Abstract
The epidermal growth factor receptor (EGFR) and Notch signaling pathways have antagonistic roles during epidermal differentiation and carcinogenesis. The molecular mechanisms regulating the crosstalk between EGFR and Notch during epidermal transformation are largely unknown. We found enhanced EGFR-dependent signaling, proliferation and oncogenic transformation caused by loss of presenilins (PS), the catalytic components of γ-secretase that generates the Notch1 intracellular domain (NICD). The underlying mechanism for abnormal EGFR signaling in PS-deficient cells involves γ-secretase-independent transcriptional upregulation of the E3 ubiquitin ligase Fbw7. Fbw7α, which targets NICD for degradation, regulates positively EGFR by affecting a proteasome-dependent ubiquitination step essential for constitutive degradation and stability of EGFR. To investigate the pathological relevance of this findings in vivo, we generated a novel epidermal conditional PS-deficient (ePS cDKO) mouse by deleting both PS in keratinocytes of the basal layer of the epidermis. The ePS cDKO mice develop epidermal hyperplasia associated with enhanced expression of both EGFR and Fbw7 and reduced NICD levels in keratinocytes. These findings establish a novel role for PS on epidermal growth and transformation by reciprocally regulating the EGFR and Notch signaling pathways through Fbw7.
Keywords: carcinogenesis, epidermis, Fbw7, Notch, γ-secretase
Introduction
Presenilins (PS: PS1 and PS2) are the catalytic components of γ-secretase, an enzymatic complex that cleaves type-I transmembrane proteins implicated in cell adhesion, differentiation, survival and proliferation (Koo and Kopan, 2004). PS/γ-secretase-mediated cleavage of the β-amyloid precursor protein has a critical role in Alzheimer's disease pathogenesis presumably by affecting the generation of β-amyloid (De Strooper, 2007). PS/γ-secretase mediates Notch1 activation by releasing the Notch1 intracellular domain (NICD) (De Strooper et al., 1999). Notch signaling inhibits keratinocyte proliferation by promoting differentiation (Lefort and Dotto, 2004). Recent findings suggest a role of loss of PS function in epidermal homeostasis. Deletion of both PS during embryogenesis induces abnormal epidermal differentiation and degeneration of hair follicles (Pan et al., 2004; Demehri et al., 2008), whereas partial loss of PS function in adulthood causes skin hyperplasia and autoimmune disease (Xia et al., 2001; Tournoy et al., 2004). The tumorigenic phenotype caused by loss of PS1 function seems to be independent of γ-secretase activity but associated with enhanced β-catenin signaling (Xia et al., 2001; Kang et al., 2002). Surprisingly, disruption of PS1/β-catenin interaction by deleting the PS1 loop domain, a region dispensable for γ-secretase activity that includes the β-catenin binding domain (Saura et al., 2000), is not sufficient to induce skin tumors (Deng et al., 2006). Its noteworthy that two PS2 mutations found in breast tumors increase cell proliferation and reduce Notch signaling indicating that they act through a loss-of-function mechanism (To et al., 2006). This suggests that alternative signaling pathways may be involved in the oncogenic phenotype caused by PS deficiency.
The epidermal growth factor receptor (EGFR) promotes proliferation of keratinocyte-derived epithelial tumors (Kalyankrishna and Grandis, 2006). EGFR maintains keratinocyte self-renewal in the basal layer of the epidermis by suppressing differentiation and promoting proliferation (Jost et al., 2000). Notably, although enhanced EGFR signaling contributes to epidermal growth and malignant transformation, Notch signaling suppresses keratinocyte proliferation (Nicolas et al., 2003). Indeed, EGFR signaling acts as a negative regulator of Notch1 transcription during suppression of keratinocyte differentiation (Kolev et al., 2008). Reduced PS1 and cleaved Notch1 and elevated EGFR levels were detected in skin papillomas of activating transcription factor-2 knockout mice (Bhoumik et al., 2008), indicating a possible mechanistic link between Notch and EGFR signaling through PS during epidermal transformation. Moreover, elevated EGFR was recently found in skin tumors of PS1- and nicastrindeficient mice (Li et al., 2007; Zhang et al., 2007). Two alternative mechanisms of EGFR regulation by PS, one involving repressed EGFR transcription by the γ-secretase-derived β-amyloid precursor protein intra-cellular domain and the other implicating altered EGFR lysosomal targeting, have been postulated (Repetto et al., 2007; Zhang et al., 2007). Despite the evidence linking PS and EGFR, it is unclear whether PS regulates cell growth and oncogenic transformation through EGFR signaling.
Fbw7, the recognition component of the Skp1-Cul1-F-box protein E3 ubiquitin ligase, acts as a tumor suppressor gene by regulating the ubiquitination and degradation of several regulators of cell division, including Notch1/4, c-Jun, cyclin E, c-Myc and mTOR (Welcker and Clurman, 2008). The Fbw7 homolog in Caenorhabditis elegans, sel-10/Cdc4, was shown to interact functionally with the PS homolog sel-12 (Wu et al., 1998), although the physiological function of Fbw7/PS interaction in mammals remains unknown. We examined the hypothesis that PS could inversely regulate EGFR and Notch signaling by affecting Fbw7. Our studies using fibroblasts and primary keratinocytes from PS-deficient mice revealed that PS negatively regulate Fbw7, which enhances EGFR expression and signaling but decreases NICD levels. These results suggest a PS-dependent crosstalk between EGFR and Notch signaling through Fbw7 during cell growth and transformation.
Results
Abnormal cell proliferation and transformation in PS-deficient cells depend on EGFR signaling
Recent evidences indicate that PS negatively regulate EGFR (Repetto et al., 2007; Zhang et al., 2007), although the relevance of this finding on epidermal tumorigenesis is still unclear. To investigate this issue, we analyzed cell growth and oncogenic transformation in immortalized skin fibroblasts from control (PS1/PS2) and PS double knockout (PS–/–) mice. Control and PS–/– fibroblasts grew similarly in standard culture conditions (10% fetal bovine serum). By contrast, PS–/– fibroblasts grew very rapidly in low and very low serum conditions (0.1–1% fetal bovine serum), suggesting that loss of PS increases the sensitivity of cells to growth factors (Figure 1a). Stable expression of hPS1 reduced the abnormal proliferation rate of PS–/– fibroblasts arguing against side effects of the transformed cell lines (Figure 1a). In order to discern the specific growth factor receptor(s) involved in the proliferative phenotype, we next performed a pharmacological drug screening. Inhibition of EGFR with PD168393 and PD153035, but not inhibiting ErbB2 (AG825), PDGFR/VEGFR (SU4312), IGFR (picropodophyllin) or FGFR (SU5402), at concentrations that lower ERK1/2 signaling (≥10 × IC50; not shown), significantly decreased the number and density of PS–/– fibroblasts (Figure 1b). Analysis of anchorage-independent growth in soft agar, an assay used previously to study EGFR-dependent oncogenic phenotypes (Di Fiore et al., 1987), revealed larger and higher number of colonies formed by PS–/– fibroblasts compared with control, PS1–/– and PS2–/– fibroblasts, a phenotype that was suppressed by inhibiting EGFR signaling (Figure 1c and data not shown). These results indicate that abnormal proliferation and transformation caused by loss of PS were dependent on EGFR signaling.
Figure 1.
Increased proliferation and oncogenic transformation of PS-deficient fibroblasts depends on EGFR signaling. (a) Control (PS1/PS2), PS–/–and PS–/–; PS1 embryonic fibroblasts were cultured for 0–72 h in the presence of 1 or 10% of fetal bovine serum (FBS). Data are presented as mean±s.e.m (n 3), *P<0.05; **P<0.001. (b) PS1/PS2 and PS–/– fibroblasts were containing cultured in media low serum (0.1% FBS) for 24 h before counting or culturing in the presence of vehicle or specific inhibitors of EGFR (PD168393, 2 μm; PD153035, 1 μm), ErbB2 (AG825, 20 μm), PDGFR/VEGFR (SU4312, 10 μm), IGFR (picropodophyllin, PPP, 10 mm) or FGFR (SU5402, 5 μm) for an additional 24 h. Data are presented as mean±s.e.m. *P<0.05. (c) Colony formation assays in control and PS–/– fibroblasts were performed in soft-agar plates in the presence or absence of PD168393 (2 μm). The number of colonies present on three plates was quantified after 15 days. This experiment is representative of three independent experiments. Data are presented as mean±s.e.m of three dishes. *P<0.05, **P<0.001.
Epidermal hyperplasia and enhanced EGFR signaling in epidermal conditional PS knockout mice
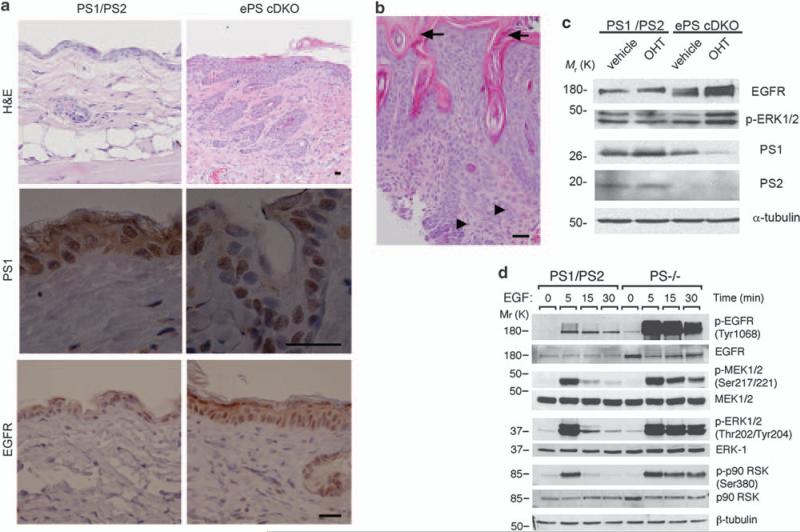
To investigate the role of EGFR signaling on epidermal proliferation and transformation caused by loss of PS function in adulthood while avoiding possible epidermal developmental effects (Shen et al., 1997; Pan et al., 2004), we generated an inducible epidermal conditional PS-deficient mouse (ePS cDKO). Mice carrying PS1 alleles flanked by loxP sites (PS1 f/f) in a PS2–/– background (Saura et al., 2004) were mated to K14-Cre-ERT2 mice expressing the tamoxifen-dependent Cre-ERT2 recombinase, under the control of the human keratin 14 (K14) promoter (Indra et al., 1999; Li et al., 2000), to generate mice bearing two PS1 floxed (f) and PS2 null alleles and expressing Cre-ERT2 in basal keratinocytes of the epidermis. Tamoxifen administration to such mice induced efficient PS1 ablation selectively in epidermal keratinocytes, thus generating mice lacking PS1 and PS2 in epidermis (Supplementary Figure S1). Mutant mice were viable, fertile and they did not exhibit phenotypic abnormalities until 2.5–3 months of age. At this age, ePS cDKO mice showed multiple skin lesions including epidermal hyperplasia and epidermal layer disorganization (Figure 2a). Interestingly, we found histological features of squamous cell carcinomas, including epidermal hyperplasia, hyperkeratosis, invasive growth pattern and mitotic cells, in the neck skin of ePS cDKO mice (Figure 2b). Immunohisto-chemical and biochemical analyses revealed upregulation of EGFR (~2-fold) coinciding with deletion of both PS in skin and primary keratinocytes from ePS cDKO mice (Figures 2a and c). As K14 promoter is specifically active in basal proliferating keratinocytes (Vasioukhin et al., 1999), PS1 was detected in some cells of the suprabasal epidermal layer in ePS cDKO mice. Sustained phosphorylation of EGFR (Tyr 1068), MEK1/2 (Ser 217/221), ERK1/2 (Thr 202/Tyr 204) and p90 RSK (Ser 380) were observed in PS-deficient fibroblasts and primary keratinocytes (Figures 2c and d). These results suggested that epidermal proliferation and transformation caused by loss of PS were associated with increased EGFR levels and signaling.
Figure 2.
PS deficiency induces epidermal hyperplasia and increased EGFR signaling in skin. (a) Paraffin-embedded sections of dorsal skin of 2.5-month-old control (PS1/PS2: PS1f/f; PS2+/–) and ePS cDKO (PS1f/f; PS2–/– ; K14-Cre-ERT2) mice were stained with hematoxylin and eosin (H&E) or processed for immunohistochemistry. H&E staining shows epidermal hyperplasia, abnormal keratinocyte proliferation and disorganization of the epidermal layer in back skin of ePS cDKO mice (× 10). Staining for PS1 (× 100) and EGFR (× 40) reveal absence of cytoplasmic and membrane PS1 expression accompanied by increased EGFR staining in epidermal keratinocytes of ePS cDKO mice. Scale bar: 30 μm. (b) H&E staining of neck skin from a 3-month-old ePS cDKO mouse showing hyperkeratosis (arrows), disorganized keratinocyte hyperplasia with infiltrating squamous epithelial cells and mitotic cells (arrowheads) consistent with histological features of squamous cell carcinomas. Scale bar: 30 μm. (c) Immunoblots showing reduced PS1 and PS2 levels and increased EGFR and phosphorylated ERK1/2 (Thr202/Tyr204) in 4-hydroxy-tamoxifen (OHT; 200 nm)-treated differentiated primary keratinocytes from ePS cDKO (PS1f/f; PS2–/– K14-Cre-ERT2) mice compared with vehicle or 4-hidroxy-tamoxifen-treated controls (PS1f/f; PS2+/–). (d) Western blotting showing increased EGFR levels and sustained EGF-induced phosphorylation of EGFR, MEK, ERK1/2 and p90RSK in PS–/– fibroblasts. The images are representative of at least three experiments.
Presenilins regulate ubiquitination-dependent EGFR degradation
We next studied the mechanisms involved in PS/γ-secretase-dependent regulation of EGFR signaling by focusing on EGFR transcription and degradation. Inhibition of γ-secretase with N-[N-(3,5-difluorophenacetyl-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) resulted in a time-dependent accumulation of β-amyloid precursor protein C-terminal fragments similar to PS–/– fibroblasts, whereas EGFR was essentially unchanged (Figure 3a). Analysis of EGFR mRNA levels by quantitative real-time reverse transcription–PCR showed unchanged EGFR transcripts in PS–/– cells (PS1/PS2: 1±0.2-fold vs PS–/–: 1±0.1-fold; n = 4–5), whereas activation of EGFR promoter was similar in control (9.3±0.5 U) and PS–/–(7.2±0.7 U) fibroblasts (P>0.05). These results suggest that regulation of EGFR expression is largely independent of γ-secretase-dependent transcription. Experiments performed with the protein synthesis inhibitor cycloheximide showed that the half-life of EGFR was significantly increased in the absence (control: ~16 h vs PS–/–: ~48 h) or presence (control: ~1 h vs PS–/– ~3 h) of ligand in PS–/– fibroblasts (Figures 3b and c) (Stoscheck and Carpenter, 1984). Consistent with a role of the lysosomal pathway on EGFR degradation, NH4Cl inhibited the degradation of EGFR in unstimulated or stimulated control and PS–/– fibroblasts, whereas the proteasome inhibitor lactacystin blocked efficiently EGFR degradation only in control fibroblasts (Figure 3c; data not shown). Under these conditions lactacystin inhibited the proteasome-dependent degradation of β-catenin (data not shown). These results indicate that efficient degradation of EGFR depends on proteasome function (Longva et al., 2002) and EGFR degradation was independent of the proteasome in the absence of PS.
Figure 3.
Presenilins regulate ubiquitination and proteasome-dependent EGFR degradation. (a) Control and PS–/– fibroblasts were cultured in the absence or presence of the γ-secretase inhibitor N-[N-(3,5-difluorophenacetyl-l-alanyl]-S-phenylglycine t-butylester (DAPT) for 0–24 h. Western blots of cell lysates revealed unchanged EGFR but higher accumulation of β-amyloid precursor protein C-terminal fragments (APPCTFs), a PS/γ-secretase substrate. β-Actin was used as a loading control. (b) EGFR turnover was analyzed in cells cultured in the presence of cycloheximide (CHX) for 0–48 h. Exposure of blots was adjusted to reflect similar levels of EGFR in both cell types at time 0. The percentage of EGFR normalized to β-actin levels of three experiments was quantified in control (white symbols) and PS–/–(black symbols) fibroblasts (bottom). (c) Degradation of EGFR was assessed in the presence of EGF plus vehicle, NH4Cl or lactacystin for the indicated times. NH4Cl inhibited EGFR degradation in control and PS–/– fibroblasts, whereas lactacystin blocked EGFR degradation only in control fibroblasts. (d) Lysates from control and PS–/– fibroblasts were immunoprecipitated with α-ubiquitin antibody and/or analyzed directly by immunoblotting as indicated. The ubiquitinated EGFR/EGFR ratio was significantly increased in PS–/– fibroblasts. Data are presented as mean±s.d. of four experiments. *P<0.05. (e) PS1/ PS2 and PS–/– fibroblasts were cultured in the absence (c) or presence of EGF on ice for 15 min and then chased at 37 °C for 0–10 min. Lysates were immunoprecipitated with α-ubiquitin antibody (top) and/or analyzed directly by western blotting with EGFR or p-EGFR antibodies (bottom).
EGFR ubiquitination is not required for internalization (Huang et al., 2007) but ubiquitination and proteasome-dependent deubiquitination are required for efficient lysosomal sorting and degradation of EGFR (Longva et al., 2002; Alwan et al., 2003; Huang et al., 2006). We then tested the hypothesis that PS could regulate EGFR stability by affecting its ubiquitination and/or internalization. In control fibroblasts, low levels of ubiquitinated EGFR were detected in the absence of ligand, whereas EGF treatment increased ubiquitinated EGFR levels (Figures 3d and e). Interestingly, ubiquitination of EGFR in PS–/– fibroblasts was significantly increased (~2-fold) in the absence of ligand (Figure 3d), whereas EGF induced deubiquitination of multiubiquitinated EGFR species (Mosesson et al., 2003) (Figure 3e). Indeed, sustained ubiquitination of EGFR, but not multiubiquitination, coincided with increased phosphorylation and stability of EGFR (Figure 3e). The EGFR is cycled through stages of ubiquitination and internalization into early endosomes, and either recycled back to the cell surface or directed to lysosomes for degradation (Marmor and Yarden, 2004). Time course analysis of cell surface EGFR by biotinylation or EGFAlexa488 labeling revealed unchanged EGFR internalization rate during 15–60 min of ligand treatment in the absence of PS, whereas impaired EGFR recycling and higher accumulation of intracellular EGFR were evident in PS–/–mentary fibroblasts at 60–180 min (Supple-Figure S2). Thus, loss of PS stabilizes EGFR by affecting a proteasomal-dependent ubiquitination/ deubiquitination step essential for efficient EGFR trafficking and degradation.
Presenilins negatively regulate the expression and activity of Fbw7
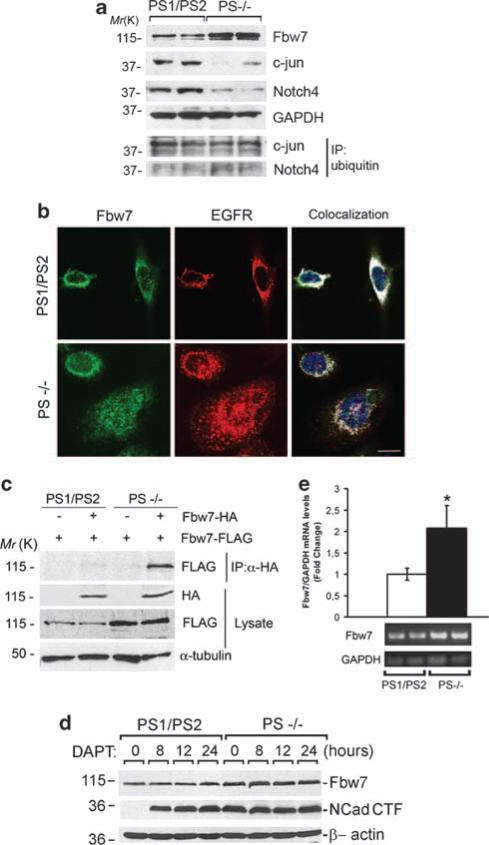
The C. elegans PS homolog sel-12 was previously shown to interact with the ubiquitin ligase sel-10/Cdc4/Fbw7 (Wu et al., 1998), although the biological function of this interaction remains unknown. We reasoned that PS could regulate EGFR ubiquitination and stability by affecting Fbw7 levels and/or activity. Consistent with this idea, we found a marked increase of Fbw7 coinciding with reduced levels and increased ubiquitination/total protein ratio of its targets c-jun and C-terminal Notch4 in PS–/– fibroblasts (Figure 4a) (Mao et al., 2004). Fbw7 staining was observed in perinuclear and nuclear compartments of control fibroblasts. Colocalization imaging analyses revealed that EGFR highly colocalizes with Fbw7 in perinuclear compartments of control fibroblasts (76±13% of Fbw7/EGFR colocalization, n = 40–50 cells). By contrast, we found an increase in Fbw7, especially in the nucleus of PS–/– fibroblasts, that highly colocalized with cytosolic and nuclear endogenous EGFR (Fbw7/EGFR colocalization: cytosol, 42±5%, nucleus, 52±24%). As Fbw7 activity depends on its dimerization (Welcker and Clurman, 2007), we examined the formation of Fbw7 dimers in the presence or absence of PS. Coexpression of Fbw7-Flag and Fbw7-HA plasmids resulted in higher steady-state levels of Fbw7 monomers and dimers in PS–/– fibroblasts (Figure 4c). Inhibition of γ-secretase with N-[N-(3,5-difluorophenacetyl-l-alanyl]-S-phenylglycine t-butyl ester did not affect Fbw7 levels, although N-cadherin C-terminal fragments accumulated in a time-dependent manner (Figure 4d). Consistent with PS-dependent regulation of Fbw7 transcription, we found a significant increase in Fbw7 mRNA transcripts in PS–/– fibroblasts (PS1/PS2: 1.0±0.1 vs PS–/–: 2.1±0.4-fold; P<0.05) (Figure 4e). These results showed that PS negatively modulates Fbw7 expression and activity by regulating its transcription.
Figure 4.
Presenilins negatively regulate the expression of Fbw7 independently of γ-secretase activity. (a) Immunoblot analysis showing increased expression of endogenous Fbw7 and decreased expression of its targets c-jun and C-terminal Notch-4 in PS–/– fibroblasts. Immunoblots at the bottom show immunoprecipitation (IP) of ubiquitinated c-jun and C-terminal Notch-4. (b) Confocal microscopy analysis showing increased expression of endogenous Fbw7 (green) and EGFR (red) especially in the nucleus (Hoescht, blue) and/or cytosol of PS–/– fibroblasts, respectively. Colocalization of Fbw7 and EGFR is shown in white. Fbw7 staining was absent in the presence of the blocking peptide (1 μg/ml; not shown). Scale bar: 10 μm. (c) Coimmunoprecipitation assays were performed in fibroblasts co-transfected with Flag-Fbw7 and HA-Fbw7 or empty vector. Total Fbw7 monomers and Flag-Fbw7/HA-Fbw7 complexes are increased in the absence of PS. (d) Inhibition of γ-secretase with N-[N-(3,5-difluorophenacetyl-l-alanyl]-S-phenylglycine t-butyl ester (DAPT) for 0–24 h resulted in a time-dependent accumulation of N-cadherin C-terminal fragments but unchanged Fbw7 levels in control fibroblasts. (e) Levels of Fbw7 mRNA analyzed by semiquantitative reverse transcription–PCR in control and PS–/– fibroblasts (n = 7). Values were normalized to GAPDH levels. Data represent mean±s.e.m. *P<0.05
Fbw7 positively regulates EGFR stability and signaling
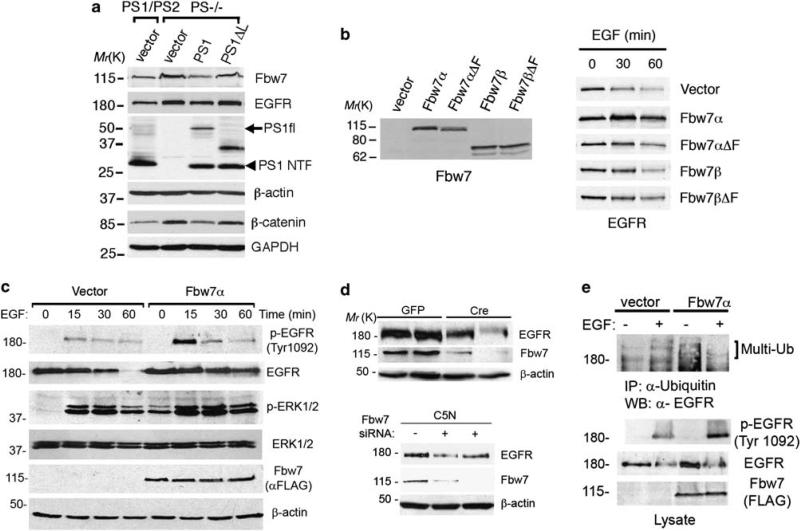
The previous results indicated that loss of PS positively regulates Fbw7 expression, which in turn could stabilize EGFR. Indeed, elevated Fbw7 was associated with higher EGFR levels in PS–/– fibroblasts, whereas stable expression of PS1, but not a PS1Δloop mutant that lacks the β-catenin binding domain (Saura et al., 2000), significantly decreased Fbw7 and EGFR levels (Figure 5a). This is an interesting finding because the PS1Δloop mutant maintains high cytosolic β-catenin levels (β-catenin/GAPDH: PS1/PS2: 1±0.1, PS–/–: 2±0.1, PS–/–;PS1: 1.3±0.1, PS–/–;PS1Δloop: 1.8±0.2; n = 4) (Figure 5a). Isoform-specific degradation of substrates by Fbw7 has been recently reported (Grim et al., 2008). Consistent with isoform- and E3 ubiquitin ligase-dependent EGFR regulation by Fbw7, Fbw7α but not Fbw7β or the loss of function mutants lacking the F-box domain (Fbw7α/β ΔF) (Welcker et al., 2004), stabilized EGFR levels and signaling in 293T cells (Figures 5b and c). In support of a role of Fbw7 in EGF stabilization, genetic inactivation of Fbw7 by using Fbw7 small interfering RNA or Cre recombinase-mediated recombination (Mao et al., 2004) significantly decreased the levels of EGFR in epithelial cells and embryonic fibroblasts (Figure 5d). Overexpression of Fbw7α in 293T cells enhanced EGFR ubiquitination and caused a rapid deubiquitination of the multi-ubiquitinated species similar to that found in PS–/–cells (Figure 5e). Together, these results indicate that Fbw7α acts as a positive regulator of EGFR stability and signaling.
Figure 5.
The ubiquitin ligase Fbw7 stabilizes EGFR expression and signaling. (a) Stable pools of PS1/PS2 or PS–/– fibroblasts expressing human PS1, PS1Δloop or empty vector were selected and generated as described in Materials and methods section. Expression of PS1 but not PS1Δloop in PS–/– fibroblasts reduces the levels of Fbw7 and EGFR. Abnormal levels of β-catenin in the cytosolic fraction of PS–/– fibroblasts were decreased after PS1 expression (bottom blots). (b) 293T cells were transiently transfected with vector, Fbw7α, Fbw7β or the loss of function mutants lacking the F-box domain Fbw7αΔF and Fbw7βΔF for 24 h. Expression of Fbw7 in non-treated cells (left panel; Ab12292 antibody) or EGFR (right panel) in cells treated with EGF (150 ng/ml) for 0–60 min were detected by western blotting. (c) Levels of EGFR and phosphorylated EGFR (Tyr 1092) or ERK1/2 after treatment with EGF were examined by immunoblotting in 293T cells transiently transfected with vector or Flag-Fbw7α. Fbw7α increases phosphorylation and stability of EGFR and enhances sustained ERK1/2 phosphorylation. (d) Top: Cre recombinase-mediated deletion of Fbw7 reduced EGFR levels in two independent Fbw7 f/f fibroblast lines. Bottom: Epithelial C5N cell lines stably expressing pSuper Fbw7 small interfering RNAs (+) showed reduced expression of EGFR compared with pSuper (–) transfected cells. (e) 293T cells were transiently transfected with vector or Flag-Fbw7α and incubated in the absence (–) or presence (+)of EGF on ice for 15 min. Immunoprecipitation/western blotting revealed increased phosphorylation and ubiquitination of EGFR in cells expressing Fbw7.
Inactivation of PS enhances EGFR-dependent signaling and cell transformation by upregulating Fbw7
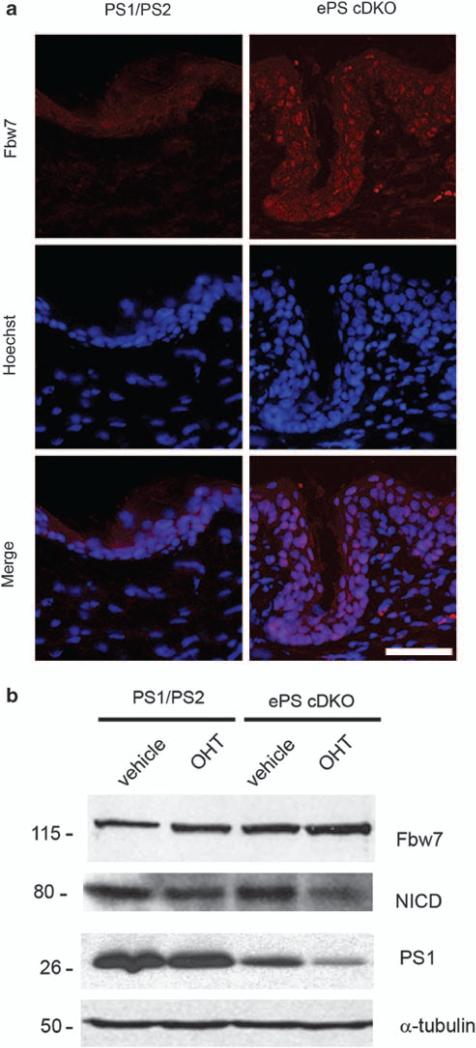
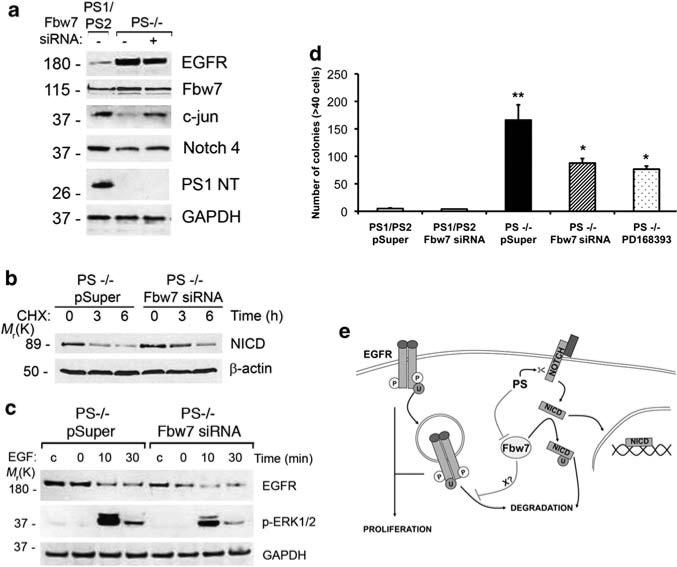
To assess whether the above findings apply in conditions of epidermal transformation, we analyzed Fbw7 expression in the epidermis of ePS cDKO mice. Immunohistochemical analyses of skin sections revealed a substantial increase in Fbw7, especially in the nucleus of epidermal cells in ePS cDKO mice (Figure 6a). We detected upregulation of Fbw7 associated with enhanced EGFR and decreased NICD levels in tamoxifen-treated primary keratinocytes of ePS cDKO mice (Figures 2c and 6b). To test the possibility that loss of PS could enhance epidermal proliferation by increasing EGFR signaling through Fbw7, we inactivated Fbw7 by using Fbw7 small interfering RNAs (Mao et al., 2004). Compared with PS–/– fibroblasts (1.5±0.2), stable expression of Fbw7 small interfering RNA reduced the level of Fbw7 mRNAs by 50% (1.10±0.04), which were similar to vector-transfected control fibroblasts (1.00±0.05). Consistent with a role of PS in regulating EGFR stability through Fbw7, Fbw7 silencing in PS–/– fibroblasts diminished EGFR while increased c-jun and C-terminal Notch4 and decreased the turnover rate of NICD (Figures 7a and b). Moreover, Fbw7 inactivation decreased significantly EGFR expression and stability as well as ERK1/2 signaling induced by EGF in PS–/– fibroblasts (Figure 7c). Finally, the number of PS–/–colonies formed in soft-agar was significantly reduced by silencing Fbw7, which was similar to those obtained by inhibiting EGFR (Figure 7d). These results support the idea that elevated Fbw7 caused by PS inactivation results in EGFR-mediated epidermal transformation.
Figure 6.
Elevated Fbw7 levels in keratinocytes of epidermal PS cDKO mice. (a) Analysis of endogenous Fbw7 in skin of ePS cDKO mice. Immunofluorescence images showing increased Fbw7 staining in cytosol and nucleus in epidermal cells of ePS cDKO mouse (PS1f/f;PS2–/– ; K14-Cre-ERT2) compared with control PS1/PS2 mouse (PS1f/f; PS2+/– ). Images: ×20. Scale bar: 30μm. (b) Primary keratinocytes isolated from 2-day-old littermate control (PS1f/f; PS2 +/–) and ePS cDKO (PS1f/f;PS2–/– ; K14-Cre-ERT2) pups were differentiated and treated with ethanol (vehicle) or 4-hidroxy-tamoxifen (OHT) for 72 h. Immunoblot analysis with anti-Fbw7, cleaved Notch1 and PS1NT antibodies showed an increase of Fbw7 coinciding with reduced PS1 and Notch1 intracelular domain (NICD) in tamoxifen-treated ePS cDKO keratinocytes.
Figure 7.
Presenilins regulate EGFR expression, signaling and transformation through Fbw7. (a) Control and PS–/– fibroblasts stably transfected with empty (–) or Fbw7 small interfering RNAs (siRNAs) (+) pSuper vectors were analyzed by immunoblotting to detect endogenous Fbw7, EGFR, c-jun, Notch-4 and PS1 (NTF). Fbw7 siRNA reduced the levels of Fbw7 targets as well as EGFR. (b) PS–/– fibroblasts stably expressing control or Fbw7 siRNAs were transiently transfected with NICD-Myc cDNA by using LipofectAMINE 2000 and treated with CHX for the indicated times. NICD stability was analyzed by western blotting with 9E10 antibody. (c) PS–/– fibroblasts stably expressing pSuper or pSuper-Fbw7 siRNA were cultured in the absence (c) or presence of EGF for 0–30 min. Inactivation of Fbw7 reduced EGFR and pERK1/2 levels. The images are representative of three independent experiments. (d) PS1/PS2 and PS–/– fibroblasts stably expressing pSuper or pSuper-Fbw7 siRNA (n = 3) were cultured in soft-agar and colonies were quantified at day 15. Mean values±s.e.m. are shown. *P<0.05, compared with PS–/– pSuper cells; **P<0.001, compared with PS1/PS2 pSuper cells. (e) Model depicting the role of PS on mediating the dual regulation of EGFR and NICD by the ubiquitin ligase Fbw7α. According to this model, PS mediate NICD generation and degradation through γ-secretase and Fbw7, respectively. By contrast, PS negatively modulate EGFR ubiquitination and stability by affecting Fbw7 directly or indirectly through an unknown effector (X).
Discussion
This study provides biochemical evidence that PS inversely modulate the EGFR and Notch pathways by regulating the ubiquitin ligase Fbw7. PS negatively regulates Fbw7, which in turn acts as positive and negative regulator of EGFR and Notch signaling, respectively (Figure 7e) (Gupta-Rossi et al., 2001; Wu et al., 2001). Functional genetic interactions between PS (sel-12) and Fbw7 (sel-10/Cdc4) were initially reported in C. elegans, in which loss of sel-10 activity leads to suppression of the egg-laying defective phenotype caused by sel-12 mutants (Wu et al., 1998). In mammalian cells, Fbw7 regulates PS levels and activity (Li et al., 2002), but a reciprocal effect has not been investigated. Our results show that PS act as negative regulators of Fbw7. Loss of PS function in fibroblasts and primary keratinocytes results in elevated expression of Fbw7 and decreased Fbw7 targets, including c-jun and Notch1/4. PS-dependent regulation of Fbw7 and EGFR is independent of its proteolytic activity as inhibition of γ-secretase does not affect expression of Fbw7 and EGFR. This result agrees with a recent report showing γ-secretase-independent regulation of EGFR degradation (Repetto et al., 2007). Notably, we found that loss of PS enhances Fbw7 mRNA transcripts suggesting that PS negatively modulates Fbw7 transcription. As the PS1Δloop mutant that stabilizes cytosolic β-catenin, leading to enhanced β-catenin/LEF signaling (Soriano et al., 2001), was unable to reduce Fbw7α, it is possible that β-catenin signaling may be involved in Fbw7 upregulation. To our knowledge, this is the first study showing a molecular mechanism regulating Fbw7 expression at the transcriptional level.
We show for the first time a possible crosstalk between Fbw7 and EGFR during cell growth and transformation. Several observations show that Fbw7 positively modulates EGFR stability and signaling, and that this effect is isoform- and E3 ligase activity-dependent. First, Fbw7α increased EGFR stability and signaling, whereas Fbw7β, the loss of function mutants lacking the F-box domain or the dominant mutant dnFbw7WD did not affect or reduced EGFR stability (Figure 5 and not shown). Second, EGFR was downregulated by genetic inactivation of Fbw7 in embryonic fibroblasts and epithelial cells or after expression of PS1, which decreased Fbw7, in PS–/– fibroblasts (Figures 5 and 7). Finally, Fbw7 silencing reduced abnormal EGFR-dependent anchorage-independent growth of PS-deficient fibroblasts. These findings define a new role for Fbw7 on facilitating EGFR stability and signaling in epidermal cells, but they do not clarify whether Fbw7 affects EGFR degradation directly or indirectly through an unknown effector (Figure 7e). These results contrast with the established function of Fbw7 as a tumor suppressor gene in several malignancies including epithelial tumors (Perez-Losada et al., 2005; Welcker and Clurman, 2008). Alternatively, our results indicate a crosstalk between PS and EGFR through Fbw7 on malignant transformation and skin carcinogenesis.
It is well-established that EGFR signaling promotes keratinocyte growth and transformation (Kalyankrishna and Grandis, 2006). The analysis of ePS cDKO mice allowed us to show a reciprocal regulation between EGFR and Fbw7 during progression of neoplastic epidermal hyperplasia. The enhancement of EGFR-mediated cell proliferation and transformation caused by loss of PS strongly argues for a role of abnormal EGFR signaling in PS-dependent epidermal hyperplasia. Although our results agree with recent reports showing elevated EGFR levels in skin tumors of PS1-and nicastrin-deficient mice (Li et al., 2007; Repetto et al., 2007; Zhang et al., 2007), they also provide a novel mechanism of EGFR regulation. Loss of PS affected EGFR recycling and degradation, whereas EGF-induced EGFR internalization was unaffected. Efficient lysosomal sorting and degradation of EGFR, but not internalization, require ubiquitination and proteasome-dependent deubiquitination (Longva et al., 2002; Alwan et al., 2003), although sustained multi-ubiquitination regulates receptor trafficking and degradation (Mosesson et al., 2003; Huang et al., 2006). The fact that loss of PS enhances basal ubiquitination and accelerates multi-deubiquitination of EGFR, which alters its proteasome-dependent downregulation, argues for a role of PS in regulating an ubiquitination/ deubiquitination step essential for trafficking of EGFR during the late endosomal/lysosomal pathway (Repetto et al., 2007) (Supplementary Figure S2). It is possible that non-proteolytic functions of ubiquitin, as those related with cell signaling or trafficking (Chen and Sun, 2009), may also contribute to altered EGFR trafficking and degradation.
Our results also raise the possibility that abnormal EGFR signaling in concert with stabilization of bcatenin levels (Xia et al., 2001; Tournoy et al., 2004) may contribute to tumor formation in PS-deficient mice. Deletion of the β-catenin binding domain in PS1 prevents skin tumors (Deng et al., 2006), whereas inactivation of Notch1 signaling causes skin tumors (Nicolas et al., 2003; Proweller et al., 2006), suggesting that altered EGFR and Notch signaling may participate in the oncogenic phenotype caused by PS deficiency. Indeed, an antagonistic crosstalk between the let-23/ EGFR and lin-12/Notch signaling pathways occurs during vulval precursor cell patterning in C. elegans (Shaye and Greenwald, 2002). Notably, LRBA, a sel-2 homolog that negatively regulates lin-12/Notch, promotes growth of cancer cells by regulating EGFR trafficking and signaling (Wang et al., 2004; de Souza et al., 2007). In proliferating keratinocytes and SCC, EGFR signaling was recently shown to suppress Notch signaling by negatively regulating Notch1 expression through p53 (Kolev et al., 2008). In agreement with a PS-dependent crosstalk between EGFR and Notch1 during epidermal carcinogenesis, reduced PS1 levels were recently associated with deficient Notch1 cleavage and elevated EGFR levels in skin papillomas of activating transcription factor-2 conditional knockout mice (Bhoumik et al., 2008). This scenario contrasts with the stabilization of NICD caused by Fbw7 mutations in leukemia (O'Neil et al., 2007). Besides regulating positively Notch signaling by generating the γ-secretase-derived NICD (De Strooper et al., 1999), our study shows that PS delays NICD degradation by controlling Fbw7 levels. Consistently, we found reduced NICD together with enhanced Fbw7 levels in keratinocytes from ePS cDKO mice. Nonetheless, Notch1 activation in epidermal basal keratinocytes triggers premature differentiation and reduced proliferation (Blanpain et al., 2006), whereas loss of Notch function causes deficient epidermal differentiation and tumor-igenesis (Nicolas et al., 2003; Proweller et al., 2006). Future studies will be necessary to discern the specific mechanisms regulating the crosstalk of β-catenin, Notch and EGFR signaling during epidermal transformation. In light of these findings, we propose a role of PS in regulating the crosstalk between EGFR and Notch signaling through Fbw7 during skin carcinogenesis.
Materials and methods
Plasmids and antibodies
The Flag-Fbw7α, Flag-Fbw7β, Flag-Fbw7α/βΔF, Flag-dnFbw7WD, HA-Fbw7α and pCS2 mNICD-Myc plasmids were previously described (Kopan et al., 1996; Welcker et al., 2004). The pER1-Luc plasmid contains the EGFR promoter cloned into the HindIII site of pGL3-Basic (Promega, Madison, WI, USA) (Rikiyama et al., 2003). Human PS1 and PS1Δloop were cloned into KpnI/BamHI sites of the pAG3Zeo vector. Antibodies against EGFR (1005), C-terminal Notch-4 (H-225), c-jun (H-79), c-myc (9E10), HA (F-7) and ubiquitin (P4D1) were from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against phosphorylated EGFR (Tyr 1068 and Tyr 1092), MEK1/2 (Ser 217/221), ERK1/2 (Thr 202/Tyr 204) and p90RSK (Ser 380) and cleaved-Notch1 (Val1744) were from Cell Signaling (Beverly, MA, USA). We used rabbit anti-Fbw7 (Ab12292; Abcam, Cambridge, UK), PS1NT (Calbiochem, San Diego, CA, USA), PS2 (Oncogene, Cambridge, MA, USA) and mouse N-cadherin (BD Biosciences, Franklin Lakes, NJ, USA) antibodies. Antibodies against β-catenin, GAPDH, Flag, β-tubulin (SAP.4G5) and β-actin (AC-15) were from Sigma- Aldrich (St Louis, MO, USA).
Generation of epidermal-specific PS cDKO mice
We generated PS2-null mice in which PS1 was conditionally ablated in epidermal keratinocytes by crossing homozygous floxed PS1 (f/f); PS2–/– mice (Saura et al., 2004) to K14-Cre-ERT2 transgenic mice (Li et al., 2000). PS1 f/+ ; PS2+/– K14-Cre-ERT2 mice were backcrossed to PS1 f/f; PS2–/– mice to obtain PS1 f/f; PS2 +/ (control) and PS1 f/f; PS2 –/– K14-Cre-ERT2 mice. The PS1 f/f and PS2–/– mice were generated in C57BL6/129 hybrid background, whereas K14-Cre-ERT2 mice were of a C57BL/6 background. The Cre-ERT2 transgene was detected in genomic DNA tails and dorsal skins by PCR using the following primers P156, 5′-GCCTGCATTACCGGTCGATGCAACGA-3′ and P157, 5′-GTGGCAGATGGCGC GGC AACACCATT-3′. Primary keratinocytes from 2- to 3-day-old littermate PS1 f/f; PS2 + / (control) and PS1 f/f; PS2–/– ; K14-Cre-ERT2 pups were cultured and differentiated for 1-week as previously described (Pirrone et al., 2005). For efficient recombination, 4-hidroxy-tamoxifen (Sigma-Aldrich) dissolved in acetone or ethanol was applied to skin (2 mg) or cultured keratinocytes (200 nm) for 72 h. All animal procedures were conducted in accordance with European Union guidelines on animal welfare and approved by the Institutional Ethical Committee.
Cell lines and proliferation assays
Mouse embryonic fibroblasts derived from control, PS–/–or Fbw7 f/f mouse embryos and the epithelial C5N cell lines have been previously described (De Strooper et al., 1999; Mao et al., 2004). For Fbw7 silencing, a Fbw7 small interfering RNA targeting coding region 1965–1983 of exon 10 was cloned into pSuper retrovirus vector (OligoEngine, Seattle, WA, USA) (Mao et al., 2004). After retroviral infection, cells were selected with puromycin (3 μg/ml) for 7 days. To generate stable cell pools, PS1/PS2 and PS–/–MEFs were transfected with pAG3Zeo, pAG3Zeo-PS1 or pAG3Zeo-PS1Δloop by using LipofectAMINE 2000 (Invitrogen, Carlsbad, CA, USA) and selected with zeocin (0.4 mg/ml) for 14 days (Saura et al., 2000). For proliferation assays, cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 0.1–10% fetal bovine serum and treated with vehicle or the indicated inhibitors. Anchorage-independent cell growth assay was performed by culturing 5000 cells resuspended in DMEM/0.35% agar/10% fetal bovine serum containing vehicle or PD168393 (2 μm; 4 h) onto a layer of 0.5% agar for 15 days (Di Fiore et al., 1987). Experiments were performed in triplicate.
Biochemical analyses
For signaling assays, cells were serum-starved for 3 h and incubated with serum-deprived DMEM containing EGF (50– 150 ng/ml) (R&D Systems, Minneapolis, MN, USA) at 37 °C for the indicated times. Cells were lysed with RIPA-DOC buffer (50 mm Tris–HCl, pH 7.4, 150 mm NaCl, 2,5 mm EDTA, 1% NP40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm Na3VO4, 50 μm NaF, 1 mm phenylmethylsulfonyl fluoride) supplemented with protease and phosphatase inhibitors (Sigma-Aldrich). EGFR turnover was analyzed in MEF in the presence or absence of EGF (150 ng/ml) and/or cycloheximide (25 μg/ml) plus NH4Cl (10 mm) or lactacystin (50 μm). EGFR stability was assayed in 293T cells transiently transfected with the indicated Fbw7 plasmids. Ubiquitination assays were performed in serum-starved cells incubated with vehicle or EGF (150 ng/ml) in DMEM for 15 min at 4 °C. Cells were washed, chased in DMEM at 37 °C for 0–30 min and lysed in RIPA-DOC buffer. Protein lysates (1 mg) were precipitated with P4D1 antibody or immunoglobulin-G plus protein A-sepharose. Immunoblotting was performed on 7.5– 12.5% SDS–polyacrylamide gel electrophoresis as previously described (Saura et al., 2000).
Immunohistological and immunofluorescence analyses
Deparaffinized mouse dorsal skins sections (5–10 mm) were stained with hematoxylin and eosin or peroxide inactivated and/or pretreated with antigen retrieval citrate (Biogenex, San Ramon, CA, USA). Sections were stained with antibodies against PS1NT (1:100) or EGFR (1:250) and processed for avidin-biotin immunoperoxidase by using the Vectastain Elite ABC kit (Vector Laboratories, Burlingame, CA, USA). For immunofluorescence staining, sections were pretreated with antigen retrieval citrate, stained with anti-Fbw7 antibody (1:250), anti-rabbit Alexa Fluor 594 antibody (1:300; Invitrogen) and Hoechst (1:10 000) as previously described (Saura et al., 2004). For Fbw7 and EGFR immunostaining, cells were permeabilized (0.1% saponin in phosphate-buffered saline), blocked and incubated with rabbit anti-Fbw7 (1:100) and mouse aEGFR (1:200; clone 225) plus anti-mouse biotinylated (1:100) antibodies before incubation with streptavidin-Cy3 (1:400) and anti-rabbit Alexa Fluor488 (1:400) and Hoechst. EGFR internalization was analyzed by incubating cells with EGF-Alexa488 (150 ng/ml; Invitrogen) at 4 °C for 15 min to avoid internalization (time 0) and chased for 15–180 min at 37 °C. Images were obtained using a Leica TCS SP5 AOBS confocal microscope and analyzed with MetaMorph 6.1 software (Universal Imaging, West Chester, PA, USA). For colocalization analysis, the number of double pixels for EGFR/Fbw7 relative to total EGFR pixels was counted for each cell field (7–8 cells per field) by using ImageJ 1.43k.
Quantitative real-time reverse transcription–PCR assays
Total RNA was isolated using the RNAeasy mini kit (Qiagen, Valencia, CA, USA) and reverse transcribed using the Super-Script II Reverse Transcriptase kit (Invitrogen). Quantitative reverse transcription–PCR was performed with specific primers and the QuantiMix EASY SYG kit (Biotools, Madrid, Spain) in an ABI PRISM 7900 Sequence Detector (Applied Biosystems, Foster City, CA, USA). Values were normalized to glyceraldehyde-3-phosphate dehydrogenase. Data analysis was performed by the ΔΔCt method using the SDS software v.2.1.
Statistical analysis
Statistical analysis was performed using the two-tailed Student's t-test or one-way analysis of variance (Super-ANOVA program). Comparisons among multiple groups were examined by Scheffé's S post hoc test. Data represent the mean±s.d. or s.e.m. Differences with P<0.05 were considered to be significant.
Supplementary Material
Acknowledgements
We thank B De Strooper for providing the immortalized PS fibroblasts, J Shen for PS1 (f/f); PS2–/– mice, G Thinakaran for the pAG3zeo-PS1Δloop plasmid, R Kopan for the mNICD-myc plasmid, BE Clurman for the Fbw7α/β-Flag cDNAs and A Johnson for the pER1-luc plasmid. We thank S Aznar-Benitah for histological expertise advice and E Martín and M Castillo for technical assistance. We thank Servei de Microscopia and Servei de Genoómica de la UAB for the excellent technical support. This study was supported by grants from Fundacioó Maratoó-TV3 (050710), Spanish Ministerio de Ciencia e Innovación (SAF2007-64115, CIBERNED CB06/05/0042 and Programa Ramón y Cajal) and 6th Framework Programme of the European Union (Marie Curie International Reintegration Grant IRG-014860). VRR received a doctoral fellowship from Generalitat de Catalunya.
Abbreviations
- PS
presenilins
- NICD
Notch1 intracellular domain
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc)
References
- Alwan HA, van Zoelen EJ, van Leeuwen JE. Ligand-induced lysosomal epidermal growth factor receptor (EGFR) degradation is preceded by proteasome-dependent EGFR de-ubiquitination. J Biol Chem. 2003;278:35781–35790. doi: 10.1074/jbc.M301326200. [DOI] [PubMed] [Google Scholar]
- Bhoumik A, Fichtman B, DeRossi C, Breitwieser W, Kluger HM, Davis S, et al. Suppressor role of activating transcription factor 2 (ATF2) in skin cancer. Proc Natl Acad Sci USA. 2008;105:1674–1679. doi: 10.1073/pnas.0706057105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry W, Pasolli H, Fuchs E. Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 2006;20:3022–3035. doi: 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Sun LJ. Nonproteolytic functions of ubiquitin in cell signaling. Mol Cell. 2009;33:275–286. doi: 10.1016/j.molcel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- de Souza N, Vallier LG, Fares H, Greenwald I. SEL-2, the C. elegans neurobeachin/LRBA homolog, is a negative regulator of lin-12/Notch activity and affects endosomal traffic in polarized epithelial cells. Development. 2007;134:691–702. doi: 10.1242/dev.02767. [DOI] [PubMed] [Google Scholar]
- De Strooper B. Loss-of-function presenilin mutations in Alzheimer disease. EMBO Rep. 2007;8:141–146. doi: 10.1038/sj.embor.7400897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, et al. A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- Demehri S, Liu Z, Lee J, Lin MH, Crosby SD, Roberts CJ, et al. Notch-deficient skin induces a lethal systemic B-lymphoproliferative disorder by secreting TSLP, a sentinel for epidermal integrity. PLoS Biol. 2008;6:e123. doi: 10.1371/journal.pbio.0060123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Tarassishin L, Kallhoff V, Peethumnongsin E, Wu L, Li YM, et al. Deletion of presenilin 1 hydrophilic loop sequence leads to impaired γ-secretase activity and exacerbated amyloid pathology. J Neurosci. 2006;26:3845–3854. doi: 10.1523/JNEUROSCI.5384-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Fiore PP, Pierce JH, Fleming TP, Hazan R, Ullrich A, King CR, et al. Overexpression of the human EGF receptor confers an EGF-dependent transformed phenotype to NIH 3T3 cells. Cell. 1987;51:1063–1070. doi: 10.1016/0092-8674(87)90592-7. [DOI] [PubMed] [Google Scholar]
- Grim J, Gustafson M, Hirata R, Hagar A, Swanger J, Welcker M, et al. Isoform- and cell cycle-dependent substrate degradation by the Fbw7 ubiquitin ligase. J Cell Biol. 2008;181:913–920. doi: 10.1083/jcb.200802076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta-Rossi N, Le Bail O, Gonen H, Brou C, Logeat F, Six E, et al. Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J Biol Chem. 2001;276:34371–34378. doi: 10.1074/jbc.M101343200. [DOI] [PubMed] [Google Scholar]
- Huang F, Goh LK, Sorkin A. EGF receptor ubiquitination is not necessary for its internalization. Proc Natl Acad Sci USA. 2007;104:16904–16909. doi: 10.1073/pnas.0707416104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Kirkpatrick D, Jiang X, Gygi S, Sorkin A. Differential regulation of EGF receptor internalization and degradation by multiubiquitination within the kinase domain. Mol Cell. 2006;21:737–748. doi: 10.1016/j.molcel.2006.02.018. [DOI] [PubMed] [Google Scholar]
- Indra A, Warot X, Brocard J, Bornert JM, Xiao J-H, Chambon P, et al. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ERT and Cre-ERT2 recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost M, Kari C, Rodeck U. The EGF receptor—an essential regulator of multiple epidermal functions. Eur J Dermatol. 2000;10:505–510. [PubMed] [Google Scholar]
- Kalyankrishna S, Grandis JR. Epidermal growth factor receptor biology in head and neck cancer. J Clin Oncol. 2006;24:2666–2672. doi: 10.1200/JCO.2005.04.8306. [DOI] [PubMed] [Google Scholar]
- Kang DE, Soriano S, Xia X, Eberhart CG, De Strooper B, Zheng H, et al. Presenilin couples the paired phosphorylation of β-catenin independent of axin: implications for β-catenin activation in tumorigenesis. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- Kolev V, Mandinova A, Guinea-Viniegra J, Hu B, Lefort K, Lambertini C, et al. EGFR signalling as a negative regulator of Notch1 gene transcription and function in proliferating keratinocytes and cancer. Nat Cell Biol. 2008;10:902–911. doi: 10.1038/ncb1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo EH, Kopan R. Potential role of presenilin-regulated signaling pathways in sporadic neurodegeneration. Nat Med. 2004;10(Suppl):S26–S33. doi: 10.1038/nm1065. [DOI] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort K, Dotto GP. Notch signaling in the integrated control of keratinocyte growth/differentiation and tumor suppression. Semin Cancer Biol. 2004;14:374–386. doi: 10.1016/j.semcancer.2004.04.017. [DOI] [PubMed] [Google Scholar]
- Li J, Pauley AM, Myers RL, Shuang R, Brashler JR, Yan R, et al. SEL-10 interacts with presenilin 1, facilitates its ubiquitination, and alters A-beta peptide production. J Neurochem. 2002;82:1540–1548. doi: 10.1046/j.1471-4159.2002.01105.x. [DOI] [PubMed] [Google Scholar]
- Li M, Indra A, Warot X, Brocard J, Messaddeq N, Kato S, et al. Skin abnormalities generated by temporally controlled RXRalpha mutations in mouse epidermis. Nature. 2000;407:633–636. doi: 10.1038/35036595. [DOI] [PubMed] [Google Scholar]
- Li T, Wen H, Brayton C, Das P, Smithson LA, Fauq A, et al. Epidermal growth factor receptor and notch pathways participate in the tumor suppressor function of γ-secretase. J Biol Chem. 2007;282:32264–32273. doi: 10.1074/jbc.M703649200. [DOI] [PubMed] [Google Scholar]
- Longva KE, Blystad FD, Stang E, Larsen AM, Johannessen LE, Madshus IH. Ubiquitination and proteasomal activity is required for transport of the EGF receptor to inner membranes of multivesicular bodies. J Cell Biol. 2002;156:843–854. doi: 10.1083/jcb.200106056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao JH, Perez-Losada J, Wu D, Delrosario R, Tsunematsu R, Nakayama KI, et al. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature. 2004;432:775–779. doi: 10.1038/nature03155. [DOI] [PubMed] [Google Scholar]
- Marmor MD, Yarden Y. Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene. 2004;23:2057–2070. doi: 10.1038/sj.onc.1207390. [DOI] [PubMed] [Google Scholar]
- Mosesson Y, Shtiegman K, Katz M, Zwang Y, Vereb G, Szollosi J, et al. Endocytosis of receptor tyrosine kinases is driven by monoubiquitylation, not polyubiquitylation. J Biol Chem. 2003;278:21323–21326. doi: 10.1074/jbc.C300096200. [DOI] [PubMed] [Google Scholar]
- Nicolas M, Wolfer A, Raj K, Kummer J, Mill P, van Noort M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- O'Neil J, Grim J, Strack P, Rao S, Tibbitts D, Winter C, et al. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to γ-secretase inhibitors. J Exp Med. 2007;204:1813–1824. doi: 10.1084/jem.20070876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Lin MH, Tian X, Cheng HT, Gridley T, Shen J, et al. γ-Secretase functions through Notch signaling to maintain skin appendages but is not required for their patterning or initial morphogenesis. Dev Cell. 2004;7:731–743. doi: 10.1016/j.devcel.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Perez-Losada J, Mao JH, Balmain A. Control of genomic instability and epithelial tumor development by the p53-Fbxw7/Cdc4 pathway. Cancer Res. 2005;65:6488–6492. doi: 10.1158/0008-5472.CAN-05-1294. [DOI] [PubMed] [Google Scholar]
- Pirrone A, Hager B, Fleckman P. Primary mouse keratinocyte culture. Methods Mol Biol. 2005;289:3–14. doi: 10.1385/1-59259-830-7:003. [DOI] [PubMed] [Google Scholar]
- Proweller A, Tu L, Lepore JJ, Cheng L, Lu MM, Seykora J, et al. Impaired Notch signaling promotes de novo squamous cell carcinoma formation. Cancer Res. 2006;66:7438–7444. doi: 10.1158/0008-5472.CAN-06-0793. [DOI] [PubMed] [Google Scholar]
- Repetto E, Yoon I-S, Zheng H, Zhang DE. Presenilin-1 regulates epidermal growth factor receptor turnover and signaling in the endosomal-lysosomal pathway. J Biol Chem. 2007;282:31504–31516. doi: 10.1074/jbc.M704273200. [DOI] [PubMed] [Google Scholar]
- Rikiyama T, Curtis J, Oikawa M, Zimonjic DB, Popescu N, Murphy BA, et al. GCF2: expression and molecular analysis of repression. Biochim Biophys Acta. 2003;1629:15–25. doi: 10.1016/s0167-4781(03)00156-8. [DOI] [PubMed] [Google Scholar]
- Saura CA, Choi SY, Beglopoulos V, Malkani S, Zhang D, Shankaranarayana Rao BS, et al. Loss of presenilin function causes impairments of memory and synaptic plasticity followed by age-dependent neurodegeneration. Neuron. 2004;42:23–36. doi: 10.1016/s0896-6273(04)00182-5. [DOI] [PubMed] [Google Scholar]
- Saura CA, Tomita T, Soriano S, Takahashi M, Leem JY, Honda T, et al. The nonconserved hydrophilic loop domain of presenilin (PS) is not required for PS endoproteolysis or enhanced Aβ42 production mediated by familial early onset Alzheimer's disease-linked PS variants. J Biol Chem. 2000;275:17136–17142. doi: 10.1074/jbc.M909624199. [DOI] [PubMed] [Google Scholar]
- Shaye DD, Greenwald I. Endocytosis-mediated downregulation of LIN-12/Notch upon Ras activation in Caenorhabditis elegans. Nature. 2002;420:686–690. doi: 10.1038/nature01234. [DOI] [PubMed] [Google Scholar]
- Shen J, Bronson RT, Chen DF, Xia W, Selkoe DJ, Tonegawa S. Skeletal and CNS defects in presenilin-1 deficient mice. Cell. 1997;89:629–639. doi: 10.1016/s0092-8674(00)80244-5. [DOI] [PubMed] [Google Scholar]
- Soriano S, Kang DE, Fu M, Pestell R, Chevallier N, Zheng H, et al. Presenilin 1 negatively regulates beta-catenin/T cell factor/lymphoid enhancer factor-1 signaling independently of β-amyloid precursor protein and notch processing. J Cell Biol. 2001;152:785–794. doi: 10.1083/jcb.152.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoscheck CM, Carpenter G. Down regulation of epidermal growth factor receptors: direct demonstration of receptor degradation in human fibroblasts. J Cell Biol. 1984;98:1048–1053. doi: 10.1083/jcb.98.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- To MD, Gokgoz N, Doyle TG, Donoviel DB, Knight JA, Hyslop PS, et al. Functional characterization of novel presenilin-2 variants in human breast cancers. Oncogene. 2006;25:3557–3564. doi: 10.1038/sj.onc.1209397. [DOI] [PubMed] [Google Scholar]
- Tournoy J, Bossuyt X, Snellinx A, Regent M, Garmyn M, Serneels L, et al. Partial loss of presenilins causes seborrheic keratosis and autoimmune disease in mice. Hum Mol Genet. 2004;13:1321–1331. doi: 10.1093/hmg/ddh151. [DOI] [PubMed] [Google Scholar]
- Vasioukhin V, Degenstein L, Wise B, Fuchs E. The magical touch: genome targeting in epidermal stem cells induced by tamoxifen application to mouse skin. Proc Natl Acad Sci USA. 1999;96:8551–8556. doi: 10.1073/pnas.96.15.8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Gamsby JJ, Highfill SL, Mora LB, Bloom GC, Yeatman TJ, et al. Deregulated expression of LRBA facilitates cancer cell growth. Oncogene. 2004;23:4089–4097. doi: 10.1038/sj.onc.1207567. [DOI] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Division. 2007;2:7. doi: 10.1186/1747-1028-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M, Clurman BE. FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat Rev Cancer. 2008;8:83–93. doi: 10.1038/nrc2290. [DOI] [PubMed] [Google Scholar]
- Welcker M, Orian A, Jin J, Grim JE, Harper JW, Eisenman RN, et al. The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc Natl Acad Sci USA. 2004;101:9085–9090. doi: 10.1073/pnas.0402770101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Hubbard EJ, Kitajewski JK, Greenwald I. Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein, and SEL-12 presenilin. Proc Natl Acad Sci USA. 1998;95:15787–15791. doi: 10.1073/pnas.95.26.15787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lyapina S, Das I, Li J, Gurney M, Pauley A, et al. Sel-10 is an inhibitor of Notch signaling that targets Notch for ubiquitin-mediated protein degradation. Mol Biol Cell. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Qian S, Soriano S, Wu Y, Fletcher AM, Wang XJ, et al. Loss of presenilin 1 is associated with enhanced β-catenin signaling and skin tumorigenesis. Proc Natl Acad Sci USA. 2001;98:10863–10868. doi: 10.1073/pnas.191284198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y-W, Wang R, Liu Q, Zhang H, Liao F-F, Xu H. Presenilin/γ-secretase-dependent processing of β-amyloid precursor protein regulates EGF receptor expression. Proc Natl Acad Sci USA. 2007;104:10613–10618. doi: 10.1073/pnas.0703903104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.