory gate in E. coli RapA. Structure PDB code 3DMQ [49].
Abundance and localisation of chromatin remodelling complexes
In addition to their diversity, remodelling complexes are surprisingly highly abundant nuclear components (table 1). In fact, the ATPase subunit of Swi/Snf is by far the least abundant of the recognised nucleosome-active remodelling complexes in S. cerevisiae and its less extensive roles may have facilitated its historical identification. The combined abundance of the Snf2 subfamily members Sth1 and Snf2 equates to approximately 1 enzyme for every 34 nucleosomes, or less than one per gene, and may correlate with an occasional requirement for these enzymes to undertake specific activities such as nucleosome ejection. The chromatin organizing enzymes Isw1, Isw2 and Chd1 are considerably more abundant at 1 enzyme for every 16 nucleosomes, perhaps reflecting a more general role for these enzymes in nucleosome spacing by sliding. The Fun30, Ino80 and Swr1 enzymes likely to have functions relating to histone exchange have overall abundance sufficient to equate to one for every 5 nucleosomes. The reason for requiring an average of more than one histone exchanger per gene is not yet apparent. It may be that in addition to performing functions relating to the directed incorporation of histone variants, some of these enzymes could have a destabilizing effect on chromatin by removing histone dimers, for example during the transit of polymerases.
Accessory subunits in multi-protein complexes and flanking domains in the Snf2 family ATPase polypeptide frequently encode subunits known to interact with chromatin, often with specificity for post-translational modifications or histone variants. This has led to the suggestion that a basic property of chromatin remodellers is that they recognise covalent histone modifications [10]. As discussed above, one function for histone recognition interactions may be to provide an allosteric regulatory mechanism to activate the remodeller in presence of its substrate [39]. A second function may be the need for the remodelling complex to maintain affinity with the nucleosome substrate.
However, the most widely recognised function for chromatin recognition domains in remodellers is to target remodelling complexes to sites of action. In this respect a potential problem is the fact that most histone binding domains have only modest affinity for epitopes. This would be anticipated to result in significant non-specific interactions with chromatin not bearing the appropriate modification. However, the localization of histone modification is often diffuse rather than punctuate meaning that the local concentration of epitopes in specific regions of the nucleus may be sufficient to generate a localised enrichment in enzyme (see accompanying review by Erdel and Rippe [52]). As some remodelling enzymes contain epitopes that are capable of recognising similar modifications [53], and many modifications share similar distributions [54], there is the potential for multiplicity and redundancy of remodelling complexes associated with large scale processes [52] such as the repair of DNA double strand breaks [55], in establishing higher order chromatin structures [56], or in transcription [57].
Translocation by Snf2 family ATPase acting on nucleosomes
Significance of the remodelling mechanism
The pathway of chromatin remodelling has great functional significance because of its implications for the exposure of DNA sequences, the organisation of the genome, and the exchange of histone proteins. Firstly, chromatin packaging generally obscures DNA so local recruitment of remodellers is required to facilitate access for genome-active processes. Secondly, genome-wide localisation shows that nucleosomes are very uniformly spaced despite the diversity of underlying DNA sequences. Deletion analysis in S. cerevisiae reveals that the spacing activity is contributed redundantly by Isw1, Isw2 and Chd1 [58], but can be locally manipulated by the effect of specialised sequences on remodelling [59]. Similar observations have been made in Schizosaccharomyces pombe for the role of related Mit1 family member [60]. Thirdly, remodelling has the potential to destabilise histone-DNA contacts that provide the link between histone post-translational modifications and bound DNA sequences [56]. The remodelling mechanism must be structurally conservative to avoid erasing such epigenetic information.
Chromatin remodelling as a nucleosome response
The molecular mechanism of remodelling has been the subject of hypothesis for many years, possibly because the mechanical parallels are intuitively accessible and because biochemical details of remodelling enzymes and chromatin substrates have been limiting.
Nucleosomes are the repeating molecular subunit of chromatin and therefore likely to be the direct substrate for chromatin remodelling. A number of different outcomes have been proposed as an end result of remodelling on nucleosomes [10], principally the repositioning of the histone octamer relative to DNA (sliding), replacement of part or all of the octamer (exchange) or removal of all or part of the histone octamer (ejection). It is also formally possible that the canonical nucleosome structure could be reconfigured to a stable alternative (switching), but this remains somewhat controversial [61-63].
Fundamentally ATP-dependent chromatin remodelling is an enzymatic process with the remodeller accelerating the rate of change between a substrate and product state. In contrast to textbook enzymology where individual covalent bonds are manipulated at localised sites, chromatin remodelling involves non-covalent process on a 200 kDa substrate. This can lead to confusions of scale because the end product of remodelling such as a slid nucleosome may be the result of a large number of stepwise turnovers of the ATPase enzyme itself. A destabilised nucleosome is not the ‘transition state’ of an individual enzyme cycle, it is the consequence of multiple enzyme cycle products building up on the nucleosome. This link between ATPase cycles of the remodelling enzyme and nucleosome outcomes is usually what is implied by “mechanism of remodelling”. It depends on how the ATPase cycle products are applied to the nucleosome, and the response of the nucleosome.
Remodelling complex structure and substrate binding
A crucial element in the mechanism of remodelling is the dynamic potential of the multiple of weak interactions within the nucleosome structure itself, and how the chromatin remodeller directs these along a specific pathway. Compositional and structural information gathered for chromatin remodellers (tables 1, 3) suggests two general classes by which remodelling enzymes might engage with the nucleosome substrate.
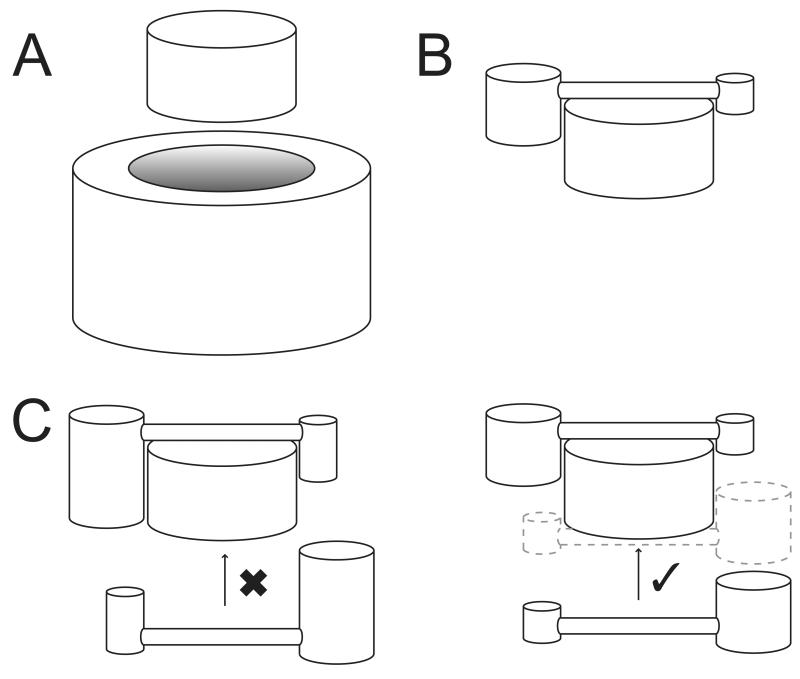
Firstly, a large remodelling machinery could envelop the entire nucleosome to control its dynamic properties (Fig. 6A). Since the nucleosome is a 200 kDa complex and volume scales with r3 for a simple sphere, this implies that an enveloping structure twice the radius of a nucleosome will have approximately 8 times its mass. Many multi-protein remodelling complexes are in the range of 1-2 MDa and EM image reconstructions are consistent with the ability to surround substrate nucleosomes [19].
Fig 6.
Possible binding orientations for remodellers on nucleosomes. (A) Enveloping of nucleosome by remodeller. (B) Cantilevering of remodeller across nucleosome. (C) Examples of monomeric dimeric and dimeric remodeller binding to nucleosome by blocking (left) or enabling (right) of symmetric second site.
Secondly, a simplified remodelling machinery could cantilever across the nucleosome (Fig. 6B) as a minimal alternative to envelopment. The non-ATPase region of D. melanogaster ISWI describes a long cylinder-like structure of 100Å length and 20Å diameter composed of HAND, SANT and SLIDE domains [64] that would be of appropriate size for such a spanning capability, and its length is conserved across Iswi subfamily proteins [26]. Arrangements equivalent to a cantilever have been modelled [65].
One feature of both enveloping and cantilever complexes is that they achieve “template commitment” to allow multiple ATPase cycles while retaining interactions with transiently destabilised nucleosomes as effects are accumulated [66, 67]. A second feature of the stabilising interactions provided by the remodeller is that they will constrain the motions of the malleable nucleosome that could otherwise flex in different ways under an applied force. This feature may therefore be crucial to enabling the remodelling process to follow a specific mechanistic pathway.
Multiple remodellers or multiple nucleosomes
Although diagrams such as those in figures 6A and 6B show a single remodeller engaged with a single nucleosome, the dyad symmetry of the nucleosome implies chromatin remodellers should bind as dimers. Indeed there is evidence that the ATPase subunits of some enzymes are dimeric [68, 69], or bind to nucleosomes as dimers [23, 67]. Alternatively, it is possible that remodeller association creates asymmetry, for example by blocking binding of a second enzyme (Fig. 6C). Asymmetry is observed in some RSC structures [17].
It is also possible that one chromatin remodelling complex could work on a dinucleosome substrate. Nucleosomes are typically found in genomes at high densities meaning that following repositioning an encounter with a neighbour is a distinct possibility. It has been proposed that collisions between adjacent nucleosomes could act as a stage in the disassembly of nucleosomes [70-72]. Conversely, enzymes that act to space nucleosomes may stabilise chromatin. In this case, in order to prevent collisions a means of sensing the adjacent nucleosome is required. Possible binding arrangements could include cooperation between remodellers on adjacent nucleosomes, or binding of a single remodeller to span adjacent linkers. However, in the simplest case contacts with linker DNA adjacent to a nucleosome enzyme complex are important for full activity so when adjacent nucleosomes interfere with these linker DNA contacts, movement in the direction of the adjacent nucleosome would be reduced.
Mechanism of nucleosome dynamics
Snf2 translocation on nucleosomes
When a remodelling complex is bound to a nucleosomal substrate the core Snf2 ATPase motor provides a double-stranded DNA translocase which can move directionally on the DNA duplex (see accompanying review by Croquette et al [73]). Template commitment suggests that the complex also maintains contact with the histone components throughout the remodelling process. Termination will occur when the remodelling complex can no longer act on the nucleosome, for example because it has been disrupted or reached a position where necessary flanking DNA is not available due to proximity with another nucleosome or some other barrier.
The capability of the Snf2 translocase for processive and directional movement is demonstrated by single molecule observations showing rapid development of induced torsion, and biochemical experiments showing blockage by hairpins, or single stranded gaps [68, 74-77]. Other SF2 double-stranded DNA translocases are observed to have apparent ‘kinetic’ step sizes distinguished by rate-determining steps down to 3-4 bp, and distinct ‘mechanical’ step sizes of 3-11 bp per ATP hydrolysis cycle [78]. Some estimates of step sizes for Snf2 family proteins have been relatively large [76] but the advent of more sophisticated detection techniques has led to progressively smaller steps with pauses every few base pairs being detected [67]. Further studies will be required to determine whether movements of several bases can be broken down into single base steps and to establish whether this applies to all Snf2 related enzymes.
The step size is of great interest as it has the potential to influence the amount of rotation generated during the remodelling process. A series of recent observations support the association of the ATPase region with nucleosomal DNA at superhelical location 2 (SHL ±2). This includes evidence that DNA gaps appear to block the action of Snf2 and Iswi subfamily remodelers when introduced at this location [76, 77, 79] and directed crosslinking consistent with an interaction of the ATPase at SHL ±2 [65, 80]. This location coincides with an important structural feature within the nucleosome; the apparent high stability across the region between superhelical locations SHL −1.5 and SHL +1.5. Stability is reflected in the high uniformity and reduced dynamics of the region in crystal structures on multiple DNA sequences [81], increased number of histone-DNA contacts in the region [82], and histone SIN mutations reducing DNA contacts which also accelerate nucleosome sliding [83]. This has been taken to suggest that that Snf2 related enzymes target a region of the nucleosome that is rate limiting for dynamics.
It is possible that DNA sequences may affect the outcome of remodelling [84] either by affecting the opportunity for engagement by Snf2-related enzymes or the response of the nucleosome to forces applied by the remodeller.
A dynamic histone octamer?
Most commonly proposed mechanisms indirectly imply that DNA is being remodelled across a static histone octamer surface. However, recent interest in nucleosome dynamics has accumulated evidence that various parts of the histone octamer may readily flex and change their binding to DNA. For example, the most external turns of DNA are known to be readily released in the process of site exposure [85, 86], and H2A-H2B dimers can be displaced [87] such that they even become exchangeable at a significant rate during remodelling [88]. Tetramers of H3 and H4 have been observed to adopt conformations that differ from those observed within octamer and nucleosome structures [89-91]. It is possible that a concerted pathway occurs during remodelling involving rearrangements in the histone octamer that weaken histone-DNA contacts altering the energetics of DNA passage across its surface. The repertoire of mechanisms proposed for remodelling may be able to be expanded from the widely discussed twist defect and bulge diffusion models and variations on them (reviewed in [92] and accompanying review by Blossey and Schiessel [93]).
Conclusion
The biochemistry of chromatin remodelling remains a highly energetic and fruitful field. The complexity of understanding the behaviours of dynamic mechanical enzymes on dynamic mechanical substrates poses significant demands on biochemical techniques more suited to homogenous and stable molecules. Likewise, mechanistic thinking has been coloured by conceptual models of rigid bodies that hide details of local structure and malleability.
The growing sophistication of both experimental and conceptual analyses is therefore crucial to a full understanding. The biochemistry of the cell involves a number of fundamental processes for which one universal and highly conserved solution has evolved due to the complexity of factors involved. It appears that directed alteration of chromatin structure by Snf2 family enzymes is such a process. They provide the means to an end required as a consequence of high-density chromatin packaging in eukaryotes.
Acknowledgements
AF is supported by a Science Foundation Ireland and the Health Research Board of Ireland. TOH is supported by Wellcome Trust Senior Fellowship 064414.
References
- 1.Stern M, Jensen R, Herskowitz I. Five SWI genes are required for expression of the HO gene in yeast. J Mol Biol. 1984;178:853–868. doi: 10.1016/0022-2836(84)90315-2. [DOI] [PubMed] [Google Scholar]
- 2.Nasmyth K, Stillman D, Kipling D. Both positive and negative regulators of HO transcription are required for mother-cell-specific mating-type switching in yeast. Cell. 1987;48:579–587. doi: 10.1016/0092-8674(87)90236-4. [DOI] [PubMed] [Google Scholar]
- 3.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–858. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abrams E, Neigeborn L, Carlson M. Molecular analysis of SNF2 and SNF5, genes required for expression of glucose-repressible genes in Saccharomyces cerevisiae. Mol Cell Biol. 1986;6:3643–3651. doi: 10.1128/mcb.6.11.3643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sternberg PW, Stern MJ, Clark I, Herskowitz I. Activation of the yeast HO gene by release from multiple negative controls. Cell. 1987;48:567–577. doi: 10.1016/0092-8674(87)90235-2. [DOI] [PubMed] [Google Scholar]
- 6.Neigeborn L, Rubin K, Carlson M. Suppressors of SNF2 mutations restore invertase derepression and cause temperature-sensitive lethality in yeast. Genetics. 1986;112:741–753. doi: 10.1093/genetics/112.4.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kruger W, Peterson CL, Sil A, Coburn C, Arents G, Moudrianakis EN, Herskowitz I. Amino acid substitutions in the structured domains of histones H3 and H4 partially relieve the requirement of the yeast SWI/SNF complex for transcription. Genes Dev. 1995;9:2770–2779. doi: 10.1101/gad.9.22.2770. [DOI] [PubMed] [Google Scholar]
- 8.Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6:2288–2298. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 9.Winston F, Carlson M. Yeast SNF/SWI transcriptional activators and the SPT/SIN chromatin connection. Trends Genet. 1992;8:387–391. doi: 10.1016/0168-9525(92)90300-s. [DOI] [PubMed] [Google Scholar]
- 10.Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- 11.Foury F, Goffeau A. Genetic control of enhanced mutability of mitochondrial DNA and gamma-ray sensitivity in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1979;76:6529–6533. doi: 10.1073/pnas.76.12.6529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cairns BR, Schlichter A, Erdjument-Bromage H, Tempst P, Kornberg RD, Winston F. Two functionally distinct forms of the RSC nucleosome-remodeling complex, containing essential AT hook, BAH, and bromodomains. Mol Cell. 1999;4:715–723. doi: 10.1016/s1097-2765(00)80382-2. [DOI] [PubMed] [Google Scholar]
- 13.Vary JC, Jr., Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol Cell Biol. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 15.Luger K, Rechsteiner TJ, Flaus AJ, Waye MM, Richmond TJ. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol. 1997;272:301–311. doi: 10.1006/jmbi.1997.1235. doi: 10.1006/jmbi.1997.1235. [DOI] [PubMed] [Google Scholar]
- 16.Asturias FJ, Chung WH, Kornberg RD, Lorch Y. Structural analysis of the RSC chromatin-remodeling complex. Proc Natl Acad Sci U S A. 2002;99:13477–13480. doi: 10.1073/pnas.162504299. doi: 10.1073/pnas.162504299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Leschziner AE, Saha A, Wittmeyer J, Zhang Y, Bustamante C, Cairns BR, Nogales E. Conformational flexibility in the chromatin remodeler RSC observed by electron microscopy and the orthogonal tilt reconstruction method. Proc Natl Acad Sci U S A. 2007;104:4913–4918. doi: 10.1073/pnas.0700706104. doi: 10.1073/pnas.0700706104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skiniotis G, Moazed D, Walz T. Acetylated histone tail peptides induce structural rearrangements in the RSC chromatin remodeling complex. J Biol Chem. 2007;282:20804–20808. doi: 10.1074/jbc.C700081200. doi: 10.1074/jbc.C700081200. [DOI] [PubMed] [Google Scholar]
- 19.Chaban Y, Ezeokonkwo C, Chung WH, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leschziner AE, Lemon B, Tjian R, Nogales E. Structural studies of the human PBAF chromatin-remodeling complex. Structure. 2005;13:267–275. doi: 10.1016/j.str.2004.12.008. doi: 10.1016/j.str.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 21.Dechassa ML, Zhang B, Horowitz-Scherer R, Persinger J, Woodcock CL, Peterson CL, Bartholomew B. Architecture of the SWI/SNF-nucleosome complex. Mol Cell Biol. 2008;28:6010–6021. doi: 10.1128/MCB.00693-08. doi: 10.1128/MCB.00693-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kasten MM, Clapier CR, Cairns BR. SnapShot: Chromatin remodeling: SWI/SNF. Cell. 2011;144:310, e311. doi: 10.1016/j.cell.2011.01.007. doi: 10.1016/j.cell.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 23.Racki LR, Yang JG, Naber N, Partensky PD, Acevedo A, Purcell TJ, Cooke R, Cheng Y, Narlikar GJ. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 25.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hargreaves DC, Crabtree GR. ATP-dependent chromatin remodeling: genetics, genomics and mechanisms. Cell Res. 2011 doi: 10.1038/cr.2011.32. doi: 10.1038/cr.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahel D, Horejsi Z, Wiechens N, Polo SE, Garcia-Wilson E, Ahel I, Flynn H, Skehel M, West SC, Jackson SP, et al. Poly(ADP-ribose)-dependent regulation of DNA repair by the chromatin remodeling enzyme ALC1. Science. 2009;325:1240–1243. doi: 10.1126/science.1177321. doi: 10.1126/science.1177321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brzeski J, Jerzmanowski A. Deficient in DNA methylation 1 (DDM1) defines a novel family of chromatin-remodeling factors. J Biol Chem. 2003;278:823–828. doi: 10.1074/jbc.M209260200. doi: 10.1074/jbc.M209260200. [DOI] [PubMed] [Google Scholar]
- 30.Law MJ, Lower KM, Voon HP, Hughes JR, Garrick D, Viprakasit V, Mitson M, De Gobbi M, Marra M, Morris A, et al. ATR-X syndrome protein targets tandem repeats and influences allele-specific expression in a size-dependent manner. Cell. 2010;143:367–378. doi: 10.1016/j.cell.2010.09.023. doi: 10.1016/j.cell.2010.09.023. [DOI] [PubMed] [Google Scholar]
- 31.Gorbalenya AE, Koonin EV. Helicases: amino acid sequence comparisons and structure-function relationships. Current opinion in structural biology. 1993;3:419–429. [Google Scholar]
- 32.Singleton MR, Dillingham MS, Wigley DB. Structure and mechanism of helicases and nucleic acid translocases. Annu Rev Biochem. 2007;76:23–50. doi: 10.1146/annurev.biochem.76.052305.115300. doi: 10.1146/annurev.biochem.76.052305.115300. [DOI] [PubMed] [Google Scholar]
- 33.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling. Curr Opin Genet Dev. 2001;11:148–154. doi: 10.1016/s0959-437x(00)00172-6. [DOI] [PubMed] [Google Scholar]
- 34.Eberharter A, Becker PB. ATP-dependent nucleosome remodelling: factors and functions. J Cell Sci. 2004;117:3707–3711. doi: 10.1242/jcs.01175. doi: 10.1242/jcs.01175. [DOI] [PubMed] [Google Scholar]
- 35.Thoma NH, Czyzewski BK, Alexeev AA, Mazin AV, Kowalczykowski SC, Pavletich NP. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 36.Richmond E, Peterson CL. Functional analysis of the DNA-stimulated ATPase domain of yeast SWI2/SNF2. Nucleic Acids Res. 1996;24:3685–3692. doi: 10.1093/nar/24.19.3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 38.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hauk G, McKnight JN, Nodelman IM, Bowman GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–723. doi: 10.1016/j.molcel.2010.08.012. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szerlong H, Hinata K, Viswanathan R, Erdjument-Bromage H, Tempst P, Cairns BR. The HSA domain binds nuclear actin-related proteins to regulate chromatin-remodeling ATPases. Nat Struct Mol Biol. 2008;15:469–476. doi: 10.1038/nsmb.1403. doi: 10.1038/nsmb.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pufall MA, Graves BJ. Autoinhibitory domains: modular effectors of cellular regulation. Annu Rev Cell Dev Biol. 2002;18:421–462. doi: 10.1146/annurev.cellbio.18.031502.133614. doi: 10.1146/annurev.cellbio.18.031502.133614. [DOI] [PubMed] [Google Scholar]
- 42.Trotter KW, Fan HY, Ivey ML, Kingston RE, Archer TK. The HSA domain of BRG1 mediates critical interactions required for glucocorticoid receptor-dependent transcriptional activation in vivo. Mol Cell Biol. 2008;28:1413–1426. doi: 10.1128/MCB.01301-07. doi: 10.1128/MCB.01301-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Minsky A, Ghirlando R, Reich Z. Nucleosomes: a solution to a crowded intracellular environment? J Theor Biol. 1997;188:379–385. doi: 10.1006/jtbi.1997.0525. doi: 10.1006/jtbi.1997.0525. [DOI] [PubMed] [Google Scholar]
- 44.Sandman K, Reeve JN. Archaeal histones and the origin of the histone fold. Curr Opin Microbiol. 2006;9:520–525. doi: 10.1016/j.mib.2006.08.003. doi: 10.1016/j.mib.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 45.San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- 46.Sikorski TW, Buratowski S. The basal initiation machinery: beyond the general transcription factors. Curr Opin Cell Biol. 2009;21:344–351. doi: 10.1016/j.ceb.2009.03.006. doi: 10.1016/j.ceb.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laine JP, Egly JM. When transcription and repair meet: a complex system. Trends Genet. 2006;22:430–436. doi: 10.1016/j.tig.2006.06.006. doi: 10.1016/j.tig.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 48.Sukhodolets MV, Cabrera JE, Zhi H, Jin DJ. RapA, a bacterial homolog of SWI2/SNF2, stimulates RNA polymerase recycling in transcription. Genes Dev. 2001;15:3330–3341. doi: 10.1101/gad.936701. doi: 10.1101/gad.936701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shaw G, Gan J, Zhou YN, Zhi H, Subburaman P, Zhang R, Joachimiak A, Jin DJ, Ji X. Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription. Structure. 2008;16:1417–1427. doi: 10.1016/j.str.2008.06.012. doi: 10.1016/j.str.2008.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yawn B, Zhang L, Mura C, Sukhodolets MV. RapA, the SWI/SNF subunit of Escherichia coli RNA polymerase, promotes the release of nascent RNA from transcription complexes. Biochemistry. 2009;48:7794–7806. doi: 10.1021/bi9004123. doi: 10.1021/bi9004123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nishino T, Komori K, Tsuchiya D, Ishino Y, Morikawa K. Crystal structure and functional implications of Pyrococcus furiosus hef helicase domain involved in branched DNA processing. Structure. 2005;13:143–153. doi: 10.1016/j.str.2004.11.008. doi: 10.1016/j.str.2004.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Erdel F, Rippe K. ISWI chromatin remodellers in mammalian cells - where, when and why? FEBS Journal. 2011 doi: 10.1111/j.1742-4658.2011.08282.x. [DOI] [PubMed] [Google Scholar]
- 53.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J Mol Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu CL, Kaplan T, Kim M, Buratowski S, Schreiber SL, Friedman N, Rando OJ. Single-nucleosome mapping of histone modifications in S. cerevisiae. PLoS biology. 2005;3:e328. doi: 10.1371/journal.pbio.0030328. doi: 10.1371/journal.pbio.0030328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van Attikum H, Gasser SM. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle. 2005;4:1011–1014. doi: 10.4161/cc.4.8.1887. [DOI] [PubMed] [Google Scholar]
- 56.Korber P, Becker PB. Nucleosome dynamics and epigenetic stability. Essays Biochem. 2010;48:63–74. doi: 10.1042/bse0480063. doi: 10.1042/bse0480063. [DOI] [PubMed] [Google Scholar]
- 57.Koerber RT, Rhee HS, Jiang C, Pugh BF. Interaction of transcriptional regulators with specific nucleosomes across the Saccharomyces genome. Mol Cell. 2009;35:889–902. doi: 10.1016/j.molcel.2009.09.011. doi: 10.1016/j.molcel.2009.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xella B, Goding C, Agricola E, Di Mauro E, Caserta M. The ISWI and CHD1 chromatin remodelling activities influence ADH2 expression and chromatin organization. Mol Microbiol. 2006;59:1531–1541. doi: 10.1111/j.1365-2958.2005.05031.x. doi: 10.1111/j.1365-2958.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 59.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 60.Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–257. doi: 10.1038/nsmb.1741. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 61.Shukla MS, Syed SH, Montel F, Faivre-Moskalenko C, Bednar J, Travers A, Angelov D, Dimitrov S. Remosomes: RSC generated non-mobilized particles with approximately 180 bp DNA loosely associated with the histone octamer. Proc Natl Acad Sci U S A. 2010;107:1936–1941. doi: 10.1073/pnas.0904497107. doi: 10.1073/pnas.0904497107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 63.Floer M, Wang X, Prabhu V, Berrozpe G, Narayan S, Spagna D, Alvarez D, Kendall J, Krasnitz A, Stepansky A, et al. A RSC/nucleosome complex determines chromatin architecture and facilitates activator binding. Cell. 2010;141:407–418. doi: 10.1016/j.cell.2010.03.048. doi: 10.1016/j.cell.2010.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grune T, Brzeski J, Eberharter A, Clapier CR, Corona DF, Becker PB, Muller CW. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 65.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. Embo J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Blosser TR, Yang JG, Stone MD, Narlikar GJ, Zhuang X. Dynamics of nucleosome remodelling by individual ACF complexes. Nature. 2009;462:1022–1027. doi: 10.1038/nature08627. doi: 10.1038/nature08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Strohner R, Wachsmuth M, Dachauer K, Mazurkiewicz J, Hochstatter J, Rippe K, Langst G. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 69.Awad S, Ryan D, Prochasson P, Owen-Hughes T, Hassan AH. The Snf2 Homolog Fun30 Acts as a Homodimeric ATP-dependent Chromatin-remodeling Enzyme. Journal of Biological Chemistry. 2010;285:9477–9484. doi: 10.1074/jbc.M109.082149. doi: DOI 10.1074/jbc.M109.082149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boeger H, Griesenbeck J, Kornberg RD. Nucleosome retention and the stochastic nature of promoter chromatin remodeling for transcription. Cell. 2008;133:716–726. doi: 10.1016/j.cell.2008.02.051. doi: 10.1016/j.cell.2008.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat Struct Mol Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dechassa ML, Sabri A, Pondugula S, Kassabov SR, Chatterjee N, Kladde MP, Bartholomew B. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lavelle C, Praly E, Bensimon D, Le Cam E, Croquette V. Nucleosome remodelling machines and other molecular motors observed at single molecule level. FEBS Journal. 2011 doi: 10.1111/j.1742-4658.2011.08280.x. [DOI] [PubMed] [Google Scholar]
- 74.Whitehouse I, Flaus A, Cairns BR, White MF, Workman JL, Owen-Hughes T. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature. 1999;400:784–787. doi: 10.1038/23506. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 75.Langst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- 76.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 77.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat Struct Mol Biol. 2005;12:747–755. doi: 10.1038/nsmb973. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 78.Lohman TM, Tomko EJ, Wu CG. Non-hexameric DNA helicases and translocases: mechanisms and regulation. Nat Rev Mol Cell Biol. 2008;9:391–401. doi: 10.1038/nrm2394. doi: 10.1038/nrm2394. [DOI] [PubMed] [Google Scholar]
- 79.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 80.Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tan S, Davey CA. Nucleosome structural studies. Current opinion in structural biology. 2011;21:128–136. doi: 10.1016/j.sbi.2010.11.006. doi: 10.1016/j.sbi.2010.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Davey CA, Sargent DF, Luger K, Maeder AW, Richmond TJ. Solvent mediated interactions in the structure of the nucleosome core particle at 1.9 a resolution. J Mol Biol. 2002;319:1097–1113. doi: 10.1016/S0022-2836(02)00386-8. doi: 10.1016/S0022-2836(02)00386-8. [DOI] [PubMed] [Google Scholar]
- 83.Flaus A, Rencurel C, Ferreira H, Wiechens N, Owen-Hughes T. Sin mutations alter inherent nucleosome mobility. Embo J. 2004;23:343–353. doi: 10.1038/sj.emboj.7600047. doi: 10.1038/sj.emboj.7600047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.van Vugt JJ, de Jager M, Murawska M, Brehm A, van Noort J, Logie C. Multiple aspects of ATP-dependent nucleosome translocation by RSC and Mi-2 are directed by the underlying DNA sequence. Plos One. 2009;4:e6345. doi: 10.1371/journal.pone.0006345. doi: 10.1371/journal.pone.0006345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Polach KJ, Widom J. Mechanism of protein access to specific DNA sequences in chromatin: a dynamic equilibrium model for gene regulation. J Mol Biol. 1995;254:130–149. doi: 10.1006/jmbi.1995.0606. doi: 10.1006/jmbi.1995.0606. [DOI] [PubMed] [Google Scholar]
- 86.Li G, Levitus M, Bustamante C, Widom J. Rapid spontaneous accessibility of nucleosomal DNA. Nat Struct Mol Biol. 2005;12:46–53. doi: 10.1038/nsmb869. doi: 10.1038/nsmb869. [DOI] [PubMed] [Google Scholar]
- 87.Bohm V, Hieb AR, Andrews AJ, Gansen A, Rocker A, Toth K, Luger K, Langowski J. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq1279. doi: 10.1093/nar/gkq1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bowman A, Ward R, El-Mkami H, Owen-Hughes T, Norman DG. Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling. Nucleic Acids Res. 2010;38:695–707. doi: 10.1093/nar/gkp1003. doi: 10.1093/nar/gkp1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sekulic N, Bassett EA, Rogers DJ, Black BE. The structure of (CENP-A-H4)(2) reveals physical features that mark centromeres. Nature. 2010;467:347–351. doi: 10.1038/nature09323. doi: 10.1038/nature09323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 92.Flaus A, Owen-Hughes T. Mechanisms for nucleosome mobilization. Biopolymers. 2003;68:563–578. doi: 10.1002/bip.10323. doi: 10.1002/bip.10323. [DOI] [PubMed] [Google Scholar]
- 93.Blossey R, Schiessel H. The dynamics of the nucleosome: thermal effects, external forces, and ATP. FEBS Journal. 2011 doi: 10.1111/j.1742-4658.2011.08283.x. [DOI] [PubMed] [Google Scholar]
- 94.Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 95.Smith CL, Horowitz-Scherer R, Flanagan JF, Woodcock CL, Peterson CL. Structural analysis of the yeast SWI/SNF chromatin remodeling complex. Nat Struct Biol. 2003 doi: 10.1038/nsb888. [DOI] [PubMed] [Google Scholar]
- 96.Cairns BR, Lorch Y, Li Y, Zhang M, Lacomis L, Erdjument-Bromage H, Tempst P, Du J, Laurent B, Kornberg RD. RSC, an essential, abundant chromatin-remodelling complex. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 97.Chaban Y, Ezeokonkwo C, Chung W-H, Zhang F, Kornberg RD, Maier-Davis B, Lorch Y, Asturias FJ. Structure of a RSC-nucleosome complex and insights into chromatin remodeling. Nat Struct Mol Biol. 2008;15:1272–1277. doi: 10.1038/nsmb.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Gelbart ME, Rechsteiner T, Richmond TJ, Tsukiyama T. Interactions of Isw2 chromatin remodeling complex with nucleosomal arrays: analyses using recombinant yeast histones and immobilized templates. Mol Cell Biol. 2001;21:2098–2106. doi: 10.1128/MCB.21.6.2098-2106.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tran HG, Steger DJ, Iyer VR, Johnson AD. The chromo domain protein chd1p from budding yeast is an ATP-dependent chromatin-modifying factor. Embo J. 2000;19:2323–2331. doi: 10.1093/emboj/19.10.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. Journal of Biological Chemistry. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. doi: DOI 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mizuguchi G, Shen X, Landry J, Wu WH, Sen S, Wu C. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science. 2004;303:343–348. doi: 10.1126/science.1090701. [DOI] [PubMed] [Google Scholar]
- 102.Krogan NJ, Keogh MC, Datta N, Sawa C, Ryan OW, Ding H, Haw RA, Pootoolal J, Tong A, Canadien V, et al. A Snf2 family ATPase complex required for recruitment of the histone H2A variant Htz1. Mol Cell. 2003;12:1565–1576. doi: 10.1016/s1097-2765(03)00497-0. [DOI] [PubMed] [Google Scholar]
- 103.Kobor MS, Venkatasubrahmanyam S, Meneghini MD, Gin JW, Jennings JL, Link AJ, Madhani HD, Rine J. A Protein Complex Containing the Conserved Swi2/Snf2-Related ATPase Swr1p Deposits Histone Variant H2A.Z into Euchromatin. Plos Biology. 2004;2:E131. doi: 10.1371/journal.pbio.0020131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Luk E, Ranjan A, FitzGerald PC, Mizuguchi G, Huang Y, Wei D, Wu C. Stepwise Histone Replacement by SWR1 Requires Dual Activation with Histone H2A.Z and Canonical Nucleosome. Cell. 2010;143:725–736. doi: 10.1016/j.cell.2010.10.019. doi: DOI 10.1016/j.cell.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Neves-Costa A, Will WR, Vetter AT, Miller JR, Varga-Weisz P. The SNF2-Family Member Fun30 Promotes Gene Silencing in Heterochromatic Loci. Plos One. 2009;4 doi: 10.1371/journal.pone.0008111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 107.Papamichos-Chronakis M, Watanabe S, Rando OJ, Peterson CL. Global Regulation of H2A.Z Localization by the INO80 Chromatin-Remodeling Enzyme Is Essential for Genome Integrity. Cell. 2011;144:200–213. doi: 10.1016/j.cell.2010.12.021. doi: DOI: 10.1016/j.cell.2010.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Clever B, Interthal H, SchmuckliMaurer J, King J, Sigrist M, Heyer WD. Recombinational repair in yeast: Functional interactions between Rad51 and Rad54 proteins. Embo Journal. 1997;16:2535–2544. doi: 10.1093/emboj/16.9.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Shah PP, Zheng XZ, Epshtein A, Carey JN, Bishop DK, Klein HL. Swi2/Snf2-Related Translocases Prevent Accumulation of Toxic Rad51 Complexes during Mitotic Growth. Mol Cell. 2010;39:862–872. doi: 10.1016/j.molcel.2010.08.028. doi: DOI 10.1016/j.molcel.2010.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Klein HL. RDH54, a RAD54 homologue in Saccharomyces cerevisiae, is required for mitotic diploid-specific recombination and repair and for meiosis. Genetics. 1997;147:1533–1543. doi: 10.1093/genetics/147.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chi P, Kwon Y, Moses DN, Seong C, Sehorn MG, Singh AK, Tsubouchi H, Greene EC, Klein HL, Sung P. Functional interactions of meiotic recombination factors Rdh54 and Dmc1. DNA Repair. 2009;8:279–284. doi: 10.1016/j.dnarep.2008.10.012. doi: DOI 10.1016/j.dnarep.2008.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Johnson RE, Henderson ST, Petes TD, Prakash S, Bankmann M, Prakash L. Saccharomyces-Cerevisiae Rad5-Encoded DNA-Repair Protein Contains DNA Helicase and Zinc-Binding Sequence Motifs and Affects the Stability of Simple Repetitive Sequences in the Genome. Mol Cell Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Blastyak A, Pinter L, Unk I, Prakash L, Prakash S, Haracska L. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. doi: DOI 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Guzder SN, Sung P, Prakash L, Prakash S. Yeast Rad7-Rad16 complex, specific for the nucleotide excision repair of the nontranscribed DNA strand, is an ATP-dependent DNA damage sensor. J Biol Chem. 1997;272:21665–21668. doi: 10.1074/jbc.272.35.21665. [DOI] [PubMed] [Google Scholar]
- 115.Reed SH, Akiyama M, Stillman B, Friedberg EC. Yeast autonomously replicating sequence binding factor is involved in nucleotide excision repair. Genes Dev. 1999;13:3052–3058. doi: 10.1101/gad.13.23.3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. Plos Genet. 2007;3:2439–2449. doi: 10.1371/journal.pgen.0030228. doi: ARTN e228 DOI 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhang ZM, Buchman AR. Identification of a member of a DNA-dependent ATPase family that causes interference with silencing. Mol Cell Biol. 1997;17:5461–5472. doi: 10.1128/mcb.17.9.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vangool AJ, Verhage R, Swagemakers SMA, Vandeputte P, Brouwer J, Troelstra C, Bootsma D, Hoeijmakers JHJ. Rad26, the Functional Saccharomyces-Cerevisiae Homolog of the Cockayne-Syndrome-B Gene Ercc6. Embo Journal. 1994;13:5361–5369. doi: 10.1002/j.1460-2075.1994.tb06871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Reid J, Svejstrup JQ. DNA damage-induced Def1-RNA polymerase II interaction and Def1 requirement for polymerase ubiquitylation in vitro. Journal of Biological Chemistry. 2004;279:29875–29878. doi: 10.1074/jbc.C400185200. doi: DOI 10.1074/jbc.C400185200. [DOI] [PubMed] [Google Scholar]
- 120.Poon D, Campbell AM, Bai Y, Weil PA. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. J Biol Chem. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 121.Auble DT, Hansen KE, Mueller CG, Lane WS, Thorner J, Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 122.Gumbs OH, Campbell AM, Weil PA. High-affinity DNA binding by a Mot1p-TBP complex: implications for TAF-independent transcription. Embo J. 2003;22:3131–3141. doi: 10.1093/emboj/cdg304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Ghaemmaghami S, Huh WK, Bower K, Howson RW, Belle A, Dephoure N, O’Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 124.Fan L, Arvai AS, Cooper PK, Iwai S, Hanaoka F, Tainer JA. Conserved XPB core structure and motifs for DNA unwinding: implications for pathway selection of transcription or excision repair. Mol Cell. 2006;22:27–37. doi: 10.1016/j.molcel.2006.02.017. doi: 10.1016/j.molcel.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 125.Sengoku T, Nureki O, Nakamura A, Kobayashi S, Yokoyama S. Structural basis for RNA unwinding by the DEAD-box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]