Abstract
Cancer classification is an important step in biomarker identification. Developing machine learning methods that correctly predict cancer subtypes/types can help in identifying potential cancer biomarkers. In this commentary, we presented ensemble classification approach and compared its performance with single classification approaches. Additionally, the application of cancer classification in identifying biomarkers for drug design was discussed.
Keywords: cancer classification, gene expression, ensemble, biomarker, drug design
INTRODUCTION
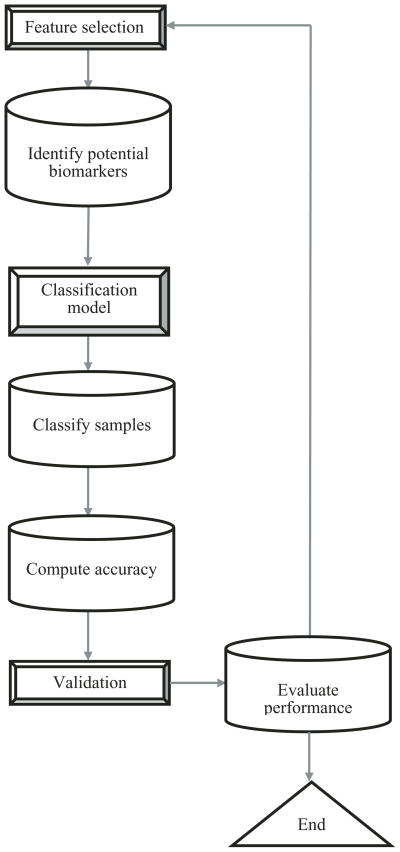
Cancer is one of the leading causes of death worldwide. As a result, developing drugs that can target cancer cells is significant. Biomarker identification using gene expression data provides an approach to identify potential candidates for cancer diagnosis [Xiong et al., 2001]. The advent of DNA microarrays that simultaneously measure the expression of thousands of genes [Harrington et al., 2000] have led to the rise in computational methods to analyze and process high throughput data. Cancer classification using gene expression data could be used to distinguish different cancer subtypes or types. Additionally, cancer classification can prove valuable by helping in the upstream screening process to identify potential biomarkers. Using feature selection methods, genes that are expressed differentially among different cancer subtypes could be suggested as potential biomarkers by evaluating whether high classification accuracy is achieved with the subset of genes. Xiong et al. [2001] proposed a general framework for a feature selection method based on classification to identify cancer biomarkers. To measure the goodness of the different feature subsets generated, Xiong et al. [2001] used the number of cancer samples incorrectly classified (false positives and false negatives) generated from a classification model to quantify the performance of the selected features. Figure 1 shows a generalized approach for identifying biomarkers. The first step identifies potential biomarkers through a feature selection method. These biomarkers could be either oncogenes or tumor suppressor genes involved in tumor initiation or progression. Next, these genes are used as features in developing a model that can predict the class to which a sample belongs. Using classification models and their accuracy in correctly predicting the different cancer subtypes, the performance of the features (e.g., genes) as potential biomarkers could be evaluated. Note that one could also skip an explicit feature selection step and use a classification model, such as decision trees, that inherently contains the feature selection step. These biomarkers could serve as potential targets for drug design that could be tested further experimentally. For example, in a previous study, Armstrong et al. [2002] classified acute lymphoblastic leukemia based on the gene expression data and identified a target gene FLT3 that was later shown experimentally to be a potential drug target [Armstrong et al., 2002, 2003].
Fig. 1.
Flowchart of biomarker identification process.
The heterogeneous nature of cancers and the need to identify better molecular biomarkers for the different subtypes increase the need for efficient and accurate machine learning methods that can correctly predict or classify cancer subtypes. Many methods have been proposed to classify cancers based on gene expression data. However, there is no general approach that has been proposed to specifically predict or classify cancer subtypes. Thus far, several research papers have used an ensemble approach for predicting or classifying cancers independent of their subtypes. We propose that an ensemble classification approach would be appropriate for classifying different cancer subtypes. Thus far, an ensemble of decision trees [Tan and Gilbert, 2003] and neural networks [Liu et al., 2004] have been used to classify cancers. Both studies found that the ensemble approach often increased the accuracy and robustness in classifying the cancer from the normal samples or between cancers based on their gene expression levels. In this commentary, a generalized, as well as specific, ensemble classification approaches that can be used to classify or predict cancer subtypes or types based on gene expression are presented. Additionally, the potential of identifying molecular biomarkers based on the classification models of cancers is discussed.
Classification of cancers faces many challenges that still need to be addressed. First, the expression levels of many genes are not differentially expressed across training samples. From a machine learning point of view, these genes should not be used as features for a classification model since they are weak distinguishers between cancer types. Many statistical methods such as T-statistics, F-statistics, and empirical Bayesian models have been used to detect differentially expressed genes across tumor samples. However, it is noteworthy that genes that are not differentially expressed could also be involved in cancers. Another problem in cancer classification is the background and system noise that result from systematic variations in the microarray experiments. This challenge can be tackled by normalizing the expression level of genes. Finally, the small number of tumor samples relative to the number of genes poses another challenge, namely high dimensionality, since the number of genes corresponds to the dimension of the feature space. For a classification model to perform relatively well and avoid the problem of high dimensionality, one could reduce the dimensionality of the feature space while maintaining the biological information in the cancer data set. Linear discriminant analysis could be used to reduce the dimensionality. This method can reduce the dimension of the feature space while maintaining the separation between the different cancer types [Wang et al., 2010].
ENSEMBLE LEARNING
The motivation behind building an ensemble model is to divide the training data into multiple data sets where each set exhibits different characteristics and properties. By building an ensemble model, the diversity can be incorporated through the use of different training parameters, different types of classifiers, or feature sets. By achieving an ensemble of diverse classifiers, a set of classifiers that do not result in the same incorrectly classified cancer samples is reached; the incorrectly classified cancer samples of the set of classifiers in the ensemble model should be uncorrelated to achieve diversity. On the other hand, building a set of classifiers such that they result in the same incorrectly classified cancer samples is no different than building a single classifier. It has been shown that ensemble methods perform better than single classifier models as long as the diversity of the base classifiers is maintained [Dietterich, 2000]. Therefore, creating a set of diverse specialized models could lead to a final model with better prediction accuracy as compared with a single classifier model. Additionally, it is easier to optimize a set of simple classifiers as compared with a single complex classifier. Ensemble models have been shown to be useful in many applications; however, its effectiveness as with any classification approach relies on the properties of the data and the diversity of the classifiers.
Building an Ensemble Model
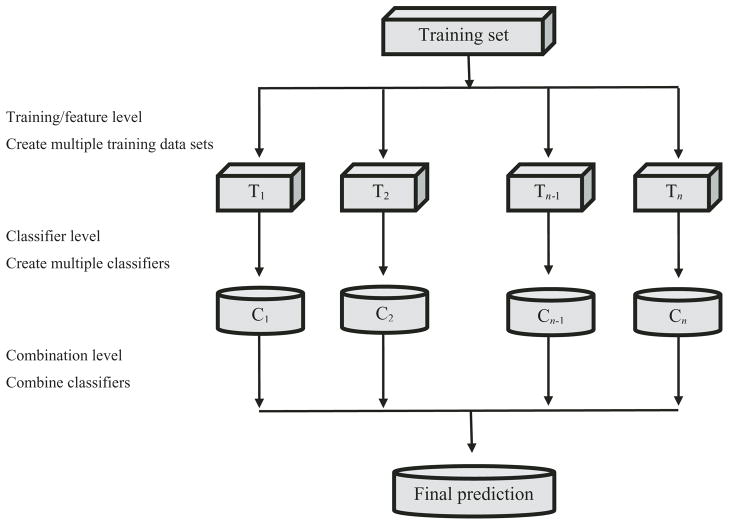
The process of ensemble learning is composed of three steps (Fig. 2). First, a set of training samples are constructed from the cancer data. Next, classification models are constructed for each set of training samples in the ensemble, which are then built into a network of models. Then each cancer test sample is classified using the network of models. Finally, the set of classifiers are integrated using a combination method that results in a single prediction of the class to which the cancer sample belongs.
Fig. 2.
General overview of an ensemble model.
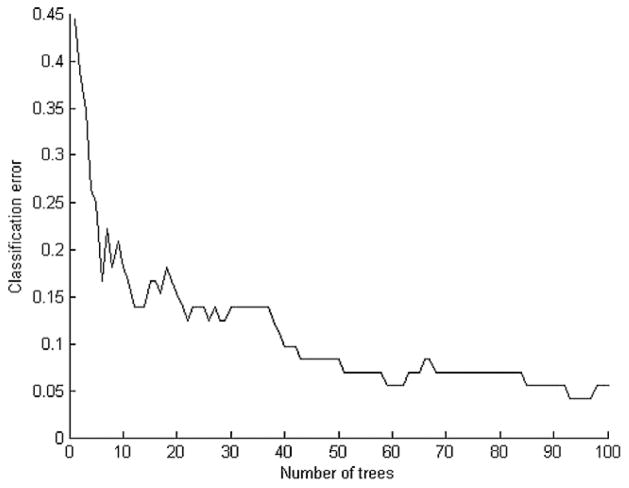
To illustrate the advantage of an ensemble approach as compared with a single classifier model, we used a leukemia gene expression data set that contains 72 samples distributed among two classes (47 samples of acute lymphoblastic leukemia and 25 samples of acute myeloid leukemia) [Golub et al., 1999]. An eightfold cross-validation method was used to compute the number of cancer samples incorrectly classified. An ensemble of three k-nearest neighbors with different initial k-values (1, 3, and 5) was compared with a single k-nearest neighbor with k = 1. As shown in Table 1, the prediction accuracy of an ensemble of the three k-nearest neighbors (93.67%) is higher than a single 1-nearest neighbor classifier (89.64%). Additionally, a single decision tree was compared with an ensemble of decision trees. As shown in Figure 3, as the ensemble number of decision trees increases, the classification error generally decreases. The prediction accuracy increased from 56% to 83% to 90% to 94% as the number of decision tress increased from one to 10 to 40 to 100. This demonstrates that an ensemble classification approach performs better than a single classification approach. It is important to note that one may risk overfitting the model if the number of decision trees is increased too much, as in the case from 40 to 90, whereas only a slight increase is achieved in the prediction accuracy.
TABLE 1.
Prediction Accuracy of a Single Classifier versus Ensemble of Classifiers
| Classifier | Prediction accuracy (%) |
|---|---|
| One k-nearest neighbor | 89.64 |
| Ensemble of three k-nearest neighbors | 93.67 |
| One decision tree | 56 |
| Ensemble of 10 decision trees | 83 |
| Ensemble of 40 decision trees | 90 |
| Ensemble of 100 decision trees | 94 |
Fig. 3.
Ensemble of decision trees.
General Approach
There are four different levels that need to be considered when building an ensemble model. The first is the training level where multiple data sets are constructed from the training data. Sampling either with or without replacement can be used to construct different subsets of training data. The second is the model or classifier level where a set of similar or different classification models are constructed. Constructing a set of similar classifiers (i.e., decision trees) is usually accompanied by different training parameters for each classifier to introduce diversity into the ensemble model. Similarly, using different classification methods can also bring diversity into the ensemble model. The third is at the feature level where random or disjoint feature sets are used for each classifier. The last is at the combination level where different combination rules can be applied to integrate the predictions made by the set of classifiers. For instance, an ensemble of five classifiers generates five predictions for a particular cancer sample. At the combination level, these five predictions are combined according to a combination rule to arrive upon a single final prediction. A simple majority vote algorithm could be used where the set of classifiers have equal weights. This method checks the most frequent occurrence of a cancer type. Alternatively, a weighted algorithm method could be applied where each model has a certain weight based on its accuracy in correctly predicting the cancer types. As there is no single ensemble model that universally outperforms other ensemble approaches, the effectiveness of an ensemble approach on a cancer data set is still largely dependent on the characteristics of the data as well as the level of diversity present in the individual classifiers.
Specific Approach
Different ensemble techniques have been proposed in machine learning and pattern recognition communities. Two methods that have been applied frequently are bagging and boosting. Bagging is one of the earliest and simplest ensemble methods. It has been shown to have generally good performance as compared with using a single classifier. Valentini et al. [2004] showed that a bagged ensemble of support vector machines applied on cancer data achieved a higher or equal prediction accuracy as compared with using a single support vector machine classifier. The core of the algorithm is the multiple training subsets constructed from the training data by randomly sampling with replacement. The different subsets are then trained by an ensemble of classifiers of the same type. Finally, a majority vote algorithm is used to predict the cancer type to which a cancer sample belonged. A bagging approach is usually associated with unstable learning classifiers. Unstable learning classifiers are classifiers, whereby small changes in the training data lead to different classification models or prediction accuracy. It is known that decision trees are unstable supervised learning methods because any change in the training patterns yields a drastic change in the decision tree structure. Many variants of the bagging approach have been introduced. Random forest combines the idea of bagging with the random selection of features to form an ensemble of decision trees. Two advantages of cancer classification through the use of random forests are as follows: overfitting that could occur as a result of the number of genes exceeding the number of samples is avoided since the number of growing decision trees increases [Breiman, 2001]; and the feature selection is inherent in the method, therefore an explicit gene selection step is not required.
Boosting is another ensemble meta-algorithm technique that can be used for cancer classification. The general idea behind boosting is that it improves the performance of a set of weak supervised learners to the level of a strong one. Boosting builds multiple data sets by resampling the training data based on a weight vector that contains the probability of each training sample being chosen. In boosting, each consecutive classifier examines the misclassified instances of the previous classifiers and attempts to address them. Specifically, the weights are recomputed for each consecutive classifier in such a way that the weights of the misclassified training samples are increased, while the weights of the correctly classified training samples are decreased. This way, a set of classifiers can be built where each classifier is more likely to select training samples that have been misclassified in the previous classifiers. Finally, the predictions of the set of classifiers are combined using the majority vote algorithm. A more generalized version of boosting is adaptive boost (adaboost) proposed by Freund and Schapire [1997]. Adaboost uses the boosting approach and weighted majority voting at the combination level. In adaboost, the weight of every component classifier is dependent and inversely correlated with the training error.
As mentioned previously, the performance of a classification model is largely dependent on the features selected; the latter could be potential biomarkers. It is known that there is a strong relationship between co-expressed genes and protein pairs. The reason behind this claim is that genes coding for proteins that have an interaction share a higher similarity than proteins that do not interact. Chuang et al. [2007] used a protein network approach to identify potential biomarkers from subnetworks constructed from the protein interaction databases. A higher prediction accuracy was achieved when classifying metastatic and nonmetastatic tumors using a subnetwork instead of individual genes as biomarkers. Here, we propose a method that combines the bagging approach with a joint feature set selection method based on a protein network. The selection of feature sets is a two-step process. First, among the different types of cancer, the most differentially expressed genes are selected. These differentially expressed genes are distributed among a set of classifiers in such a way that every classifier contains at least one differentially expressed gene. The second step involves adding more features to our set of classifiers in order to enhance the capability of the model in correctly classifying or predicting the cancer samples. The selection of features is based on the network constructed from protein–protein interactions. The core idea is to build a network of models where each model is composed of at least one differentially expressed gene and its k-nearest neighbors in a protein–protein interaction network, where k is a predefined threshold. After building an ensemble of feature sets, classification is performed using multiclass support vector machines on the different models to predict the cancer type. The idea behind this ensemble model is to build a set of classifiers where each classifier has a number of genes that likely perform similar biological functions or share a similar pathway. By taking the k-nearest neighbors of a differentially expressed gene, we anticipate that a number of diverse specialized models and an increase in the accuracy of cancer type prediction could be achieved. This ensemble approach could help provide more specific and better hypothesis of the gene pathways and cellular processes that relate to a cancer subtype, i.e., target a pathway/process rather than one gene or interaction.
APPLICATION OF CANCER CLASSIFICATION IN IDENTIFYING BIOMARKERS FOR DRUG DESIGN
The development of drugs that target cancer cells is a challenging pharmaceutical task. Cancer classification based on gene expression has the potential to assist the drug design process by aiding in the identification of potential biomarkers [Clarke et al., 2004]. Drug design is a lengthy process that passes through many developmental, as well as clinical testing, stages. Therefore, developing more efficient methods of identifying biomarkers for different cancer subtypes that can be verified experimentally could enhance the drug pipeline.
The ensemble classification approach could provide a method of identifying biomarkers for different cancer subtypes and thus could lead to potential targets. Ensemble classification can detect genes that work as a group in determining a cancer subtype. Specifically, if an ensemble model achieves high prediction accuracy, then the selected features could be considered as important biomarkers. The candidates selected using this approach may not necessarily be successful targets in cancer treatment because of the complexities involved in tumor initiation and progression. However, an optimistic hypothesis could be generated stating that if the genes that distinguish cancer subtypes are identified, they could be potential biomarkers for the subtypes and could be investigated further experimentally.
An ensemble classification model can be used to help predict biomarkers. Having prior information about potential biomarkers that contribute to distinguishing the different cancer subtypes could be useful in building the classification model. Next, one could investigate whether the features can accurately distinguish between the different cancer subtypes. Although Xiong et al. [2001] used a single classification model to identify the biomarkers, an ensemble classification approach could also have been used for quantifying the performance of the features selected. Ensemble classification approach is a more stable and accurate classification approach if diversity is a major aspect of the data set than using a single classifier as shown in Dietterich [2000] and Valentini et al. [2004] as well as in our analysis (Table 1 and Fig. 3).
CONCLUSION
Ensemble modeling could be very useful in cancer classification as it outperforms single classifiers and enhances prediction accuracy. However, a major issue that remains to be addressed is the fact that no single ensemble model can outperform all other ensemble models in cancer classification. This is due to the different ways one can construct an ensemble classifier, the diversity of the network of models built, and the characteristics of the cancer data sets. Nevertheless, ensemble modeling offers improved prediction accuracy and could further aid in the drug design process of cancer subtypes, given the heterogeneity of cancers, which ensemble classification appears amenable to modeling.
Acknowledgments
This research was supported in part by the National Institutes of Health (R01GM079688 and 1R01GM089866) and the National Science Foundation (CBET 0941055 and DBI 0701709).
References
- Armstrong SA, Kung AL, Mabon ME, Silverman LB, Stam RW, Den Boer ML, Pieters R, Kersey JH, Sallan SE, Fletcher JA, et al. Validation of a therapeutic target identified by gene expression based classification. Cancer Cell. 2002;3:173–183. doi: 10.1016/s1535-6108(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Armstrong SA, Staunton JE, Silverman LB, Pieters R, Den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet. 2002;30:41–47. doi: 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- Breiman L. Random forests. Mach Learn. 2001;45:5–32. [Google Scholar]
- Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol Syst Biol. 2007;3:1–10. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke PA, Poele RT, Workman P. Gene expression microarray technologies in the development of new therapeutic agents. Eur J Cancer. 2004;40:2560–2591. doi: 10.1016/j.ejca.2004.07.024. [DOI] [PubMed] [Google Scholar]
- Dietterich TG. Ensemble methods in machine learning. In: Kittler J, Roli F, editors. Proceedings of the First International Workshop on Multiple Classifier Systems. London, UK: Springer-Verlag; 2000. pp. 1–15. [Google Scholar]
- Freund Y, Schapire ER. A decision-theoretic generalization of on-line learning and an application to boosting. J Comput Syst Sci. 1997;55:119–139. [Google Scholar]
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, et al. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- Harrington CA, Rosenow C, Retief J. Monitoring gene expression using DNA microarrays. Curr Opin Microbiol. 2000;3:285–291. doi: 10.1016/s1369-5274(00)00091-6. [DOI] [PubMed] [Google Scholar]
- Liu B, Cui Q, Jiang T, Ma S. A combinational feature selection and ensemble neural network method for classification of gene expression data. BMC Bioinformatics. 2004;5:136–147. doi: 10.1186/1471-2105-5-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan AC, Gilbert D. Ensemble machine learning on gene expression data for cancer classification. Appl Bioinformatics. 2003;2(Suppl 3):75–83. [PubMed] [Google Scholar]
- Valentini G, Muselli M, Ruffino F. Cancer recognition with bagged ensembles of support vector machines. Neurocomputing. 2004;56:461–466. [Google Scholar]
- Wang SL, You HZ, Lei YK, Li XL. Performance comparison of tumor classification based on linear and non-linear dimensionality reduction methods. Adv Intell Comput Theories Appl. 2010;6215:291–300. [Google Scholar]
- Xiong M, Fang Z, Zhao J. Biomarker identification by feature wrappers. Genome Res. 2001;11:1878–1887. doi: 10.1101/gr.190001. [DOI] [PMC free article] [PubMed] [Google Scholar]