Abstract
Small interfering RNAs (siRNAs) guide RNA-induced silencing complexes (RISC) to target mRNAs for sequence-specific silencing. A fundamental aspect of this highly coordinated process is a guide strand–specific loading of the siRNA duplex into the RISC for the accurate target recognition, which is currently dictated by certain duplex parameters such as thermodynamics. Here, we show that minor changes in the overhang structure have profound effects on the extent to which the individual strands of the siRNA duplex participate in RNAi activity. We demonstrate that the two strands of the siRNA are similarly eligible for assembly into RISC for the siRNAs with symmetric overhangs, whereas those with asymmetric RNA/deoxythymidine dinucleotide (dTdT) overhangs exhibit a distinct preference in favor of a strand with an RNA overhang that drives a mature RISC affinity to the desired target. We believe that this additional determinant provides a plausible and simple approach for improving the strand selection, thereby considerably increasing a specificity of target silencing.
Introduction
RNA interference (RNAi) is an evolutionarily conserved mechanism of post-transcriptional gene silencing that triggers messenger RNA (mRNA) cleavage in a sequence-specific manner. The RNAi technology has been harnessed by researchers for the basic research as well as for the development of new therapies for disease. Prototypical small interfering RNA (siRNA) duplexes, originally described by Elbashir and colleagues [1], are 21-nt double-stranded RNAs with 2-nt symmetric 3′-overhangs, suggestive of processing by an RNase III–type enzyme such as Dicer [2]. The two strands of the siRNA duplex are first assembled into Argonaute proteins as double-stranded RNAs (pre-RNA-induced silencing complex (RISC) and one strand is discarded by unwinding or cleavage to form a mature RISC [3]. This discarded strand is called the passenger strand (or sense strand), whereas the other strand finally retained in mature RISC is called the guide strand (or antisense strand). Our current knowledge of the specific selection bias of the guide strand is governed by the relative thermodynamic asymmetry of the first one to four bases at each end of the precursor duplexes, as a strand whose 5′ end is less stable is favored for incorporation into the mature RISC [4,5], which is the hallmark of microRNA (miRNA) species.
However, this attribute alone is clearly not sufficient to eliminate an inappropriate loading of the passenger strand that generates an active RISC, as is revealed by a genome-wide scale profiling, in which an unintended and extensive off-target silencing mediated by the passenger strand is widespread [6], which might lead to toxic phenotypes and cause serious problems in therapeutic applications [7]. In addition, since a transcriptome-wide profiling of endogenous small RNA populations in living cells has become widely employed [8], it has become apparent that a substantial proportion of even miRNA passenger strand species indeed accumulates to distinct and characteristic levels in Argonaute proteins [9,10]. Taken together, these findings argue that conventional factors, such as thermodynamic stability, do not entirely circumvent possible off-target effects by the passenger strand. Therefore, the extra determinants that promote the efficient strand selection of the desired strand are apparently necessary.
A deoxythymidine dinucleotide (dTdT) overhang has been a common industry standard for classical siRNAs to reduce cost and confer increased nuclease resistance [1]. However, decreased gene silencing potency of siRNAs with the dTdT overhang was observed, due to a shorter in vivo duration than those with RNA overhang [11]. In addition, we previously reported that asymmetric siRNAs (asiRNAs) solely composed of RNA result in more efficient silencing than those with dTdT overhang [12]. We found that a strand with RNA overhang assembles into a human Argonaute 2 (hAGO2)-RISC more efficiently than a counterpart with dTdT overhang, because of an increased affinity toward hAGO2 [12]. In this respect, we modified a conventional siRNA (19+2) with an asymmetric RNA/dTdT overhang, in which one strand is composed of RNA overhang and the other of dTdT overhang. By introducing this asymmetric 3′ overhang structure, it was possible to drive a mature RISC affinity to the stand complementary to the desired target, thus considerably reducing off-target silencing by the unwanted strand.
Materials and Methods
siRNAs
Chemically synthesized RNAs were purchased from Bioneer and annealed according to the manufacturer's protocol. Detailed sequence information of siRNAs is provided in each figure.
Cell culture and siRNA transfection
A549 (CCL-185) and HeLa (CCL-2) cell lines were purchased from ATCC and maintained at 37°C in Dulbecco's modified Eagle's medium (DMEM) (Gibco) supplemented with 10% (v/v) fetal bovine serum (Gibco). Cells were routinely subcultured to maintain exponential growth. For siRNA transfection, cells were plated on 12-well plates 24 hours before transfection at 30%–50% confluence in complete medium. Lipofectamine RNAiMAX (for A549 transfection) was used for siRNA transfection following the manufacturer's protocol (Invitrogen). The final concentrations of siRNAs transfected are provided in each figure.
Plasmids
DNA oligonucleotides including target sequences corresponding to the siRNA duplexes in Fig. 1 and Supplementary Fig. S1 (Supplementary Data are available online at www.liebertpub.com/nat; see below for the DNA sequence) were cloned into the SpeI and HindIII sites of the pMIR-REPORT luciferase vector (Ambion) The sequences for each if the oligonucleotides were as below.
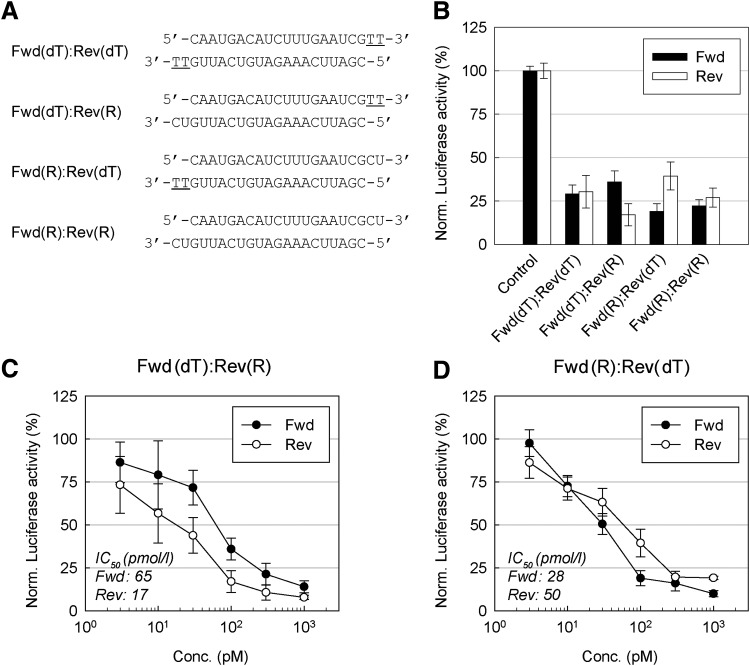
FIG. 1.
Effects of the small interfering RNA (siRNA) 3′-overhangs on the RNA interference (RNAi) activity of each strand. (A) The structures of the siRNA duplexes with symmetric or asymmetric overhangs. Underlined letters indicate DNA. (B) Reporter gene knockdown activities of the siRNAs. A549 cells were transfected with each firefly luciferase reporter construct, either with the forward (Fwd) or reverse (Rev) strand target shown in (A); Renilla luciferase expressing pRL-SV40 vector and 0.1 nM of siRNAs were used. Luciferase activity was measured 24 hours after transfection. Luciferase activities relative to mock transfection condition (reagent only) are presented as bar graphs. (C–D) Target gene knockdown activities of Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT) duplexes. Diverse concentration of duplexes, reporter construct (Fwd or Rev) and pRL-SV40 vector were cotransfected into A549 cells for 24 hours and target gene knockdown activity was analyzed as described in (B). Half maximal inhibitory concentration (IC50) values for each strand are shown in the insets. All data in the graphs represent mean±standard deviation (SD) values of three independent experiments. dT, deoxythymidine; R, RNA.
Fig. 1 Fwd target:
5′-CTAGTAGTACAGCGATTCAAAGATGTCATTGTCTC-3′
5′-AGCTGAGACAATGACATCTTTGAATCGCTGTACTA-3′
Fig. 1 Rev target:
5′-CTAGGAGACAATGACATCTTTGAATCGCTGTACTA-3′
5′-AGCTTAGTACAGCGATTCAAAGATGTCATTGTCTC-3′
Supplementary Fig. S1C Fwd target:
5′-CTAGCTCATGGGTGGAATCATATTGGA-3′
5′-AGCTTCCAATATGATTCCACCCATGAG-3′
Supplementary Fig. S1C Rev target:
5′-CTAGTCCAATATGATTCCACCCATGAG-3′
5′-AGCTCTCATGGGTGGAATCATATTGGA-3′
Luciferase knockdown analysis
Cells were plated on 24-well plates at 50% confluence in complete medium. After 24 hours, pMIR-REPORT based plasmids containing siRNA target sites, pRL-SV40 (1 ng, Promega), and siRNAs were cotransfected using Lipofectamine 2000 reagent following the manufacturer's protocol (Invitrogen). Twenty-four hours later, luciferase activity was analyzed using the Dual Luciferase Reporter Assay System (Promega). Results were quantified with the 20/20n Luminometer (Turner Biosystems). Dose response analysis was performed using Sigma Plot 11 (Systat Software, Inc.) fitting the curves to the “four parameter logistic curve” fit, and half maximal inhibitory concentration (IC50) was calculated for each siRNA.
Quantitative real-time polymerase chain reaction
Total RNA was extracted from cells using an Isol-RNA lysis reagent (5 Prime) according to manufacturer's instructions. Total RNA (500 ng) was used as a template for complementary DNA (cDNA) synthesis using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) according to the manufacturer's protocol. Aliquots (1/20) of each cDNA reaction were analyzed by quantitative real-time reverse transcription polymerase chain reaction using the StepOne Real-Time PCR System (Applied Biosystems). Gene-specific primers were mixed with SYBR Premix Ex Taq (Takara). In the analysis of the siRNA target gene knockdown efficiency, the target genes and an internal control mRNA (TUBA1A) levels were determined using the relative standard curve quantitation method. The primer sequences for each gene are:
Regulatory factor X–associated ankyrin-containing protein (RFXANK)-forward: 5′-CGTGACGTGGACATCAACA-3′
RFXANK-reverse: 5′- CCTGTTGCACTTTCCGGTAT-3′
Nuclear receptor 2C2-associated protein (NR2C2AP)-forward: 5′-CAGCTGCAGATCCAGTTTCA-3′
NR2C2AP-reverse: 5′-CACTTCAGCAGCTGGTATGG-3′
TUBA1A-forward: 5′-GACCAAGCGTACCATCCAGT-3′
TUBA1A-reverse: 5′-CACGTTTGGCATACATCAGG-3′
Cell lysate preparation
To prepare cell lysates expressing FLAG-tagged hAGO2 proteins, HEK293T cells were cultured in DMEM (Welgene) supplemented with 10% (v/v) fetal bovine solution (Welgene) at 37°C in 5% CO2. HEK293T cells grown on 10-cm dishes were transfected with 10 μg of pcDNA3.1-FLAG-hAGO2 using the calcium phosphate method. Cells were washed three times with cold PBS, pH 7.4, and collected by centrifugation at 6,000 rpm at 4°C for 5 minutes. The cell pellets were then resuspended in three packed-cell volumes of hypotonic lysis buffer (25 mM HEPES-NaOH, pH 7.4, 10 mM NaCl, 5 mM DTT, 0.1% Tween-20 and 1× EDTA-free Protease Inhibitor cocktail) and clarified by centrifugation at 17,000 rpm at 4°C for 30 min. The supernatant was flash frozen in liquid nitrogen and immediately stored at −80°C in single-use aliquots.
In vitro small RNA loading assay
Ten nanomoles (f.c.) of radiolabeled duplexes (5′-32P-radiolabeled RNA strand annealed to unlabeled phosphorylated partner strand) were incubated at 37°C for 1 hour in 40 μL of in vitro RNAi reaction. Ten percent of reaction was removed for input fraction and the rest of the sample was incubated with anti-FLAG M2 Affinity Gel (Sigma-Aldrich) for 2 hours with gentle rocking at 4°C. The resin was then washed four times with 10 bead volumes of wash buffer containing 25 mM HEPES-NaOH, pH 7.4, 300 mM NaCl, 1.5 mM MgCl2 and 1x protease inhibitor cocktail (Roche). RNA samples were then phenol extracted, ethanol precipitated, and resolved by 15% native polyacrylamide gel electrophoresis at 4°C. Phosphorimaging was performed using a BAS-2500 image analyzer (Fujifilm), and signal intensities were quantified using MultiGauge (Fujifilm).
In vitro target RNA cleavage assay
For generation of target RNAs, CTFG-17 target mRNAs were in vitro transcribed and radiolabeled at the 5′-cap by guanylyltransferase using the mScript mRNA production system (Epicentre) according to the manufacturers instruction, followed by polyacrylamide gel electrophoresis purification. Target RNA cleavage assays were performed essentially as described previously [13]. Briefly, the reaction was incubated at 30°C using 10 nM siRNA, 2.5 nM cap-radiolabeled target RNA, 1 mM ATP, 1 mM MgCl2, 0.1 μg/μL creatine kinase, 25 mM creatine phosphate, and 0.4 U/μL RNasin (Promega). siRNA duplexes were preincubated with lysate at 30°C for 20 minutes before the addition of cap-labeled target RNA. Phosphorimaging was performed using a BAS-2500 image analyzer (Fujifilm), and signal intensities were quantified using MultiGauge (Fujifilm).
In vitro RISC assembly assays (Gel-shift assay)
In vitro RISC assembly assays were performed essentially as described previously [14,15]. Briefly, each reaction consisted of 5 μL of lysate, 3 μL of 40× reaction mix [16], 1 μL of 100 nM 5′-32P-radiolabeled small RNA duplex, and 1 μL of lysis buffer. After incubation at the indicated time points, the reaction was terminated with 2 μL of heparin mix (60 mM KCl, 3 mM MgCl2, 3% PEG8000, 8% glycerol and 4 mg/mL heparin). The RISCs were resolved by vertical native gel electrophoresis at 10W for 1.5 hour at 4°C. Phosphorimaging was performed using a BAS-2500 image analyzer (Fujifilm), and signal intensities were quantified using MultiGauge (Fujifilm).
Results and Discussion
Effect of 3′-overhang structures of siRNA duplexes on the RNAi activity
To investigate the effect of the siRNA 3′-overhang on the RNAi activity of each strand, we designed four siRNA duplexes that share the same target sequence, but with different overhang types. Both strands of Fwd(dT)–Rev(dT) and Fwd(R)–Rev(R) duplexes have dTdT and RNA overhang, respectively. In contrast, Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT) duplexes consist of strands with different overhangs, dTdT and RNA (Fig. 1A). We designed two reporter vectors that harbor firefly luciferase in both orientations [i.e., forward (Fwd) and reverse (Rev)] within the 3′-untranslated region (Supplementary Fig. S1). Note that we here use the terms “forward strand” and “reverse strand” of each siRNA because the guide and passenger strand will vary depending on the reporter vector analyzed. We cotransfected A549 cells with each siRNA, firefly luciferase construct, and Renilla luciferase vector as a transfection control. Firefly luciferase versus Renilla luciferase activities were analyzed 24 hours after transfection (Fig. 1B).
The siRNAs with symmetric overhangs, such as Fwd(dT)–Rev(dT) and Fwd(R)–Rev(R), displayed a comparable RNAi activity for both of the targets. Regardless of strand and target combinations, 70%–78% knockdown was observed. However, the siRNAs with asymmetric overhangs, such as Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT), showed a distinct preference for one of two targets. As shown in Fig. 1B, the Fwd(dT)–Rev(R) duplex triggered a strong suppression (83% knockdown) of the reverse target (pMIR-REPORT-Rev), but moderate suppression (64% knockdown) of the forward target (pMIR-REPORT-Fwd). In contrast, the Fwd(R)–Rev(dT) duplex showed a reversed pattern of the target silencing (i.e., 81% and 61% knockdown against the forward and reverse target, respectively). We further performed IC50 analysis of each strand of the siRNAs with asymmetric overhangs, which showed a 3.8-fold (Fwd: 65, Rev: 17) and 1.8-fold (Fwd: 28, Rev: 50) higher activity of the strands with RNA overhang than those with dTdT overhang (Fig. 1C, D). We also compared IC50 values of siRNAs with asymmetric and symmetric overhangs. The strands with RNA overhangs from asymmetric siRNAs [Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT)] showed a similar activity to their counterpart from a symmetric siRNA [Fwd(R)–Rev(R)] (Fig. 1C, D and Supplementary Fig. S2B). Meanwhile, the strands with dTdT overhangs from asymmetric siRNAs [Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT)] exhibited a lower activity than their counterpart from a symmetric siRNA [Fwd(dT)–Rev(dT)] (Fig. 1C, D and Supplementary Fig. S2A). It was important to note that, for the siRNAs with asymmetric overhangs [Fwd(dT)–Rev(R) and Fwd(R)–Rev(dT)], we observed a substantial difference in IC50 values (e.g., Fwd: 65, Rev: 17) (Fig. 1C), whereas those with symmetric overhangs [Fwd(R)–Rev(R) and Fwd(dT)–Rev(dT)] had little or no differences (e.g., Fwd: 32, Rev: 30) (Supplementary Fig. S2A, B).
We designed an additional set of siRNAs and reporter vectors to examine whether the different overhang formats generally affect the strand selectivity for target gene silencing (Fig. S1C). As expected, the Fwd(dT)–Rev(R) duplex showed a relatively higher RNAi activity for the reverse target (79% knockdown) than the forward target (70% knockdown). For the Fwd(R)–Rev(dT) duplex, we observed a dramatic difference in RNAi activity of each strand. While Fwd(R) strand showed a strong suppression of the forward target (83% knockdown), Rev(dT) strand merely achieved 22% knockdown for the reverse target (Supplementary Fig. S1D). Conclusively, these results indicate that the strand with an RNA overhang appears to be preferentially processed into the guide strand during the RNAi pathway.
siRNAs with asymmetric RNA/dTdT overhangs provide a higher degree of specificity
To biochemically investigate an important step that may lead to a preferential silencing of the siRNAs with asymmetric overhangs observed in reporter experiments, we established a minimal in vitro system for assessing the individual contribution of each siRNA with different overhang structures for the strand selection. We synthesized eight different small-RNA duplex substrates (For the detailed compositions of each small-RNA duplex, please see Fig. 2A). Each small-RNA duplex, which contained a radiolabeled 5′-phosphate (32P) on either the forward or reverse strand and a non-radiolabeled 5′-phosphate on the other strand, was incubated in cytoplasmic extracts expressing FLAG-tagged hAGO2, the core catalytic components of the RISC, and then immunoprecipitated with anti-FLAG antibody conjugated beads. Immunoprecipitated proteins were removed by phenol extraction and recovered 32P-radiolabeled small RNAs were analyzed by native gel electrophoresis to determine how much portion of individual single-stranded RNAs were matured from their immature siRNA duplexes (Fig. 2B). For the siRNAs with asymmetric RNA/dTdT overhangs (dT–R or R–dT), no matter if it is forward- or reverse-strand radiolabeled, were selective for the strands with RNA overhangs, with a 2- to3-fold greater strand selection efficiency (i.e., 59.5% vs. 28.6% and 26.8% vs. 73.1%) than those with symmetric overhangs (R–R or dT–dT), which clearly explains the previous results of the preferential target silencing obtained with reporter experiments (Fig. 2B).
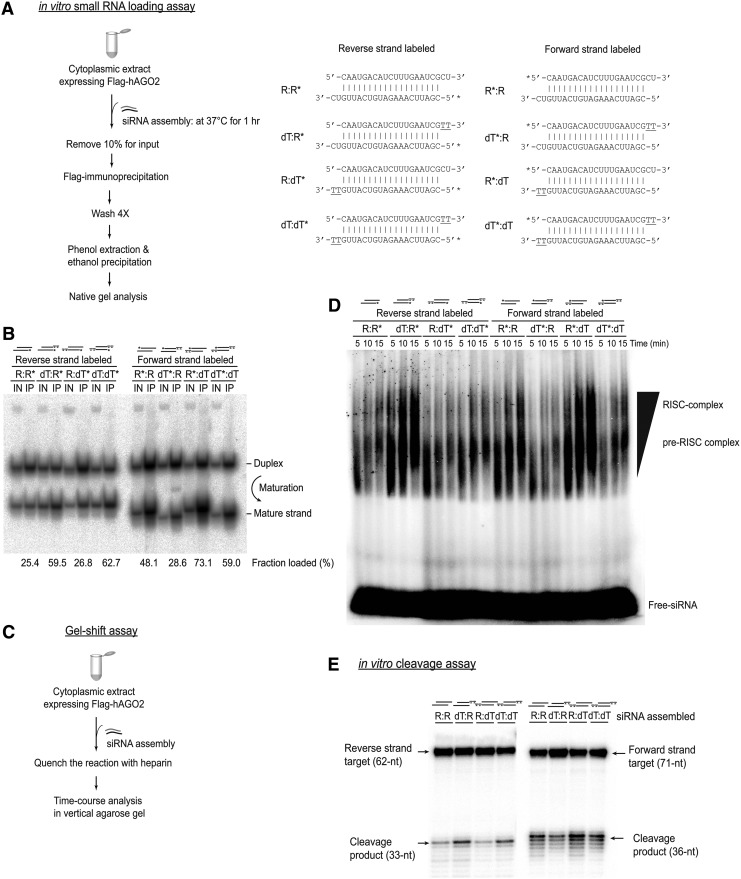
FIG. 2.
Distinct loading preferences of the strands of siRNAs with asymmetric RNA/dTdT overhangs. (A) Schematic description of in vitro loading assay. For example, R:R* duplex contains a non-radiolabeled 5′-phosphate on the forward strand and radiolabeled 5′-phosphate on the reverse strand. Others are denoted in a similar way. (B) Synthetic duplexes carrying one labeled strand were incubated with lysates from HEK 293T cells expressing FLAG-hAGO2. hAGO2 complexes were sequentially immunopurified (IP) and analyzed along with the 10% input (IN) relative to the IP lanes were loaded. Numbers beneath image are the relative fraction loaded (%), calculated as: [(Mature)/(Duplex+Mature)]×100. (C) Schematic description of gel-shift assay. (D) Time-course analysis of the mature- and pre-RISCs formed in lysates using 5′-32P-radiolabeled siRNA duplex variants. Complexes assembled at the indicated time points were resolved on a native gel system. (E) Lysates from HEK 293T cells expressing FLAG-tagged hAGO2 were incubated with siRNA duplexes for the indicated time points. Cleavage reactions were analyzed by 12% urea polyacrylamide gel electrophoresis and detected by autoradiography. HEK, human embryonic kidney; hAGO2, human Argonaute 2; RISC, RNA-induced silencing complex.
Strands with RNA overhangs from asymmetric siRNAs assembled a higher amount of mature RISCs
Because the immunoprecipitation assay by itself does not distinguish between an immature duplex in Argonaute and a functionally matured RISC, we performed a gel-shift assay to further investigate whether these mature strands are indeed incorporated into a mature RISC in vitro [14,15] (Fig. 2C). Time-course assembly of each different siRNA duplex showed that strands with RNA overhangs (dT–R* and R*–dT) formed a much higher levels of the RISCs than those with dTdT overhangs (dT*–R and R–dT*) when the overhangs are asymmetric (RNA/dTdT) (Fig. 2D). The accumulation of fast-migrating, high-molecular-weight complexes derived from the strands with RNA overhangs (dT–R* and R*–dT) diagnose a high degree of functionally matured RISC formation, as compared to those with dTdT overhangs (dT*–R and R–dT*) (Fig. 2D). In contrast, the siRNAs with symmetric overhangs (R–R*, R*–R or dT–dT*, dT*–dT) exhibited a weak or no strand selectivity.
We next sought to determine whether these functionally matured RISCs trigger a higher efficacy of hAGO2-mediated target cleavage catalysis. Incubation of either a 5′-32P-cap-radiolabeled forward or reverse strand target mRNA in cytoplasmic extracts, containing exogenously assembled four different siRNA variants (R–R, dT–R, R–dT, and dT–dT), produced 5′-cleavage products at the positions expected for hAGO2-catalyzed cleavage (Fig. 2E). This suggests that different 3′-overhang structures do not have any influence on the cleavage site position, indicating that the siRNAs with asymmetric RNA/dTdT overhangs enter the canonical RNAi pathway as well. As expected, the dT–R duplex displayed a considerably higher efficacy of target cleavage than a counterpart R–dT duplex for the reverse strand target, whereas an inverse correlation was observed for the forward strand target (Fig. 2E). Together, we demonstrated in several different contexts that by simply introducing the asymmetric RNA/dTdT overhang drastically shifted the balance in favor of a strand with an RNA overhang. We concluded that siRNA duplexes exhibited a specific dependence on the 3′-overhang, and those with an asymmetric overhang showed a biased preference for the strand with an RNA overhang, which efficiently assembled into the mature RISC and thereby triggers a selective target silencing of the desired strand.
Selective suppression of endogenous transcript by siRNAs with asymmetric overhangs
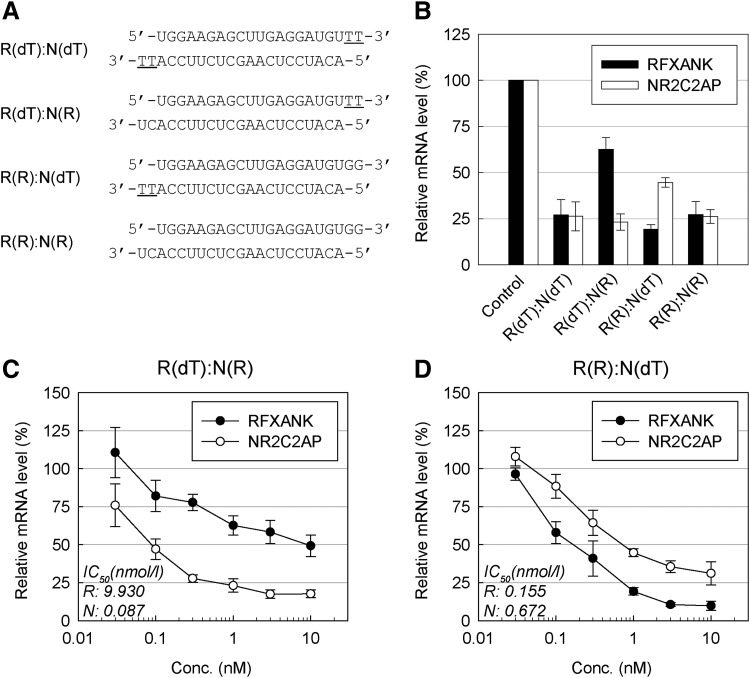
Our results demonstrate that the siRNA duplex with an asymmetric overhang confers the improved intended strand-silencing potency. We went further by applying our findings to direct a strand-specific knockdown of natural antisense transcript (NAT) pair genes. NATs are endogenous RNA molecules transcribed from the opposite DNA strand to other transcripts (sense transcripts) and overlap in part with sense transcripts [17]. From the identified human NATs by other group [18], RFXANK and NR2C2AP NAT pair was selected, and the corresponding siRNAs targeting the overlap sequence were designed and examined (Fig. 3A and Supplementary Fig. S1B). As expected, we observed a strand-specific target gene silencing of the asymmetric overhang duplexes. For the R(dT)–N(R) duplex, consisting of RFXANK targeting strand (R) with a dTdT overhang and NR2C2AP targeting strand (N) with an RNA overhang, 37.4% and 76.9% knockdown was achieved for RFXANK and NR2C2AP respectively (Fig. 3B). In contrast, the R(R)–N(dT) duplex exhibited a reversed pattern of target silencing (i.e., 80.8% and 55.3% knockdown of RFXANK and NR2C2AP respectively) (Fig. 3B).
FIG. 3.
Effects of the siRNA 3′-overhangs on the RNAi activity for the endogenous sense and anti-sense transcripts. (A) The structures of the siRNA duplexes with symmetric or asymmetric overhangs. R and N represent RFXANK and NR2C2AP transcript targeting strands, respectively. Underlined letters indicate DNA. (B) Endogenous gene knockdown activities of the siRNAs. Each siRNA in (A) was transfected into A549 cells (at 1 nM of final concentration) for 24 hours, and target mRNA levels were analyzed by quantitative real-time reverse transcription polymerase chain reaction. The values plotted as “relative target mRNA level” on the y-axis in the figure were calculated as the target mRNA level divided by levels of TUBA1A (control) mRNA, and the mRNA levels relative to the mock transfection control (reagent only) are presented. (C–D) Target gene knockdown activities of R(dT)–N(R) and R(R)–N(dT) duplexes. Diverse concentrations of duplexes were transfected into A549 cells for 24 hours and target gene knockdown activity was analyzed as described in (B). IC50 values for each strand are shown in the insets. All data in the graphs represent mean±SD values of three independent experiments.
The IC50 comparison of the siRNAs with asymmetric and symmetric overhangs showed that the strands with RNA overhangs from R(dT)–N(R) and R(R):N(dT) exhibited a similar activity to those from R(R)–N(R) (Fig. 3C, D and Supplementary Fig. S2D). The strands with dTdT overhangs from asymmetric siRNAs, however, showed an inferior activity to those from a symmetric siRNA [R(dT)–N(dT)] (Fig. 3C, D and Supplementary Fig. S2C). IC50 analysis also confirmed that siRNAs with asymmetric overhangs displayed a strong selection bias in favor of the strands with RNA overhangs (Fig. 3C, D). Especially, R(dT)–N(R) duplex displayed a dramatic preference (∼100-fold) for NR2C2AP target compared with RFXANK, indicating that the siRNA with an asymmetric RNA/dTdT overhang has a high potential for specific silencing of NAT pairs.
Influence of overhang structures on strand selection and silencing efficiency
Several previous studies have suggested that siRNAs with asymmetric overhangs had enhanced efficacy relative to siRNAs with symmetric overhangs [19–22]. For example, Sano et al. showed that siRNAs with a unilateral 2-nt 3′-overhang on the guide strand are more effective than those with 3′-overhang at both ends [21]. The main difference between ours and previously reported siRNA structures is that ours has 3′-overhang for both strands (bilateral overhang), which conforms to the conventional siRNA structure (19+2).
We further compared and evaluated RNAi potencies of ours and some possible end structure variants—asymmetric siRNAs: RNA/blunt, DNA/blunt, and RNA/dTdT and symmetric siRNAs: RNA/RNA, dTdT/dTdT, and blunt/blunt (Supplementary Fig. S3A). On the whole, the siRNAs with asymmetric overhangs [Fwd(B)–Rev(R), Fwd(B)–Rev(dT), and Fwd(dT)–Rev(R)] displayed a strong strand-selection bias, whereas those with symmetric overhangs [Fwd(R):Rev(R) and Fwd(dT):Rev(dT)] or non-overhang [Fwd(B):Rev(B)] showed weak or no strand selectivity (Supplementary Fig. S3B). Although the siRNA with RNA/dTdT overhang was less efficient than those with unilateral overhangs (RNA/blunt and DNA/blunt), the greatest advantage of our siRNA is that it maintains the most widely used siRNA structure, consisting of 19 bp double-stranded RNA with 2-nt 3′-overhangs of both strands (19+2), which is especially familiar and convenient for nonexpert users.
The biased strand selection from asymmetric siRNA structures have suggested that strand shortening from the 3′ end of the passenger strand could provide a valuable approach to reducing off-target effects [20–22]; however, such structures might be unfamiliar to many researchers not specialized in this field. In this sense, we believe that the most valuable and strongest point of our siRNA is that it can be adopted easily by nonexpert users. This design feature may provide a feasible approach for nonexpert to rationally designing an efficient RNAi trigger and be beneficial from the point of view of a large-scale manufacturing. We previously reported that a mature RISC formation was largely dependent on the affinity between hAGO2 and the 3′-end structure of the guide strand [12]. Specifically, the guide strand with an RNA overhang incorporates the mature RISC more efficiently, presumably because of an increased affinity towards the PAZ domain of hAGO2 [12]. These observations led us to investigate the effect of the siRNA with an asymmetric RNA/dTdT overhang. Surprisingly, our collective results demonstrate that the siRNA with an asymmetric RNA/dTdT overhang exhibits a distinct preference in favor of a strand with an RNA overhang, thereby considerably increasing a specificity of target silencing. We postulate that the hAGO2-PAZ preferentially binds to the 3′ end of the strand with an RNA overhang and this anchored strand remain associated with hAGO2 to form a mature RISC, whereas the partner strand with a dTdT overhang is less preferred.
Our results demonstrate that our siRNA structure can be applied to specific repression of NAT target pairs. By using the siRNA with an asymmetric RNA/dTdT overhang, we observed up to 100-fold enhanced silencing activity for the intended target based on IC50 value. Our siRNA structure does not require any chemical modification or introduction of mismatches to maximize the target silencing. Along with the fact that the siRNA with a symmetric overhang readily targets both sense and antisense strand, it should also be noted that, by combining the strands with different 3′-overhang, targeting a specific transcript or both transcripts can be adjustable. Considering widespread NATs in mammalian genomes and their functions, including chromatin remodeling and transcriptional interference [23], we believe that our siRNA structure could be a useful tool to investigate the regulatory roles of NATs.
In conclusion, we describe a simple and novel approach of biasing siRNA strand selection from the conventional siRNA structure (19+2) with an asymmetric RNA/dTdT overhang, which requires relatively minimal input from nonexpert users. Nowadays, siRNAs can be routinely ordered from a variety of commercial vendors, and the practical process to design and manufacture the siRNAs needed for a given experiment should be straightforward and rather unsophisticated, especially for industrial-scale production. A simple alteration of overhang structure does not require any intricate chemical modification within an internal 19-bp duplex structure, which therefore provides researchers with a simple and low-cost approach for a large-scale automated synthesis of highly functional siRNAs.
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (Ministry of Education, Science, and Technology; no. 2012R1A2A2A01045528) and the Next-Generation BioGreen 21 Program (no. PJ00820603 and no. PJ00801101), Rural Development Administration, Republic of Korea. This work was also supported by a Global Research Laboratory grant from the Ministry of Education, Science, and Technology of Korea (no. 2008-00582). SWH was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (NRF-2013R1A1A2059693).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K. and Tuschl T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411:494–498 [DOI] [PubMed] [Google Scholar]
- 2.Kim VN. (2005). MicroRNA biogenesis: Coordinated cropping and dicing. Nat Rev Mol Cell Biol 6:376–385 [DOI] [PubMed] [Google Scholar]
- 3.Kawamata T. and Tomari Y. (2010). Making RISC. Trends Biochem Sci 35:368–376 [DOI] [PubMed] [Google Scholar]
- 4.Schwarz DS, Hutvagner G, Du T, Xu Z, Aronin N. and Zamore PD. (2003). Asymmetry in the assembly of the RNAi enzyme complex. Cell 115:199–208 [DOI] [PubMed] [Google Scholar]
- 5.Khvorova A, Reynolds A. and Jayasena SD. (2003). Functional siRNAs and miRNAs exhibit strand bias. Cell 115:209–216 [DOI] [PubMed] [Google Scholar]
- 6.Jackson AL, Bartz SR, Schelter J, Kobayashi SV, Burchard J, Mao M, Li B, Cavet G. and Linsley PS. (2003). Expression profiling reveals off-target gene regulation by RNAi. Nat Biotechnol 21:635–637 [DOI] [PubMed] [Google Scholar]
- 7.Fedorov Y, Anderson EM, Birmingham A, Reynolds A, Karpilow J, Robinson K, Leake D, Marshall WS. and Khvorova A. (2006). Off-target effects by siRNA can induce toxic phenotype. RNA 12:1188–1196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruby JG, Stark A, Johnston WK, Kellis M, Bartel DP. and Lai EC. (2007). Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17:1850–1864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT. and Lai EC. (2008). The regulatory activity of microRNA star species has substantial influence on microRNA and 3′ UTR evolution. Nat Struct Mol Biol 15:354–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamura K, Liu N. and Lai EC. (2009). Distinct mechanisms for microRNA strand selection by Drosophila Argonautes. Mol Cell 36:431–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strapps WR, Pickering V, Muiru GT, Rice J, Orsborn S, Polisky BA, Sachs A. and Bartz SR. (2010). The siRNA sequence and guide strand overhangs are determinants of in vivo duration of silencing. Nucleic Acids Res 38:4788–4797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong SW, Park JH, Yun S, Lee CH, Shin C. and Lee DK. (2014). Effect of the guide strand 3′-end structure on the gene silencing potency of asymmetric siRNA. Biochem J 461:427–434 [DOI] [PubMed] [Google Scholar]
- 13.Martinez J, Patkaniowska A, Urlaub H, Luhrmann R. and Tuschl T. (2002). Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110:563–574 [DOI] [PubMed] [Google Scholar]
- 14.Kawamata T. and Tomari Y. (2011). Native gel analysis for RISC assembly. Methods Mol Biol 725:91–105 [DOI] [PubMed] [Google Scholar]
- 15.Tomari Y, Du T, Haley B, Schwarz DS, Bennett R, Cook HA, Koppetsch BS, Theurkauf WE. and Zamore PD. (2004). RISC assembly defects in the Drosophila RNAi mutant armitage. Cell 116:831–841 [DOI] [PubMed] [Google Scholar]
- 16.Haley B, Tang G. and Zamore PD. (2003). In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30:330–336 [DOI] [PubMed] [Google Scholar]
- 17.Faghihi MA. and Wahlestedt C. (2009). Regulatory roles of natural antisense transcripts. Nat Rev Mol Cell Biol 10:637–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li YY, Qin L, Guo ZM, Liu L, Xu H, Hao P, Su J, Shi Y, He WZ. and Li. YX. (2006). In silico discovery of human natural antisense transcripts. BMC Bioinformatics 7:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hohjoh H. (2004). Enhancement of RNAi activity by improved siRNA duplexes. FEBS Lett 557:193–198 [DOI] [PubMed] [Google Scholar]
- 20.Sun X, Rogoff HA. and Li CJ. (2008). Asymmetric RNA duplexes mediate RNA interference in mammalian cells. Nat Biotechnol 26:1379–1382 [DOI] [PubMed] [Google Scholar]
- 21.Sano M, Sierant M, Miyagishi M, Nakanishi M, Takagi Y. and Sutou S. (2008). Effect of asymmetric terminal structures of short RNA duplexes on the RNA interference activity and strand selection. Nucleic Acids Res 36:5812–5821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang CI, Yoo JW, Hong SW, Lee SE, Kang HS, Sun X, Rogoff HA, Ban C, Kim S, Li CJ. and Lee DK. (2009). Asymmetric shorter-duplex siRNA structures trigger efficient gene silencing with reduced nonspecific effects. Mol Ther 17:725–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li K. and Ramchandran R. (2010). Natural antisense transcript: A concomitant engagement with protein-coding transcript. Oncotarget 1:447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.