Abstract
Single cell analysis and cell sorting has enabled the study of development, growth, differentiation, repair and maintenance of “liquid” tissues and their cancers. The application of these methods to solid tissues is equally promising, but several unique technical challenges must be addressed. This report illustrates the application of multidimensional flow cytometry to the identification of candidate stem/progenitor populations in non-small cell lung cancer and paired normal lung tissue. Seventeen paired tumor/normal lung samples were collected at the time of surgical excision and processed immediately. Tissues were mechanically and enzymatically dissociated into single cell suspension and stained with a panel of antibodies used for negative gating (CD45, CD14, CD33, glycophorin A), identification of epithelial cells (intracellular cytokeratin), and detection of stem/progenitor markers (CD44, CD90, CD117, CD133). DAPI was added to measure DNA content. Formalin fixed paraffin embedded tissue samples were stained with key markers (cytokeratin, CD117, DAPI) for immunofluorescent tissue localization of populations detected by flow cytometry. Disaggregated tumor and lung preparations contained a high proportion of events that would interfere with analysis, were they not eliminated by logical gating. We demonstrate how inclusion of doublets, events with hypodiploid DNA, and cytokeratin+ events also staining for hematopoietic markers reduces the ability to quantify epithelial cells and their precursors. Using the lung cancer/normal lung data set, we present an approach to multidimensional data analysis that consists of artifact removal, identification of classes of cells to be studied further (classifiers) and the measurement of outcome variables on these cell classes. The results of bivariate analysis show a striking similarity between the expression of stem/progenitor markers on lung tumor and adjacent tumor-free lung.
Key terms: non-small cell lung cancer, lung stem cells, flow cytometry data analysis, solid tissue, collagenase
Flow cytometry is an analytical technique designed to make measurements on single cells in suspension. As instrumentation, reagents and analytical tools have improved, flow cytometry has been applied brilliantly to the study of heterogeneous “liquid” tissues such as blood and bone marrow, as well as other easily dissociated solid tissues such as lymph node and spleen. Much of what has been discovered in the fields of immunology and hematology is the direct result of the ability to study such heterogeneous tissues at the level of the single cell. Classical hematologists were able to discover differentiation pathways and make inferences about the identity of hematopoietic progenitor cells by grouping cells on the basis of their morphological features and affinity for dyes (1). These techniques reached their limitations when cells of different function but identical morphology (e.g., CD4 and CD8 T lymphocytes) could not be distinguished, and when important cell populations, inferred by biological experiments, evaded detection because of their low frequency (e.g., definitive hematopoietic stem cells). These problems (2,3) and many others, have yielded to flow cytometry because of its robust analytical capabilities, and also because it has been a preparative method since the inception of the fluorescence activated cell sorter (4,5).
Flow cytometry has the potential to advance the study of solid tissue differentiation, maintenance and repair in the same way, but its application has been challenging since it was first used to characterize ovarian carcinoma(6), pancreas (7), and hepatocytes (8). Many of the difficulties that early investigators faced are still problematic: How to tease strongly adhered cells into single cell suspension while maintaining viability; how to quantify the selection bias that occurs when some cell types survive the process better than others; how to determine which markers and functions are perturbed by the process of disaggregation, and when such loss is irreversible; how to identify and eliminate cellular debris and other sources of artifact from the analysis; and finally, having identified discrete cell populations, how to understand this information in the context of whole tissue architecture? This report will attempt to address these problems using the detection of a panel of putative stem/progenitor markers in non-small cell lung carcinoma and normal adjacent lung tissue as an example.
The hyaluronic acid receptor CD44, a ubiquitous adhesion molecule, was early implicated in tumor metastasis (9) and is the principal marker proposed by Clarke and coworkers to identify tumorigenic breast cancer cells (10). In normal stem cell biology, CD44 has been proposed to play a role in the migration and homing of mesenchymal stem cells (11). CD90 appears to be a lineage-independent adult tissue stem cell marker and was first described on primitive human BM stem cells (12). It is also expressed on oval cells of the liver (13), and on perivascular stem cells (14) which are closely related to mesenchymal stem cells (15). We have proposed CD90 as a principal cancer stem/progenitor cell marker in a variety of epithelial cancers and demonstrated its presence on cytokeratin+/ABCG2+ cells in lung, ovarian, gastric and breast cancer (16,17). CD45−/CD90+ cells have been detected in liver tumors and in the circulation of liver cancer patients (18). When CD44 and CD90 are coexpressed on cytokeratin+ cells, they mark highly tumorigenic cells in breast cancer. As few as 50 cells directly sorted from clinical isolates are capable of tumor formation when coinjected with irradiated tumor (16,19) or adipose-derived stromal cells (20). Further, in liver cancer, CD44+/CD90+ cells demonstrated a more aggressive phenotype than their CD44 negative/CD90+ counterpart and form metastatic lesions in the lungs of immunodeficient mice (18). Like CD90, the type III tyrosine kinase CD117 (KIT, stem cell factor receptor) marks stem/progenitor cells in a variety of tissues, but is also present in terminally differentiated cells such as mast cells (21). Activating mutations in CD117 are implicated in gastrointestinal stromal tumors (GISTs) (22), and myeloid leukemia (23). In normal stem cell biology, CD117 was recently used to identify and sort-purify human lung stem cells capable of regenerating bronchioles, alveoli, and pulmonary vessels (24). In the bone marrow CD133 marks hematopoietic stem/progenitor cells (25). It is present on human prostate epithelial basal cells (26) and has also been implicated on putative cancer stem cells in a variety of tumors (27–29), and most recently in poor risk lung cancer (30).
Methods
Annotated Methods
An annotated version of this methods section with commentary is available as an online Supplement 1 (Supporting Information) to this article.
Tissue Procurement and Transport
Non-small cell lung cancer samples and paired adjacent normal lung tissue were obtained from 17 patients at the time of surgical resection of the tumor. Specimens were collected under protocols approved by the University of Pittsburgh Internal Review Board (UPCI 99–053, 020391, 0503126, 07090247). The tissues were immediately immersed in sterile heparinized tissue culture medium (sodium heparin, 10 U/mL) and transported to the laboratory on an ice pack in a cooler.
Tissue Processing
After the tissue is accessioned, it is weighed, photographed, a physical description is recorded, and a sample is taken for formalin fixation and paraffin embedding. A schematic diagram of tissue processing workflow is provided in Supporting Information Figure 1, and the expected cell recovery of several tissue types prepared by mechanical dissociation (scalpels and screens) and collagenase digestion is shown in Supporting Information Table S1.
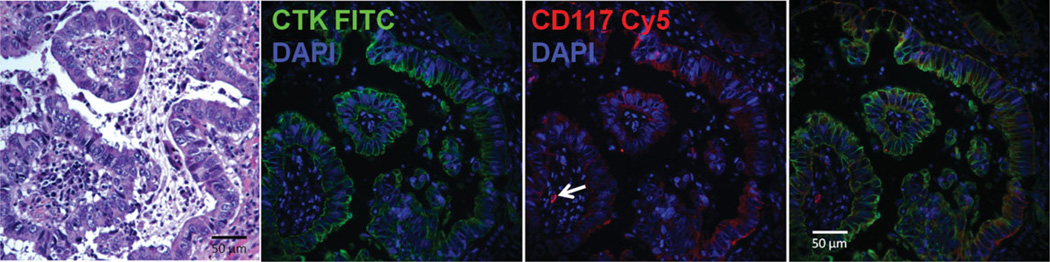
Figure 1.
Immunofluorescent staining of non-small cell lung cancer. FFPE sections were stained with hematoxylin/eosin (left panel) or for cytokeratin (green), CD117 expression (red), or DNA content (blue). These key markers, which are also used in the flow cytometry panel, allow us to assess the quality of the tumor specimen and provide information about the histological location of marker+ cells. In this specimen the great majority of cytokeratin+ cells express CD117. This information helps us confirm that enzymatic digestion has not reduced the proportion of CD117 cells when the same tumor is prepared for flow cytometry. The arrow (center panel) shows a solitary cytokeratin negative CD117+ mast cell. The presence of mast cells serves as a CD117 positive control for both immunohistostaining and flow cytome-try in CD117 negative tumors.
In the present study single cell suspensions were prepared from malignant lesions and tumor-free adjacent lung tissue as previously described (31). Briefly, tumors and lung tissue were minced with paired scalpels and digested with type I collagenase (0.4% in RPMI 1640 medium, Cat. No. C-0130, Sigma Chemicals, St. Louis MO) and DNase (350 KU/mL, Sigma Chemicals, St. Louis MO, Cat. No. D-5025) and disaggregated through 100 mesh stainless steel screens. Undigested tissue clumps were subjected to repeated rounds of digestion. Viable cells were separated from erythrocytes and debris on a Ficoll-Hypaque gradient (Histopaque 1077, Sigma Chemicals). Erythrocytes were lysed using an ammonium chloride lysing solution without fixative (Beckman-Coulter, Cat No. IM3630d). The complete laboratory procedure for tissue disaggregation is provided as in online Supplement 2 (Supporting Information).
Histology and Immunohistostaining
Normal and tumor tissues were fixed for 24 hours in neutral buffered formalin (Sigma Cat. No. F5554). Paraffin sections (5–6 µm) were prepared from embedded tissues. Tissue sections were heated (60°C, 20 min), deparaffinized (3 washes in xylenes), rehydrated by successive washes in absolute ethanol, 90% ethanol, 75% ethanol and deionized water and rinsed twice in Dako wash buffer (Dako). Antigen retrieval was performed at 125°C for 20 min in pH 9.0-EDTA buffer (Dako). After 2 washes in Dako wash buffer, the tissue sections were incubated for 1 hour in a blocking solution (PBS, 5% goat serum, 0.05% Tween 20) to reduce nonspecific antibody binding. Immunofluorescent staining was performed using CD117 (1:400 (35.7µg/mL),Dako Cat. No. A4502, polyclonal). The primary rabbit antibody was substituted by Dako Universal Negative Control for Rabbit Antibodies (ready to use, Dako Cat.No.N1699). Primary antibody and control were incubated for overnight at 4°C. Tissue sections were washed twice using DAKO Wash Buffer prior to applying biotinylated secondary goat anti-rabbit antibody (1:500 (1.52 µg/mL), Dako Cat. No. E0432) for 1 hour at room temperature. Tissue sections were washed twice with Dako wash buffer and incubated with streptavidin-Cy3 (1:500 (2µg/mL),Sigma, Cat. No. 6402) for 30 minutes at room temperature. Slides were washed again and tissue sections were incubated with Alexa 488-conjugated anti-pan-cytokeratin (1:200 (2.5µg/mL), clone AE1-AE-3, eBiosciences Cat. No. 53–9003–80) antibody for 1 hour at room temperature. Stained tissue sections were washed again twice in Dako wash buffer and nuclear staining was attained through 10minute incubation with DAPI (7.15µM Invitrogen, Cat. No. D1306). Slides were washed twice in PBS-A and mounted in Prolong Gold anti-fade reagent (Invitrogen, Cat. No. P36934). Immunofluorescent staining was observed and photographed using an epi-fluorescence microscope (Nikon Eclipse TE 2000-U).
Flow Cytometric Staining
Non-specific binding of fluorochrome-conjugated antibodies was minimized by preincubating pelleted cell suspensions for 5 minutes with neat decomplemented (56°C, 30 minutes) mouse serum (5 µL) (17). Prior to intracellular cytokera-tin staining, cells were stained for surface markers (2 µL each added to the cell pellet, 15–30 minutes on ice; CD44-PE (Beckman-Coulter, Cat No. A32537), CD90-biotin (BD, Cat. No. 555594), Streptavidin-ECD (Beckman Coulter, Fullerton, CA Cat. No. IM3326), CD14-PECy5 (Beckman-Coulter, Cat. No. IM2640U), CD33-PECy5 (Beckman-Coulter, Cat. No. IM2647U), Glycophorin A-PECy5 (BD Biosciences, Cat.No.559944), CD133-APC (Miltenyi Biotech Cat. No. 130–090–854), CD117-PC7 (Beckman Coulter, Cat. No. IM3698), CD45-APCCy7 (BD, Cat. No. 348805)), and fixed with 2% methanol-free formaldehyde (Polysciences, Warrington, PA). Cells were then permeabilized with 0.1% saponin (Beckman Coulter) in phosphate buffered saline with 0.5% human serum albumin (10 minutes at room temperature), cell pellets were incubated with 5 µL of neat mouse serum for 5 minutes, centrifuged and decanted. The cell pellet was disaggregated and incubated with 2 µL of anti-pan cytokeratin-FITC (Beckman Coulter, Cat. No. IM2356) for 30 minutes. Cell pellets were diluted to a concentration of 10 million cells/400 µL of staining buffer and DAPI (Life Technologies, Grand Island NY, Cat. D1306) was added 10 minutes before sample acquisition, to a final concentration of 7.7µg/mL and 40 µL/ 106 cells (17).
Sample Acquisition
Multi-dimensional flow cytometric acquisition was performed using a 10-color Gallios cytometer (Beckman Coulter, Miami FL). An effort was made to acquire a total of 1.8 million events per sample at rates not exceeding 10,000 events/ second. For DAPI staining, PMT gain was optimized for linear (cell cycle) detection of 2N cells (tissue lymphocytes). The cytometer was calibrated to predetermined photomultiplier target channels prior to each use using SpectrAlign beads (DAKO, Cat. No. KO111) and 8-peak Rainbow Calibration Particles (Spherotech, Libertyville, IL, Cat. No. RCP-30–5A). Offline compensation and analyses were performed using VenturiOne software designed for multidimensional rare event problems (Applied Cytometry, Dinnington, Sheffield, UK). Spectral compensation matrices were calculated for each experiment using single-stained mouse IgG capture beads (Becton Dickinson, Cat. No. 552843) for each tandem antibody and hard stained beads (Calibrite, BD) for single molecule dyes (Becton Dickinson, FITC, PE (Cat. No. 349502), APC (Cat. No. 340487)).
Results
Immunofluorescent Staining of a Primary Lung Adenocarcinoma
In order to determine the histologic location of stem/progenitor marker positive cells in primary adenocarcinoma of the lung, we prepared FFPE sections and stained for histology and the expression of key markers used in the flow cytometry panel. Cytokeratin, which identifies epithelial cells, and DAPI, which stains nuclei, were used in combination with other key markers. Figure 1 shows expression of CD117 on cytokeratin+ tumor cells from a primary adenocarcinoma of the lung. In this specimen most tumor cells, which are distinguished from normal cells by their histologic features, express CD117. CD117 was also detected on solitary cytokeratin negative mast cells. Other markers used in this study (CD44, CD90, α-SMA, Ki67) and validated for FFPE sections and flow cytometry, but not shown here, are detailed in Supporting Information Methods.
Artifacts of Tissue Digestion
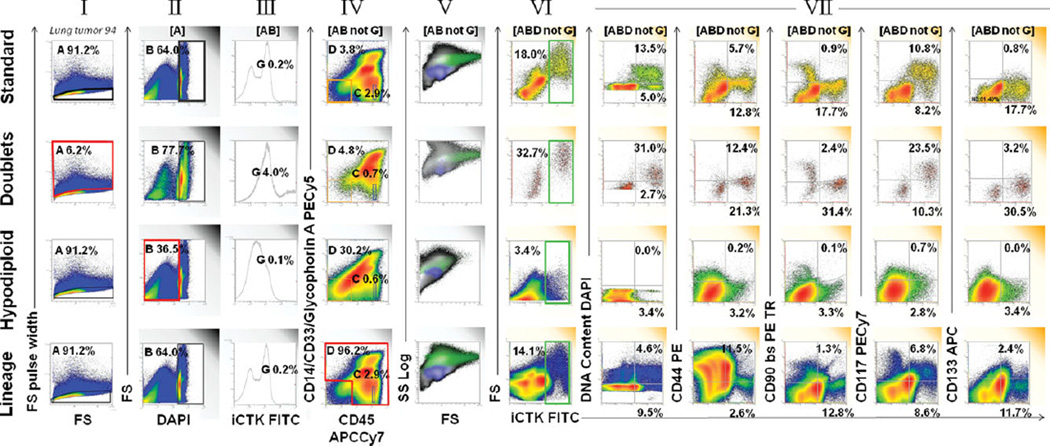
In order to detect rare events in disaggregated lung tumor and adjacent tissue, we removed several potential sources of noise and artifact (Fig. 2). For “doublet discrimination” (Fig. 2, row 1 column I) forward scatter pulse area was plotted versus forward scatter pulse width. Next (Fig. 2, II) forward scatter was plotted versus DAPI fluorescence to eliminate events with <2N DNA content. The events to the far left of the histogram are subcellular debris. The events smearing leftward from the 2N peak are early apoptotic cells that have begun to degrade their DNA or hypodiploid tumor cells. Figure 2, column III is a one parameter histogram of cytokeratin-FITC fluorescence, used to eliminate the last 10 channels with saturating FITC fluorescence (not able to be spectrally compensated). Events with saturating FITC fluorescence represented only 0.2% of “clean” events, but spilled over into the PE and PE-Texas red channels as false positive events if not removed. The remaining rows of Figure 2 illustrate the properties of the events that were eliminated during artifact removal. The details are presented in the figure legend.
Figure 2.
Strategies for artifact removal in analysis of disaggregated solid tissue. The top row shows our standard protocol for artifact removal prior to detection of ploidy and four stem/progenitor markers measured on a primary nonsmall cell lung cancer sample. The first four panels (left to right) show the steps in artifact removal: (I) Eliminate doublets and cell clusters by forward scatter pulse analysis; (II) Eliminate events with hypodiploid DNA or no detectable DNA; (III) Eliminate saturating events (last 10 channels); (IV) Use a dump gate to eliminate cells that stain for markers not present on the population of interest. Next (V) forward versus side scatter is shown on ″cleaned″ events in a color precedence density plot [blue = CD45bright lymphocytes (IV), green = cytokeratin+(VI)]. The next five plots (VII) show individual features versus intracellular cytokeratin. This tumor had high forward scatter, a high proportion of aneuploid cytokeratin+ cells, scant population of CD44, CD90, and CD133+ cells and a prominent population of CD117+ cells. The second row shows the properties of event doublets and clusters (I red region, eliminated from the top analysis). Doublets expressed greater cytokeratin, were of higher light scatter and had greater DNA content than singlet cells, indicating that most were undigested tumor cell clusters. The third row shows the properties of hypodiploid events (II, red region). These bind antibody, but the proportions are very different from that of cells with 2N DNA. The bottom row shows the staining of all events outside the lineage negative gate (IV, red region). There is a population that appears to stain for cytokeratin, but the proportion of aneuploid cells is greatly reduced, making it unlikely that most are genuine tumor cells. These apparently cytokeratin+ events have populations that streak into the ″positive″ regions for many of the stem/progenitor regions.
A dump gate was used to eliminate events that are known to be outside the domain of events of interest. It has the advantage of also removing events that bind antibody nonspecifically, as well as events with autofluorescence at the detection wavelength. The use of the dump gate (Fig. 2, column IV) requires some explanation. In earlier iterations of this panel we simply used CD45 versus cytokeratin to identify and eliminate CD45+ (hematopoietic cells). The sporadic appearance of a puzzling cytokeratin+/CD45+ population led us to add a myeloid/erythroid lineage cocktail to clarify this issue.
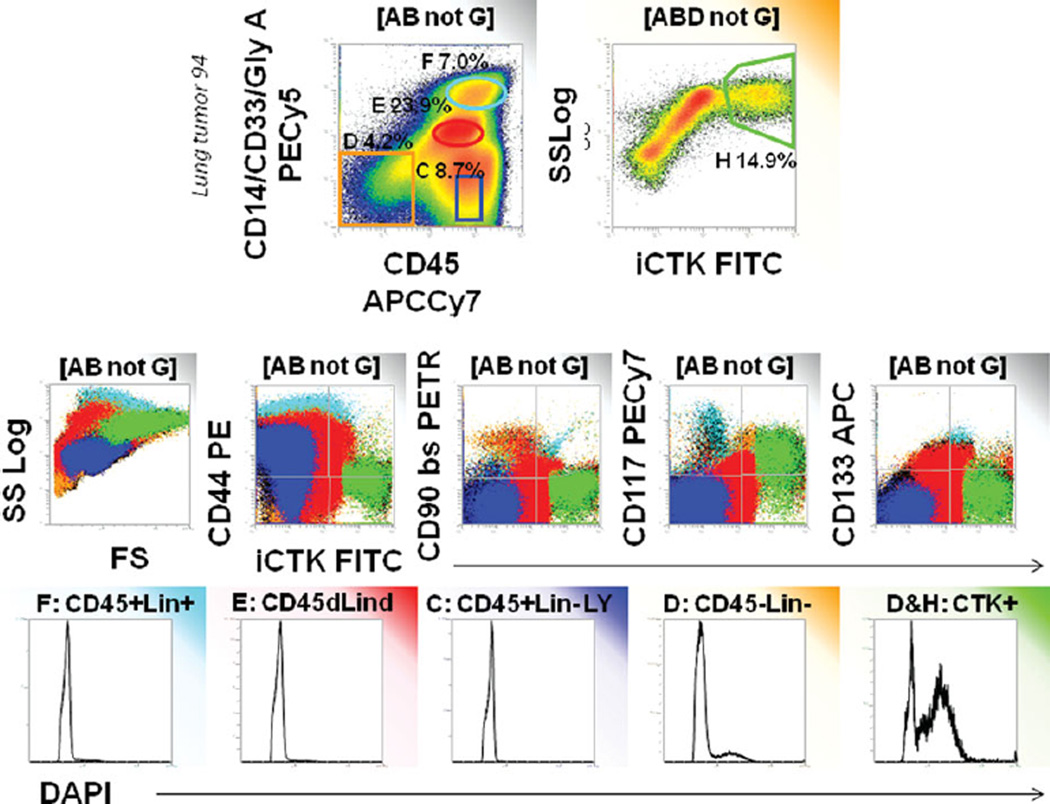
Figure 3 provides an investigation of the populations that we eliminate using this 2-parameter dump gate. Color-event gating is used to identify the CD45−/lineage- population (orange), cytokeratin+ events (green), and 3 major populations outside the CD45−/lineage- gate: lymphocytes (blue) and two myeloid populations (red, turquoise). Although the two populations staining for myeloid markers appear to spread into cytokeratin+ events, analysis of DNA staining shows that they are diploid and therefore probably not tumor cells. In contrast, CD45−/lineage- cells (orange) have a discernible population with DNA >2N, which is even more prominent in CD45−/lineage-/cytokeratin+ cells (green). We investigated the apparent CD45+/cytokeratin+ population further using imaging flow cytometry, which revealed monocytoid cells with cytokeratin+ cytoplasmic inclusions (Supporting Information Fig. S2). We conclude that heme lineage+/cytokeratin+ events should be excluded from analysis.
Figure 3.
Characteristics of populations eliminated by CD45/heme lineage dump gate. Color-eventing is used to trace four major populations observed in the CD45 by heme lineage histogram. All events are gated as described in Figure 1 The majority of aneuploid cells are contained within the nonhematopoietic gate used as a first step in analysis (D, orange). Aneuploid cells are further concentrated when events within D are further gated on cytokeratin+. Lymphocytes (C, blue) are predictably cytokeratin negative, diploid, CD44+ and negative for stem/progenitor markers. Monocytes (E, red) are diploid and appear to be cytokeratin dim to bright, but isotype controls reveal the dim population to be negative (not shown). Cytokeratin bright monocytes have cytokeratin+ cytoplasmic inclusions (Supporting Information Fig. S1). Granulocytes and mast cells (F, turquoise). Granulocytes are diploid, autofluorescent, and negative for cytokeratin and stem/ progenitor markers. Mast cells are CD117+ and CD44+.
Choosing Classifiers and Outcomes for Multidimensional Flow Cytometry Data Analysis
In the present example stem/progenitor marker expression (CD44, CD90, CD117, CD133) is compared on tumor cells and cells from adjacent normal lung on paired samples for 17 patients. After limiting the analysis to nonhematopoietic cells, we classified cells based on cytokeratin expression (epithelial versus nonepithelial or pre-epithelial) and ploidy (2N versus >2N), yielding four classes of cells on which to examine outcomes (stem/progenitor markers, and light scatter, Fig. 4). The reason for using ploidy as a classifier is that in tumor samples, we could be certain that the majority of aneuploid cells were of bona fide tumor origin (as opposed to normal stromal or epithelial cells). It should be noted that the converse is not true; all 2N cells are not normal and pseudodiploid tumor cells are well documented (32,33). Had our question or hypothesis been different, we may have chosen to use ploidy as an outcome rather than as a classifier. Tumor infiltrating lymphocytes, identified by CD45 expression, were used as internal standards defining 2N DNA and lymphoid (i.e. small cell) light scatter.
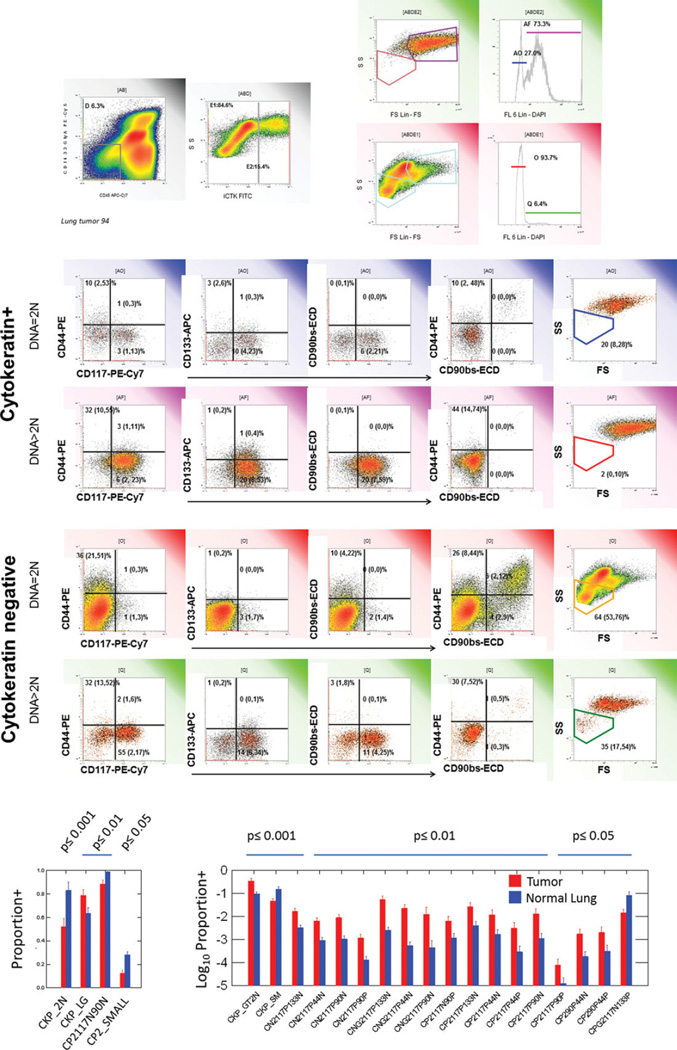
Figure 4.
Identification of classifiers and outcomes. The top panels show identification of cytokeratin+ (E2) and cytokeratin negative (E1) cells among nonhematopoietic (D) cells. These were further subdivided in diploid and aneuploid populations, creating four classes on which to measure outcomes (stem/progenitor markers, light scatter). The region percents listed are mean values, parentheses indicate lower and upper 95% confidence intervals. The same analysis was performed on normal lung samples (Supporting Information Fig. S3). Bottom panels: Bivariate comparison of tumor and normal lung. Eighty-six quantitative variables extracted from analysis were compared between tumor and normal lung samples. Log normally distributed variables were log transformed prior to analysis. Only statistically significant comparisons are shown (uncorrected P-values, Student′s 2-tailed test, pooled variance). Bars represent mean values, error bars = standard error of the mean.
Data Exploration
Figure 4 also illustrates the combination of a histogram array and data table, in which a representative sample is chosen to illustrate the analytical strategy, but the region statistics are based on the entire study population (17 patients). For the outcome variables, mean values, with 95% confidence intervals in parentheses, are provided to facilitate comparisons.
Scanning Figure 4 it is apparent that the majority of cells with >2N DNA (aneuploid/proliferating cells) are in the cytokeratin+ population and the majority of small cells are in the cytokeratin negative population. Inspection of the cytokeratin+ population reveals that cells bearing the stem/ progenitor associated markers CD44, CD117 and CD133 are more prevalent among cells with >2N DNA. Although there is some marker coexpression, CD44, CD117 and CD133+ populations are largely distinct. Although CD117 is the most prominent stem/progenitor marker in this series, some care must be taken in the analysis. Tumor samples in our dataset dichotomized on the basis of CD117 expression and the example shown is CD117+, illustrating the difficulty in choosing a truly representative sample.
The cytokeratin negative population is more interesting than might have been assumed a priori. Particularly in the subset with >2N DNA, there is a prominent population of CD117+ cells, unlikely to be mast cells because of their ploidy. There is also a small but prominent population of low light scatter cells, which again, because of aneuploidy may represent small undifferentiated (i.e. cytokeratin negative) tumor cells. Finally, among the 2N population there is a robust population of cells coexpressing CD44 and CD90, most likely identifying tumor-associated mesenchymal stromal cells, an important component of the tumor niche.
An analogous graphic representing analysis of adjacent grossly normal lung tissue is shown in Supporting Information Figure S3. Cells with >2N DNA content are scant. Cytokeratin+ diploid cells include a small population of CD117+ cells, reported to be normal lung stem cells (24), as well as CD117−/CD133+ cells, absent or reduced in lung tumors. Mesenchymal stromal cells (CD44+/CD90+) are present and low light scatter cells are prominent in the diploid cytokeratin negative population.
From a total of 86 quantitative variables extracted from the data by conventional analysis (gates and regions), at total of 22 were significantly different (p ≤ 0.05) between tumor and normal lung in an uncorrected bivariate comparison (Fig. 4, bottom panel). Like all multivariate cytometry problems, rigorous comparison of tumor and normal lung is complicated by the fact that there are more variables than observations, many of the variables are heavily correlated, and multiple comparisons increase the risk of chance association. This problem is thoroughly treated, using this dataset, in a companion article (34). However, even a simple bivariate analysis reveals that 3 of the largest effects in the data set (p = 0.001) are those identifiable by morphology and simple immunohistochemistry: 1) CKP_2N = cytokeratin+/euploid (tumors have less); 2) CKP>2N = cytokeratin+/aneuploid (tumors have more); 3) CKP_SM = cytokeratin+/lymphoid light scatter (tumors have less). The fourth variable CN2117P133N = cytokeratin negative/diploid/CD133 negative, does not correspond to a known population, as mast cells were removed from the analysis by gating on CD45 negative events.
Discussion
FFPE sections (Fig. 1) are critical to the interpretation of flow cytometry performed on digested tissues. These preparations provide a histologic context for key markers used in flow cytometry and provide a standard by which single cells suspensions may be evaluated for selection bias. The principal disadvantages of immunohistostaining, the small number of cells that can be evaluated, the subjective nature of analysis, and the technical difficulties associated with polychromatic staining, are easily overcome by flow cytometry. In the present data set, immunofluorescent staining provided several important cues for interpretation of flow cytometric data: 1) Cytokeratin negative CD117+ cells are mast cells; 2) Some morphologically identifiable tumor cells are cytokeratin negative; 3) The dichotomy observed by flow cytometry between patients with CD117+ and CD117− tumors was confirmed; and 4) Tissue processing for flow cytometry results in overre-presentation of hematopoietic cells, especially lymphocytes. In previous studies, combining immunohistostaining with flow cytometry allowed us to localize the cytokeratin+/CD44+/ CD90+ population observed by flow cytometry to the invasive edge of breast tumors (19), and to determine the histologic location of CD45−/CD146−/CD31−/CD34+ adipose stem cells (35).
Among the first flow cytometric applications for disaggregated tumors was the study of tumor infiltrating immune cells (36). Compared to the study of epithelial tissues, which are complex vascularized structures in which cells are organized by avid adhesion to extracellular matrices and each other, tissue infiltrating immune cells are weakly associated and readily recovered as viable single cells. Tumors can be challenging to dissociate because they may be hardened by fibrosis and contain necrotic areas. However, careful observation and sequential digestion of tumor tissue will actually yield more cells per gram than normal tissue (Supporting Information Table S1).
Even the most careful tissue digestion will result in undigested tissue clumps, apoptotic cells, dead cells and subcellular debris. All of these will interfere with analysis and interpretation unless identified in the data set and removed by logical gating. The methods described here have previously been used for adipose tissue (35), normal breast and breast cancer tissues (19). Stem/progenitor populations are rare in most tissues and require special considerations for their detection and enumeration (37,38). Most importantly, it is necessary to examine a sufficient number of “clean” events to yield an appropriate number of analyzable events. A population of 100 cells in an analytical region will give a Poisson counting coefficient of variation of 10%, as originally worked out by Student for the hemocytometer (39). To take an example from a recently published article describing multipotential adult human lung stem cells (24), which were present at a frequency of 1/24,000, a minimum of 2.4 million events (post artifact removal) must be acquired to attain a counting CV = 10%. Dealing with such large data sets requires specialized analytical software designed for parallel processing. Several packages are available for offline analysis. A hierarchical approach to data analysis, such as the artifact removal, classifier, outcome method described here helps to focus data exploration and analysis. However, the problem of more quantifiable features (i.e. analytical regions) than cases, with many variables highly correlated, is inherent in multidimensional cytometry data, and argues ultimately for an automated approach to data analysis. In its simplest form, this entails applying modern multivariate statistical techniques (40,41) to the results of conventional gate/region type analyses such as those described here. Eventually, it may be possible to replace manual gate/region-based analysis with automated cluster-finding algorithms, but this can be a double-edged problem if attaining complete objectivity requires us to relinquish a wealth of a priori knowledge concerning the biological constraints imposed on marker expression.
In this data set, three of the four most significant distinguishing features identified by bivariate analysis involved a combination of morphology (light scatter), cytokeratin expression, and DNA content, features long used to identify tumor cells. Prior to analysis of stem/progenitor marker expression on nonhematopoietic cells, we chose to identify four classifier populations on the basis of cytokeratin expression and DNA content. In tumor samples, cytokeratin1 cells with >2N DNA are clearly tumor cells, but this does not exclude the possibility of cytokeratin negative or pseudodi-ploid tumor cells. Similarly normal lung airway cells have a proliferative (and therefore >2N) component (Supporting Information Fig. S3).
After subsetting the data on the basis of cytokeratin expression and DNA content, we found a striking similarity between stem/progenitor marker patterns in tumor and adjacent tumor-free lung. The conservation of expression patterns suggests that these proteins may play important functional roles in both tumor and the normal tissues (24). Similarly, we (17) and others (42–44) have demonstrated that constitutive self-protection mediated by ABC transporter activity in normal tissue stem cells can be retained or re-expressed in a subset of malignant cells. These data support the interpretation that airway stem cells and their malignant counterparts share at least some of these growth factor receptors and adhesion molecules, as has been demonstrated in colon cancer and normal colon (45). For example, CD44/CD90 expression on cytokeratin negative cells is consistent with mesenchymal stem cells in normal tissue, but in metastatic cancer, CD44/CD90 coexpression on cytokeratin positive cells (19) may signal epithelial to mesenchymal transition (46).
Taken together, our finding that tumor cells share stem/ progenitor and adhesion markers with tumor-free chronically injured lung tissue is consistent with the hypothesis that the self-renewing, self-protected tumorigenic cell can take the form of a stem-progenitor hybrid in aggressive epithelial neoplasms such as lung cancer (17). Combining stem-like self-renewal and protection with high proliferative capacity, they need not be rare to exploit mechanisms employed by normal tissue stem cells for their renewal and survival.
Supplementary Material
Acknowledgments
The authors would like to acknowledge our clinical collaborators James D. Luketich and Adam M. Brufsky, as well as Dr. Ludovic Zimmerlin, James Arbore and E. Michael Meyer for their assistance in the development of the methods presented here.
Grant sponsor: Department of Defense; Grant numbers: BC032981, BC044784; Grant sponsor: Production Assistance for Cellular Therapy (PACT); Grant number: #N01-HB-37165; Grant sponsor: UPCI Cytometry Facility; Grant number: CCSG P30CA047904; Grant sponsors: Commonwealth of Pennsylvania, the Hillman Foundation, the Glimmer of Hope Foundation.
Footnotes
Additional Supporting Information may be found in the online version of this article.
Literature Cited
- 1.Ehrlich P. Methodologische Beitra¨ge zur Physiologie und Pathologie der verschiedenen Formen der Leukocyten. Z Klin Med. 1879;1:5553–5560. [Google Scholar]
- 2.Reinherz EL, Kung PC, Goldstein G, Schlossman SF. Further characterization of the human inducer T cell subset defined by monoclonal antibody. J Immunol. 1979;123:2894–2996. [PubMed] [Google Scholar]
- 3.Spangrude GJ, Heimfeld S, Weissman IL. Purification and characterization of mouse hematopoietic stem cells. Science. 1988;241:58–62. doi: 10.1126/science.2898810. [DOI] [PubMed] [Google Scholar]
- 4.Fulwyler MJ. Electronic separation of biological cells by volume. Science. 1965;150:910–911. doi: 10.1126/science.150.3698.910. [DOI] [PubMed] [Google Scholar]
- 5.Hulett HR, Bonner WA, Barrett J, Herzenberg LA. Cell sorting: automated separation of mammalian cells as a function of intracellular fluorescence. Science. 1969;166:747–749. doi: 10.1126/science.166.3906.747. [DOI] [PubMed] [Google Scholar]
- 6.Bast RC, Jr, Feeney M, Lazarus H, Nadler LM, Colvin RB, Knapp RC. Reactivity of a monoclonal antibody with human ovarian carcinoma. J Clin Invest. 1981;68:1331–1337. doi: 10.1172/JCI110380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabinovitch A, Russell T, Shienvold F, Noel J, Files N, Patel Y, et al. Preparation of rat islet B-cell-enriched fractions by light-scatter flow cytometry. Diabetes. 1982;31:939–943. doi: 10.2337/diacare.31.11.939. [DOI] [PubMed] [Google Scholar]
- 8.Morin O, Patry P, Lafleur L. Heterogeneity of endothelial cells of adult rat liver as resolved by sedimentation velocity and flow cytometry. J Cell Physiol. 1984;119:327–334. doi: 10.1002/jcp.1041190311. [DOI] [PubMed] [Google Scholar]
- 9.Haynes BF, Liao HX, Patton KL. The transmembrane hyaluronate receptor (CD44): Multiple functions, multiple forms. Cancer Cells. 1991;3:347–350. [PubMed] [Google Scholar]
- 10.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhu H, Mitsuhashi N, Klein A, Barsky LW, Weinberg K, Barr ML, Demetriou A, Wu GD. The role of the hyaluronan receptor CD44 in mesenchymal stem cell migration in the extracellular matrix. Stem Cells. 2006;24:928–935. doi: 10.1634/stemcells.2005-0186. [DOI] [PubMed] [Google Scholar]
- 12.Craig W, Kay R, Cutler RL, Lansdorp PM. Expression of Thy-1 on human hematopoietic progenitor cells. J Exp Med. 1993;177:1331–1342. doi: 10.1084/jem.177.5.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herrera MB, Bruno S, Buttiglieri S, Tetta C, Gatti S, Deregibus MC, Bussolati B, Camussi G. Isolation and characterization of a stem cell population from adult human liver. Stem Cells. 2006;24:2840–2850. doi: 10.1634/stemcells.2006-0114. [DOI] [PubMed] [Google Scholar]
- 14.Zannettino ACW, Paton S, Arthur A, Khor F, Itescu S, Gimble JM, Gronthos S. Multipotential human adipose-derived stromal stem cells exhibit a perivascular phenotype in vitro and in vivo. J Cellular Physiol. 2008;214:413–421. doi: 10.1002/jcp.21210. [DOI] [PubMed] [Google Scholar]
- 15.Young HE, Steele TA, Bray RA, Hudson J, Floyd JA, Hawkins K, Thomas K, Austin T, Edwards C, Cuzzourt J, et al. Human reserve pluripotent mesenchymal stem cells are present in the connective tissues of skeletal muscle and dermis derived from fetal, adult, and geriatric donors. Anatomical Record. 2001;264:51–62. doi: 10.1002/ar.1128. [DOI] [PubMed] [Google Scholar]
- 16.Donnenberg VS, Luketich JD, Landreneau RJ, DeLoia JA, Basse P, Donnenberg AD. Tumorigenic epithelial stem cells and their normal counterparts. Ernst Schering Foundation Symp Proc. 2006;5:245–263. doi: 10.1007/2789_2007_054. [DOI] [PubMed] [Google Scholar]
- 17.Donnenberg VS, Landreneau RJ, Donnenberg AD. Tumorigenic stem and progenitor cells: Implications for the therapeutic index of anti-cancer agents. J Control Release. 2007;122:385–391. doi: 10.1016/j.jconrel.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang ZF, Ngai P, Ho DW, Yu WC, Ng MNP, Lau CK, Li MLY, Tam KH, Lam CT, Poon RTP, et al. Identification of local and circulating cancer stem cells in human liver cancer. Hepatology. 2008;47:919–928. doi: 10.1002/hep.22082. [DOI] [PubMed] [Google Scholar]
- 19.Donnenberg VS, Donnenberg AD, Zimmerlin L, Landreneau RJ, Bhargava R, Wetzel RA, Basse P, Brufsky AM. Localization of CD44 and CD90 positive cells to the invasive front of breast tumors. Cytometry B Clin Cytom. 2010;78B:287–301. doi: 10.1002/cyto.b.20530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zimmerlin L, Donnenberg AD, Rubin JP, Basse P, Landreneau RJ, Donnenberg VS. Regenerative therapy and cancer: in vitro and in vivo studies of the interaction between adipose-derived stem cells and breast cancer cells from clinical isolates. Tissue Eng Part A. 2011;17:1–2. 93–106. doi: 10.1089/ten.tea.2010.0248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iemura A, Tsai M, Ando A, Wershil BK, Galli SJ. The c-kit ligand, stem cell factor, promotes mast cell survival by suppressing apoptosis. Am J Pathol. 1994;144:321–328. [PMC free article] [PubMed] [Google Scholar]
- 22.Sarlomo-Rikala M, Kovatich AJ, Barusevicius A, Miettinen M. CD117: A sensitive marker for gastrointestinal stromal tumors that is more specific than CD34. Mod Pathol. 1998;11:728–734. [PubMed] [Google Scholar]
- 23.Moore MAS. Converging pathways in leukemogenesis and stem cell self-renewal. Exp Hematol. 2005;33:719–737. doi: 10.1016/j.exphem.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 24.Kajstura J, Rota M, Hall SR, Hosoda T, D’Amario D, Sanada F, Zheng H, Ogorek B, Rondon-Clavo C, Ferreira-Martins J, et al. Evidence for human lung stem cells. N Engl J Med. 2011;364:1795–1806. doi: 10.1056/NEJMoa1101324. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Handgretinger R, Gordon PR, Leimig T, Chen X, Buhring HJ, Niethammer D, Kuci S. Biology and plasticity of CD133+ hematopoietic stem cells. Ann N Y Acad Sci. 2003;996:141–151. doi: 10.1111/j.1749-6632.2003.tb03242.x. [DOI] [PubMed] [Google Scholar]
- 26.Rizzo S, Attard G, Hudson DL. Prostate epithelial stem cells. Cell Proliferation. 2005;38:363–374. doi: 10.1111/j.1365-2184.2005.00356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, Henkelman RM, Cusi-mano MD, Dirks PB. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 28.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445:106–110. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 29.Maitland NJ, Collins AT. Prostate cancer stem cells: A new target for therapy. J Clin Oncol. 2008;26:2862–2870. doi: 10.1200/JCO.2007.15.1472. [DOI] [PubMed] [Google Scholar]
- 30.Woo T, Okudela K, Mitsui H, Yazawa T, Ogawa N, Tajiri M, Yamamoto T, Rino Y, Kitamura H, Masuda M. Prognostic value of CD133 expression in stage I lung adeno-carcinomas. Int J Clin Exp Pathol. 2010;4:32–42. [PMC free article] [PubMed] [Google Scholar]
- 31.Donnenberg VS, Meyer EM, Donnenberg AD. Measurement of multiple drug resistance transporter activity in putative cancer stem/progenitor cells. In: Yu J, editor. Methods in Molecular Biology. Volume 568 Cancer Stem Cells. New York: Humana Press, Springer; 2009. pp. 261–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abeln EC, Corver WE, Kuipers-Dijkshoorn NJ, Fleuren GJ, Cornelisse CJ. Molecular genetic analysis of flow-sorted ovarian tumour cells: Improved detection of loss of heterozygosity. Br J Cancer. 1994;70:255–262. doi: 10.1038/bjc.1994.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Navin N, Kendall J, Troge J, Andrews P, Rodgers L, McIndoo J, Cook K, Stepansky A, Levy D, Esposito D, et al. Tumour evolution inferred by single-cell sequencing. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Normolle DP, Donnenberg VS, Donnenberg AD. Statistical classification of multivar-iate flow cytometric data analyzed by manual gating: Stem, progenitor, and epithelial marker expression in nonsmall cell lung cancer and normal lung. Cytometry A. 2013;83A:150–160. doi: 10.1002/cyto.a.22240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmerlin L, Donnenberg VS, Pfeifer ME, Meyer EM, Peault B, Rubin JP, Donnenberg AD. Stromal vascular progenitors in adult human adipose tissue. Cytometry A. 2010;77A:22–30. doi: 10.1002/cyto.a.20813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ferry BL, Flannery GR, Robins RA, Lawry J, Baldwin RW. Phenotype of cytotoxic effector cells infiltrating a transplanted, chemically induced rat sarcoma. Immunology. 1984;53:243–250. [PMC free article] [PubMed] [Google Scholar]
- 37.Zimmerlin L, Donnenberg VS, Donnenberg AD. Rare event detection and analysis in flow cytometry: Bone marrow mesenchymal stem cells, breast cancer stem/progenitor cells in malignant effusions, and pericytes in disaggregated adipose tissue. In: Hawley TS, Hawley RG, editors. Flow Cytometry Protocols, 3rd ed. Volume 699, Methods in Molecular Biology. New York, N.Y: Humana Press; 2011. [DOI] [PubMed] [Google Scholar]
- 38.Donnenberg AD, Donnenberg VS. Rare-event analysis in flow cytometry. Clin Lab Med. 2007;27:627–652. doi: 10.1016/j.cll.2007.05.013. viii. [DOI] [PubMed] [Google Scholar]
- 39.Student [Gossett WS] On the error of counting with a haemacytometer. Biometrika. 1907;5:351–360. [Google Scholar]
- 40.Zou H, Hastie T. Regularization and variable selection via the elastic net. J Royal Statist Soc B. 2005;67:301–320. [Google Scholar]
- 41.Breiman L. Random forests. Machine Learn. 2001;45:5–32. [Google Scholar]
- 42.Szotek PP, Pieretti-Vanmarcke R, Masiakos PT, Dinulescu DM, Connolly D, Foster R, Dombkowski D, Preffer F, MacLaughlin DT, Donahoe PK. Ovarian cancer side population defines cells with stem cell-like characteristics and Mullerian Inhibiting Substance responsiveness. Proc Natl Acad Sci USA. 2006;103:11154–11159. doi: 10.1073/pnas.0603672103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engelmann K, Shen H, Finn OJ. MCF7 side population cells with characteristics of cancer stem/progenitor cells express the tumor antigen MUC1. Cancer Res. 2008;68:2419–2426. doi: 10.1158/0008-5472.CAN-07-2249. [DOI] [PubMed] [Google Scholar]
- 44.Harris MA, Yang H, Low BE, Mukherje J, Guha A, Bronson RT, Shultz LD, Israel MA, Yun K. Cancer stem cells are enriched in the side population cells in a mouse model of glioma. Cancer Res. 2008;68:10051–10059. doi: 10.1158/0008-5472.CAN-08-0786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dalerba P, Kalisky T, Sahoo D, Rajendran PS, Rothenberg ME, Leyrat AA, Sim S, Okamoto J, Johnston DM, Qian D, et al. Single-cell dissection of transcriptional heterogeneity in human colon tumors. Nat Biotechnol. 2011;29:1120–1127. doi: 10.1038/nbt.2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mani SA, Guo W, Liao M-J, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.