Abstract
Multifocal plane microscopy (MUM) has made it possible to study subcellular dynamics in 3D at high temporal and spatial resolution by simultaneously imaging distinct planes within the specimen. MUM allows high accuracy localization of a point source along the z-axis since it overcomes the depth discrimination problem of conventional single plane microscopy. An important question in MUM experiments is how the number of focal planes and their spacings should be chosen to achieve the best possible localization accuracy along the z-axis. Here, we propose approaches based on the Fisher information matrix and report spacing scenarios called strong coupling and weak coupling which yield an appropriate 3D localization accuracy. We examine the effect of numerical aperture, magnification, photon count, emission wavelength and extraneous noise on the spacing scenarios. In addition, we investigate the effect of changing the number of focal planes on the 3D localization accuracy. We also introduce a new software package that provides a user-friendly framework to find appropriate plane spacings for a MUM setup. These developments should assist in optimizing MUM experiments.
OCIS codes: (100.2000) Digital image processing, (180.2520) Fluorescence microscopy, (110.3055) Information theoretical analysis, (180.6900) Three-dimensional microscopy
1. Introduction
The development of multifocal plane microscopy (MUM) in the recent past has made it possible to track the 3D dynamics in live cells at high temporal and spatial resolution by simultaneously imaging different focal planes within the sample [1–7]. MUM overcomes the depth discrimination problem of conventional single plane microscopy and thereby allows high accuracy localization of subcellular structures and single molecules along the z-axis [3,8–10]. MUM has been used to study different biological problems [11–14]. For instance, in [11] MUM is used to understand the effect of tubulovesicular transport carriers on intracellular trafficking pathways within 3D cellular environments. Furthermore, in [13] MUM is utilized to study the 3D dynamics of single molecules in live epithelial cells.
One of the important questions in the design of MUM experiments concerns selecting the appropriate spacings between the focal planes. Varying the plane spacing changes the image profiles of the object of interest (e.g. a point source) at the focal planes. An implication of changes in the image profiles is that the accuracy for localizing the object as it moves along the z-axis will be affected [3, 14]. Another implication is that the object of interest may become difficult to detect in the acquired image. For instance, the image profile at a focal plane far from the object of interest will be diffusely spread out, making the object both difficult to localize and difficult to detect with respect to that focal plane. As a consequence, for a given MUM setup the choice of the plane spacing determines whether a point source can be localized with a consistent level of accuracy, and whether it can be continuously detected by the imaging system, as it moves along the z-axis.
In addition to the plane spacing, selecting the appropriate number of focal planes to cover a sample of a certain thickness is also of importance in the design of MUM experiments. Different authors have used different numbers of planes to cover their desired viewing and tracking depth, i.e. a range along the z-axis over which the particle is detectable by the imaging system. For instance, in [12] a 4-plane MUM setup, covering a 2 μm depth, is utilized to study the dynamics of tubulovesicular transport containers. In [13] a 4-plane MUM setup providing a 10 μm viewing and tracking depth is used to study the 3D single molecule dynamics in live epithelial cells. More recently, in [15] a 9-plane MUM setup is developed that provides a viewing and tracking depth of 2.25 – 18 μm. Increasing the number of focal planes can enhance the viewing and tracking depth. However, in fluorescence microscopy experiments, regardless of the number of focal planes, a specific number of photons is collected from the sample per acquisition. This fixed number of photons is then split among multiple focal planes. Hence, each plane detects fewer photons when the number of planes is increased. A poorer localization accuracy might therefore be obtained when using a large number of planes. This is due to the fact that decreasing the number of detected photons at each focal plane worsens the localization accuracy of the MUM setup [14, 16].
In this paper, we address the above concerns by investigating the practical localization accuracy measure (PLAM) for a MUM setup. The PLAM provides the best possible accuracy (standard deviation) with which an isolated single molecule can be localized, and it is calculated using the Fisher information matrix (FIM) [16–18]. The latter represents the amount of information the data provides about an unknown parameter [19]. For our analysis, we consider two design requirements that are typically encountered while setting up MUM experiments. The first requirement is to achieve a relatively constant PLAM along the z-axis such that the 3D location of a subcellular structure or single molecule can be estimated with the same level of accuracy across the viewing and tracking depth. The second requirement is to allow for a relatively large viewing and tracking depth across the sample in order to cover the z-range over which the cellular process of interest occurs. Taking into account these design considerations, we provide guidelines to set up appropriate MUM experiments for different applications. Aside from the number of planes and their spacings, a variety of other imaging parameters such as photon count, system magnification and the numerical aperture of the objective lens also influence the PLAM. Here, we also examine the effect of these parameters on the plane spacing.
In practice, the calculation of the PLAM is computationally expensive. Thus, without appropriate software determining the appropriate number of focal planes and their spacing can be a complicated and time consuming procedure. We therefore also introduce a new software module called MUMDesignTool that calculates and plots the PLAM along the z-axis and provides a user-friendly framework for finding the appropriate number of planes and plane spacings for a MUM setup. The results of the paper can be reproduced using the MUMDesignTool.
2. Simulations
The results of this paper are calculated using a new software module, the MUMDesignTool, developed in the MATLAB environment (The MathWorks Inc., Natick, MA). The software is available at http://www.wardoberlab.com/ and its detailed description can be found in Section 4.8. Using this software we model an isolated single molecule that is imaged by a pixelated detector in the presence of background and readout noise [16]. A Poisson process models the background effect and a Gaussian process models the readout noise of the detector (see Appendix A for more details). The pixel dimensions are 13 μm × 13 μm. We assume the point spread function (PSF) is given by the Born and Wolf model [20]. The emission wavelength is assumed to be the same for all focal planes. The refractive index noil of the immersion oil is 1.515. The tube length L of the microscope is 160 mm. The remaining parameters are given below each figure.
3. Theory
3.1. Behavior of the PLAM and the Fisher information matrix for a MUM setup
In this section, we briefly review the concepts concerning the PLAM and define several terms which are necessary for our later discussions. For all of our analysis we assume that a single molecule is modeled as a point source. We denote the best possible localization accuracy that can be achieved for estimating the x, y and z coordinates of the single molecule by x0-PLAM, y0-PLAM and z0-PLAM (axial-PLAM), respectively. The PLAMs are calculated using the well-known Cramér-Rao lower bound (CRLB) [19,21], which is specified in terms of the inverse of the FIM (see Appendix A). The FIM represents the amount of information the data provides about an unknown parameter of interest which in the current case pertains to the 3D location of the single molecule [19, 22]. We refer to the amount of information about the x, y and z coordinates of the single molecule as x0-FIM, y0-FIM and z0-FIM (axial-FIM). Large numerical values of x0-FIM, y0-FIM and axial-FIM, which correspond to small values of x0-PLAM, y0-PLAM and axial-PLAM, respectively, specify a better localization accuracy.
The FIM and therefore the PLAM depend on a variety of imaging parameters such as the numerical aperture of the objective lens, the magnification, the emission wavelength, the photon count and the z-position of the point source. The behavior of the PLAM as a function of the mentioned parameters can be explained through the mathematical expression of the FIM (see Appendix A). This paper is primarily devoted to the investigation of the effect of these parameters on the PLAM, in the context of MUM, with an emphasis on the effect of the z-position of the point source with respect to the focal planes.
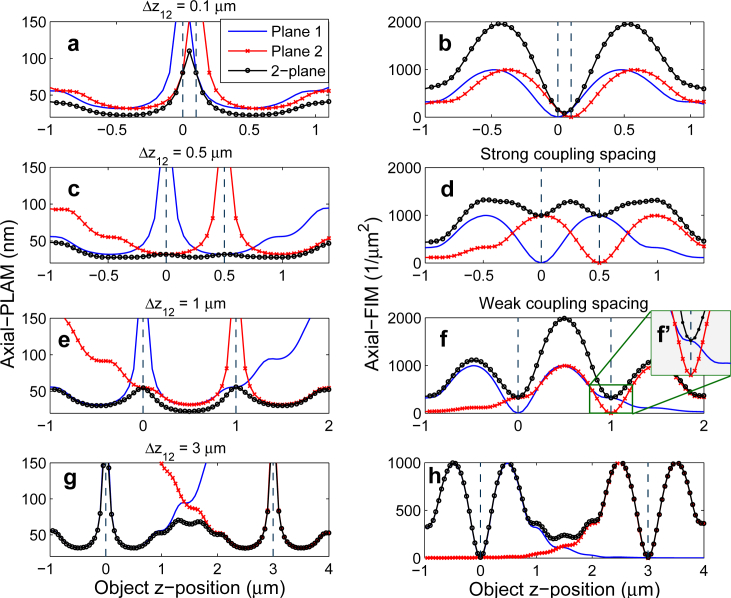
Figure 1(a) shows the axial-FIM for a conventional single plane microscopy setup with a standard infinity-corrected (i.e. design) focal plane as a function of the z-position of the point source (“Plane 1”). For ease of reference, we denote this and its corresponding axial-PLAM by axial-FIM1 and axial-PLAM1, respectively. As can be seen, the axial-FIM1 is zero when the object is at the focal plane, implying that the data does not provide any information about the z-location of the point source; we refer to this spot as the zero of the axial-FIM1. A consequence of the zero of the axial-FIM1 is the depth discrimination problem, implying that there is a high uncertainty in estimating the z-position of the point source when it is located near the focal plane [3,18]. This problem is illustrated in Fig. 1(b) (“Plane 1”), which shows the axial-PLAM1 increases without bound as the point source approaches Plane 1.
Fig. 1.
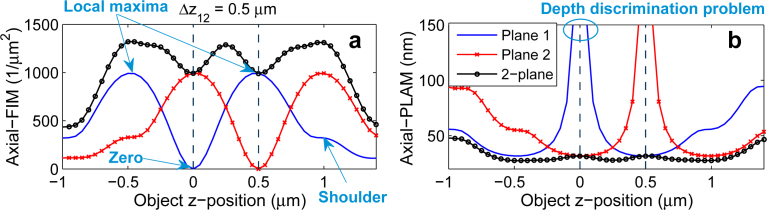
The behavior of the axial-FIM and the axial-PLAM for conventional single plane microscopy and MUM. (a) The axial-FIM and (b) the axial-PLAM for a 2-plane MUM setup and two conventional single plane setups (Plane 1 and Plane 2) as a function of the z-position of the point source. The zero, local maxima and shoulders of the axial-FIM of Plane 1, and the depth discrimination problem can be seen in panels (a) and (b), respectively. The results are calculated for a 100x, NA 1.3 objective lens. The plane spacing (Δz12) is 0.5 μm, the photon count is 250 photons/plane and the emission wavelength is 520 nm. The background level is 1 photon/pixel/plane and the standard deviation of the readout noise is 2 e−/pixel. The ROI size is 11 × 11 pixels.
By moving the point source away from the focal plane the axial-FIM1 increases and at some z-position the axial-FIM1 reaches a peak which we refer to as the local maximum of the axial-FIM1. Further moving the point source away from the focal plane gradually decreases the axial-FIM1 and at some z-position the axial-FIM1 has a bump which we refer to as the shoulder of the axial-FIM1. The described behavior of the axial-FIM1 is symmetric with respect to the z-position of the focal plane due to the axial symmetry of the Born and Wolf 3D PSF, which has been used to calculate the FIM [20].
Figure 1(b) also shows the axial-PLAM for a 2-plane MUM setup along the z-axis. For brevity, we refer to this and its corresponding axial-FIM as axial-PLAMMUM and axial-FIMMUM, respectively. As can be seen, the axial-PLAMMUM is relatively constant along the z-axis including at the focal planes when compared to the axial-PLAM for a conventional (single-plane) microscope (i.e. “Plane 1”). This implies that MUM overcomes the depth discrimination problem of conventional microscopy and allows high accuracy z-localization. This is due to the fact that the axial-FIMMUM is the sum of the axial-FIMs of the individual planes (i.e. axial-FIM1 and axial-FIM2) because of the independence of data acquisition at each focal plane (see Fig. 1(a)) [3, 14]. Hence, the axial-FIMMUM is nonzero for a range of z-positions including at the focal planes.
3.2. Graphical interpretation of the design of focal plane spacing for a MUM setup
We now give a graphical interpretation for the design of the focal plane spacing for a MUM setup. Our design objective is to obtain an appropriate level of the axial-PLAM for the MUM system. Due to the fact that the PLAMMUM is specified in terms of the inverse of the FIMMUM, minimizing the axial-PLAMMUM is closely related to maximizing the axial-FIMMUM (see Appendix A for a more detailed discussion of the relationship of the axial-PLAM to the axial-FIM). We carry out the main steps of the focal plane spacing design by investigating the axial-FIM as this will prove to be a very convenient criterion due to two important properties of the axial-FIM. First, as pointed out earlier, the axial-FIMMUM is the sum of the axial-FIMs for the planes that make up the MUM system (see also Appendix A). It is this additive property of the axial-FIMs that makes it very convenient to define the focal plane spacing problem in terms of the axial-FIMs rather than directly through the axial-PLAMs. Hence, as shown in Fig. 1(a), the axial-FIMMUM curve along the z-axis is obtained by adding the axial-FIM curves of the individual focal planes (i.e. axial-FIM1 and axial-FIM2 for the 2-plane configuration of Fig. 1). The second property of the axial-FIM which we need is that, to a good approximation, the graphs of axial-FIMs of the different focal planes are simply translated versions of the graph of the axial-FIM for the design focal plane along the z-axis (see Fig. 1(a)). We should note that this property is based on the assumption that different focal planes have similar experimental conditions, e.g. the photon count and extraneous noise, as would be the case if identical detectors are used and the emission light is equally split amongst the detectors for the different planes. This second property in particular implies that changing the position of a focal plane with respect to the other planes amounts to a corresponding translation of the graph of the axial-FIM.
These two properties immediately provide the basis for a graphical interpretation of the design process for focal plane spacing. The design process can therefore be thought of as shifting the graphs of the axial-FIMs such that their sum, i.e. the axial-FIM of the MUM configuration, has the desired values for the range of z-positions for the point source that are of interest. Building on the idea of shifting and adding the similar FIMs for the different focal planes, in Appendix B we develop a fast approach for the calculation of the FIMMUM that also accounts for focal planes with different photon counts.
4. Results
4.1. Strong and weak coupling spacings: constant z-localization accuracy along the z-axis
An important requirement in the design of MUM experiments is to achieve a constant axial-PLAMMUM along the z-axis as it allows estimating the axial location of an object with a constant level of accuracy across the viewing and tracking depth. Here, as a first step we investigate the influence of different plane spacings on the axial-PLAMMUM assuming that the number of focal planes is two. In the subsequent sections, we will study MUM setups with more than two focal planes. The behavior of the axial-PLAM of a 2-plane MUM setup for different plane spacings is illustrated in the left-hand side plots of Fig. 2, where it is shown that altering the plane spacing changes the flatness of the curve. When the plane spacing is small (Δz12 = 0.1 μm), there is a significant variation in the axial-PLAMMUM value between the focal planes, i.e. the axial-PLAMMUM varies from 22 nm to 110 nm over the z-range of [−0.6, 1.1] μm (see Fig. 2(a)).
Fig. 2.
The effect of plane spacing on the axial-PLAM and the axial-FIM for a MUM setup. The left-hand (right-hand) side plots show the axial-PLAM (axial-FIM) for a 2-plane MUM setup as a function of the z-position of the point source. The figure also shows the axial-FIMs and axial-PLAMs of focal planes 1 and 2. The plane spacings (Δz12) are 0.1, 0.5, 1 and 3 μm from top to bottom. (c) and (d) show the strong coupling spacing whereas (e) and (f) show the weak coupling spacing. (f’) shows the shoulder of the axial-FIM1 more clearly. The results are calculated for a 100x, NA 1.3 oil immersion objective lens where the photon count is 250 photons/plane and the emission wavelength is 520 nm. The background level is 1 photon/pixel/plane and the standard deviation of the readout noise is 2 e−/pixel. The ROI size is 11 × 11 pixels.
By increasing the plane spacing, the axial-PLAMMUM becomes more constant along the z-axis and a certain spacing (Δz12 = 0.5 μm) yields a relatively flat curve (see Fig. 2(c)). For this spacing, the axial-PLAMMUM varies from 28 nm to 31 nm over the same z-range of [−0.6, 1.1] μm. This relatively constant axial-PLAMMUM is achieved when the local maximum of the axial-FIM of the second plane (axial-FIM2) falls on the zero of the axial-FIM of the first plane (axial-FIM1), as shown in Fig. 2(d). We refer to this spacing as the strong coupling spacing. Further increasing the plane spacing worsens the flatness of the curve (see Fig. 2(e)). Hence, we set the largest acceptable plane spacing to be the case where the shoulder of the axial-FIM2 overlaps with the zero of the axial-FIM1, which can be seen in Figs. 2(f) and 2(f’). We refer to this spacing as the weak coupling spacing. For this spacing, the axial-PLAMMUM varies from 23 nm to 54 nm over the z-range of [−0.6, 1.1] μm, as can be seen in Fig. 2(e) (note that the variation also remains unchanged over the larger z-range of [−1, 2] μm).
A very large plane spacing (Δz12 = 3 μm) results in significant variations in the curve, with large axial-PLAMMUM values both between the focal planes and at the focal planes, i.e. the axial-PLAMMUM varies from 31 nm to 381 nm over the z-range of [−0.6, 1.1] μm (see Fig. 2(g)). The latter is due to the fact that the axial-FIMs of two distantly spaced focal planes make small contributions to the axial-FIMMUM at the focal planes (see Fig. 2(h)). Hence, the axial-PLAMMUM at each focal plane is large. In other words, two distantly spaced focal planes are similar to two separate conventional microscopy setups.
4.2. Lateral-PLAM and constant x- and y-localization accuracy along the z-axis
Aside from a constant z-localization accuracy, achieving constant x- and y-localization accuracy is also of importance in the context of 3D tracking. Here, we analyze the behavior of the x0-PLAM and y0-PLAM for a MUM setup along the z-axis. For this purpose, we define the lateral-PLAM as the square root of the sum of (x0-PLAMMUM)2 and (y0-PLAMMUM)2. The lateral-PLAM quantifies the best possible accuracy for the lateral localization of a particle. A large value for the lateral-PLAM at a certain z-position implies a poor lateral localization accuracy at that z-position.
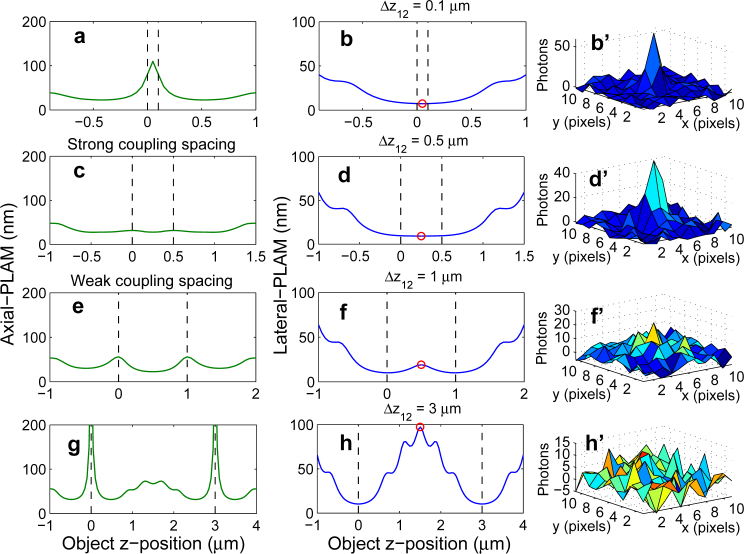
Figure 3 shows the axial-PLAM and the corresponding lateral-PLAM for a 2-plane MUM setup for different plane spacings. All imaging conditions are the same as those used for Fig. 2. When the focal planes are located close to one another (Δz12 = 0.1 μm), the lateral-PLAM varies from 7.3 nm to 32.6 nm over the z-range of [−0.3, 0.8] μm (see Fig. 3(b)). Adjusting the plane spacing based on the strong coupling spacing (Fig. 3(c)) and the weak coupling spacing (Fig. 3(e)) provides relatively constant lateral-PLAMs. More specifically, for the strong coupling spacing the lateral-PLAM varies from 9.2 nm to 13.7 nm over the z-range of [−0.3, 0.8] μm (Fig. 3(d)), whereas for the weak coupling spacing it varies from 10.1 nm to 19 nm over the same z-range (Fig. 3(f)). For a large plane spacing (Δz12 = 3 μm), the lateral-PLAM varies significantly, i.e. from 10.3 nm to 45.1 nm, over the same z-range (see Fig. 3(h)) which implies that the lateral location of the particle cannot be estimated with a constant level of accuracy.
Fig. 3.
The behavior of the lateral-PLAM. The left and middle columns show the axial-PLAM and the lateral-PLAM, respectively, for a 2-plane MUM setup as a function of the z-position of the point source. The plane spacings (Δz12) are 0.1, 0.5, 1 and 3 μm from top to bottom. The right column shows the mesh plots of the simulated images of point sources located at z-positions shown by the red circles on the design plane. The simulation parameters are identical to those used in Fig. 2.
4.3. Large viewing depth for qualitative imaging applications
In the previous sections, we analyzed the effect of plane spacing on the 3D localization accuracy of a point source along the z-axis and discussed spacing scenarios for quantitative 3D tracking using MUM. In some imaging applications, however, the objective is the qualitative 3D visualization of events. An example of such applications is the trafficking of receptors from sorting endosomes to the plasma membrane [12]. A key requirement in such cases is that the particle/structure is continuously detectable in the acquired data as it moves within the sample. Here, by making use of the lateral-PLAM, we investigate how the plane spacing affects the visual identifiability of a particle in a MUM setup.
By definition, a large value of the lateral-PLAM predicts poor lateral localization accuracy, which can also be interpreted as high uncertainty in visually detecting the particle in the acquired image. Figure 3 (middle panels) shows the behavior of the lateral-PLAM for different plane spacings for a 2-plane MUM setup. For a small plane spacing (Δz12 = 0.1 μm), the numerical value of the lateral-PLAM at the midpoint between the focal planes is relatively small, i.e. 7.3 nm. Correspondingly, the point source can be clearly visually identified in a MUM image as shown in the mesh plot (Fig. 3(b’), where only the image from the first focal plane is shown). As the plane spacing increases, the numerical value of the lateral-PLAM varies significantly such that at a certain plane spacing the point source becomes barely detectable in the image. For example, for a plane spacing of 1 μm that corresponds to the weak coupling spacing, the lateral-PLAM for a point source at z = 0.5 μm is 19 nm and at this position the particle is marginally visible in the image (see Fig. 3(f’)). For larger plane spacings the particle can no longer be identified in the image (Fig. 3(h’)). This is due to the fact that as the plane spacing increases, for certain z-positions (especially near the midpoint between the focal planes), the distance between the particle and each focal plane becomes so large that the particle is significantly out of focus and is therefore undetectable in the image.
Thus, from the above discussion we see that the weak coupling scenario provides a guideline for adjusting the plane spacing to achieve a large viewing range and helps in designing MUM setups, for example, to observe a particular cellular process that occurs over a large z-range.
4.4. Effects of numerical aperture, magnification and emission wavelength on the spacing scenarios
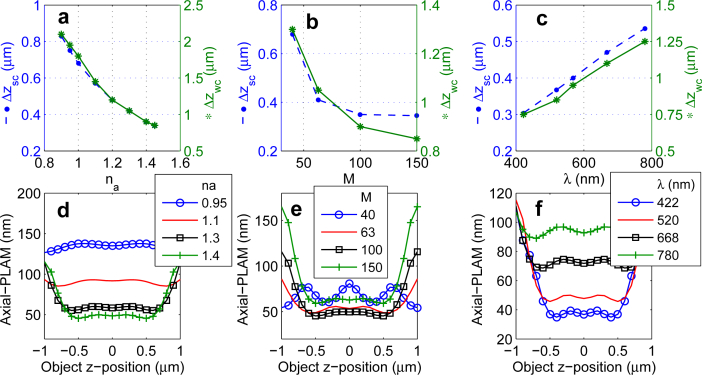
The FIM of a MUM setup depends on a second group of parameters such as the numerical aperture, magnification and emission wavelength. Therefore, in this section we investigate how these factors affect the plane spacings. We first study the effect of numerical aperture. Figure 4(a) shows the effect of changing the numerical aperture on the strong and weak coupling spacings, where we observe an inverse dependence of the spacings on the numerical aperture assuming that all other parameters remain the same. For instance, the strong coupling spacing increases from Δzsc = 0.35 μm to Δzsc = 0.75 μm (i.e. a 114% elongation) when the numerical aperture decreases from 1.4 to 0.95. This implies that a low NA objective supports relatively constant 3D localization accuracy over a larger viewing and tracking depth than a high NA objective. This result is not surprising considering the fact that given a low NA objective, the PSF has a broader profile in the xz-plane and/or the yz-plane. The broader profile leads to a stretched axial-FIM along the z-axis for each focal plane and thereby it is expected that a low NA objective leads to a more constant axial-PLAMMUM along the z-axis. However, the numerical value of the axial-PLAMMUM for the low NA objective along the z-axis is typically higher than that of a high NA objective (Fig. 4(d)). For example, for a 1.1 NA objective, the axial-PLAMMUM varies from 85 nm to 93 nm over the z-range of [−0.8, 0.8] μm. Over the same z-range, the numerical value of the axial-PLAMMUM for a 1.4 NA objective varies from 45 nm to 77 nm. This is also an expected result given the fact that use of a low NA objective yields a broader image profile in the xy-plane, which translates to a poorer localization accuracy. We note that an analogous behavior is also observed for the weak coupling spacing (not shown). The high numerical value of the axial-PLAMMUM for the 0.95 NA objective can be reduced in part by collecting more photons from the sample due to the inverse dependence of the PLAM on the photon count. Thus, an immediate implication of this result is that depending on the requirements for the viewing and tracking depth and the desired level of 3D localization accuracy, it is necessary to make a careful choice of experimental parameters (e.g. numerical aperture of the objective lens) and imaging conditions (photon count/signal from the sample).
Fig. 4.
The effect of changing imaging parameters on the spacing scenarios. A plot of the strong coupling spacing (Δzsc) and weak coupling spacing (Δzwc) versus (a) the numerical aperture na, (b) magnification M and (c) emission wavelength λ. (d), (e) and (f) show the axial-PLAM for a 2-plane MUM setup as a function of the z-position of the point source for different numerical apertures, magnifications and emission wavelengths, respectively. In (d), (e) and (f), the plane spacings are adjusted based on the strong coupling scenario. The photon count is 250 photons/plane. The ROI size is 32 × 32 pixels. The magnification is 100, the emission wavelength is 520 nm and the numerical aperture is 1.4. The background level and the standard deviation of the readout noise are 2.5 photons/pixel/plane and 8 e−/pixel, respectively.
We next examine the effect of magnification on the spacing scenarios. Analogous to the behavior with respect to the numerical aperture, the strong and weak coupling spacings increase with decreasing values of magnification assuming all other parameters are the same (Fig. 4(b)). More specifically, as the magnification decreases from 150x to 40x the strong coupling spacing increases from 0.35 μm to 0.68 μm. However, as shown in Fig. 4(e), the behavior of the axial-PLAMMUM as a function of z-position for different magnification values exhibit a distinct behavior. As the magnification increases from 40x to 63x, the numerical value of the axial-PLAMMUM for a z-range of [−0.8, 0.8] μm varies from 60 nm to 81 nm, and from 49 nm to 59 nm, respectively. This implies that with increasing magnification values, one can expect relatively constant z-localization accuracy over a certain z-range. However, this behavior is true only up to a certain point. For instance, for a magnification of 150x the axial-PLAMMUM value exhibits greater variation, i.e. from 59 nm to 108 nm over the z-range of [−0.8, 0.8] μm when compared to a 100x magnification for which the axial-PLAMMUM varies from 46 nm to 77 nm over the same z-range. This is due to the fact that at very high magnifications, the image of the point source is spread out over such a large number of pixels that the number of photons detected from the point source at each pixel becomes relatively small compared to the readout noise. This results in the observed variation in the axial-PLAMMUM, which depends not only on the total number of detected photons but also on the spatial distribution of the detected photons over the pixels [12, 14].
We last study the behavior of the spacing scenarios as a function of the emission wavelength. Increasing the emission wavelength increases the strong and weak coupling spacings (see Fig. 4(c)). As a consequence, a fluorophore with a large emission wavelength can provide a relatively constant level of accuracy across a larger viewing and tracking depth (see Fig. 4(f)). Similar to the discussion regarding the numerical aperture, the PSF given a large emission wavelength has a broader profile in the xz-plane (or yz-plane) and thereby one would expect a large emission wavelength to yield a more constant axial-PLAMMUM along the z-axis.
4.5. Effects of photon count and extraneous noise on the spacing scenarios
We next investigate the dependence of the strong and weak coupling spacings on the photon count and readout noise. In the presence of extraneous noise, altering the photon count changes the strong and weak coupling spacings (see Fig. 5(a)) as it alters the local maxima and shoulders of the axial-FIMs of the individual focal planes. More specifically, increasing the photon count from 100 photons to 8000 photons increases the strong coupling spacing from 0.34 μm to 0.44 μm (i.e. a 29% elongation). In addition, decreasing the readout noise also increases the strong and weak coupling spacings (see Fig. 5(b)). For example, reducing the standard deviation of the readout noise from 10 e−/pixel to 1 e−/pixel increases the strong coupling spacing from 0.36 μm to 0.46 μm (i.e. a 28% enhancement). The effect of background noise on the strong and weak coupling spacings is similar to the effect of readout noise and hence is omitted for brevity.
Fig. 5.
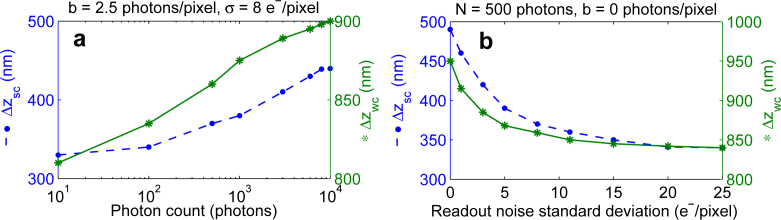
The effect of photon count and extraneous noise on the spacing scenarios. A plot of the strong coupling spacing (Δzsc) and weak coupling spacing (Δzwc) as a function of (a) the photon count (N) and (b) the standard deviation of the readout noise (σ). The ROI size is 32 × 32 pixels. The magnification is 100, the emission wavelength is 520 nm and the numerical aperture is 1.4. In (a), the background level and the standard deviation of the readout noise are 2.5 photons/pixel and 8 e−/pixel, respectively. In (b), the photon count and the background level are 500 photons and 0 photons/pixel, respectively.
It is important to note that the effect of changing the photon count and extraneous noise on the strong and weak coupling spacings is not significant when compared to the effect of magnification, numerical aperture and emission wavelength. An intuitive explanation of this behavior is as follows. In the absence of extraneous noise, changing the photon count only scales the FIM (due to the linear dependence of the FIM on the photon count) and therefore does not change the locations of the local maxima and shoulders of the axial-FIM (for details see Eq. (2) in Appendix A). In the presence of extraneous noise, however, there is a nonlinear dependence of the FIM on the photon count. This nonlinear dependence, in addition to scaling the FIM, causes the observed slight change in the positions of the local maxima and shoulders of the axial-FIM.
4.6. Spacing scenarios for MUM setups with more than 2 focal planes
In the previous sections, we described scenarios for adjusting the plane spacing for a 2-plane MUM setup that provide a relatively constant 3D localization accuracy along the z-axis. We next extend the spacing scenarios to MUM setups with more than two focal planes. The strong coupling spacing is obtained when the focal planes are positioned in such a way that the local maximum of the axial-FIM of a given plane overlaps with the zero of the axial-FIM of the adjacent plane. Figures 6(a) and 6(b) show the axial-PLAM and the lateral-PLAM, respectively, for a 4-plane MUM setup with planes adjusted based on the strong coupling spacing. For the given set of imaging conditions, the axial-PLAMMUM and the lateral-PLAM vary from 23 nm to 30 nm and from 9.8 nm to 13 nm, respectively, over the z-range of [−0.2, 1.5] μm. This implies that the extension of the strong coupling spacing provides a relatively constant 3D localization accuracy along the z-axis.
Fig. 6.
The strong and weak coupling spacings for a 4-plane MUM setup. (a) The axial-PLAM and (b) the lateral-PLAM as a function of the z-position of the point source for the strong coupling spacing for a 4-plane MUM setup. The planes are placed at 0, 0.45, 0.9 and 1.35 μm. (c) and (d) show the same for the weak coupling spacing for a 4-plane MUM setup where the planes are located at 0, 1, 2 and 3 μm. The magnification is 100, the numerical aperture is 1.3, the photon count is 250 photons/plane and the ROI size is 11 × 11 pixels. The emission wavelength is 520 nm. The background level and the standard deviation of the readout noise are 20 photons/pixel/plane and 3 e−/pixel, respectively.
The weak coupling spacing can be defined in an analogous way, i.e. the shoulder of the axial-FIM of one plane is selected to overlap with the zero of the axial-FIM of the adjacent plane. Figures 6(c) and 6(d) show that the axial-PLAMMUM and the lateral-PLAM for the extension of the weak coupling spacing are relatively constant along the z-axis as well. More specifically, the axial-PLAMMUM and the lateral-PLAM vary from 26 nm to 68 nm and from 11.6 nm to 22 nm, respectively, over the larger z-range of [−0.4, 3.4] μm.
4.7. Increasing the number of focal planes within a specific z-range does not necessarily enhance the localization accuracy along the z-axis
An important question in the design of MUM experiments concerns the appropriate number of focal planes that are required to cover a sample of a certain thickness. To address this concern, we next study the effect of changing the number of focal planes on the 3D localization accuracy of a MUM setup along the z-axis. Figure 7(a) shows the axial-PLAM for MUM setups with 2 to 8 focal planes over the range of [−1, 1] μm, where the planes are placed based on the strong coupling spacing. We assume that the total photon count is fixed and is split equally among the focal planes. The axial-PLAM of the 2-plane setup has relatively small numerical values along the z-axis (see Fig. 7(a)). However, the values vary significantly (i.e, they vary from 34 nm to 67 nm over the z-range of [−1, 1] μm). By increasing the number of planes from 2 to 3, the numerical values of the axial-PLAMMUM increase while the curve becomes more flat (i.e. the axial-PLAMMUM varies from 37 nm to 56 nm over the same z-range).
Fig. 7.
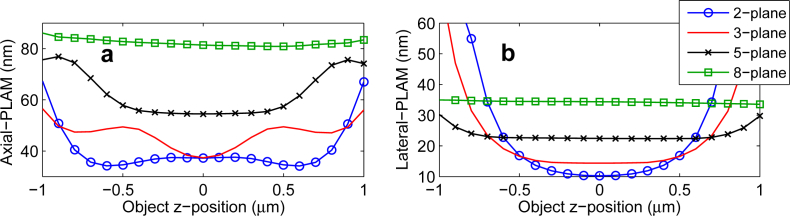
The effect of changing the number of focal planes on the 3D localization accuracy. (a) The axial-PLAM and (b) the lateral-PLAM for a MUM setup with different numbers of focal planes over a range of [−1, 1] μm. The magnification is 100, the numerical aperture is 1.3 and the ROI size is 11 × 11 pixels. The emission wavelength is 520 nm. The focal planes are located based on the strong coupling spacing. The total photon count is 1000 photons and is split equally among the focal planes. The background level and the standard deviation of the readout noise are 25 photons/pixel/plane and 8 e−/pixel, respectively.
Further increasing the number of focal planes, e.g. to 5 or 8, yields more flat axial-PLAMMUM curves along the z-axis. For instance, for an 8-plane setup the axial-PLAM values vary from 81 nm to 86 nm over the same z-range. However, the numerical value of the axial-PLAM for a MUM setup with a large number of planes is consistently greater than that for a MUM setup with a small number of planes. A consistently large axial-PLAMMUM value implies a poor localization accuracy along the z-axis. Figure 7(b) shows that the effect of changing the number of focal planes on the lateral-PLAM is analogous to its effect on the axial-PLAMMUM.
An important implication of this behavior is that one can achieve a relatively constant 3D localization accuracy across the viewing and tracking depth by increasing the number of focal planes. However, as the number of focal planes increases, the numerical values of the localization accuracy become consistently large along the z-axis. The reason for this behavior is that by placing a large number of focal planes, the emitted light from the point source is split among the different planes such that the number of photons detected from the point source at each plane becomes relatively small when compared to the readout noise. This results in large numerical values in the axial-PLAMMUM and the lateral-PLAM which depend on the total number of detected photons per plane [14].
4.8. MUMDesignTool
The MUMDesignTool is a new software module developed in the MATLAB environment based on an object-oriented programming methodology. This software provides a graphical user interface to calculate and plot the 3D localization accuracy for MUM setups with up to 10 focal planes. In addition, the MUMDesignTool provides two working modes called “rapid” (noise-free) and “precise” (corrupted by background, stochastic signal amplification and readout noise). The rapid mode allows the fast calculation of the 3D localization accuracy for a MUM setup by representing the FIM of the MUM setup in terms of the unit photon count FIM (uFIM) of the design focal plane (see Appendix B). This fast calculation is possible by assuming that the magnification is constant at different focal planes and that the data is devoid of extraneous noise sources. As a result, the rapid mode provides the ability to change the plane spacing, the number of focal planes and the photon count percentages in real-time, and to simultaneously visualize the FIM and PLAM. On the other hand, the precise mode of the MUMDesignTool calculates the 3D localization accuracy without considering the assumptions made in the rapid mode (see Appendix A).
In addition, the MUMDesignTool is capable of exporting the results and acquisition parameters as image and text files, respectively. An approach to designing the plane spacing using this package is to first use the rapid mode to interactively change the plane spacing and the number of planes and visualize the behavior of the PLAM. This helps to find a candidate for one of the introduced spacing scenarios. The next step is then to run the precise mode with the extraneous noise parameters, and the spacings found in the rapid mode to ensure that the designed spacings are appropriate in the presence of noise.
5. Conclusion
In this paper, we developed design approaches for adjusting the plane spacing for a MUM setup called the strong and weak coupling spacings. The strong coupling spacing is designed for quantitative imaging experiments which require relatively constant 3D localization accuracy along the z-axis. The weak coupling spacing, on the other hand, was primarily intended for qualitative imaging experiments where the continuous visualization of particles over a large viewing and tracking depth is of interest. We investigated the effect of imaging parameters such as the numerical aperture and system magnification on these spacing scenarios. In addition, we showed that placing both a small number of focal planes and a large number of focal planes in a specific z-range do not necessarily provide an appropriate 3D localization accuracy along the z-axis. We also introduced the MUMDesignTool that helps to determine the appropriate plane spacings for a MUM setup.
Acknowledgments
This work was supported in part by the National Institutes of Health ( R01 GM085575).
Appendix A: FIM and PLAM for a MUM setup
Here, we express the FIM for a MUM setup for a general single molecule microscopy experiment. Let Θ ⊆ ℝ3 be an open parameter space and let θ = (x0, y0, z0) ∈ Θ be the vector of unknown parameters representing the 3D location of a single molecule in the object space with respect to the design focal plane, i.e. the standard infinity-corrected focal plane in conventional single plane microscopy. Consider a pixelated detector {C1,...,CKpix} which consists of Kpix pixels, where Ck ⊆ ℝ2, k = 1,...,Kpix, denotes the area occupied by the kth pixel. Assume that the pixels are disjoint. Suppose that we have Kpln focal planes where the first plane is equivalent to the design focal plane. It has been shown that the photon counts detected by the pixels of the nth plane due to a single molecule axially located at z0 are realizations of independent Poisson random variables with expected values [16, 22]
| (1) |
where Nn is the expected number of detected photons on the nth infinite detector plane (i.e. ℝ2) due to the single molecule such that , with Ntot denoting the total number of detected photons due to the single molecule on an arbitrarily positioned infinite detector plane (i.e. ℝ2). Note that Ntot is independent of the number of focal planes. The term Ck,n denotes the kth pixel at the nth focal plane. In addition, Mn is the lateral magnification at the nth focal plane, Δz1n is the distance between the design focal plane and the nth focal plane in the object space (with Δz11 = 0) and qz0 is the image function [14, 16].
The image function describes the image of a stationary single molecule on the detector at unit lateral magnification when the single molecule is located on the z-axis in the object space. Here, we assume that the image function is given by the Born and Wolf 3D PSF (for more information see [14, 20]). Moreover, it has been previously shown, under geometrical optics, that the lateral magnification for a focal plane that is shifted by a distance of Δz1n from the design focal plane is given by [23, 24]
where noil is the refractive index of the immersion oil and L is the tube length of the microscope.
For a practical microscopy setup where the acquired data is corrupted by extraneous noise sources, the expression of the FIM for the parameter-vector θ at the nth focal plane is given by [16, 22, 25]
| (2) |
where νθ,n(k) = μθ,n(k) + bk,n with bk,n, k = 1,..., Kpix, n = 1,..., Kpln, denoting the photon count due to the background noise at pixel Ck and plane n. The term ψn(k) is the so-called noise coefficient that depends on the type of detector [25]. In the absence of readout noise, ψn(k) = 1 for all k = 1,..., Kpix, n = 1,..., Kpln [16]. In the presence of readout noise and when using charge coupled device (CCD) and complementary metal oxide semiconductor (CMOS) detectors, the noise coefficient is given by [16]
where ηk,n and denote the mean and the variance of the readout noise, respectively, for k = 1,...,Kpix and n = 1,..., Kpln. The expression of the noise coefficient in the presence of stochastic signal amplification and readout noise, i.e. when using an electron-multiplying CCD (EMCCD) detector, is omitted for brevity but can be found in [25].
Since the data acquisition in each focal plane is independent of the data acquisition in the other planes, the FIM of a MUM setup is the sum of the FIMs of the individual planes [14] and we have
| (3) |
For the current 3D localization problem, the FIM I(θ) for any given focal plane or for the MUM setup is a 3 × 3 matrix. The main diagonal elements of this 3 × 3 matrix provide information about the x, y and z coordinates of the single molecule and we refer to them as the x0-FIM, y0-FIM and z0-FIM (axial-FIM), respectively. According to the Cramér-Rao inequality [21], the covariance matrix of any unbiased estimator θ̂ of an unknown parameter θ is always greater than or equal to the inverse FIM, i.e. cov(θ̂) ≥ I−1(θ) [16, 19]. Therefore, the square roots of the main diagonal elements of the inverse FIM provide lower bounds for the accuracy (standard deviation) with which the x, y and z coordinates of the single molecule can be estimated. We denote these lower bounds by x0-PLAM, y0-PLAM and z0-PLAM (axial-PLAM), respectively. It is important to note that the PLAM has been previously validated by comparing it with the standard deviation of the estimated locations of single molecules in actual microscopy experiments (see e.g. [14, 26]).
The precise mode of the MUMDesignTool is capable of calculating the FIM and PLAM for a MUM setup in the presence of extraneous noise sources such as background, stochastic signal amplification and readout using the general expression given by Eq. (3). For this purpose, the FIM for each focal plane is calculated separately using Eq. (2).
Appendix B: approximate but fast calculation of the FIM
In Section 3.2, we discussed a graphical interpretation for the design of the focal plane spacing for a MUM system. Building on the same idea and by making additional assumptions, here we develop an approach for the fast calculation of the FIM for a MUM setup. We also discuss the additional assumptions and the resulting properties of the FIM that are exploited for the design process. This fast approach is also used in the rapid mode of the MUMDesignTool. In this mode of the software, the design process can be carried out graphically in real-time as it avoids the often very time-consuming calculations of the full computations.
From Eq. (1) we have μθ,n(k) = Nnμ̃θ,n(k). Assuming that the data is devoid of extraneous noise sources, νθ,n(k) = μθ,n(k) and ψn(k) = 1 for k = 1,..., Kpix, n = 1,..., Kpln. This assumption will prove to be useful for the fast calculation of the FIM for focal planes with different expected number of detected photons. Hence, from Eq. (2) it follows
We refer to Ĩn(θ) = Ĩn(x0, y0, z0), θ = (x0, y0, z0) ∈ Θ as the unit photon count FIM (uFIM) of the nth plane which is a function of Mn and Δz1n, n = 1,..., Kpln. We now assume that the lateral magnification is the same for all focal planes, i.e. we suppose Mn = M1 for all n = 2,...,Kpln. Using this assumption it follows that Ĩn(θ) = Ĩn(x0, y0, z0) = Ĩ1(x0, y0, z0 −Δz1n), n = 2,..., Kpln. Combining this result with Eq. (3), we define the fast FIMMUM as
which is a weighted sum of the uFIM of the design focal plane evaluated at different z-positions. The above equation implies that, in this approximation setting, obtaining the FIM for a MUM setup for a range of z-positions only requires one calculation that is the uFIM of the design focal plane (i.e. Ĩ1) for the range of z-positions. On the other hand, from Eq. (3) obtaining the FIM for a MUM setup for the same range of z-positions using the precise approach requires Kpln × a calculations, where Kpln calculations are needed for the FIMs of the individual planes (i.e. I1,..., IKpln) and a is the number of configurations to be tried when varying the plane spacing.
As a consequence, the rapid mode of the MUMDesignTool, which uses the above approximate approach, can significantly speed up the design procedure for the plane spacing for a MUM setup that is otherwise very time consuming using the precise method. However, it is important to note that the PLAM obtained using the rapid mode is valid under the assumptions that the observed data is devoid of extraneous noise and that the lateral magnification is the same for all focal planes. In a practical situation where the data is corrupted by extraneous noise, we recommend verifying the results of the fast calculations by performing the precise calculations (which correspond to the noisy case). If there is a significant discrepancy between the results of the rapid mode and the results of the precise mode, the plane spacing designed using the rapid mode (i.e. the candidate spacing) should be fine tuned. The fine tuning can be performed by slightly increasing and/or decreasing the candidate spacing and then rerunning the precise mode. The results of the precise mode for these slightly different spacings can then be compared to find the desired spacing (e.g. the spacing that provides the flattest axial-PLAM curve).
References and links
- 1.Prabhat P., Ram S., Ward E. S., Ober R. J., “Simultaneous imaging of different focal planes in fluorescence microscopy for the study of cellular dynamics in three dimensions,” IEEE Trans. Nanobiosci. 3, 237–242 (2004). 10.1109/TNB.2004.837899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard P. M., Greenaway A. H., “Simultaneous multiplane imaging with a distorted diffraction grating,” Appl. Opt. 38, 6692–6699 (1999). 10.1364/AO.38.006692 [DOI] [PubMed] [Google Scholar]
- 3.Ram S., Chao J., Prabhat P., Ward E. S., Ober R. J., “Overcoming the depth discrimination barrier in widefield microscopes: 3D single molecule tracking with high axial accuracy,” Proc. SPIE 6862, 68620O (2008). 10.1117/12.763097 [DOI] [Google Scholar]
- 4.Dehmelt L., Bastiaens P. I. H., “Spatial organization of intracellular communication: insights from imaging,” Nature Rev. Mol. Cell Biol. 11, 440–452 (2010). 10.1038/nrm2903 [DOI] [PubMed] [Google Scholar]
- 5.Liu S., Kromann E., Krueger W., Bewersdorf J., Lidke K., “Three dimensional single molecule localization using a phase retrieved pupil function,” Opt. Express 21, 29462–29487 (2013). 10.1364/OE.21.029462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jesacher A., Roider C., Ritsch-Marte M., “Enhancing diffractive multi-plane microscopy using colored illumination,” Opt. Express 21, 11150–11161 (2013). 10.1364/OE.21.011150 [DOI] [PubMed] [Google Scholar]
- 7.Wolter S., Loschberger A., Holm T., Aufmkolk S., Dabauvalle M., van de Linde S., Sauer M., “rapidSTORM: accurate, fast open-source software for localization microscopy,” Nat. Methods 9, 1040–1041 (2012). 10.1038/nmeth.2224 [DOI] [PubMed] [Google Scholar]
- 8.Badieirostami M., Lew M. D., Thompson M. A., Moerner W. E., “Three-dimensional localization precision of the double-helix point spread function versus astigmatism and biplane,” Appl. Phys. Lett. 97, 161103 (2010). 10.1063/1.3499652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kirshner H., Aguet F., Sage D., Unser M., “3-D PSF fitting for fluorescence microscopy: implementation and localization application,” J. Microsc. 249, 13–25 (2013). 10.1111/j.1365-2818.2012.03675.x [DOI] [PubMed] [Google Scholar]
- 10.Dalgarno P. A., Dalgarno H. I. C., Putoud A., Lambert R., Paterson L., Logan D. C., Towers D. P., Warburton R. J., Greenaway A. H., “Multiplane imaging and three dimensional nanoscale particle tracking in biological microscopy,” Opt. Express 18, 877–884 (2010). 10.1364/OE.18.000877 [DOI] [PubMed] [Google Scholar]
- 11.Gan Z., Ram S., Ober R. J., Ward E. S., “Using multifocal plane microscopy to reveal novel trafficking processes in the recycling pathway,” J. Cell Sci. 126, 1176–1188 (2013). 10.1242/jcs.116327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prabhat P., Gan Z., Chao J., Ram S., Vaccaro C., Gibbons S., Ober R. J., Ward E. S., “Elucidation of intracellular recycling pathways leading to exocytosis of the Fc receptor, FcRn, by using multifocal plane microscopy,” Proc. Natl. Acad. Sci. USA 104, 5889–5894 (2007). 10.1073/pnas.0700337104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ram S., Kim D., Ober R. J., Ward E. S., “3D single molecule tracking with multifocal plane microscopy reveals rapid intracellular transferrin transport at epithelial cell barriers,” Biophys. J. 103, 1594–1603 (2012). 10.1016/j.bpj.2012.08.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ram S., Prabhat P., Chao J., Ward E. S., Ober R. J., “High accuracy 3D quantum dot tracking with multifocal plane microscopy for the study of fast intracellular dynamics in live cells,” Biophys. J. 95, 6025–6043 (2008). 10.1529/biophysj.108.140392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abrahamsson S., Chen J., Hajj B., Stallinga S., Katsov A. Y., Wisniewski J., Mizuguchi G., Soule P., Mueller F., Darzacq C. D., Darzacq X., Wu C., Bargmann C. I., Agard D. A., Dahan M., Gustafsson M. G. L., “Fast multicolor 3D imaging using aberration-corrected multifocus microscopy,” Nat. Methods 10, 60–63 (2013). 10.1038/nmeth.2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ober R. J., Ram S., Ward E. S., “Localization accuracy in single-molecule microscopy,” Biophys. J. 86, 1185–1200 (2004). 10.1016/S0006-3495(04)74193-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abraham A. V., Ram S., Chao J., Ward E. S., Ober R. J., “Quantitative study of single molecule location estimation techniques,” Opt. Express 17, 23352–23373 (2009). 10.1364/OE.17.023352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ram S., Ward E. S., Ober R. J., “How accurately can a single molecule be localized in three dimensions using a fluorescence microscope?” Proc. SPIE 5699, 426–435 (2005). 10.1117/12.587878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kay S. M., Fundamentals of Statistical Signal Processing: Estimation Theory (Prentice Hall PTR, Upper Saddle River, NJ, 1993). [Google Scholar]
- 20.Born M., Wolf E., Principles of Optics, 7th ed. (Cambridge University Press, Cambridge, UK, 2002). [Google Scholar]
- 21.Rao C. R., Linear Statistical Inference and its Applications (John Wiley and Sons, New York, 1965). [Google Scholar]
- 22.Snyder D. L., Miller M. I., Random Point Processes in Time and Space, 2nd ed. (Springer Verlag, New York, USA, 1991). 10.1007/978-1-4612-3166-0 [DOI] [Google Scholar]
- 23.Chao J., Ram S., Ward E. S., Ober R. J., “A comparative study of high resolution microscopy imaging modalities using a three-dimensional resolution measure,” Opt. Express 17, 24377–24402 (2009). 10.1364/OE.17.024377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tao L., Nicholson C., “The three-dimensional point spread functions of a microscope objective in image and object space,” J. Microsc. 178, 267–271 (1995). 10.1111/j.1365-2818.1995.tb03604.x [DOI] [PubMed] [Google Scholar]
- 25.Chao J., Ward E. S., Ober R. J., “Fisher information matrix for branching processes with application to electron-multiplying charge-coupled devices,” Multidim. Sys. Sig. Proc. 23, 349–379 (2012). 10.1007/s11045-011-0150-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ram S., Prabhat P., Ward E. S., Ober R. J., “Improved single particle localization accuracy with dual objective multifocal plane microscopy,” Opt. Express 17, 6881–6898 (2009). 10.1364/OE.17.006881 [DOI] [PMC free article] [PubMed] [Google Scholar]