Abstract
To generate γ-retroviral vectors for stable conjoint expression of artificial microRNAs (amiR) and therapeutic genes in primary human lymphocytes, and to identify the design parameters that are key for successful vector generation. Gamma-retroviral vectors were designed to co-express both amiRs and a linked reporter gene, truncated CD34 (tCD34). Artificial miRs based on microRNAs miR-16, miR-142, miR-146b, miR-150, miR155, and miR-223 were inserted into sites within the intron of the vector and tested for tCD34 expression by flow cytometry (FACS). Different constructs were assembled with amiRs targeted to knockdown expression of suppressor of cytokine signaling 1 (SOCS1) or programmed cell death 1 (PDCD1, PD-1). Three of the six amiRs maintained tCD34 expression. Expansion of primary human T cells transduced with these amiR vectors, as well as transgene expression, were equivalent to control engineered T cells over a 40-day period. Knockdown of SOCS1 RNA and PD-1 expression by FACS was shown to vary between constructs, dependent on either the specific short interfering RNA sequence used in the amiR, or the microRNA backbone and location in the vector intron. Gamma-retroviral vectors that both efficiently knockdown endogenous gene expression and maintain linked transgene production can be produced, but empirical vector evaluations were best suited for optimal construct analysis.
Introduction
The discovery of RNA interference and the subsequent finding of microRNAs (miRs) led to new understandings of cellular regulation and novel opportunities to manipulate gene expression [1–4]. While transfection of short interfering RNA (siRNA) can be used to rapidly screen for the phenotype of target gene knockdowns, long-term manipulation of gene expression requires sustained delivery of the interfering RNA through integrating gene transfer vectors. The most common design for these vectors has been to use strong RNA polymerase III (PolIII) promoters to drive the expression of short hairpin RNA (shRNA) where the interfering RNA is expressed as a stem loop structure that mimics the precursor microRNA (pre-miRNA) [5–8]. Most recently, several groups have gone a step further to mimic natural miRNA biology by producing artificial constructs that are analogous to the primary miRNA (pri-miRNA) [9–12]. These artificial miRNAs (amiRs) can be transcribed by RNA PolII promoters, as are most natural miRNAs.
Significant advances have been made in the last decade in the ability to transduce primary cells for human gene therapy applications [13,14]. The successful application of these techniques in T cell adoptive cell transfer gene therapy trials for cancer has led to long-term clinical responses in diverse malignancies [15]. Using retroviral vectors (both γ-retroviral and lentiviral vectors have been used) to deliver tumor antigen targeting receptors based on T cell receptors (TCR) or chimeric antigen receptors are particularly attractive approaches for cancer immunotherapy [15–18]. The target cell for these cancer gene therapy trials were mature T cells, which have a number of regulator feedback systems that limit uncontrolled immune responses in healthy individuals [19,20]. While the expression of these T cell feedback regulatory networks are essential in the control of autoimmunity, they may limit the in vivo effectiveness of T cells genetically engineered to express anti-tumor antigen receptors. Herein, we describe γ-retroviral vector design principles for the co-expression of an RNAi inducing moiety along with a model anti-tumor antigen receptor.
Materials and Methods
Artificial miR vector design
The MSGV1 (MSCV-based splice-gag) γ-retroviral vector is similar to the MFG vector with a long terminal repeat derived from the murine stem cell virus (MSCV) and the murine leukemia virus (MLV) extended packaging signal (ψ+), and the splice acceptor region of the POL gene immediately upstream of the ENV start codon, which is located within an NcoI site [21,22]. In MSGV2, the NcoI site is removed and replaced with a multiple cloning site. The truncated CD34 (tCD34) gene was made using polymerase chain reaction (PCR) as described [23] with the addition of a 5′ NotI site and 3′ EcoRI site, and inserted into MSGV2 using these sites to produce MSGV2-tCD34. amiR sequences were synthesized (Blue Heron Biotech) based on available pri-miR sequences for miR-16, miR-142, miR-146b, miR-150, miR-155, and miR-223. The 5′ end of each amiR was synthesized with restriction enzyme sites BglII-BsrGI-XhoI-SpeI and with 3′ addition of XhoI-BglII-SpeI-BsrGI. The sequences from commercially derived (Thermo Scientific and Qiagen) siRNA or shRNAs were substituted for the stem region of each microRNA. For example, the anti-PD-1 amiR based on miR-223 (lowercase sequence to follow) and siRNA SA3 (uppercase sequence to follow) has this sequence: cctttctctctctttccctctagggtcacatctcccaggatgatctcacttccccacagaagctcttggcctggcctcctgcagtgccacgctcCGGAGAGCTTCGTGCTAAACTggacactccatgtggtagagAGTTTAGCACGAAGCTCTCCGagtgcggcacatgcttaccagctctaggccagggcagatgggatatgacgaatggactgccagctggatacaaggatgctca. The complete sequence of each miR backbone is shown in Table 1. amiR sequences were inserted into the BsrGI or BglII sites within the intron of MSGV2-tCD34. All vectors were confirmed by enzyme digestion and DNA sequencing.
Table 1.
Artificial microRNA Backbone Sequences
| 5′ Sequence | |
|---|---|
| miR-16 | cctcaaaaatacaaggatctgatcttctgaagaaaatatatttctttttattcatagctcttatgatagcaatgtcagcagtgcct |
| miR-142 | acaaggagggctggggggctcttggagcaggagtcaggaggcctgggcagcctgaagagtacacgccgacggacagacagacagtgcagtcacc |
| miR-146b | ttactcatcctgggaacgggagacgattcacagaagaaagcatgcaagagcagcgtccaggctgaaagaactttggccacctggcac |
| miR-150 | ggacctgggtataaggcagggactgggcccacggggaggcagcgtccccgaggcagcagcggcagcggcggctcctctccccatggccctg |
| miR-155 | acaaaccaggaaggggaaatctgtggtttaaattctttatgcctcatcctctgagtgctgaaggcttgctgtaggctgtatgctg |
| miR-223 | cctttctctctctttccctctagggtcacatctcccaggatgatctcacttccccacagaagctcttggcctggcctcctgcagtgccacgctc |
| 3′ Sequence | |
|---|---|
| miR-16 | agtaaggttgaccatactctacagttgtgttttaatgtatattaatgttactaatgtgttttcagttttattgatagtcttttcagtatt |
| miR-142 | tgagtgtactgtgggcttcggagatcacgccactgctgccgcccgctgcccgccaccatcttcctcggcgctcggggacctcgtgtg |
| miR-146b | tgcccggcagtgctacaacatcaatgccaaggccgtggggcagctgatggtttgggctcccaacttcccagccaggtgcttctgcag |
| miR-150 | ggacctggggaccccggcaccggcaggccccaaggggtgaggtgagcgggcattgggacctcccctccctgtactcccatct |
| miR-155 | gtgtatgatgcctgttactagcattcacatggaacaaattgctgccgtgggaggatgacaaagaagcatgagtcaccctgctgg |
| miR-223 | agtgcggcacatgcttaccagctctaggccagggcagatgggatatgacgaatggactgccagctggatacaaggatgctca |
| Loop | |
|---|---|
| miR-16 | ttaagattctaaaattatct |
| miR-142 | aacagcactggaggg |
| miR-146b | gtgagctctagcaa |
| miR-150 | ctgggctcagacc |
| miR-155 | ttttgcctccaactga |
| miR-223 | ggacactccatgtggtagag |
All nucleotide sequences are shown in 5′ to 3′ orientation.
miR, microRNA.
Retroviral vector preparation and transduction
To generate γ-retrovirus, 293 GP cells, which stably express GAG and POL proteins, were transfected as previously described [24,25]. In brief, 9 μg of vector DNA and 4 μg of RD114 envelope plasmid DNA were mixed with lipofectamine 2000 (Life Technologies) in antibiotic free medium and incubated at room temperature for 20 minutes. The mixture was applied to 293GP cells that had been plated the prior day on a 100-mm2 poly-lysine –coated plate (Becton Dickinson). After 6 hours of incubation, the medium was replaced with Dulbecco's modified Eagle's medium (Life Technologies) with 10% fetal bovine serum and the viral supernatants were harvested 48 hours later. The peripheral blood lymphocytes (PBL) used in this study were obtained from healthy donors and metastatic melanoma patients seeking treatments at the Surgery Branch, National Cancer Institute. All primary human cells were obtained under Institutional Review Board reviewed and approved clinical protocols. Briefly, PBL were collected by leukapheresis, and lymphocytes were separated by Ficoll/Hypaque cushion centrifugation, washed in Hank's balanced salt solution and resuspended at a concentration of 1×106/mL in AIM-V® medium (Life Technologies) supplemented with 300 IU/mL interleukin-2, 50 ng/mL of an agonistic anti-CD3 antibody, and 5% heat-inactivated human serum type AB (Valley Biomedical). Two days later, 2×106 total cells were transduced at 0.5×106 cells/mL with retroviral supernatant spun onto RetroNectin (Takara Bio) coated non-tissue culture treated 6-well plates as described by the manufacturer. Transduced cells were allowed to expand in AIM-V media as above, without OKT-3. HEK293 and 293GP cells were purchased from ATCC.
Endpoint and real-time PCR
For the detection of integrated viral sequences in the genomic DNA of transduced lymphocytes, we utilized an end-point polymerase chain reaction (PCR) assay, whereby integration of an amiR-containing vector results in amplification of a ∼350-bp band. Integration of a vector that does not contain any amiR yields a ∼100-bp band instead. Genomic DNA was isolated, from transduced lymphocytes cultured during one week after transduction, using the DNeasy Blood and Tissue kit (Qiagen). Oligonucleotide primers flanking the BglII and BsrGI cloning sites within the intronic region of the vector were synthesized by Integrated DNA Technologies. PCR reactions were performed using the PCR Supermix (Life Technologies in a GeneAmp PCR System 9700 thermal cycler (Life Technologies) following the instructions provided by the supplier. For real-time reverse-transcription PCR (RT-PCR), total RNA was isolated using RNeasy Mini Kit (Qiagen) and reverse transcribed using the ThermoScript RT-PCR system (Life Technologies). All RT-PCR reactions were performed using an ABI 7500 FAST real-time PCR system instrument (Life Technologies). All TaqMan probes and reagents were purchased directly from Applied Biosystems (Life Technologies) and TaqMan β-actin control reagents kit was used for normalization.
Fluorescence activated cell sorting analysis
Transduced T cells were stained with a phycoerythrin-labeled anti-CD34 and with allophycocyanin-labeled anti-PD-1 (BD Pharmingen). Cells were analyzed using a FACScanto II flow cytometer with CellQuest software (BD Biosciences) or FlowJo software (Tree Star, Inc).
Results
Evaluation of amiR vector design using a vector expressing tCD34
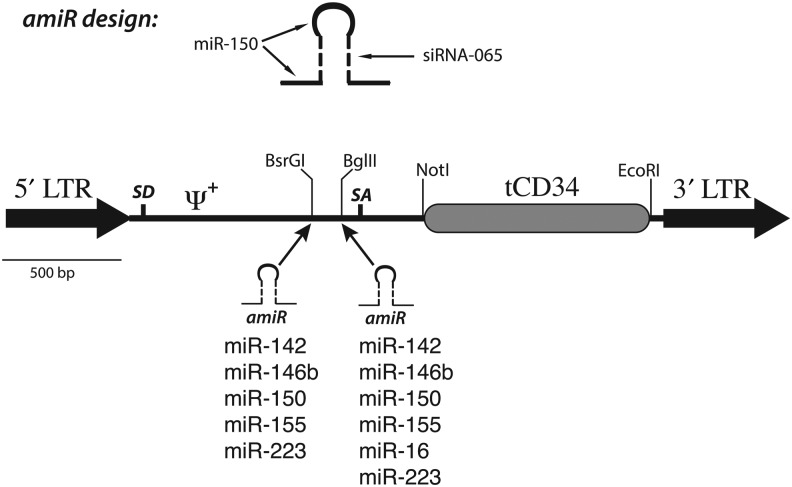
A γ-retroviral vector (MSGV2–tCD34) was designed to express a truncated version of the human hematopoietic progenitor protein CD34 (tCD34) in a vector containing the Moloney murine leukemia virus (MoMLV) extended packaging signal and the naturally occurring intronic elements from the MoMLV ENV gene, similar in design to the MFG class of vectors (Fig. 1) [21]. tCD34 has a number of advantages for use as cell surface marker, the most attractive being the availability of certified reagents for clinical applications to enrich for CD34+ cells [23,26]. The tCD34 gene was flanked by restriction enzyme sites permitting the rapid substitution of alternative coding sequences for potentially therapeutic genes, such as anti-tumor antigen receptors.
FIG. 1.
Artificial microRNA–truncated CD34 (amiR–tCD34) vector design. Shown in the top diagram is the design principle for amiRs, which contain the 5′, loop, and 3′ sequences from a primary microRNA (miR) transcript with the stem sequence being derived from a small interfering RNA (siRNA). The MSGV2–tCD34 vector is shown in the bottom diagram with the location of the insertion sites for the amiR sequences. LTR, long terminal repeat; SD, splice donor; ψ+, extended packaging sequence; SA, splice acceptor.
The MSGV2-tCD34 vector backbone was used as the platform for the delivery of artificial microRNAs (amiRs) designed to knockdown expression of endogenous genes.
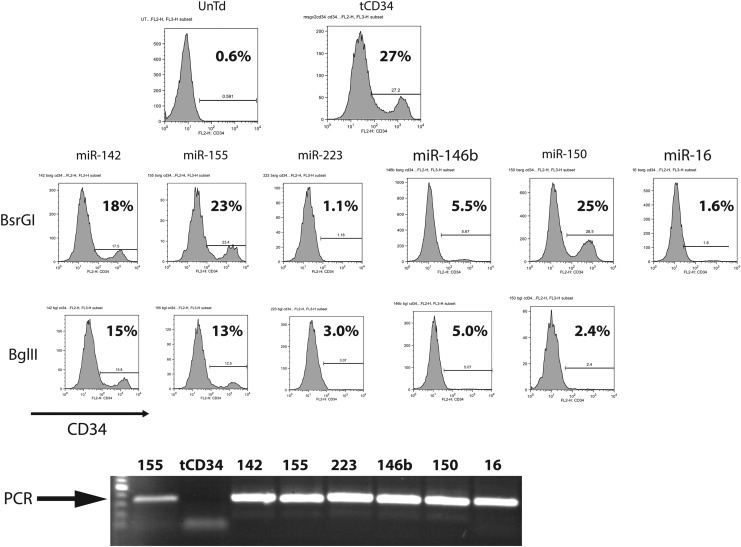
As backbones for the amiR sequences we chose microRNA previously shown to be abundantly expressed in human lymphocytes, specifically miR-142, miR-146b, miR-150, miR-155, miR-16, and miR-223 [27–30]. Using a representative siRNA sequence targeted to TIPE2, we constructed 11 individual vectors with the amiRs inserted into one of two restriction enzyme sites within the intron of the tCD34 vector (Fig. 1). We chose to directly engineer primary human T cells, as established T cell lines cannot correctly model the diversity of gene expression found in random human donor lymphocytes. Retroviral vector supernatant was prepared from each construct and used to transduce activated human T cells. Two days later we assayed for transduction efficiency by fluorescence activated cell sorting analysis (FACS) analysis of cells expressing tCD34. As shown in Fig. 2, the T cells transduced with the parental vector produced 27% tCD34+ cells, while expression of tCD34 in the amiR vectors varied from 1.1% to 25%. Interestingly, three of the amiR vector designs, using miR-223, miR-146b, and miR-16 backbones yielded poor tCD34 expression when inserted into either restriction enzyme site. This observation was reproducible with independently produced batches of vector supernatant.
FIG. 2.
Screening of amiR backbones. Shown in the top three rows are flow cytometry (FACS) plots of transduced primary human T cells stained for the cell surface expression of CD34, with percent positive cells as shown. Rows labeled BsrGI and BglII indicate which restriction enzyme site within the MSGV2–tCD34 vector was used to insert the amiR. The bottom row shows a photograph of the polymerase chain reaction (PCR) reaction products of DNA extracted from transduced T cells and subject to amplification using primers on either side of the BglII and BsrGI sites. Data are representative of four independent transductions using different donor lymphocytes.
To determine if the lack of tCD34 expression in transduced human PBL was caused by the lack of transduction, we extracted DNA from transduced lymphocytes and used PCR to determine the presence of integrated proviral DNA. The specific PCR primers used were in the intron and flanked the amiR insertion region (on either side of the BglII and BsrGI sites). A PCR product was observed for each amiR vector-transduced PBL culture (Fig. 2), regardless of tCD34 expression, indicating that the amiR sequence was associated with the proviral integrate and we did not observe any gross rearrangements of the amiR sequence by this assay. These results suggest that the lack of tCD34 expression in these specific vectors was due to post-transcriptional events.
The purpose of this experiment was to screen vector designs for subsequent experiments targeting relevant cellular targets, and thus all subsequent vectors were based on amiR sequences from mirR-142, miR-150, and miR-155. The reason for the lack of tCD34 expression in some vector designs was not investigated further but possibly involved negative influences on vector mRNA stability, processing, or translational efficiency.
amiR vectors targeting SOCS1
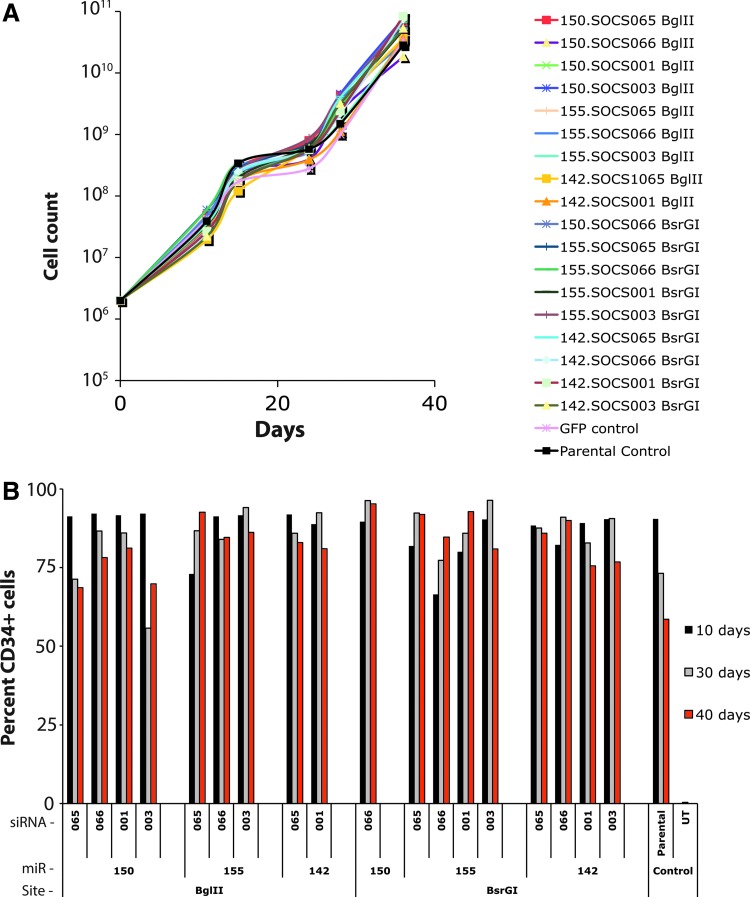
As a first gene of potential interest to manipulate in T cells we chose to target suppressor of cytokine signaling 1 (SOCS1). Eight siRNA or shRNAs targeting SOCS1 were initially evaluated for their ability to knockdown SOCS1 mRNA in 293T and PBL with the four most active sequences used to form the stems of amiRs based on microRNAs 142, 150, and 155 (data not shown). A total of 18 different vectors were assembled and used to transduce PBL. Vector nomenclature was as follows: microRNA. siRNA restriction enzyme (e.g., 150.065 BglII was a vector using the miR-150 backbone with a stem from siRNA 065 inserted into the BglII site). Because previous reports had demonstrated that high-level expression of shRNAs could affect in vitro cell viability, we monitored growth of the SOCS1-amiR vector transduced PBL cultures for 40 days (Fig. 3A). No significant differences in SOCS1-amiR vector transduced cell growth curves were observed compared to control cells, including untransduced cells, green fluorescent protein (GFP) vector transduced, and an unrelated amiR. We also determined whether there were any changes in tCD34 gene expression over time and again did not observe any significant differences in tCD34 expression over time versus the parental tCD34 vector (Fig. 3B).
FIG. 3.
Cell viability and transgene expression of amiR vector transduced primary T cells. (A) Eighteen different amiR vectors targeting suppressor of cytokine signaling 1 (SOCS1) were used to transduce primary human T cells along with control vectors and maintained in cell culture for 40 days. Shown is the cell count over time. (B) The same 18 vector-transduced cell cultures were assayed for tCD34' gene expression by FACS at days 10, 30, and 40 post transduction. Shown were the percent CD34 positive cells at the given time points. Columns are labeled to indicate which siRNA was used, the microRNA (miR) backbone, and the vector insertion site. Data shown were from one of two similar experiments using different donors.
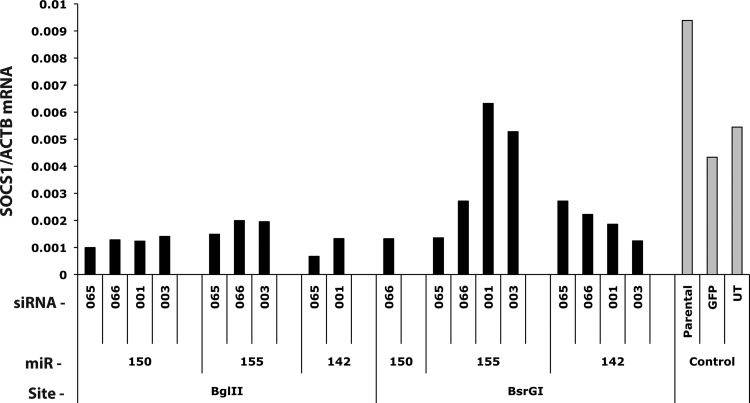
Evaluation of the activity of 18 amiR vectors targeting SOCS1 was performed by measuring RNA levels of SOCS1 in transduced T cells (Fig. 4). In comparison with control cultures, several of the constructs reduced SOCS1 RNA greater than 4-fold (e.g., 142.065 BglII), while other constructs such as 155.003 BsrGI did not reduce SOCS1 mRNA. Furthermore, we observed that the identical amiR construct could behave differently dependent on its cloning site (e.g., compare 142.065 BglII and 142.065 BsrGI), indicating the necessity to empirically evaluate all constructs for knockdown activity. In some experiments, as the one depicted in Fig. 4, SOCS1 mRNA expression in untransduced lymphocytes or lymphocytes transduced with GFP was different from that in MSGV2-tCD34-tranduced cells. This observation, however, was not consistent across different donors.
FIG. 4.
SOCS1 knockdown by amiR vectors. The amount of SOCS1 mRNA relative to beta-actin was determined in peripheral blood lymphocytes (PBL) cultures transduced with the 18 SOCS1 amiR vectors and 3 control cultures. RNA values were determined by qualitative real-time PCR. Data shown were from one of two similar experiments using different donors.
Artificial miR vectors targeting PD-1
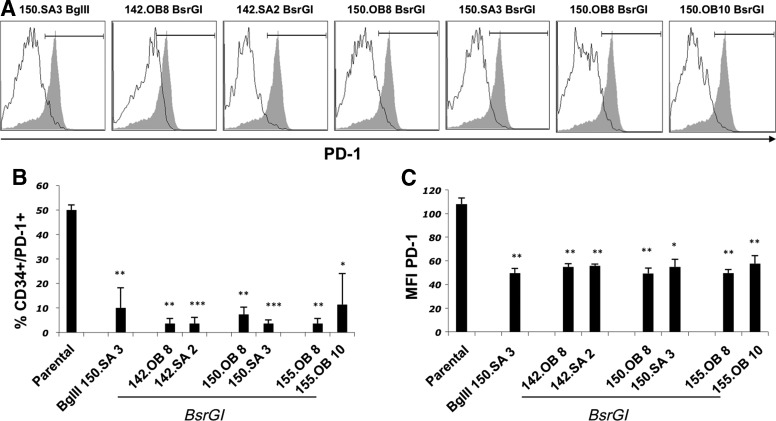
We next chose to target programmed cell death 1 (PDCD1), whose gene product is the PD-1 protein. When we sought to target PD-1, we found that endogenous PD-1 expression was only transiently expressed post T cell activation and varied greatly from donor to donor making it a difficult target to evaluate amiR activity in vitro. Therefore, we introduced, into primary T cells, the wild type PD-1 sequence expressed by a constitutive promoter and then transduced these cells with amiR vectors targeting PD-1. To determine the ability of the amiR vectors to knockdown PD-1 protein expression, seven anti-PD-1 amiR vectors were used to engineer PD-1 gene-transduced primary T cells from three independent donors and five days post-transduction, the expression of tCD34 and PD-1 on the T cell surface was determined (Fig. 5A). Each of the amiR vectors reduced PD-1 surface expression in comparison with T cells transduced with the parental tCD34 vector, both in the percentage of PD-1-postive cells (Fig. 5A) and in the mean fluorescence intensity of the PD-1 staining (Fig. 5B).
FIG. 5.
Programmed cell death protein 1 (PD-1) knockdown by amiR vectors. The percentage of PD-1+/CD34+cells was determined 5 days post transduction with seven different amiR vectors in multiple donor PBL engineered to constitutively express PD-1. (A) Histograms showing a representative example of PD-1 expression on human PBL engineered to co-express PD-1 along with the indicated amiR (solid line) or control vector (shaded histogram). Events gated on lymphoid, single, viable CD3+ cells. (B) Shown are the mean values from three independent transductions using different donors. (C) Shown are the mean fluorescence intensity values of PD-1 expression for tCD34+ PD-1+ cells (average of three independent transductions performed on different donors). Parental refers to cells transduced with the MSGV2–tCD34 vector. Paired t-test: ***p<0.001, **p<0.01, *p<0.05.
Discussion
Gamma-retroviral vectors are one of the core gene transfer technologies used in human gene therapy clinical applications and have been used extensively in trials targeting hematopoietic cells using ex vivo gene transfer procedures. They have been used in multiple trials targeting T lymphocytes for diseases as diverse as adenosine deaminase deficiency-severe combined immunodeficiency, HIV infection, cancer, and most importantly, have a long-term safety record [14,31–33]. In this report we examined the design of γ-retroviral vectors devised to express both a protein coding sequence and also an amiR sequence with the ability to knockdown target gene expression. We took an empirical approach by testing several microRNA backbones inserted into two different sites within the intron of an MFG-like vector.
An amiR is a hybrid RNA molecule with RNA sequences derived from the 5′ and 3′ flanking and loop regions of endogenous microRNAs but where the stem sequence has been replaced by an short interfering RNA (siRNA) sequence. For potential clinical applications, amiRs have a number of advantages over similar shRNA expression platforms, including the use of RNA polymerase II promoters, which can mimic natural microRNA expression; and the endogenous microRNA processing machinery does not trigger cellular self-defense mechanisms such as interferon induction [34]. While many RNAi knockdown vectors designed for general laboratory experimentation use short-hairpin designs, over-expression of shRNAs using RNA polymerase III vectors have been demonstrated to mediate toxicity via saturation of microRNA processing pathways leading to lethality in animal models [35,36]. These shRNA vectors have also been shown to be toxic to transduced human T cells [37] and we also observed this phenomenon in preliminary experiments.
Many microRNAs are naturally found within introns or other noncoding sequences, and previous publications using lentiviral vectors also reported that these genetic elements could be inserted and effectively expressed within artificial introns of gene transfer vectors [12]. By insertion into the vector intron, we theorized that the amiR insert would not interfere with the expression of tCD34, as they would be spliced out from the mature mRNA that was translated into the tCD34 protein. Interestingly, this was not the case for three of the amiRs based on miR-223, 146b, and 16, for which we did not observe tCD34 expression (Fig. 2). In the report by Amendola et al., using a lentiviral vector gene transfer platform, an amiR based on miR-223 was shown to not interfere with linked reporter gene expression when inserted into an artificial intron [12]. Thus it is possible that the differences observed in the effectiveness of the given amiR vector to maintain the linked reporter protein expression are likely to be context dependent, which reinforces the concept of empirical vector design evaluation.
SOCS gene family members are involved in the regulation of T cell differentiation and function as negative feedback elements to cytokine stimulation [38,39]. Following cytokine receptor binding, Janus kinase (JAK) is activated which in turn activates the signal transducers and activators of the transcription pathway. SOCS1 binds to JAK and inhibits further T cell activation. Therefore, inhibition of SOCS1 might enhance T cell function by lessening this feedback and potentially enhancing proliferation and effector functions. Although we assembled and tested multiple amiR vectors targeting SOCS1, we were unable to reduce SOCS1 RNA expression more than 80% (Fig. 4). This amount of SOCS1 RNA reduction may not be sufficient for a relevant biological effect and further vector optimization would be warranted.
PD-1 is a cell surface protein that is expressed on activated T cells and upon binding its ligands PD-L1 and PD-L2, inhibits T cell function, and is likely involved in limiting peripheral inflammatory responses and autoimmunity [40,41]. Most significantly, many tumors express PD-L1 and in clinical trials using anti-PD-1 monoclonal antibodies, clinical efficacy has been demonstrated in a variety of cancers [42]. We recently reported that PD-1 is down regulated in ex vivo gene modified T cells administered to cancer patients in TCR gene therapy trials [43]. This lack of significant PD-1 expression on ex vivo cultured T cells necessitated the introduction of an expression cassette constitutively expressing PD-1 in order for us to evaluate the PD-1 amiR vectors. While artificial in design, we felt that if any of the amiR vectors we able to reduce vector-mediated PD-1 expression, it would likely be possible to see reduction of PD-1 under natural T cell activation. Data presented in Fig. 5 demonstrated a near 90% reduction in PD-1 protein expression, which may merit further investigation.
The purpose of the investigations reported herein was to determine the feasibility of functional amiR insertion into the commonly used MFG-class of γ-retroviral vectors. Based on these vectors design principles, γ-retroviral vectors containing an amiR plus potentially therapeutic proteins can be readily assembled, but different combinations of amiR sequence and vector insertion site need to be taken into consideration for optimal function.
Acknowledgments
The authors would like to thank Arnold Mixon and Shawn Farid for technical support with FACS analysis. This work was supported by the intramural program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health, Bethesda Maryland.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.McManus MT. and Sharp PA. (2002). Gene silencing in mammals by small interfering RNAs. Nat Rev Genet 3:737–747 [DOI] [PubMed] [Google Scholar]
- 2.Flynt AS. and Lai EC. (2008). Biological principles of microRNA-mediated regulation: shared themes amid diversity. Nat Rev Genet 9:831–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brown BD. and Naldini L. (2009). Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet 10:578–585 [DOI] [PubMed] [Google Scholar]
- 4.Berezikov E. (2011). Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12:846–860 [DOI] [PubMed] [Google Scholar]
- 5.Silva JM, Li MZ, Chang K, Ge W, Golding MC, Rickles RJ, Siolas D, Hu G, Paddison PJ, et al. (2005). Second-generation shRNA libraries covering the mouse and human genomes. Nat Genet 37:1281–1288 [DOI] [PubMed] [Google Scholar]
- 6.Chang K, Elledge SJ. and Hannon GJ. (2006). Lessons from Nature: microRNA-based shRNA libraries. Nat Methods 3:707–714 [DOI] [PubMed] [Google Scholar]
- 7.An DS, Qin FX, Auyeung VC, Mao SH, Kung SK, Baltimore D. and Chen IS. (2006). Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther 14:494–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Manjunath N, Wu H, Subramanya S. and Shankar P. (2009). Lentiviral delivery of short hairpin RNAs. Adv Drug Deliv Rev 61:732–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du G, Yonekubo J, Zeng Y, Osisami M. and Frohman MA. (2006). Design of expression vectors for RNA interference based on miRNAs and RNA splicing. FEBS J 273:5421–5427 [DOI] [PubMed] [Google Scholar]
- 10.Boudreau RL, Martins I, and Davidson BL. (2009). Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol Ther 17:169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lebbink RJ, Lowe M, Chan T, Khine H, Wang X. and McManus MT. (2011). Polymerase II promoter strength determines efficacy of microRNA adapted shRNAs. PloS One 6:e26213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amendola M, Passerini L, Pucci F, Gentner B, Bacchetta R, and Naldini L. (2009). Regulated and multiple miRNA and siRNA delivery into primary cells by a lentiviral platform. Mol Ther 17:1039–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kay MA. (2011). State-of-the-art gene-based therapies: the road ahead. Nat Rev Genet 12:316–328 [DOI] [PubMed] [Google Scholar]
- 14.Kohn DB. (2010). Update on gene therapy for immunodeficiencies. Clin Immunol 135:247–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park TS, Rosenberg SA, and Morgan RA. (2011). Treating cancer with genetically engineered T cells. Trends Biotechnol 29:550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jorritsma A, Schotte R, Coccoris M, De Witte MA. and Schumacher TN. (2011). Prospects and limitations of T cell receptor gene therapy. Curr Gene Ther 11:276–287 [DOI] [PubMed] [Google Scholar]
- 17.Lagisetty KH. and Morgan RA. (2012). Cancer therapy with genetically-modified T cells for the treatment of melanoma. J Gene Med 14:400–404 [DOI] [PubMed] [Google Scholar]
- 18.Ramos CA. and Dotti G. (2011). Chimeric antigen receptor (CAR)-engineered lymphocytes for cancer therapy. Expert Opin Biol Ther 11:855–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldszmid RS. and Trinchieri G. (2012). The price of immunity. Nat Immunol 13:932–938 [DOI] [PubMed] [Google Scholar]
- 20.Saito T. and Yamasaki S. (2003). Negative feedback of T cell activation through inhibitory adapters and costimulatory receptors. Immunol Rev 192:143–160 [DOI] [PubMed] [Google Scholar]
- 21.Frankel TL, Zhang L, Burns WR, Zheng Z. and Morgan RA. (2011). The position of the AUG start codon in MFG-based gamma-retroviral vectors has a dramatic effect on translation-dependent protein expression. J Gene Med 13:478–486 [DOI] [PubMed] [Google Scholar]
- 22.Hughes MS, Yu YY, Dudley ME, Zheng Z, Robbins PF, Li Y, Wunderlich J, Hawley RG, Moayeri M, Rosenberg SA. and Morgan RA. (2005). Transfer of a TCR gene derived from a patient with a marked antitumor response conveys highly active T-cell effector functions. Hum Gene Ther 16:457–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fehse B, Richters A, Putimtseva-Scharf K, Klump H, Li Z, Ostertag W, Zander AR. and Baum C. (2000). CD34 splice variant: an attractive marker for selection of gene-modified cells. Mol Ther 1:448–456 [DOI] [PubMed] [Google Scholar]
- 24.Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, et al. (2009). Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114:535–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kochenderfer JN, Feldman SA, Zhao Y, XU H, Black MA, Morgan RA, Wilson WH. and Rosenberg SA. (2009). Construction and preclinical evaluation of an anti-CD19 chimeric antigen receptor. J Immunother 32:689–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Govers C, Berrevoets C, Treffers-Westerlaken E, Broertjes M. and Debets R. (2012). Magnetic-activated cell sorting of TCR-engineered T cells, using tCD34 as a gene marker, but not peptide-MHC multimers, results in significant numbers of functional CD4+ and CD8+ T cells. Hum Gene Ther Methods 23:213–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salaun B, Yamamoto T, Badran B, Tsunetsugu-Yokota Y, Roux A, Baitsch L, Rouas R, Fayyad-Kazan H, Baumgaertner P, et al. (2011). Differentiation associated regulation of microRNA expression in vivo in human CD8+ T cell subsets. J Transl Med 9:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grigoryev YA, Kurian SM, Hart T, Nakorchevsky AA, Chen C, Campbell D, Head SR, Yates JR., 3rd and Salomon DR. (2011). MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes. J Immunol 187:2233–2243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta A, Nagilla P, Le HS, Bunney C, Zych C, Thalamuthu A, Bar-Joseph Z, Mathavan S. and Ayyavoo V. (2011). Comparative expression profile of miRNA and mRNA in primary peripheral blood mononuclear cells infected with human immunodeficiency virus (HIV-1). PloS One 6:e22730. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.D'Agostino DM, Zanovello P, Watanabe T. and Ciminale V. (2012). The microRNA regulatory network in normal- and HTLV-1-transformed T cells. Adv Cancer Res 113:45–83 [DOI] [PubMed] [Google Scholar]
- 31.Rossi JJ, June CH. and Kohn DB. (2007). Genetic therapies against HIV. Nat Biotechnol 25:1444–1454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenberg SA, Restifo NP, Yang JC, Morgan RA. and Dudley ME. (2008). Adoptive cell transfer: a clinical path to effective cancer immunotherapy. Nat Rev Cancer 8:299–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Scholler J, Brady TL, Binder-Scholl G, Hwang WT, Plesa G, Hege KM, Vogel AN, Kalos M, Riley JL, et al. (2012). Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med 4:132ra153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer M, Kinkl N, Meixner A, Kremmer E, Riemenschneider M, Förstl H, Gasser T, and Ueffing M. (2009). Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene Ther 16:142–147 [DOI] [PubMed] [Google Scholar]
- 35.Grim D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F. and Kay MA. (2006). Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature 441:537–541 [DOI] [PubMed] [Google Scholar]
- 36.Mcbride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso R, et al. (2008). Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci (USA) 105:5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo HL, Chang T, Yam P, Marcovecchio PM, Li S, Zaia JA. and Yee JK. (2007). Inhibition of HIV-1 replication with designed miRNAs expressed from RNA polymerase II promoters. Gene Ther 14:1503–1512 [DOI] [PubMed] [Google Scholar]
- 38.Ilangumaran S. and Rottapel R. (2003). Regulation of cytokine receptor signaling by SOCS1. Immunol Rev 192:196–211 [DOI] [PubMed] [Google Scholar]
- 39.Palmer DC. and Restifo NP. (2009). Suppressors of cytokine signaling (SOCS) in T cell differentiation, maturation, and function. Trends Immunol. 30:592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pardoll DM. (2012). The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12:252–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Topalian SL, Drake CG. and Pardoll DM. (2012). Targeting the PD-1/B7-H1(PD-L1) pathway to activate anti-tumor immunity. Curr Opin Immunol 24:207–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, Mcdermott DF, Powderly JD, Carvajal RD, Sosman JA, et al. (2012). Safety, activity, and immune correlates of anti-PD-1 antibody in cancer. New Engl J Med 366:2443–2454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abate-Daga D, Hanada KI, Davis JL, Yang JC, Rosenberg SA. and Morgan RA. (2013). Expression profiling of TCR-engineered T cells demonstrates over-expression of multiple inhibitory receptors in persisting lymphocytes. Blood 122:1399–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]