Abstract
Research in the deep terrestrial biosphere is driven by interest in novel biodiversity and metabolisms, biogeochemical cycling, and the impact of human activities on this ecosystem. As this interest continues to grow, it is important to ensure that when subsurface investigations are proposed, materials recovered from the subsurface are sampled and preserved in an appropriate manner to limit contamination and ensure preservation of accurate microbial, geochemical, and mineralogical signatures. On February 20th, 2014, a workshop on “Trends and Future Challenges in Sampling The Deep Subsurface” was coordinated in Columbus, Ohio by The Ohio State University and West Virginia University faculty, and sponsored by The Ohio State University and the Sloan Foundation’s Deep Carbon Observatory. The workshop aims were to identify and develop best practices for the collection, preservation, and analysis of terrestrial deep rock samples. This document summarizes the information shared during this workshop.
Keywords: deep biosphere, deep subsurface, drilling, contamination, shale, deep life
INTRODUCTION
It has been estimated that up to 25 × 1029 bacterial cells are present in the terrestrial subsurface, potentially accounting for 40–60% of all bacterial cells on Earth (Whitman et al., 1998; McMahon and Parnell, 2014). The depth limit for life on Earth is unknown, but likely tied to upper temperature limits and the availability of water in terrestrial systems. Microorganisms have been detected in 3.6 km deep groundwater accessed via South African gold mines (Moser et al., 2003), in sub-sea floor sediments (Schrenk et al., 2010), and at almost 4 km beneath ice sheets in Lake Vostok (Priscu et al., 1999). However, given the extent of the deep biosphere, the majority of potential habitats remain almost completely unexplored (Edwards et al., 2012). As such, a series of wide-ranging research questions remain unanswered: what controls the subsurface microbial abundance (McMahon and Parnell, 2014)? What is the taxonomic diversity of these systems (Teske and Sorensen, 2007)? What microbial metabolisms are active across diverse chemical and physical conditions (Orsi et al., 2013)? How do cells survive exceedingly low fluxes of energy and nutrients that lead to extremely slow doubling times, and bring to question the energy requirements for cellular maintenance and repair (Hoehler and Jorgensen, 2013)? How do taxonomically similar microorganisms appear in seemingly isolated deep environments across the Earth (L’Haridon et al., 1995)? How are microorganisms impacted when human activity alters these deep subterranean and oceanic environments? These outstanding questions emphasize the importance of continued deep subsurface research, in both terrestrial and marine systems.
SAMPLE COLLECTION AND CONTAMINATION ASSESSMENT
Recovering material from the subsurface generally requires drilling technologies to reach suitable depths, although in some instances pre-existing infrastructure may be used for sample collection (e.g., South African gold mines). A number of drilling techniques including hollow-stem auger coring, cable-tool coring, and rotary sonic are suitable for shallow sampling in unconsolidated sediments (Kieft, 2010). While these techniques can be used without drilling fluids, thus limiting potential contamination of recovered materials, they are not suitable for recovery of deeper rock and sediments. For accessing deeper materials (> 300 m), rotary drilling is generally used in conjunction with added drilling fluids. This contrasts with drilling in marine sediments, where surrounding ocean water can be used as the drilling fluid. Such fluids are frequently muds (bentonite, and other organic constituents), although formation waters, foams and gasses can be substituted in some instances. Although these fluids are essential to seal the borehole, to cool and lubricate the drill bit, and to adjust density and viscosity with the borehole, they can support extremely high densities of microorganisms, and must be carefully managed when acquiring microbiological samples (Beeman and Suflita, 1989). Preserving in situ geochemical and microbiological signatures during recovery of core material is technically challenging. Returning sediments to the surface from deep locations can take significant time, during which such signatures may change. Rock and sediments exhibiting high porosity and permeability may be particularly at risk to these changes. Technologies are currently being developed to design a freeze-shoe sampler that would enable the freezing of sediment and rock cores in situ during recovery, and thus prevent microbiological and geochemical shifts.
Samples and measurements can be obtained at multiple points during and after borehole drilling. In situ pore water chemistry can be estimated during drilling, via the use of devices that enable “probe-at-the-bit” measurements (Hall et al., 2008). Once a well has been developed, U-tube borehole fluid samplers can be used to remove the drilling fluid or monitor its dilution over time with ground water flow and then collect true formation fluids and gasses at near in situ conditions (Freifeld, 2009; Stotler et al., 2011). Further, geochemical conditions and microbial community structures for specific fractures can be determined through the use of packers that isolate those fractures within a borehole for sampling (Haveman et al., 1999; Shimizu et al., 2006; Purkamo et al., 2013). Finally, significant understanding of the mineralogy and geochemistry of a subsurface environment can be derived from the effective utilization of well log data (Onstott et al., 1998).
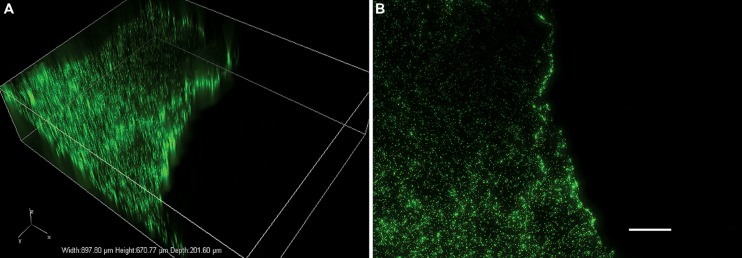
When solid rock and sediment matrices are recovered, a primary concern focuses around potential contamination issues. Contamination can occur at several points during the drilling and coring process. Sources include: surface water used during drilling, air contamination of the mud tanks, additives to the drilling fluid, contaminated surfaces of the mud pumps, core barrels and drill bits, and contamination from overlying formations and groundwater via a process known as drilling drag-down. Due to the extremely low biomass in deep subsurface formations, special care must be taken to minimize microbial contamination as even a small quantity of exogenous bacteria can mask indigenous biomarkers and compromise cultivation and enrichment efforts. Common methods to assess the extent of contamination include the use of chemical, microbiological and particle tracers, with multiple, redundant, tracers recommended to ensure sample integrity (Russell et al., 1992; Kieft et al., 2007). Typical tracers include visual markers such as fluorescein and rhodamine B (Russell et al., 1992; Wandrey et al., 2010), and chemical tracers such as perfluorocarbons (McKinley and Colwell, 1996; Smith et al., 2000; House et al., 2003; Pfiffner et al., 2008; Santelli et al., 2010) and perdeuterated n-octacosane (nC28; Agouron Institute Drilling Project, 2014). Some of these tracers may be added throughout drilling (e.g., fluorescein), some may be changed as suites at discrete formational/depth changes (e.g., perfluorocarbons), while others are applied onto bits and core barrels prior to drilling (e.g., perdeuterated n-octacosane). Fluorescent microspheres 0.5– 1.0 μm diameter can be used as a proxy for bacterial cells, and quantified by microscopy (Kallmeyer et al., 2006; Kieft et al., 2007; Stroes-Gascoyne et al., 2007; Pfiffner et al., 2008; Santelli et al., 2010; Cardace et al., 2013; Yanagawa et al., 2013; Figure 1). Fluorescent microspheres can be added to the drilling fluid but become cost-prohibitive and impractical in the deep subsurface as large volumes of fluids are needed; instead microspheres may be deployed in the core catcher in a plastic bag that ruptures as core material enters the core barrel (Kieft et al., 2007; Pfiffner et al., 2008; Mason et al., 2010; Yanagawa et al., 2013). Microbiological tracers (e.g., active microbial cells) have also been used to assess penetration of microorganisms into core material (Zhang et al., 2005). Finally, total organic carbon (TOC) measurements in recovered material can be a surrogate for contamination from the carboxymethyl cellulose (CMC) component of drilling mud (Wandrey et al., 2010).
FIGURE 1.
Example of fluorescent microspheres (green dots) on shale showing contaminated and uncontaminated regions. The 2.5 by 4 cm shale was exposed to an aqueous solution containing 0.5 μm Fluoresbrite yellow-green microspheres (Polysciences Inc., Warrington, PA, USA) at a concentration of 3.64 × 108 particles/mL. Images were obtained using a Nikon Eclipse Ti inverted microscope at 100x total magnification (10x objective) and NIS Elements v. 4.00.07 software. Image (A) shows a volumetric composite of captured Z-stack images over a depth of 201.60 μm; image (B) shows the same data as a composite maximum-intensity projection. Scale bar = 100 μm.
A rigorous assessment of contamination includes the sub-sampling of all materials coming in contact with cores before, during and after all operational steps (e.g., drilling operations, core retrieval, and sample processing). Example samples for contamination analyses include swabs from surfaces used for drilling, coring, or paring; samples of drilling muds and return cuttings, especially when new formations are encountered; samples of drill bit lubricants; and swabs/samples from core liners (Kieft et al., 2007; Pfiffner et al., 2008). Additionally, the collection of sample “blanks” at multiple points throughout drilling and sampling processing allows for the detection of environmental contaminants (e.g., air, moisture, gloves, glovebag) to distinguish from native microorganisms. Once cores have been analyzed for tracers and potential contamination has been documented, they must be immediately subsampled prior to geochemical and microbiological analyses. Common paring and disaggregation/size reduction methods utilize core extrusion (Russell et al., 1992; Kieft et al., 2007), a hammer and/or chisel, (Pfiffner et al., 2008) circular saws with hydraulic crushing, (Santelli et al., 2010) or mortar and pestle/ball mill (Herrera and Cockell, 2007) depending upon the lithology and subsequent analyses. Field samples must be immediately preserved using appropriate methods to retain competency for subsequent microbial and geochemical analyses. Where non-culturing approaches are to be used, rapid freezing is generally ideal to capture microbial community structures from molecular biomarkers (e.g., nucleic acids, proteins, and lipids). If culturing approaches are to be applied, samples should be maintained at in situ pressures and temperatures or refrigerated and used as soon as possible to prevent outgrowth of organisms (Haldeman et al., 1994; Brockman et al., 1998).
MEASURING MICROBIAL BIOMASS, ACTIVITY, AND COMMUNITY STRUCTURE
A number of techniques can be leveraged to determine microbial community structure, function, biomass concentration, and activity in the deep biosphere. DNA-based analyses are tractable in these environments, and can range from single gene biomarker studies to shotgun community genomic investigations that inform microbial community structure and functional potential (Zhang et al., 2005; Chivian et al., 2008; Wrighton et al., 2012, 2013; Dong et al., 2014). Catalyzed reported deposition fluorescent in situ hybridization (CARD-FISH) has been used in some environments to identify active cells, while demonstrating that DNA is sufficiently intact to hybridize with primers and probes (Hoehler and Jorgensen, 2013). Recently, amino acid-based racemization rates have been used to constrain potential depth limits and temperatures for microbial activity (Onstott et al., 2013). RNA-based analyses for microbial activity are challenging in low-biomass deep terrestrial environments. Messenger RNA signatures may change during sample recovery, although the ability to sample at some deep subsurface locations (e.g., South African gold mines) may enable the rapid preservation of recovered biomass. As discussed earlier, mechanisms for freezing samples during the coring process may offer another solution to preserving signatures that would otherwise change rapidly.
Similarly, lipid biomarker profiles can be determined from intact polar lipids (PLs) or their derived fatty acid methyl esters to provide estimates of biomass and determine the relative abundance of taxonomic groups, including Eukaryotes, Bacteria, and Archaea (Fredrickson et al., 1995; White et al., 1996; White and Ringelberg, 1997; Schubotz et al., 2009). Polar lipid analyses can be used to estimate total biomass and the proportion of viable versus dead cells (Balkwill et al., 1988; Findlay et al., 1989; Fredrickson et al., 1995; White and Ringelberg, 1997). In conjunction with taxonomic identification, these analyses can be used to infer microbial phenotypic states as they relate to environmental conditions, and are therefore very useful for subsurface studies. For example, enriched trans and cyclopropyl fatty acids have been used to indicate microbial responses to stress and toxicity (Heipieper et al., 1992; White and Ringelberg, 1997), while higher proportions of cyclopropyl fatty acids versus monounsaturated and saturated fatty acids have been used to indicate microbial starvation (Guckert et al., 1986; Kieft et al., 1994). Additionally, respiratory quinones have been employed to infer environmental redox potentials (Hedrick and White, 1986; White and Ringelberg, 1997) and may be useful in environments where direct geochemical measurements are difficult to obtain. Finally, isotope signatures in PLs offer a valuable indicator of microbial function in deep subsurface environments. Isotopic compositions are determined using a gas chromatograph combustion interface isotope ratio mass spectrometer (GC-C-IRMS).
Deoxyribonuclicacid- and biomarker-based analyses require the extraction of biological material from subsurface cores, a process complicated by low biomass concentration, chemical, and physical factors (Herrera and Cockell, 2007). The nature of the matrix itself, which is often characterized by low porosity, carbonate precipitates, and brine minerals and fluids, impacts the amount and quality of extractable DNA, RNA, and lipids (Nielsen and Petersen, 2000; Herrera and Cockell, 2007; Wu et al., 2009). In many systems, cells are encased in the physical matrix and this directly affects the choice of subsampling to optimize extraction efficiency. DNA is usually extracted from environmental samples by direct cell lysis, using chemical or physical lysis, or a combination of both (Zhou et al., 1996; Griffiths et al., 2000; Hurt et al., 2001; Barton et al., 2006). Although many commercial DNA extraction kits have been developed to increase extraction reproducibility and yield, it is recommended that multiple methods be tested and compared, either on actual sample material or chemically similar samples (Barton et al., 2006; Herrera and Cockell, 2007; Novinscak and Filion, 2011; Direito et al., 2012; Paulin et al., 2013). DNA sorption onto mineral surfaces is a significant problem with low-biomass samples; although blocking agents or carrier molecules have been shown to help overcome this challenge (Barton et al., 2006; Direito et al., 2012).
Microbial activity in recovered material can be measured through laboratory batch enrichments or continuous flow-through experiments. Given that both pressure and temperature increase with depth in terrestrial subsurface environments, microorganisms living in such environments must be able to tolerate, survive, and even proliferate under these conditions. Hydrostatic pressure-adapted microorganisms are known as piezophiles, and have optimal growth rates at pressures greater than 0.1 MPa, while hyperpiezophiles require pressures > 60 MPa for optimal growth (Bartlett, 2002). Growth experiments at these pressures are therefore often desirable to obtain relevant data on cultivable organisms (Zeng et al., 2009). High-pressure devices allow recovered samples to be maintained at in situ pressures and temperatures. Some of these systems require no de-pressurization following core recovery, and allow pressurized material to be subsampled and incubated under in situ conditions (Kyo et al., 1991; Parkes et al., 2009). Other pressure core samplers have been designed to recover material from depth at in situ pressures (Pettigrew, 1992), and have been effectively used in the oceanic deep subsurface (Dickens et al., 2003). These samplers require de-pressurization for subsampling. U-tube samplers can acquire fluid and microbial samples at formation pressure and are readily adaptable to subsampling (Freifeld, 2009; Stotler et al., 2011). Although rapid de-pressurization can result in cell death, slower-rates of de-pressurization do not necessarily cause lethal damage to piezophiles (Yayanos and Dietz, 1983; Chastain and Yayanos, 1991; Park and Clark, 2002). Resulting material can be re-pressurized to desired pressures using relatively simple equipment, such as modified Hungate tubes (Bowles et al., 2011) or pressure bags (Kato et al., 2008) inside high pressure stainless steel vessels, enabling microbial batch cultivation in the laboratory. Continuous flow-through high-pressure reactors have also been developed, and recently used for determining rates of anaerobic methane oxidation (AOM; Deusner et al., 2010; Zhang et al., 2011). The ability to culture microbial assemblages under environmentally representative conditions enables rate measurements for microbially catalyzed reactions to be determined (Brockman et al., 1998; Deusner et al., 2010; Tamegai et al., 2012), while enrichment of specific microbial groups can be used to obtain either pure cultures or enriched microbial consortia. Resulting biomass provides abundant material for omics-based analyses of functional potential, such as transcript expression or proteomics. In addition, cultivation at high pressures and temperatures can be used to remove contaminant species that are unlikely to tolerate such extreme conditions.
PHYSICAL AND CHEMICAL CHARACTERIZATION OF ROCK CORE AND FLUIDS
In recovered rock samples, the development of linkages between pore structure and microbial parameters is key for understanding the distribution of microbial communities. In fine-grained shale systems, source rocks have low porosities and extremely low permeabilities, on the order of nanodarcies (Javadpour, 2009; Sondergeld et al., 2010). Understanding the microstructural controls on porosity and permeability has implications for the nature of biodiversity in such systems, in that it governs the movement of cells and chemicals within the rock. Conversely, other host rocks such as sandstone and carbonate systems can have higher porosities and permeabilities, with greater potential for microbial and chemical transport through fractures and matrix pores in such formations (Fredrickson et al., 1997; Dong et al., 2014). Currently, a range of new imaging technologies can be used in concert with more conventional characterization methods (Bryndzia and Braunsdorf, 2014). The advent of focused ion beam – scanning electron microscopic (FIB – SEM) techniques now allows us to image pore networks in the rock matrix (Curtis et al., 2012). This 3-dimensional method is part of a broader suite of instruments that image rock samples including X-ray and neutron computed tomography – XCT and NCT (Perfect et al., 2014). Recently, the application of Small and ultra-small angle neutron scattering (USANS) has proved a valuable complement for the analysis of porosity and pore connectivity at the nanometer to the centimeter scale (Anovitz et al., 2013; Jin et al., 2013). Sample preparation for X-ray and neutron tomography of native rock core only requires the material to be sized (length and diameter) according to the desired resolution of the instrument – e.g., imaging the pore features < 1 micron requires samples ranging from a few to 10’s of mm3. Neutron scattering is conducted on ~150 micron-thick polished sections pressure impregnated with epoxy and mounted on 1 × 2 inch quartz slides (Anovitz et al., 2013). For higher resolution assessment down to the nanoscale by dual beam-FIB, a small chip or core roughly a few mm’s on a side or diameter, respectively, is used (Curtis et al., 2012).
Given the highly heterogeneous nature of pore networks and fractures in many rock types and samples, determining spatial aspects of microbial activity is important, yet technically difficult. In a novel experimental setup, microautoradiography techniques were applied to core material to determine spatial locations for microbial sulfate reduction. In situ 35S-sulfate reduction was monitored using freshly fractured cores wrapped in silver foil (Russell et al., 1992; Fredrickson et al., 1997; Krumholz et al., 1997). 35S-sulfide was retained on the foil and offered a two-dimensional image of discrete pockets of microbial activity that could be mapped to physical and chemical characterizations of the core. These activity measurements can be directly related to cation and anion analyses of pore waters trapped in cores using crush and leach methods and correcting for drilling fluid contamination using tracers. Stable N-isotope analyses have become sensitive enough to obtain the N and O isotopic compositions of pore water trapped in rock with only 1% porosity (Silver et al., 2012). Formation gas composition and its isotopic signatures provide valuable information on whether methanogenesis is taking place within the formation and is typically monitored during drilling. The pore water gas compositions of cores can be measured by quickly transferring intact cores into evacuated leak-tight cylinders. The cores then degas into the cylinders and are sampled for gas composition and even for noble gas dating of the pore water (Lippmann-Pipke et al., 2011).
Other chemical signatures in rock matrices can be analyzed using isotope analysis tools to infer carbon pools, or determine potential microbial activity. The carbon isotopic composition of TOC is an excellent indicator for determining the source and type of organic matter, given that diagenetic alterations or removal of organic matter pools does not significantly affect the δ13C of bulk organic matter in sedimentary rocks, particularly in black shales (Meyers, 1994; Bekker et al., 2008; Young et al., 2008; Ader et al., 2009; Jiang et al., 2012). Conversely, N and S isotope fractionations in such media are closely associated with microbial processes like sulfate reduction and denitrification (Berner, 1978; Gautier, 1986; Beier and Hayes, 1989; Altabet, 2006; Quan et al., 2008). The bulk δ13C and δ34S also serve as excellent tracers for microbial oxidation of methane via sulfate reduction as this process leaves the sulfate pool enriched in 34S while adding light 12C to the total carbon pool (Kemp and Thode, 1968; Jahnke et al., 1995; Freeman, 2001; Seal, 2006). The C, N, and S isotopes are conservative and do not get altered by exposure of the core to air and mild temperature changes during storage. However, as with previously described analysis techniques, microbial contamination from drilling fluids and muds and their potential to impact paleoenvironmental signals is best avoided by using material from inner portions of recovered cores.
FUTURE CHALLENGES
Despite advances in deep biosphere sampling techniques, and development of high-resolution molecular analysis tools, a range of challenges still exists in understanding these environments. How representative are collected samples? How can samples be better preserved for downstream analyses? Advances are currently being made in sample collection efforts, with development of freeze-shoe samplers that can freeze sediment cores in situ. Perhaps the greatest challenge is developing a predictive understanding of microbial processes in such environments based on a limited number of expensive, difficult to collect vertical borehole samples. Characterization will require greater linkages between biogeochemical, geophysical, mineralogical, and microbiological data, and the presentation of these results in a regional and global context. Additional research is also needed to determine the effects of engineered activities (e.g., hydraulic fracturing, geologic CO2 sequestration) on shale and other rock-hosted biodiversity, as the scale of these activities has the potential to promote significant change within the terrestrial subsurface. Using best practices for the collection, preservation, and analysis of biological and chemical signatures of these samples is key to advancing our understanding of the deep biosphere.
WORKSHOP PARTICIPANTS
David R. Cole (Coorganizer), Paula J. Mouser (Coorganizer), Shihka Sharma (Coorganizer), Michael J. Wilkins (Coorganizer), Kelly C. Wrighton (Coorganizer), Jennifer F. Biddle, Elizabeth H. Denis, Jim K. Fredrickson, Thomas L. Kieft, Tullis C. Onstott, Lee Peterson, Susan M. Pfiffner, Tommy J. Phelps, Matthew O. Schrenk.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
REFERENCES
- Ader M., Macouin M., Trindade R., Hadrien M., Yang Z., Sun Z., et al. (2009). A multilayered water column in the Ediacaran Yangtze platform? Insights from carbonate and organic matter paired δ13C. Earth Planet. Sci. Lett. 288 213–227 10.1016/j.epsl.2009.09.024 [DOI] [Google Scholar]
- Agouron Institute Drilling Project. (2014). Sampling Protocols. Available at: http://agourondrillingproject.wordpress.com/sampling-protocols/ [Google Scholar]
- Altabet M. A. (2006). “Isotopic tracers of the marine nitrogen cycle: present and past,” in The Handbook of Environmental Chemistry Vol. 2: Marine Organic Matter: Biomarkers, Isotopes and DNA (Berlin: Springer; ) 251–293 [Google Scholar]
- Anovitz L. M., Cole D. R., Rother G., Allard L. F., Jackson A. J., Littrell K. C. (2013). Diagenetic changes in macro- to nano-scale porosity in the St. Peter Sandstone: an (ultra) small angle neutron scattering and backscattered electron imaging analysis. Geochim. Cosmochim. Acta 102 280–305 10.1016/j.gca.2012.07.035 [DOI] [Google Scholar]
- Balkwill D., Leach F., Wilson J., Mcnabb J., White D. (1988). Equivalence of microbial biomass measures based on membrane lipid and cell wall components, adenosine triphosphate, and direct counts in subsurface aquifer sediments. Microb. Ecol. 16 73–84 10.1007/BF02097406 [DOI] [PubMed] [Google Scholar]
- Bartlett D. H. (2002). Pressure effects on in vivo microbial processes. Biochim. Biophys. Acta 1595 367–381 10.1016/S0167-4838(01)00357-0 [DOI] [PubMed] [Google Scholar]
- Barton H. A., Taylor N. M., Lubbers B. R., Pemberton A. C. (2006). DNA extraction from low-biomass carbonate rock: an improved method with reduced contamination and the low-biomass contaminant database. J. Microbiol. Methods 66 21–31 10.1016/j.mimet.2005.10.005 [DOI] [PubMed] [Google Scholar]
- Beeman R. E., Suflita J. M. (1989). Evaluation of deep subsurface sampling procedures using serendipitous microbial contaminants as tracer organisms. Geomicrobiol. J. 7 223–233 10.1080/01490458909377868 [DOI] [Google Scholar]
- Beier J., Hayes J. (1989). Geochemical and isotopic evidence for paleoredox conditions during deposition of the Devonian-Mississippian New Albany Shale, southern Indiana. Geol. Soc. Am. Bull. 101 774–782 [DOI] [PubMed] [Google Scholar]
- Bekker A., Holmden C., Beukes N., Kenig F., Eglinton B., Patterson W. (2008). Fractionation between inorganic and organic carbon during the Lomagundi (2.22–2.1 Ga) carbon isotope excursion. Earth Planet. Sci. Lett. 271 278–291 10.1016/j.epsl.2008.04.021 [DOI] [Google Scholar]
- Berner R. A. (1978). Sulfate reduction and the rate of deposition of marine sediments. Earth Planet. Sci. Lett. 37 492–498 10.1016/0012-821X(78)90065-1 [DOI] [Google Scholar]
- Bowles M. W., Samarkin V. A., Joye S. B. (2011). Improved measurement of microbial activity in deep-sea sediments at in situ pressure and methane concentration. Limnol. Oceanogr. 9 499–506 10.4319/lom.2011.9.499 [DOI] [Google Scholar]
- Brockman F. J., Li S. W., Fredrickson J. K., Ringelberg D. B., Kieft T. L., Spadoni C. M., et al. (1998). Post-sampling changes in microbial community composition and activity in a subsurface paleosol. Microb. Ecol. 36 152–164 10.1007/s002489900102 [DOI] [PubMed] [Google Scholar]
- Bryndzia L. T., Braunsdorf N. R. (2014). From source rock to reservoir – the evolution of self-sourced unconventional resource plays. Elements 10 271–276 10.2113/gselements.10.4.271 [DOI] [Google Scholar]
- Cardace D., Hoehler T., Mccollom T., Schrenk M., Carnevale D., Kubo M., et al. (2013). Establishment of the Coast Range ophiolite microbial observatory (CROMO): drilling objectives and preliminary outcomes. Sci. Dril. 16 45–55 10.5194/sd-16-45-2013 [DOI] [Google Scholar]
- Chastain R. A., Yayanos A. A. (1991). Ultrastructural changes in an obligately barophilic marine bacterium after decompression. Appl. Environ. Microbiol. 57 1489–1497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chivian D., Brodie E. L., Alm E. J., Culley D. E., Dehal P. S., Desantis T. Z., et al. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322 275–278 10.1126/science.1155495 [DOI] [PubMed] [Google Scholar]
- Curtis M. E., Sondergeld C. H., Ambrose R. J., Rai C. S. (2012). Microstructural investigation of gas shales in two and three dimensions using nanometer-scale resolution imaging. Am. Assoc. Pet. Geol. Bull. 96665–677 [Google Scholar]
- Deusner C., Meyer V., Ferdelman T. G. (2010). High-pressure systems for gas-phase free continuous incubation of enriched marine microbial communities performing anaerobic oxidation of methane. Biotechnol. Bioeng. 105 524–533 10.1002/bit.22553 [DOI] [PubMed] [Google Scholar]
- Dickens G. R., Schroeder D., Hinrichs K.-U. and the Leg 201 Scientific Party (2003). “The pressure core samples (PCS) on ODP leg 201: general operations and gas release,” in Proceedings of the Ocean Drilling Program, Initial Reports eds D’hondt S. L., Jorgensen B. B., Miller D. J. (College Station, TX: Ocean Drilling Program; ). Available at: http://www-odp.tamu.edu/publications/201_IR/chap_03/chap_03.htm [Google Scholar]
- Direito S. O. L., Marees A., Röling W. F. M. (2012). Sensitive life detection strategies for low-biomass environments: optimizing extraction of nucleic acids adsorbing to terrestrial and Mars analogue minerals. FEMS Microbiol. Ecol. 81 111–123 10.1111/j.1574-6941.2012.01325.x [DOI] [PubMed] [Google Scholar]
- Dong Y., Kumar C. G., Chia N., Kim P.-J., Miller P. A., Price N. D., et al. (2014). Halomonas sulfidaeris-dominated microbial community inhabits a 1.8 km-deep subsurface Cambrian Sandstone reservoir. Environ. Microbiol. 16 1695–1708 10.1111/1462-2920.12325 [DOI] [PubMed] [Google Scholar]
- Edwards K. J., Becker K., Colwell F. (2012). “The deep, dark energy biosphere: intraterrestrial life on earth,” in Annual Review of Earth and Planetary Sciences Vol. 40 ed. Jeanloz R. (Palo Alto: Annual Reviews; ) 551–568 [Google Scholar]
- Findlay R. H., King G. M., Watling L. (1989). Efficacy of phospholipid analysis in determining microbial biomass in sediments. Appl. Environ. Microbiol. 55 2888–2893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson J. K., Mckinley J. P., Bjornstad B. N., Long P. E., Ringelberg D. B., White D. C., et al. (1997). Pore-size constraints on the activity and survival of subsurface bacteria in a late cretaceous shale-sandstone sequence, northwestern New Mexico. Geomicrobiol. J. 14 183–202 10.1080/01490459709378043 [DOI] [Google Scholar]
- Fredrickson J. K., Mckinley J. P., Nierzwicki-Bauer S. A., White D. C., Ringelberg D. B., Rawson S. A., et al. (1995). Microbial community structure and biogeochemistry of Miocene subsurface sediments: implications for long-term microbial survival. Mol. Ecol. 4 619–626 10.1111/j.1365-294X.1995.tb00262.x [DOI] [Google Scholar]
- Freeman K. H. (2001). Isotopic biogeochemistry of marine organic carbon. Rev. Mineral. Geochem. 43 579–605 10.2138/gsrmg.43.1.579 [DOI] [Google Scholar]
- Freifeld B. (2009). The U-tube: a new paradigm for borehole fluid sampling. Sci. Drill. 8 41–45 10.5194/sd-8-41-2009 [DOI] [Google Scholar]
- Gautier D. L. (1986). Cretaceous shales from the western interior of North America: sulfur/carbon ratios and sulfur-isotope composition. Geology 14 225–228 [DOI] [Google Scholar]
- Griffiths R. I., Whiteley A. S., Donnell A. G., Bailey M. J. (2000). Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA- and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66 5488–5491 10.1128/AEM.66.12.5488-5491.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guckert J. B., Hood M. A., White D. C. (1986). Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl. Environ. Microbiol. 52 794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldeman D. L., Amy P. S., White D. C., Ringelberg D. B. (1994). Changes in bacteria recoverable from subsurface volcanic rock samples during storage at 4∘C. Appl. Environ. Microbiol. 60 2697–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall D. R., Leany F., Turner P., Durrand C., Lundgreen D., Wise D. (2008). “Drill bit assembly with a probe.” (Google Patents). Available at: http://www.google.com/patents/US20070114061 [Google Scholar]
- Haveman S. A., Pedersen K., Ruotsalainen P. (1999). Distribution and metabolic diversity of microorganisms in deep igneous rock aquifers of Finland. Geomicrobiol. J. 16 277–294 10.1080/014904599270541 [DOI] [Google Scholar]
- Hedrick D. B., White D. C. (1986). Microbial respiratory quinones in the environment: I. A sensitive liquid chromatographic method. J. Microbiol. Methods 5 243–254 10.1016/0167-7012(86)90049-7 [DOI] [Google Scholar]
- Heipieper H. J., Diefenbach R., Keweloh H. (1992). Conversion of cis unsaturated fatty acids to trans, a possible mechanism for the protection of phenol-degrading Pseudomonas putida P8 from substrate toxicity. Appl. Environ. Microbiol. 58 1847–1852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera A., Cockell C. S. (2007). Exploring microbial diversity in volcanic environments: a review of methods in DNA extraction. J. Microbiol. Methods 70 1–12 10.1016/j.mimet.2007.04.005 [DOI] [PubMed] [Google Scholar]
- Hoehler T. M., Jorgensen B. B. (2013). Microbial life under extreme energy limitation. Nat. Rev. Microbiol. 11 83–94 10.1038/nrmicro2939 [DOI] [PubMed] [Google Scholar]
- House C. H., Cragg B. A., Teske A. (2003). “Drilling contamination tests during ODP leg 201 using chemical and particulate tracers,” in Proceedings of the Ocean Drilling Program, Initial Reports Vol. 201 eds D’Hondt S. L., Jørgensen B. B., Miller D. J., et al. (College Station, TX: Ocean Drilling Program; ) 1–19 [Google Scholar]
- Hurt R. A., Qiu X., Wu L., Roh Y., Palumbo A. V., Tiedje J. M., et al. (2001). Simultaneous recovery of RNA and DNA from soils and sediments. Appl. Environ. Microbiol. 67 4495–4503 10.1128/AEM.67.10.4495-4503.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahnke L. L., Summons R. E., Dowling L. M., Zahiralis K. D. (1995). Identification of methanotrophic lipid biomarkers in cold-seep mussel gills: chemical and isotopic analysis. Appl. Environ. Microbiol. 61 576–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Javadpour F. (2009). Nanopores and apparent permeability of gas flow in mudrocks (shales and siltstone). J. Can. Petrol. Technol. 48 16–21 10.2118/09-08-16-DA [DOI] [Google Scholar]
- Jiang G., Wang X., Shi X., Xiao S., Zhang S., Dong J. (2012). The origin of decoupled carbonate and organic carbon isotope signatures in the early Cambrian (ca. 542–520Ma) Yangtze platform. Earth Planet. Sci. Lett. 317 96–110 10.1016/j.epsl.2011.11.018 [DOI] [Google Scholar]
- Jin L., Mathur R., Rother G., Cole D., Bazilevskaya E., Williams J., et al. (2013). Evolution of porosity and geochemistry in Marcellus Formation black shale during weathering. Chem. Geol. 356 50–63 10.1016/j.chemgeo.2013.07.012 [DOI] [Google Scholar]
- Kallmeyer J., Mangelsdorf K., Cragg B., Horsfield B. (2006). Techniques for contamination assessment during drilling for terrestrial subsurface sediments. Geomicrobiol. J. 23 227–239 10.1080/01490450600724258 [DOI] [Google Scholar]
- Kato C., Nogi Y., Arakawa S. (2008). “Isolation, cultivation, and diversity of deep-sea piezophiles,” in High-Pressure Microbiology eds Michiels C., Bartlett D. H., Aertsen A. (Washington, DC: ASM Press; ) 203–217 [Google Scholar]
- Kemp A., Thode H. (1968). The mechanism of the bacterial reduction of sulphate and of sulphite from isotope fractionation studies. Geochim. Cosmochim. Acta 32 71–91 10.1016/0016-7037(68)90088-4 [DOI] [Google Scholar]
- Kieft T. L. (2010). “Sampling the deep sub-surface using drilling and coring techniques,” in Handbook of Hydrocarbon and Lipid Microbiology ed. Timmis K. (Berlin Heidelberg: Springer; ) 3427–3441 10.1007/978-3-540-77587-4_267 [DOI] [Google Scholar]
- Kieft T. L., Phelps T. J., Fredrickson J. K. (2007). Drilling, coring, and sampling subsurface environments. Man. Environ. Microbiol. 799–817 [Google Scholar]
- Kieft T. L., Ringelberg D. B., White D. C. (1994). Changes in ester-linked phospholipid fatty acid profiles of subsurface bacteria during starvation and desiccation in a porous medium. Appl. Environ. Microbiol. 60 3292–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krumholz L. R., Mckinley J. P., Ulrich G. A., Suflita J. M. (1997). Confined subsurface microbial communities in cretaceous rock. Nature 386 64–66 10.1038/386064a0 [DOI] [Google Scholar]
- Kyo M., Tuji T., Usui H., Itoh T. (1991). Colletion, isolation and cultivation system for deep-sea microbes study: concept and design. Oceans 1 419–423 [Google Scholar]
- L’Haridon S., Reysenbacht A. L., Glenat P., Prieur D., Jeanthon C. (1995). Hot subterranean biosphere in a continental oil reservoir. Nature 377 223–224 10.1038/377223a0 [DOI] [PubMed] [Google Scholar]
- Lippmann-Pipke J., Sherwood Lollar B., Niedermann S., Stronick N., Naumann R., Vanheerden E., et al. (2011). 2 Ga old crustal neon component implies long water-rock reaction times of deep fracture water. Chem. Geol. 283 287–296 10.1016/j.chemgeo.2011.01.028 [DOI] [Google Scholar]
- Mason O. U., Nakagawa T., Rosner M., Van Nostrand J. D., Zhou J., Maruyama A., et al. (2010). First investigation of the microbiology of the deepest layer of ocean crust. PLoS ONE 5:e15399 10.1371/journal.pone.0015399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinley J. P., Colwell F. S. (1996). Application of perfluorocarbon tracers to microbial sampling in subsurface environments using mud-rotary and air-rotary drilling techniques. J. Microbiol. Methods 26 1–9 10.1016/0167-7012(96)00826-3 [DOI] [Google Scholar]
- McMahon S., Parnell J. (2014). Weighing the deep continental biosphere. FEMS Microbiol. Ecol. 87 113–120 10.1111/1574-6941.12196 [DOI] [PubMed] [Google Scholar]
- Meyers P. A. (1994). Preservation of elemental and isotopic source identification of sedimentary organic matter. Chem. Geol. 114 289–302 10.1016/0009-2541(94)90059-0 [DOI] [Google Scholar]
- Moser D. P., Onstott T. C., Fredrickson J. K., Brockman F. J., Balkwill D. L., Drake G. R., et al. (2003). Temporal shifts in the geochemistry and microbial community structure of an ultradeep mine borehole following isolation. Geomicrobiol. J. 20 517–548 10.1080/713851170 [DOI] [Google Scholar]
- Nielsen P., Petersen S. O. (2000). Ester-linked polar lipid fatty acid profiles of soil microbial communities: a comparison of extraction methods and evaluation of interference from humic acids. Soil Biol. Biochem. 32 1241–1249 10.1016/S0038-0717(00)00041-9 [DOI] [Google Scholar]
- Novinscak A., Filion M. (2011). Effect of soil clay content on RNA isolation and on detection and quantification of bacterial gene transcripts in soil by quantitative reverse transcription-PCR. Appl. Environ. Microbiol. 77 6249–6252 10.1128/AEM.00055-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onstott T. C., Magnabosco C., Aubrey A. D., Burton A. S., Dworkin J. P., Elsila J. E., et al. (2013). Does aspartic acid racemization constrain the depth limit of the subsurface biosphere? Geobiology 12 1–19 10.1111/gbi.12069 [DOI] [PubMed] [Google Scholar]
- Onstott T. C., Phelps T. J., Colwell F. S., Ringelberg D., White D. C., Boone D. R., et al. (1998). Observations pertaining to the origin and ecology of microorganisms recovered from the deep subsurface of Taylorsville Basin, Virginia. Geomicrobiol. J. 15 353–385 10.1080/01490459809378088 [DOI] [Google Scholar]
- Orsi W. D., Edgcomb V. P., Christman G. D., Biddle J. F. (2013). Gene expression in the deep biosphere. Nature 499 205–208 10.1038/nature12230 [DOI] [PubMed] [Google Scholar]
- Park C. B., Clark D. S. (2002). Rupture of the cell envelope by decompression of the deep-sea methanogen Methanococcus jannaschii. Appl. Environ. Microbiol. 68 1458–1463 10.1128/AEM.68.3.1458-1463.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkes R. J., Sellek G., Webster G., Martin D., Anders E., Weightman A. J., et al. (2009). Culturable prokaryotic diversity of deep, gas hydrate sediments: first use of a continuous high-pressure, anaerobic, enrichment and isolation system for subseafloor sediments (DeepIsoBUG). Environ. Microbiol. 11 3140–3153 10.1111/j.1462-2920.2009.02018.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulin M. M., Nicolaisen M. H., Jacobsen C. S., Gimsing A. L., Sørensen J., Bælum J. (2013). Improving Griffith’s protocol for co-extraction of microbial DNA and RNA in adsorptive soils. Soil Biol. Biochem. 63 37–49 10.1016/j.soilbio.2013.02.007 [DOI] [Google Scholar]
- Perfect E., Cheng C. L., Kang M., Bilheux H. Z., Lamanna J. M., Gragg M. J., et al. (2014). Neutron imaging of hydrogen-rich fluids in geomaterials and engineered porous media: a review. Earth Sci. Rev. 129 120–135 10.1016/j.earscirev.2013.11.012 [DOI] [Google Scholar]
- Pettigrew T. L. (1992). “The design and operation of a wireline pressure core sampler (PCS),” in Ocean Drilling Program Technical Note, No 17 (College Station, TX: A&M University; ). [Google Scholar]
- Pfiffner S. M., Onstott T. C., Ruskeeniemi T., Talikka M., Bakermans C., Mcgown D., et al. (2008). Challenges for coring deep permafrost on Earth and Mars. Astrobiology 8 623–638 10.1089/ast.2007.0159 [DOI] [PubMed] [Google Scholar]
- Priscu J. C., Adams E. E., Lyons W. B., Voytek M. A., Mogk D. W., Brown R. L., et al. (1999). Geomicrobiology of subglacial ice above Lake Vostok, Antarctica. Science 286 2141–2144 10.1126/science.286.5447.2141 [DOI] [PubMed] [Google Scholar]
- Purkamo L., Bomberg M., Nyyssönen M., Kukkonen I., Ahonen L., Kietäväinen R., et al. (2013). Dissecting the deep biosphere: retrieving authentic microbial communities from packer-isolated deep crystalline bedrock fracture zones. FEMS Microbiol. Ecol. 85 324–337 10.1111/1574-6941.12126 [DOI] [PubMed] [Google Scholar]
- Quan T. M., Van De Schootbrugge B., Field M. P., Rosenthal Y., Falkowski P. G. (2008). Nitrogen isotope and trace metal analyses from the Mingolsheim core (Germany): evidence for redox variations across the Triassic-Jurassic boundary. Global Biogeochem. Cycles 22 GB2014 10.1029/2007GB002981 [DOI] [Google Scholar]
- Russell B. F., Phelps T. J., Griffin W. T., Sargent K. A. (1992). Procedures for sampling deep subsurface microbial communities in unconsolidated sediments. Ground Water Monit. Remediat. 12 96–104 10.1111/j.1745-6592.1992.tb00414.x [DOI] [Google Scholar]
- Santelli C. M., Banerjee N., Bach W., Edwards K. J. (2010). Tapping the subsurface ocean crust biosphere: low biomass and drilling-related contamination calls for improved quality controls. Geomicrobiol. J. 27 158–169 10.1080/01490450903456780 [DOI] [Google Scholar]
- Schrenk M. O., Huber J. A., Edwards K. J. (2010). Microbial provinces in the subseafloor. Annu. Rev. Mar. Sci. 2 279–304 10.1146/annurev-marine-120308-081000 [DOI] [PubMed] [Google Scholar]
- Schubotz F., Wakeham S. G., Lipp J. S., Fredricks H. F., Hinrichs K.-U. (2009). Detection of microbial biomass by intact polar membrane lipid analysis in the water column and surface sediments of the Black Sea. Environ. Microbiol. 11 2720–2734 10.1111/j.1462-2920.2009.01999.x [DOI] [PubMed] [Google Scholar]
- Seal R. R. (2006). Sulfur isotope geochemistry of sulfide minerals. Rev. Miner. Geochem. 61 633–677 10.2138/rmg.2006.61.12 [DOI] [Google Scholar]
- Shimizu S., Akiyama M., Ishijima Y., Hama K., Kunimaru T., Naganuma T. (2006). Molecular characterization of microbial communities in fault-bordered aquifers in the Miocene formation of northernmost Japan. Geobiology 4 203–213 10.1111/j.1472-4669.2006.00077.x [DOI] [Google Scholar]
- Silver B. J., Raymond R., Sigman D. M., Prokopeko M., Sherwood Lollar B., Lacrampe-Couloume G., et al. (2012). The origin of NO3- and N2 in deep subsurface fracture water of South Africa. Chem. Geol. 294–295 51–62 10.1016/j.chemgeo.2011.11.017 [DOI] [Google Scholar]
- Smith D. C., Spivack A. J., Fisk M. R., Haveman S. A., Staudigel H. and the Leg 185 Shipboard Scientific Party. (2000). “Methods for quantifying potential microbial contamination during deep ocean coring,” in Ocean Drilling Program Technical Note, No 28 (College Station, TX: A&M University; ). 1–19 Available at: http://www-odp.tamu.edu/publications/tnotes/tn28/INDEX.HTM [Google Scholar]
- Sondergeld C. H., Newsham K. E., Comisky J. T., Rice M. C., Rai C. S. (2010). “Petrophysical considerations in evaluating and producing shale gas resources,” in Proceedings of the SPE Unconventional Gas Conference (Pittsburgh, PA: Society of Petroleum Engineers; ) 34 10.2118/131768-MS [DOI] [Google Scholar]
- Stotler R. L., Frape S. K., Freifeld B. M., Holden B., Onstott T. C., Ruskeeniemi T., et al. (2011). Hydrogeology, chemical and microbial activity measurement through deep permafrost. Ground Water 49 348–364 10.1111/j.1745-6584.2010.00724.x [DOI] [PubMed] [Google Scholar]
- Stroes-Gascoyne S., Schippers A., Schwyn B., Poulain S., Sergeant C., Simonoff M., et al. (2007). Microbial community analysis of opalinus clay drill core samples from the mont terri underground research laboratory, Switzerland. Geomicrobiol. J. 24 1–17 10.1080/01490450601134275 [DOI] [Google Scholar]
- Tamegai H., Nishikawa S., Haga M., Bartlett D. H. (2012). The respiratory system of the piezophile Photobacterium profundum SS9 grown under various pressures. Biosci. Biotech. Biochem. 8 1506–1510 10.1271/bbb.120237 [DOI] [PubMed] [Google Scholar]
- Teske A., Sorensen K. B. (2007). Uncultured archaea in deep marine subsurface sediments: have we caught them all? ISME J. 2 3–18 10.1038/ismej.2007.90 [DOI] [PubMed] [Google Scholar]
- Wandrey M., Morozova D., Zettlitzer M., Würdemann H. (2010). Assessing drilling mud and technical fluid contamination in rock core and brine samples intended for microbiological monitoring at the CO2 storage site in Ketzin using fluorescent dye tracers. Int. J. Greenh. Gas Control 4 972–980 10.1016/j.ijggc.2010.05.012 [DOI] [Google Scholar]
- White D. C., Ringelberg D. B. (1997). “Utility of the signature lipid biomarker analysis in determining in situ viable biomass, community structure, and nutritional/physiologic status of deep subsurface microbiota,” in Microbiology of the Terrestrial Deep Subsurface eds Amy P. S., Haldeman D. L. (Boca Raton, FL: CRC Press; ) 119–136 [Google Scholar]
- White D. C., Stair J. O., Ringelberg D. B. (1996). Quantitative comparisons of in situ microbial biodiversity by signature biomarker analysis. J. Ind. Microbiol. 17 185–196 10.1007/BF01574692 [DOI] [Google Scholar]
- Whitman W. B., Coleman D. C., Wiebe W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95 6578–6583 10.1073/pnas.95.12.6578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton K. C., Castelle C. J., Wilkins M. J., Hug L. A., Sharon I., Thomas B. C., et al. (2013). Metabolic interdependencies between phylogenetically novel fermenters and respiratory organisms in an unconfined aquifer. ISME J. 8 1452–1463 10.1038/ismej.2013.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrighton K. C., Thomas B. C., Sharon I., Miller C. S., Castelle C. J., Verberkmoes N. C., et al. (2012). Fermentation, hydrogen, and sulfur metabolism in multiple uncultivated bacterial phyla. Science 337 1661–1665 10.1126/science.1224041 [DOI] [PubMed] [Google Scholar]
- Wu Y., Ding N., Wang G., Xu J., Wu J., Brookes P. C. (2009). Effects of different soil weights, storage times and extraction methods on soil phospholipid fatty acid analyses. Geoderma 150 171–178 10.1016/j.geoderma.2009.02.003 [DOI] [Google Scholar]
- Yanagawa K., Nunoura T., Mcallister S., Hirai M., Breuker A., Brandt L. D., et al. (2013). The first microbiological contamination assessment by deep-sea drilling and coring by the D/V Chikyu at the Iheya North hydrothermal field in the Mid-Okinawa Trough (IODP Expedition 331). Front. Microbiol. 4:327 10.3389/fmicb.2013.00327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yayanos A. A., Dietz A. S. (1983). Death of a hadal deep-sea bacterium after decompression. Science 220 497–498 10.1126/science.220.4596.497 [DOI] [PubMed] [Google Scholar]
- Young S. A., Saltzman M. R., Bergström S. M., Leslie S. A., Chen X. (2008). Paired δ13Ccarb and δ13Corg records of upper Ordovician (Sandbian–Katian) carbonates in North America and China: implications for paleoceanographic change. Palaeogeogr. Palaeocl. 270 166–178 10.1016/j.palaeo.2008.09.006 [DOI] [Google Scholar]
- Zeng X., Birrien J.-L., Fouquet Y., Cherkashov G., Jebbar M., Querellou J., et al. (2009). Pyrococcus CH1, an obligate piezophilic hyperthermophile: extending the upper pressure-temperature limits for life. ISME J. 3 873–876 10.1038/ismej.2009.21 [DOI] [PubMed] [Google Scholar]
- Zhang G., Dong H., Xu Z., Zhao D., Zhang C. (2005). Microbial diversity in ultra-high-pressure rocks and fluids from the Chinese continental scientific drilling project in China. Appl. Environ. Microbiol. 71 3213–3227 10.1128/AEM.71.6.3213-3227.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Maignien L., Zhao X., Wang F., Boon N. (2011). Enrichment of a microbial community performing anaerobic oxidation of methane in a continuous high-pressure bioreactor. BMC Microbiol. 11:137 10.1186/1471-2180-11-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Bruns M. A., Tiedje J. M. (1996). DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 62 316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]