Summary
One envisioned function of homologous recombination (HR) is to find a template for DNA synthesis from the resected 3′-OH molecules that occur during double-strand break (DSB) repair at broken or stalled replication forks. However, the interplay between DNA synthesis and HR remains poorly understood in higher eukaryotic cells. Here, we reveal new functions for breast cancer proteins BRCA2 and PALB2 at blocked replication forks and show a role for these proteins in stimulating polymerase eta (Polη) to initiate DNA synthesis. PALB2, BRCA2 and Polη co-localize at stalled or collapsed replication forks after hydroxyurea treatment. Moreover, PALB2 and BRCA2 interact with Polη, and are required to sustain the recruitment of Polη at blocked replication forks. PALB2 and BRCA2 stimulate Polη-dependent DNA synthesis on D-loop substrates. We conclude that PALB2 and BRCA2, in addition to their functions in D-loop formation, play crucial roles in the initiation of recombination-associated DNA synthesis by Polη-mediated DNA repair.
Keywords: PALB2, BRCA2, polymerase η, DNA replication and homologous recombination
Introduction
Faithful DNA replication is essential to prevent accumulation of mutations and to maintain genome integrity. Replication forks are vulnerable to unrepaired DNA damage or secondary structures, which can lead to fork stalling or collapse. A stalled replication fork can be arrested but still has the ability to restart. In contrast, a collapsed replication fork leads the generation of replication-dependent DNA double-strand breaks (DSBs), triggering the recruitment of DSB repair machineries. Tumor suppressor proteins PALB2 (Partner and localizer of BRCA2) and BRCA2 (breast cancer type 2 susceptibility protein) are essential for active homologous recombination (HR) repair, the main mechanism of error-free homology-directed repair of DNA DSBs in mammalian cells (Xia et al., 2006). The central activity of HR is performed by RAD51 using resected DNA DSBs to invade a homologous double-strand DNA to form a displacement loop (D-loop). Defects in HR cause genomic instability and promote accumulation of cancer-promoting mutations. PALB2 and BRCA2 mutations have been associated with predisposition to breast and pancreatic cancer (Roy et al., 2012), and PALB2- or BRCA2-deficient cells are sensitive to PARP inhibitors (Bryant et al., 2005; Buisson et al., 2010). Furthermore, PALB2 (FANCN) and BRCA2 (FANCD1) are mutated in a subgroup of Fanconi Anemia (Howlett et al., 2002; Xia et al., 2007) an inherited genomic instability disorder caused by mutations in genes regulating replication-dependent removal of interstrand DNA crosslinks (Moldovan and D'Andrea, 2009).
PALB2 links BRCA1 and BRCA2 to promote efficient DNA repair by HR (Sy et al., 2009a; Xia et al., 2006; Zhang et al., 2009a; Zhang et al., 2009b). In the absence of PALB2, the recruitment of BRCA2 and RAD51 to DSBs is defective (Xia et al., 2007; Xia et al., 2006). PALB2 interacts with BRCA1 via its N-terminal coiled-coil domain (Buisson and Masson, 2012; Sy et al., 2009b; Zhang et al., 2009a; Zhang et al., 2009b) and with BRCA2 via its WD40 domain (in C-terminal) (Xia et al., 2007). The coiled-coil domain is also important for the self-interaction of PALB2. In the presence of DNA damage, PALB2 dissociates and interacts with BRCA1 to allow PALB2 localization and HR activation (Buisson and Masson, 2012). A third domain, named ChAM (Chromatin-Association Motif), is located at the center of the protein (395 to 446 amino acid) and is required for PALB2 chromatin localization (Bleuyard et al., 2012). Recently, we have shown that purified PALB2 binds D-loops preferentially and interacts directly with RAD51 to stimulate strand invasion (Buisson et al., 2010; Dray et al., 2010). At the same time, the Kowalczykowski, Heyer, and West laboratories reported the purification of full length BRCA2. Human BRCA2 promotes RAD51 filament assembly on single-strand DNA, which stimulates RAD51-mediated DNA strand exchange (Jensen et al., 2010; Liu et al., 2010; Thorslund et al., 2010).
Following D-loop formation by RAD51, the 3′tail single-stranded DNA is used as a primer for extension by polymerases. The polymerases δ, η, υ, ε, ζ, κ and REV1 have all been shown to be involved in HR although their exact functions remain unclear. Currently, only polymerases δ, η, and κ are known to extend a D-loop structure after RAD51 single-strand DNA invasion (Li et al., 2013; McIlwraith et al., 2005; Sebesta et al., 2011; Sebesta et al., 2013; Sneeden et al., 2013). In yeast, genetic evidence has shown clearly that polymerase δ is important for HR and gene conversion (Holmes and Haber, 1999; Maloisel et al., 2008) but these results have not yet been confirmed in human cells. Moreover, it has been recently shown that the subunit p12 of Polδ is degraded in S-phase by the CRL4cdt2 complex after replicative-associated DNA damage (Terai et al., 2013; Zhang et al., 2013). These results suggest that, in human cells, polymerase δ could not be essential for stalled and collapsed replication forks repair. However, further investigations will be necessary to better understand the regulation of Polδ after DNA damage and the biological significance of this observation. In DT40 cells, Polη-deficient cells showed a significant decrease in the frequency of both Ig gene conversion and HR-dependent repair of I-SceI- induced DSBs when compared to wild-type cells (Kawamoto et al., 2005). Interestingly, in human cells, overexpression of Polη and κ leads to an increase in HR frequency (Sebesta et al., 2013) while absence of Polη slightly decreases HR (Moldovan et al., 2010). Deletion of both Pol η and κ leads to a 50% decrease in HR frequency (Sebesta et al., 2013) suggesting a cooperation mechanism between these polymerases.
We hypothesized that different polymerases could be involved in recombination-associated DNA synthesis. In this study, we focused on Polη in order to better define its function in HR. Polη has been shown to co-localize and interact with RAD51 (Kannouche et al., 2001; McIlwraith et al., 2005), and several studies have shown that, contrary to Polδ and κ, Polη activity for D-loop extension does not require the loading of PCNA and RFC (Li et al., 2013; McIlwraith et al., 2005; Sebesta et al., 2011; Sebesta et al., 2013; Sneeden et al., 2013). These results suggest that polη activity could be regulated by other factors.
In humans, the deletion of Polη leads to Xeroderma pigmentosum variant (XPV), which is associated with an increase risk of skin cancer induced by UV radiation (Johnson et al., 1999; Masutani et al., 1999). Polη is a member of the Y-family DNA polymerases (with Polι, Polκ and REV1) that specialize in performing DNA damage bypass repairs involving TT-cyclobutane pyrimidine dimer (CPD) (Masutani et al., 1999), 8-oxoguanine (8-oxoG) (Haracska et al., 2000b) or O6-methylguanine (me6G) (Haracska et al., 2000a). These DNA lesions can cause collapsed replication forks, impeding lesion bypass by replicative DNA polymerases (Polε and Polδ). Although the biological role of Polη in UV-induced DNA damage repair is well understood, very little is known about the regulation of Polη activity during HR repair.
In this study, we show that Polη interacts directly with PALB2 and BRCA2 at DSBs induced by collapsed replication forks. We show that PALB2 and BRCA2 are important for (i) Polη localization to collapsed replication forks, and (ii) Polη-dependent DNA synthesis for D-loop extension. Our results establish PALB2 and BRCA2 as key regulators of the extension step after strand invasion at replication-dependent DSBs.
Results
PALB2 and BRCA2 promote the accumulation of Polη at collapsed replication forks
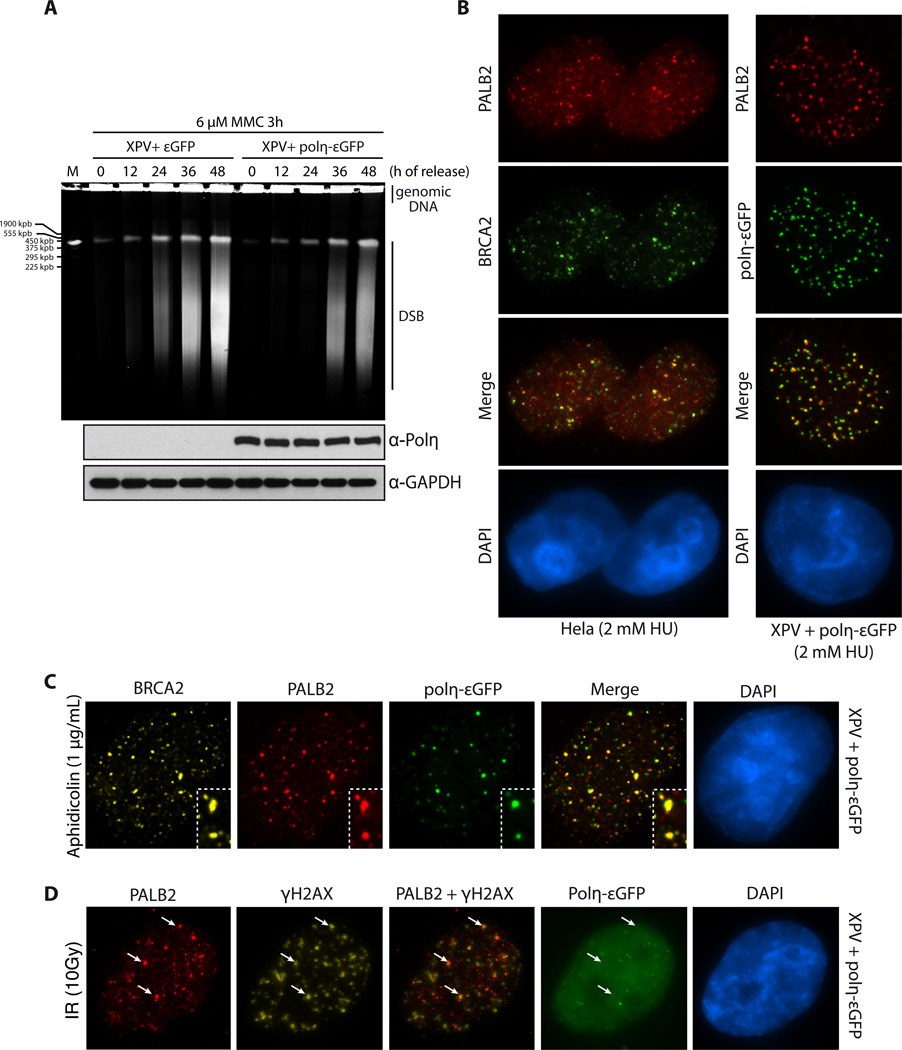
To better define the function and the regulation of Polη at DSBs, we focused on the localization and recruitment of Polη with other HR proteins in the presence of replication fork stalling agents such as hydroxyurea (HU), aphidicolin or mitomicyn C (MMC). These drugs cause collapsed replication forks after prolonged exposure (Petermann et al., 2010). First, we confirmed previously published reports showing that Polη-deficient cells present a defect in repairing DSBs (de Feraudy et al., 2007; Kawamoto et al., 2005; Moldovan et al., 2010; Sebesta et al., 2013). We used XPV cells (Polη-deficient cell line) complemented or not with εPolη. After MMC treatment, Polη-deficient XPV cells showed a stronger accumulation of DSBs compared to Polη-complemented XPV cells 24–48 hours after release into fresh medium (Fig. 1A). As a control, we also showed that these Polη-complemented XPV cells rescued UV sensitivity (Suppl. Fig. 1A). Next, we scrutinized the localization of Polη at blocked replication forks. We confirmed that Polη forms foci after MMC, HU, and aphidicolin treatment (Suppl. Fig 1B). Furthermore, PALB2, BRCA2, RAD51, PCNA, RPA and/or γ-H2AX co-localized with Polη at DSBs induced by collapsed replication forks in Hela or Polη-εGFP-complemented XPV cells (Fig. 1A, 1B and Suppl. Fig. 1C–D). We used HU and aphidicolin rather than MMC because it is very difficult to observe PALB2 and BRCA2 foci after MMC treatment owing to their smaller size. To prove that the localisation of Polη is happening at collapsed replication forks, we observed that about 70 % of cells observed displayed a co-localization between Polη and the DSB marker RAD51 (Suppl. Fig. 2A). In contrast, γ-irradiation-induced Polη foci formation was very limited, confirming previous observations (Kannouche et al., 2001), and the few foci observed did not co-localize with PALB2, RAD51 or γ-H2AX (Fig. 1C, Suppl. Fig. 2B), suggesting a role for Polη in DSB repair uniquely at blocked replication forks.
Figure 1. Polη, PALB2 and BRCA2 recruitment at replication dependent DNA double-strand breaks.
(A) Pulsed-field gel electrophoresis was used to visualize double-strand break (DSB) formation in XPV cells complemented with εGFP or Polη-εGFP, after treatment with 6 µM MMC for 3h and release for the times indicated. (B) Co-localization of PALB2 and BRCA2 or PALB2 and Polη-εGFP at DSBs induced by HU. DNA was counterstained with DAPI. (C–D) Immunofluorescence staining of the indicated DNA repair proteins at DSBs induced by aphidicolin or IR treatment in XPV cells complemented with Polη-εGFP. DNA was counterstained with DAPI. The white arrows indicate co-localization of PALB2 and γ-H2AX.
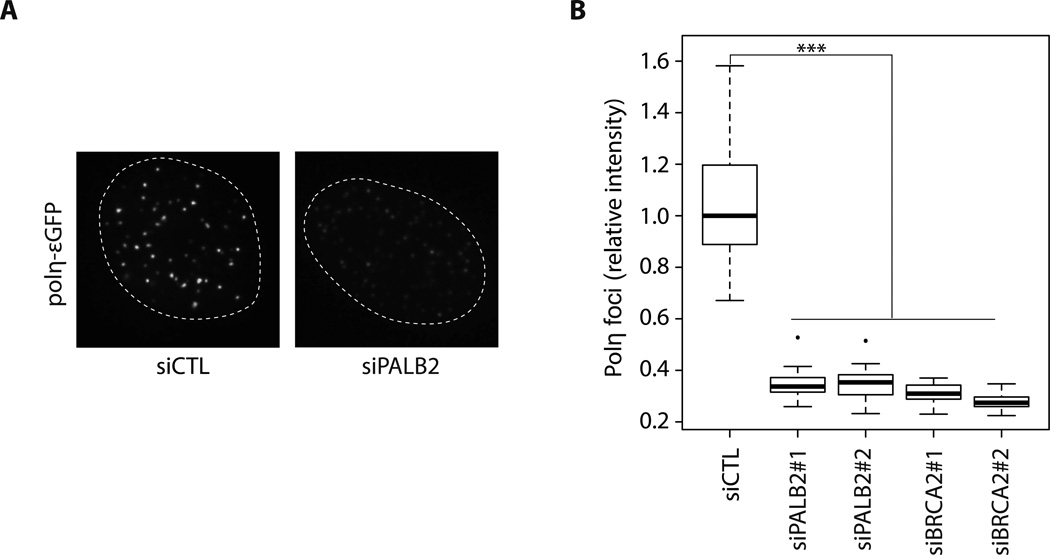
Next, we wondered whether Polη localization at collapsed replication forks is affected by HR proteins. We used siRNAs against PALB2 or BRCA2, two key regulators of RAD51 and HR activity, in Polη-εGFP-complemented XPV cells (Suppl. Fig. 2C). In PALB2 or BRCA2 knockdowns, Polη was still recruited to DSBs; however, we observed a significant decrease in the intensity of Polη foci (quantified in Fig. 2), suggesting that PALB2 and BRCA2 are important for Polη accumulation at replication-dependent DNA DSBs (see also Suppl. Fig. 2D).
Figure 2. Knockdown of PALB2 or BRCA2 decreases Polη foci formation.
(A) A representative example of Polη-εGFP foci intensity in cells transfected with a control siRNA or PALB2 siRNA. (B) Quantification of Polη-εGFP foci intensity induced by HU treatment in control, PALB2 or BRCA2 knockdowns. *** P < 0.001. The median is represented by a black line.
These results show that PALB2 and BRCA2 are required for the accumulation of Polη at DSBs triggered by collapsed replication forks, and provide evidence that PALB2 and BRCA2 could be important for Polη activity.
Purified PALB2 and BRCA2 stimulate Polη
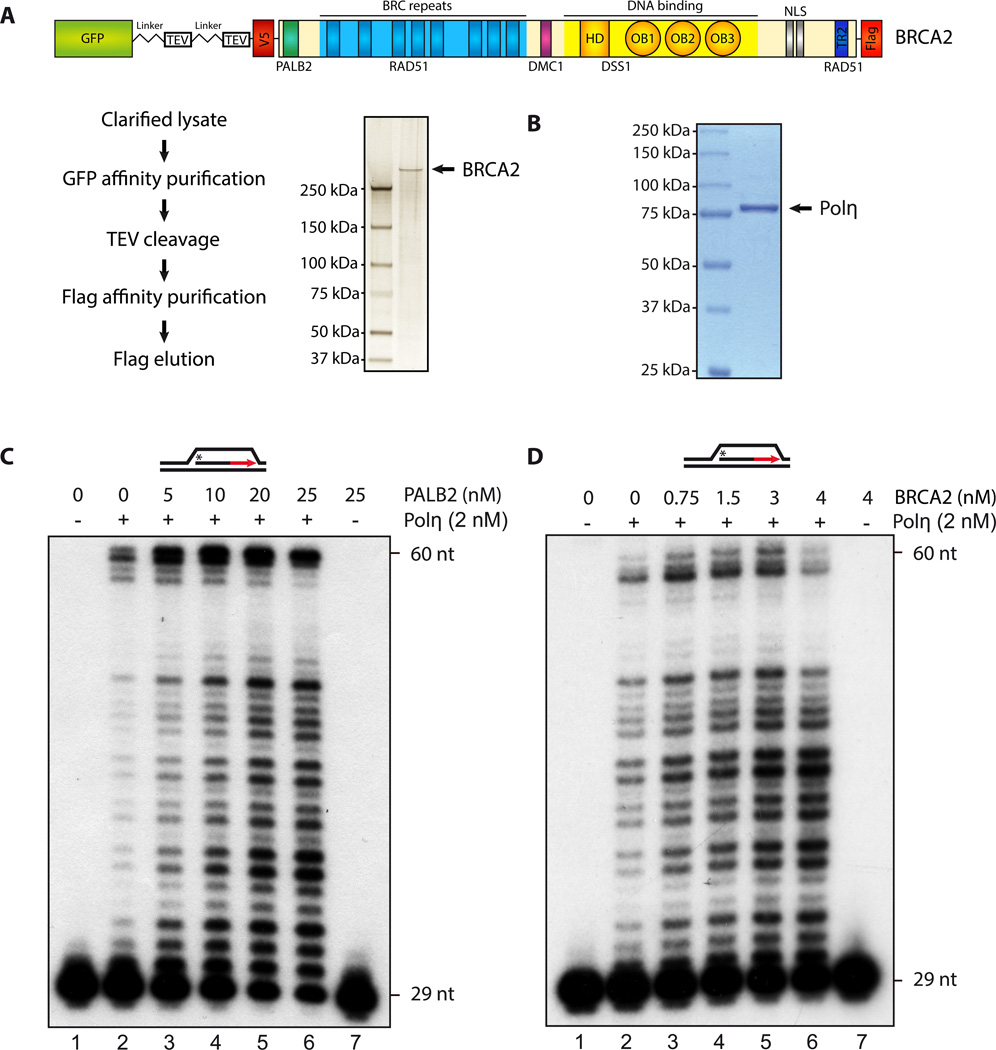
A key step in HR is the formation of a D-loop structure, characterized by the invasion of a single-strand DNA (ssDNA) into a homologous duplex DNA. We purified human BRCA2 (Fig. 3A), Polη (Fig. 3B), and PALB2 (Buisson et al., 2010) in order to see whether both PALB2 and BRCA2 could directly affect the DNA polymerization activity of Polη. First, we confirmed the ability of Polη to promote DNA synthesis on a D-loop substrate (Suppl. Fig. 3A). A control reaction showed that, at 2 nM Polη, only a few synthesis products were detected below 48 nucleotides, while DNA synthesis was complete for some of the events (60 nt) (Fig. 3C, lane 2). Then, when Polη was supplemented with increasing concentrations of purified PALB2, a strong stimulation of Polη-dependent DNA synthesis was observed, leading to intermediate- and full-length DNA products (lanes 3–6). As PALB2 is a major partner of BRCA2, it was important to test whether BRCA2 could also stimulate Polη. Purified BRCA2 also stimulated Polη, although less efficiently than PALB2. Because BRCA2 purification is technically difficult, we also used a functional BRCA2 chimera protein (termed piccolo BRCA2 or piBRCA2) to test Polη stimulation with a higher concentration of protein. We have shown previously that piBRCA2 has very similar properties to full length BRCA2 as it stimulates RAD51 D-loop formation and the accumulation of RAD51 on chromatin (Buisson et al., 2010). piBRCA2 enhanced Polη activity (Suppl. Fig. 3B). Time-course experiments revealed that DNA was synthesized progressively over a 30 minute-period (Suppl. Fig. 4A–B).
Figure 3. PALB2 and BRCA2 stimulate D-loop extension by Polymerase η.
SDS-PAGE of purified human BRCA2 (A), or Polη ((B), 1 µg) stained by silver stain or Coomassie blue, respectively. (C–D) 32P-labeled D-loop was incubated with Polη (2 nM) with the indicated amounts of purified PALB2 or BRCA2. DNA synthesis products were analyzed by denaturing PAGE.
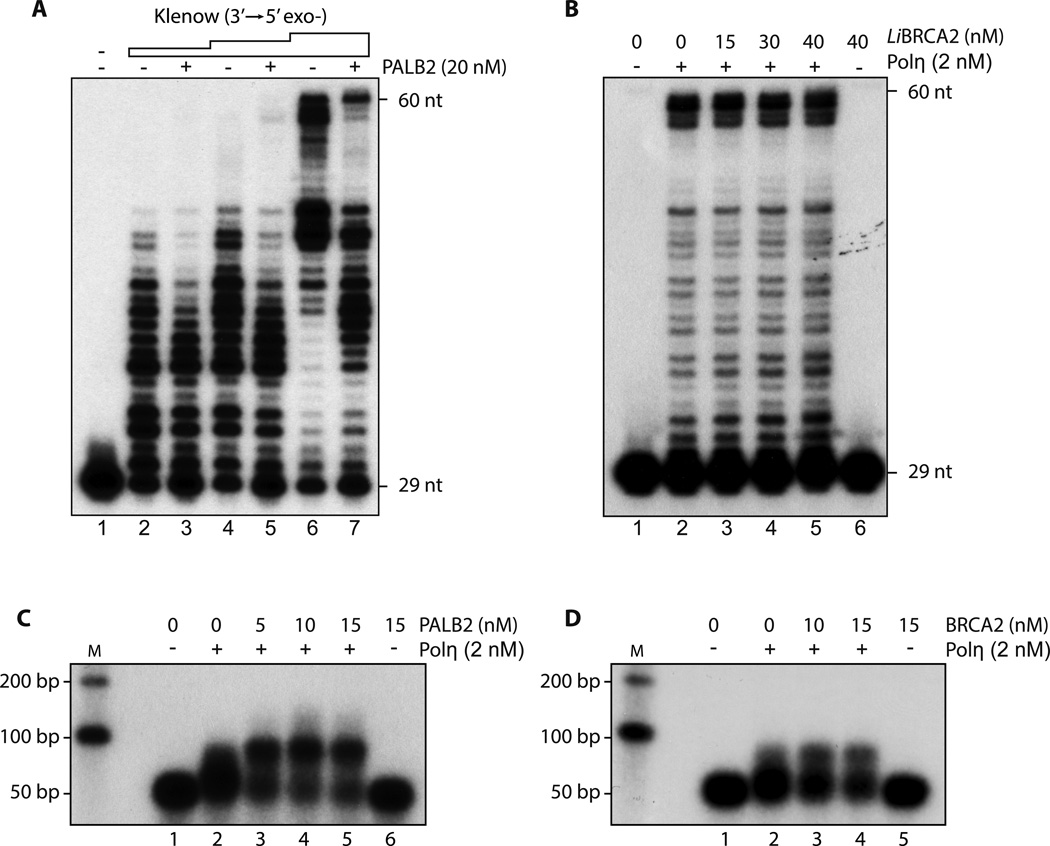
To confirm the specificity of PALB2/BRCA2 stimulation of Polη, several controls were performed. First, we used E.coli DNA polymerase I Klenow fragment (3′–5′ exo−) (Suppl. Fig. 5A). PALB2 or piBRCA2 failed to stimulate Klenow polymerase; indeed, its activity was inhibited (Fig. 4A and suppl. Fig. 5B). Second, unlike PALB2 and BRCA2, BRCA1 did not enhance Polη activity (Suppl. Fig. 6A), although BRCA1 is necessary for their recruitment to DSBs. Third, in E. coli, the single-strand binding protein, SSB, recruits DNA polymerase V (Polη homolog) to the primer end of RecA-coated DNA for DNA synthesis (Arad et al., 2008). Unlike what was observed in bacteria, human RPA protein did not activate Polη (Suppl. Fig. 6B). We then used a BRCA2 homologue from Leishmania infantum, which bears the DNA binding domain BRC7 and BRC8-like repeats. Like hBRCA2, LiBRCA2 stimulates LiRAD51 in recombination assays (Genois et al., 2012). Importantly, LiBRCA2 failed to stimulate human Polη, suggesting that the observed stimulation is specific for human BRCA2 protein (Fig. 4B). In addition, RAD52 was not able to enhance Polη-mediated DNA synthesis, suggesting that a DNA annealing activity is not required (Suppl. Fig. 6C).
Figure 4.
(A) Klenow DNA polymerase activity is not enhanced by PALB2 D-loop substrate was incubated with PALB2 (20 nM) followed by the addition of Klenow polymerase (0.25, 0.5 and 2.5 µU). DNA synthesis products were analyzed by denaturing PAGE.
(B) Leishmania infantum BRCA2 does not stimulate Polη 32P-labeled D-loop was incubated with the indicated amounts of LiBRCA2 prior to the addition of Polη (2 nM). DNA synthesis products were analyzed by denaturing PAGE.
(C–D) PALB2 and BRCA2 stimulate D-loop extension by Polη Purified D-loop substrate was incubated with increasing concentrations of PALB2 or BRCA2 following by addition of Polη. DNA synthesis products were analyzed by alkaline gel electrophoresis.
To further demonstrate that PALB2 and BRCA2 stimulate Polη DNA synthesis, we used a D-loop substrate more similar to those found in eukaryotes in vivo. The D-loop DNA substrate was purified from a RAD51-mediated D-loop assay (Suppl. Fig. 7A). Using this substrate, Polη extended the invaded ssDNA only poorly (Fig. 4C, lane 2). However, the addition of PALB2 (cf. lanes 2 and 3–5), BRCA2 (Fig. 4D, cf. lanes 2 and 3–4), or piBRCA2 (Suppl. Fig. 7B) increased Polη-mediated DNA synthesis. We removed RAD51 during the D-loop purification step to be sure that DNA synthesis was not stimulated artificially by increased D-loop formation due to RAD51 (enhanced by the presence of PALB2/BRCA2). Experiments showed that Polη is activated rapidly in the presence of PALB2 or piBRCA2 (Fig. 4C, Suppl. Fig. 7B) but that extension ceased after addition of 50–100 nucleotides. Topological constraints severely affect DNA synthesis (Li et al., 2009). Adding topoisomerase I at a concentration that transformed supercoiled plasmids into relaxed plasmids (Suppl. Fig. 8A) did not affect Polη/PALB2 or Polη/piBRCA2 DNA extension, suggesting that halted DNA synthesis is not caused by a topological constraint (Suppl. Fig. 8B–C).
Our data suggests that PALB2 and BRCA2 target Polη at the 3′-terminus for DNA synthesis. We used a primer accessibility assay to monitor whether PALB2 and BRCA2 bind the 3′-end of the primer within the D-loop structure. In this assay, PALB2 or piBRCA2 are bound to the D-loop substrate, followed by the addition of E. coli exonuclease III (Exo III) to excise the primer terminus in the 3′–5′ direction. Control reactions showed that Exo III degraded the 5′-labelled primer (Suppl. Fig. 9, lane 2). However, when PALB2 or piBRCA2 were added before Exo III, a region of 10 nucleotides from the 3′-end was protected from degradation (Suppl. Fig. 9A–B). Hence, once bound to the D-loop substrate, our results suggest that PALB2 and BRCA2 bind the 3′-end of D-loop substrates to initiate DNA synthesis by Polη.
Direct interaction between Polη and PALB2 or BRCA2 is required for stimulation of DNA synthesis
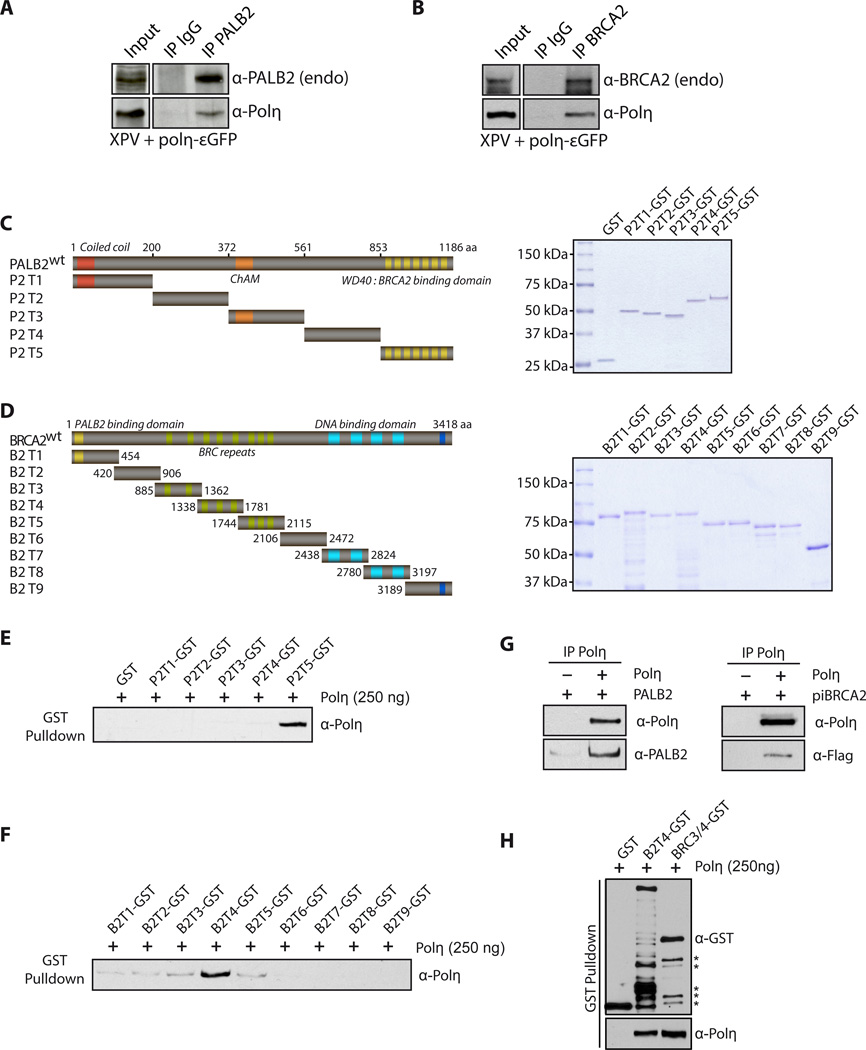
As suggested by the co-localization of PALB2 and BRCA2 with Polη in immunofluorescence studies, we found that Polη co-immunoprecipitated with endogenous PALB2 or BRCA2 in XPV-complemented cells (Fig. 5A,B). We also detected a complex between endogenous Polη, BRCA2 and PALB2 in untreated and HU-treated U2OS cells (Suppl. Fig. 10). Moreover, the stimulatory effect of PALB2 and BRCA2 on Polη suggested that these proteins could interact directly. To map the interaction regions between Polη and both PALB2 and BRCA2, we used a series of non-overlapping glutathione S-transferase (GST) fusion proteins (designated PALB2 P2-T1 to P2-T5, and BRCA2 B2-T1 to B2-T9; Fig. 5C,D) to define the regions of PALB2 and BRCA2 that interact with Polη. GST pull-down assays revealed that Polη binds the WD40 domain of PALB2 (residues 853–1186) (Fig. 5E) and truncation 4 of BRCA2 (residues 1338–1781) containing the BRC3, BRC4 and BRC5 domains (Fig. 5F). In addition, we found that both purified PALB2 and piBRCA2 bind Polη directly (Fig. 5G). Since the purification scheme of these proteins include a benzonase treatment, we infer that this interaction is not DNA mediated. As piBRCA2 bears the BRC3/4 domain and interacts directly with Polη, we purified a smaller GST fragment of truncation 4 with only BRC3/4. GST-BRC3/4 still interacts with Polη with the same affinity as truncation 4 of BRCA2 (Fig. 5H).
Figure 5. PALB2 and BRCA2 interact directly with Polη.
(A) Cell extracts from complemented XPV cells were subjected to immunoprecipitation with anti-PALB2 or (B) anti-BRCA2 antibodies. Immunoprecipitated proteins were detected by Western Blotting with the indicated antibodies.
(C) Left: Scheme of the PALB2 or (D) BRCA2 deletion variants fused to GST. Right: SDS-PAGE of the corresponding purified proteins. (E) GST alone or GST-PALB2 truncation (P2T1 to P2T5) were incubated with Polη followed by GST pulldown and detection of Polη by Western blotting. (F) GST alone or GST-BRCA2 truncations (B2T1 to B2T9) were incubated with Polη, followed by GST pulldown. The beads were washed and bound proteins were eluted with Laemmli buffer, and revealed by western blotting with the antibodies indicated. The input for PALB2 or BRCA2 truncations are shown in (C) and (D). (G-H) Co-immunoprecipitation of purified PALB2, piBRCA2, B2T4 or BRC3/4 and Polη. Asterisk: degradation products.
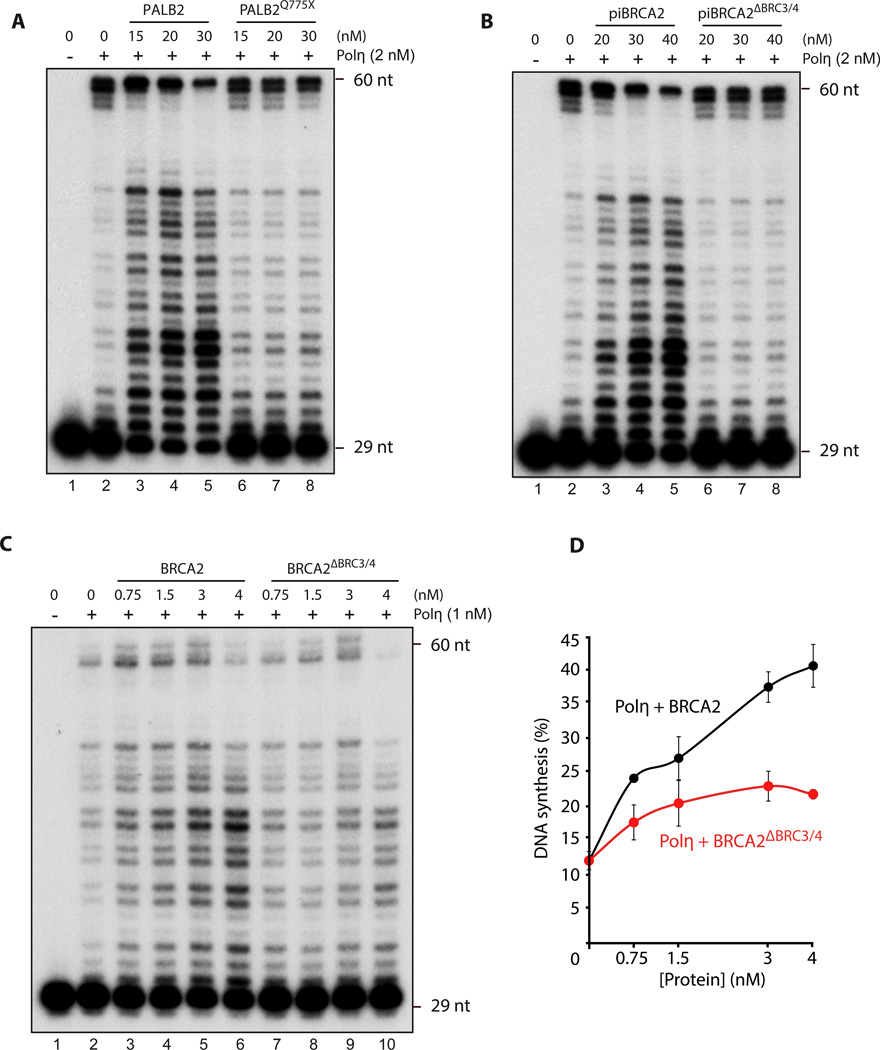
We next examined whether the Polη interaction domains within PALB2 or BRCA2 were important for Polη stimulation. We purified PALB2Q775X—a mutant of PALB2 with a nonsense codon just before the WD40 domain (Suppl. Fig. 11A–B). This mutant was found among French-Canadian women with breast cancer (Foulkes et al., 2007). Since it bears intact DNA binding domains, PALB2Q775X binds DNA similarly to full-length PALB2 (Suppl. Fig. 11C). Remarkably, PALB2Q775X completely lost its ability to enhance Polη DNA synthesis and extend D-loop oligonucleotide substrates (Fig. 6A, Suppl. Fig. 11D) or purified D-loops (Suppl. Fig. 11E). Similar experiments were also performed with piBRCA2 and BRCA2. We deleted the BRC3/4 domain from the piBRCA2 chimera or full-length BRCA2 (Suppl. Fig. 12 and 13). Unlike piBRCA2 or BRCA2, piBRCA2ΔBRC3/4 or BRCA2ΔBRC3/4 failed to enhance Polη DNA synthesis (Fig. 6B–D, Suppl. Fig. 12D–E) while both proteins displayed similar DNA binding properties (Suppl. Fig. 12C and 13C). These results show that the WD40 domain of PALB2 and the BRC3/4 repeat of BRCA2 are crucial for DNA synthesis by Polη.
Figure 6. Polη interactions domains within PALB2 or piBRCA2 are essential for Polη stimulation.
(A–B–C) 32P-labeled D-loop oligonucleotides was first incubated with indicated concentrations of PALB2, piBRCA2 with/without the Polη interacting domains following by the addition of Polη. DNA synthesis products were analyzed by denaturing PAGE. (D) quantification of the results shown in (C).
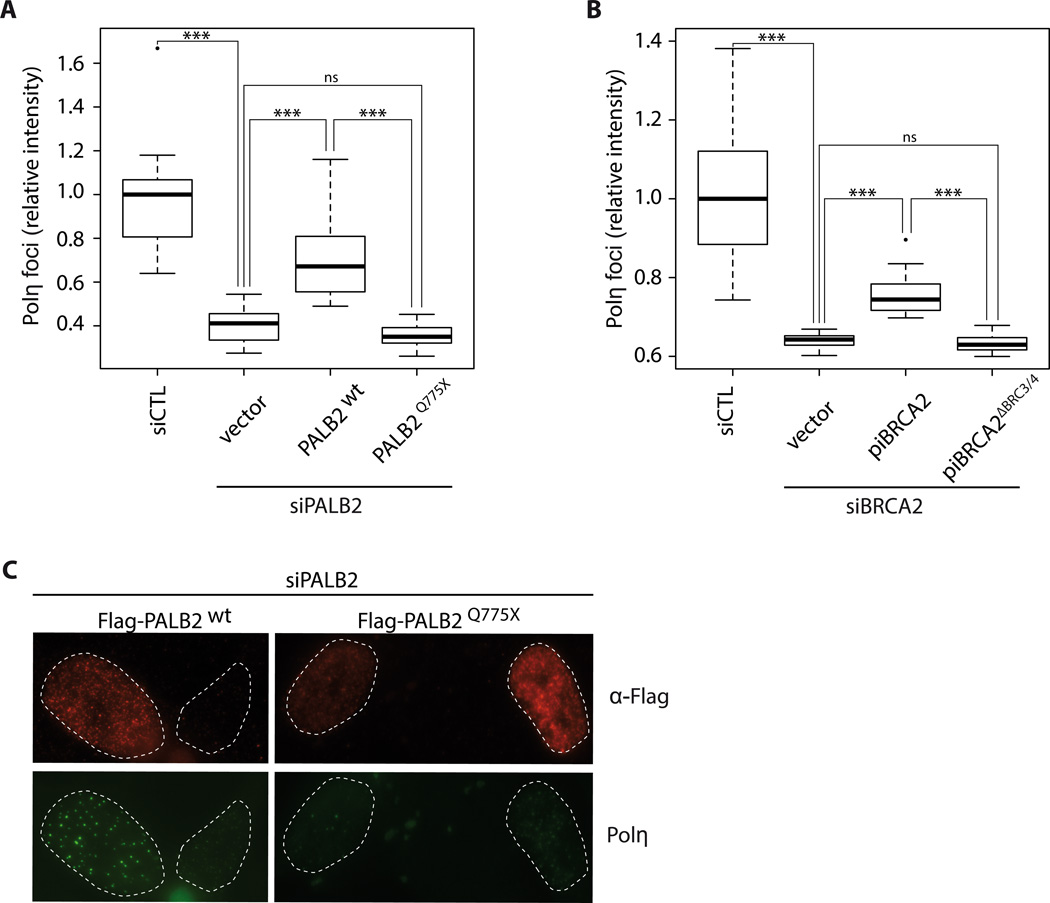
Next, we tested whether disruption of the Polη interaction domains in PALB2 and BRCA2 affected Polη foci formation in vivo after HU treatment. First, consistent with our previous data, the interaction between PALB2 and Polη was lost with a C-terminal truncation mutation containing the WD40 domain (PALB2Q775X, Suppl. Fig. 14A), while deletion of BRC3/4 in piBRCA2 abrogated the interaction with Polη (Suppl. Fig. 14B). These siRNA-resistant constructs, along with wild-type controls, were used to complement siPALB2 and siBRCA2 cells, respectively. Polη foci formation was severely affected in siPALB2 and siBRCA2 cells transfected with PALB2Q775X or piBRCA2ΔBRC3/4, compared to wild-type constructs (Fig. 7A–B–C and Suppl. Fig. 14C). These results show that the WD40 domain of PALB2 and the BRC3/4 repeat of BRCA2 are important for Polη foci formation at broken replication forks.
Figure 7. Deletion of Polη interaction domains in PALB2 and piBRCA2 affect polymerase η foci formation.
(A) quantification of Polη-εGFP foci intensity induced by HU treatment in control, and siPALB2 knockdowns complemented with vector alone, wild-type PALB2, or PALB2Q775X. (B) Quantification of Polη-εGFP foci intensity induced by HU treatment in control, and siBRCA2 knockdowns complemented with vector alone, wild-type piBRCA2, or piBRCA2ΔBRC3/4. *** P < 0.001, ns not significant between the identified groups. The median is represented by a black line. (C) Representative images of Figure 7A. XPV complemented cells knockdown for PALB2, and complemented with Flag-PALB2 wild-type, and Flag-PALB2 Q775X were subjected for immunofluorescence against Polη (green) or anti-Flag antibodies to discriminate transfected cells. siRNA PALB2 resistant constructs were used.
Discussion
Replicative DNA polymerases are often stalled by DNA lesions, and the replication blockage is released by translesion DNA synthesis and HR. How these processes are intertwined is poorly understood in higher eukaryotic cells. Our data reveal novel roles for BRCA2 and PALB2 in coordinating the DNA synthesis step of HR during the repair of replication forks.
PALB2 and BRCA2: two important players at blocked DNA replication forks
Our results show that PALB2 and BRCA2 are important for the proper localization of Polη to DSBs at replication forks and to enhance its activity for D-loop extension. Recent studies ascribe a new role to BRCA2/PALB2 tumor suppressors at broken replication forks. Unlike collapsed replication forks, which require active HR for repair (Petermann et al., 2010), RAD51 promotes replication restart independently of HR at stalled replication forks. At stalled replication forks, a possible model is that helicases such WRN, BLM or SMARCAL1 facilitate fork regression into a « Chicken Foot » structure, which is protected against MRE11 degradation by the BRCA pathway (Hashimoto et al., 2010; Petermann and Helleday, 2010; Schlacher et al., 2011; Schlacher et al., 2012). The BRCA pathway achieves this function by promoting RAD51 nucleoprotein filament formation and stabilization at stalled forks (Schlacher et al., 2012). The double-stranded end of the chicken foot structure is used by RAD51 and its mediators to promote strand invasion and D-loop formation. In contrast, prolonged replication blocks, caused mainly by a failure to restart, lead to collapsed replication forks and DSB formation. DSBs are generated by the MUS81-EME1 complex and then are repaired by RAD51-mediated HR. In both mechanisms, DNA synthesis is required and Polη action would benefit from the presence of PALB2 and BRCA2 proteins after strand invasion.
We show that both PALB2 and BRCA2 bind the 3′-end of the invading strand in the D-loop structure. Since PALB2 and BRCA2 interact with Polη, our data suggest that they target DNA Polη to the end of the invading strand, in addition to their role in stimulating RAD51 during strand invasion. We initially planned to study the function of PALB2 or BRCA2 in the presence of RAD51 in terms of their Polη stimulation activities. However, this is extremely difficult as one cannot distinguish between the RAD51 invasion activity promoted by PALB2/BRCA2 and the polη elongation activity stimulated by PALB2/BRCA2, as elongation is possible only after DNA invasion has occurred. Hence, we purified D-loop structures in order to monitor PALB2/BRCA2 activity at the elongation step without interference by RAD51. After D-loop formation, RAD51 may be an obstacle to the initiation of the elongation step in making the 3'-OH end inaccessible to DNA polymerases. RAD54 is able to eliminate RAD51 at the dsDNA and thereby release the 3'-OH (Solinger et al., 2002). Indeed, in yeast, in the absence of RAD54, Polη and δ are unable to extend the D-loop structures formed by RAD51 (Li et al., 2009).
More than one polymerase is required for effective HR
We have shown that Polη is stimulated by BRCA2 and PALB2—two regulators of RAD51 at stalled or collapsed replication forks—which is similar to what is observed in prokaryotic cells. In E.coli, DNA polymerase V, the prokaryotic homolog of Polη, catalyzes DNA synthesis on a TLS substrate in the presence of RecA (RAD51 homolog in E.coli) (Pham et al., 2002; Schlacher et al., 2005). While our data suggest that Polη-dependent DNA synthesis is enhanced specifically by BRCA2 and PALB2, we do not exclude the possibility that other DNA polymerases could be stimulated by BRCA2 and PALB2. The human genome encodes at least 13 nuclear DNA polymerases (Lange et al., 2011). Several polymerases have been identified as being important for DSB repair by HR. S. cerevisiae Polδ affects gene conversion tract length during mitotic recombination from site-specific DSBs (Maloisel et al., 2008), and both PCNA-associated DNA Polδ and ε are important for gene conversion (Holmes and Haber, 1999). Polymerases ζ, ν, and REV1 have been shown to be involved in DSB repair and their knockdown in human cells causes a significant decrease of HR (Moldovan et al., 2010; Sharma et al., 2012). In human cells, Polδ and Polη and Polδ are the only polymerases known able to extend a D-loop structure (Li et al., 2013; McIlwraith et al., 2005; Sebesta et al., 2011; Sebesta et al., 2013; Sneeden et al., 2013) The exact function of all these polymerases in DSB repair is still poorly understood and several polymerases might replace each other when required. Indeed, a recent report shows that only the double depletion of Polη and Polκ affects HR significantly (Sebesta et al., 2013). Another possibility could be that these polymerases are involved in different alternative DSB repair pathways. For instance, the DSBR (double-strand break repair) pathway is characterized by a second-end capture and double Holliday junction formation leading to crossover or non-crossover products, whereas SDSA (synthesis-dependent strand-annealing) is characterized by displacement of the extended D-loop by BLM helicase with annealing with the second extremity of the break leading to only non-crossover products (Krejci et al., 2012; Moynahan and Jasin, 2010). We hypothesize that, depending on the pathway used, different polymerases could be involved. This is consistent with recent work showing that expression of an inactive Polη led to an increase of sister chromatin exchange (Bergoglio et al., 2013)—a similar phenotype to that observed in BLM deficient cells—suggesting that Polη could be more important in the SDSA pathway. Furthermore, Pol% has been shown to be more processive than Polη when extending D-loop structures (Li et al., 2009; Sebesta et al., 2013; Sneeden et al., 2013). We hypothesize that Polη will be necessary only for short extension or for the initiation of D-loop extension, and is then substituted by Polδ for the generation of longer track length. In this regard, it is important to note that Polη is a low fidelity DNA polymerase. Thus, enabling extended DNA synthesis by Polη could lead to enhanced mutagenesis and genomic instability. Finally, we showed that Polη does not localize to DSBs provoked by IR treatment, confirming previous results (Kannouche et al., 2001). For this reason, we think that polymerase δ or κ could be the major players in D-loop extension at DSBs not associated with a replication fork. Further work will be needed to clearly understand the interplay between all these polymerases and whether PALB2 and BRCA2 are also important for their regulation. The regulation of these polymerases during repair DNA synthesis might also be different depending on species, as PALB2 and BRCA2 are absent in yeast. Hence, binding to BRCA2 and PALB2 might control the proper use of polymerases in a temporal and DNA damage-specific manner in human cells.
Conclusion
Previous studies on BRCA2 and PALB2 clearly established these proteins as essential regulators of HR through RAD51 regulation. Our findings show that PALB2 and BRCA2 are important both for polymerase η localization and DNA polymerization activity, leading us to propose a new mechanistic function for these HR mediator proteins. The effect of the PALB2 Q775X mutant on DNA synthesis was revealing, as this mutation is associated with breast cancer (Foulkes et al., 2007). The deleterious effect of this mutation might be related to the inability to interact with both Polη and BRCA2. As a result, PALB2-deficient cells undergo genome rearrangement and instability as DSBs are not properly repaired. Altogether, our results suggest that the defects observed in BRCA2- and PALB2-deficient or mutated cells, are related not only to RAD51-dependent strand invasion, but also to a deficiency in DNA synthesis.
Supplementary Material
Highlights.
-
-
PALB2, BRCA2 and Polη co-localize at stalled or collapsed replication forks
-
-
PALB2 and BRCA2 interact with Polη
-
-
PALB2 and BRCA2 stimulate Polη-dependent DNA synthesis on D-loop substrates
-
-
PALB2 and BRCA2 play crucial roles in recombination-associated DNA synthesis
Acknowledgements
We thank Elliot Drobetsky for XPV and complemented XPV cell lines, Bing Xia for providing the PALB2 antibody, and Mark Baker for discussions. We also thank Lee Zou, Helen Rothnie and Isabelle Brodeur for helpful comments on the manuscript, Eric Paquet for bioinformatic analyses and Marie-Michelle Genois for purified LiBRCA2. R.B. was a FQNRT doctoral scholar, and J.-Y.M. is a FRQS Chercheur National. This work was supported by NIH grant ES015252 (to P.S.) and the CIHR (to J.-Y.M.).
References
- Arad G, Hendel A, Urbanke C, Curth U, Livneh Z. Single-stranded DNA-binding protein recruits DNA polymerase V to primer termini on RecA-coated DNA. J Biol Chem. 2008;283:8274–8282. doi: 10.1074/jbc.M710290200. [DOI] [PubMed] [Google Scholar]
- Bergoglio V, Boyer AS, Walsh E, Naim V, Legube G, Lee MY, Rey L, Rosselli F, Cazaux C, Eckert KA, et al. DNA synthesis by Pol eta promotes fragile site stability by preventing under-replicated DNA in mitosis. The Journal of cell biology. 2013;201:395–408. doi: 10.1083/jcb.201207066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard JY, Buisson R, Masson JY, Esashi F. ChAM, a novel motif that mediates PALB2 intrinsic chromatin binding and facilitates DNA repair. EMBO Rep. 2012;13:135–141. doi: 10.1038/embor.2011.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Buisson R, Dion-Cote AM, Coulombe Y, Launay H, Cai H, Stasiak AZ, Stasiak A, Xia B, Masson JY. Cooperation of breast cancer proteins PALB2 and piccolo BRCA2 in stimulating homologous recombination. Nat Struct Mol Biol. 2010;17:1247–1254. doi: 10.1038/nsmb.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buisson R, Masson JY. PALB2 self-interaction controls homologous recombination. Nucleic acids research. 2012 doi: 10.1093/nar/gks807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Feraudy S, Limoli CL, Giedzinski E, Karentz D, Marti TM, Feeney L, Cleaver JE. Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene. 2007;26:5713–5721. doi: 10.1038/sj.onc.1210385. [DOI] [PubMed] [Google Scholar]
- Dray E, Etchin J, Wiese C, Saro D, Williams GJ, Hammel M, Yu X, Galkin VE, Liu D, Tsai MS, et al. Enhancement of RAD51 recombinase activity by the tumor suppressor PALB2. Nat Struct Mol Biol. 2010;17:1255–1259. doi: 10.1038/nsmb.1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foulkes WD, Ghadirian P, Akbari MR, Hamel N, Giroux S, Sabbaghian N, Darnel A, Royer R, Poll A, Fafard E, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genois MM, Mukherjee A, Ubeda JM, Buisson R, Paquet E, Roy G, Plourde M, Coulombe Y, Ouellette M, Masson JY. Interactions between BRCA2 and RAD51 for promoting homologous recombination in Leishmania infantum. Nucleic acids research. 2012 doi: 10.1093/nar/gks306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L. Replication past O(6)-methylguanine by yeast and human DNA polymerase eta. Molecular and cellular biology. 2000a;20:8001–8007. doi: 10.1128/mcb.20.21.8001-8007.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nature genetics. 2000b;25:458–461. doi: 10.1038/78169. [DOI] [PubMed] [Google Scholar]
- Hashimoto Y, Ray Chaudhuri A, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nature structural & molecular biology. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes AM, Haber JE. Double-strand break repair in yeast requires both leading and lagging strand DNA polymerases. Cell. 1999;96:415–424. doi: 10.1016/s0092-8674(00)80554-1. [DOI] [PubMed] [Google Scholar]
- Howlett NG, Taniguchi T, Olson S, Cox B, Waisfisz Q, De Die-Smulders C, Persky N, Grompe M, Joenje H, Pals G, et al. Biallelic inactivation of BRCA2 in Fanconi anemia. Science. 2002;297:606–609. doi: 10.1126/science.1073834. [DOI] [PubMed] [Google Scholar]
- Jensen RB, Carreira A, Kowalczykowski SC. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature. 2010;467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of Xeroderma pigmentosum. Science. 1999;285:263–265. doi: 10.1126/science.285.5425.263. [DOI] [PubMed] [Google Scholar]
- Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes & development. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto T, Araki K, Sonoda E, Yamashita YM, Harada K, Kikuchi K, Masutani C, Hanaoka F, Nozaki K, Hashimoto N, et al. Dual roles for DNA polymerase eta in homologous DNA recombination and translesion DNA synthesis. Molecular cell. 2005;20:793–799. doi: 10.1016/j.molcel.2005.10.016. [DOI] [PubMed] [Google Scholar]
- Krejci L, Altmannova V, Spirek M, Zhao X. Homologous recombination and its regulation. Nucleic acids research. 2012 doi: 10.1093/nar/gks270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nature reviews. Cancer. 2011;11:96–110. doi: 10.1038/nrc2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Holzschu DL, Sugiyama T. PCNA is efficiently loaded on the DNA recombination intermediate to modulate polymerase delta eta, zeta activities. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:7672–7677. doi: 10.1073/pnas.1222241110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Stith CM, Burgers PM, Heyer WD. PCNA is required for initiation of recombination-associated DNA synthesis by DNA polymerase delta. Molecular cell. 2009;36:704–713. doi: 10.1016/j.molcel.2009.09.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Doty T, Gibson B, Heyer WD. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol. 2010;17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloisel L, Fabre F, Gangloff S. DNA polymerase delta is preferentially recruited during homologous recombination to promote heteroduplex DNA extension. Molecular and cellular biology. 2008;28:1373–1382. doi: 10.1128/MCB.01651-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- McIlwraith MJ, Vaisman A, Liu Y, Fanning E, Woodgate R, West SC. Human DNA polymerase eta promotes DNA synthesis from strand invasion intermediates of homologous recombination. Mol Cell. 2005;20:783–792. doi: 10.1016/j.molcel.2005.10.001. [DOI] [PubMed] [Google Scholar]
- Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moldovan GL, Madhavan MV, Mirchandani KD, McCaffrey RM, Vinciguerra P, D'Andrea AD. DNA polymerase POLN participates in cross-link repair and homologous recombination. Mol Cell Biol. 2010;30:1088–1096. doi: 10.1128/MCB.01124-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham P, Seitz EM, Saveliev S, Shen X, Woodgate R, Cox MM, Goodman MF. Two distinct modes of RecA action are required for DNA polymerase V-catalyzed translesion synthesis. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11061–11066. doi: 10.1073/pnas.172197099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Christ N, Siaud N, Egashira A, Wu H, Jasin M. Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell. 2011;145:529–542. doi: 10.1016/j.cell.2011.03.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlacher K, Leslie K, Wyman C, Woodgate R, Cox MM, Goodman MF. DNA polymerase V and RecA protein, a minimal mutasome. Molecular cell. 2005;17:561–572. doi: 10.1016/j.molcel.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Schlacher K, Wu H, Jasin M. A Distinct Replication Fork Protection Pathway Connects Fanconi Anemia Tumor Suppressors to RAD51-BRCA1/2. Cancer cell. 2012;22:106–116. doi: 10.1016/j.ccr.2012.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Haracska L, Krejci L. Reconstitution of DNA repair synthesis in vitro and the role of polymerase and helicase activities. DNA repair. 2011;10:567–576. doi: 10.1016/j.dnarep.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebesta M, Burkovics P, Juhasz S, Zhang S, Szabo JE, Lee MY, Haracska L, Krejci L. Role of PCNA and TLS polymerases in D-loop extension during homologous recombination in humans. DNA repair. 2013 doi: 10.1016/j.dnarep.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Hicks JK, Chute CL, Brennan JR, Ahn JY, Glover TW, Canman CE. REV1 and polymerase zeta facilitate homologous recombination repair. Nucleic acids research. 2012;40:682–691. doi: 10.1093/nar/gkr769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneeden JL, Grossi SM, Tappin I, Hurwitz J, Heyer WD. Reconstitution of recombination-associated DNA synthesis with human proteins. Nucleic acids research. 2013 doi: 10.1093/nar/gkt192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinger JA, Kiianitsa K, Heyer WD. Rad54, a Swi2/Snf2-like recombinational repair protein, disassembles Rad51:dsDNA filaments. Molecular cell. 2002;10:1175–1188. doi: 10.1016/s1097-2765(02)00743-8. [DOI] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Chen J. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A. 2009a;106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sy SM, Huen MS, Zhu Y, Chen J. PALB2 Regulates Recombinational Repair through Chromatin Association and Oligomerization. J Biol Chem. 2009b;284:18302–18310. doi: 10.1074/jbc.M109.016717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai K, Shibata E, Abbas T, Dutta A. Degradation of p12 Subunit by CRL4Cdt2 E3 Ligase Inhibits Fork Progression after DNA Damage. J Biol Chem. 2013;288:30509–30514. doi: 10.1074/jbc.C113.505586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol. 2010;17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Dorsman JC, Ameziane N, de Vries Y, Rooimans MA, Sheng Q, Pals G, Errami A, Gluckman E, Llera J, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- Zhang F, Fan Q, Ren K, Andreassen PR. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res. 2009a;7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol. 2009b;19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhao H, Darzynkiewicz Z, Zhou P, Zhang Z, Lee EY, Lee MY. A Novel Function of CRL4Cdt2: Regulation of the subunit structure of DNA polymerase delta in response to DNA damage and during the S phase. J Biol Chem. 2013;288:29550–29561. doi: 10.1074/jbc.M113.490466. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.