Abstract
Oculocutaneous albinism (OCA) is a heterogeneous group of autosomal recessive disorders resulting from mutations of the tyrosinase (TYR) gene and presents with either complete or partial absence of pigment in the skin, hair and eyes due to a defect in an enzyme involved in the production of melanin. In this study, mutations in the TYR gene of 30 unrelated Iranian OCA1 patients and 100 healthy individuals were examined using PCR-sequencing. Additionally, in order to predict the possible effects of new mutations on the structure and function of tyrosinase, these mutations were analyzed by SIFT, PolyPhen and I-Mutant 2 software. Here, two new pathogenic p.C89S and p.H180R mutations were detected in two OCA1 patients. Moreover, the R402Q and S192Y variants, which are common non-pathogenic polymorphisms, were detected in 17.5% and 35% of the patients, respectively. The outcome of this study has extended the genotypic spectrum of OCA1 patients, which paves the way for more efficient carrier detection and genetic counseling.
Introduction
Oculocutaneous albinism (OCA) is a group of congenital heterogeneous disorders characterized by either complete or partial absence of pigment in the skin, hair and eyes because of the absence of or a defect in an enzyme involved in the production of melanin [1]. OCA's symptoms include poor visual acuity, nystagmus, iris transillumination, strabismus, photophobia, foveal hypoplasia and misrouting of the optic nerve fibers at the chiasm [2].
OCA type 1 (OCA1, MIM 203100) is the most severe form of albinism and is caused by mutations in the tyrosinase gene (TYR, MIM 606933; 11q14–q21). Other subtypes of OCA include OCA type 2 (OCA2, MIM#203200) caused by mutations in the OCA2 gene (15q11.2–q12), OCA type 3 (OCA3, MIM#203290) associated with mutations in the tyrosinase-related protein gene (TYRP1, 9p23) and OCA type 4 (OCA4, MIM#606574) because of mutations in the membrane-associated transporter gene (MATP, 5p13.3) [3]. The prevalence of different forms of OCA fluctuates widely in different populations [4], where OCA1 is the most common subtype found in Caucasians and accounts for about 50% of all cases worldwide [5], [6], while OCA2 is most common in Africa and accounts for about 30% of all cases worldwide [7].
There are two subtypes of OCA1: OCA1A and OCA1B. OCA1 is caused by a mutation causing a complete lack of tyrosinase activity, while mutations resulting in the retainment of some enzyme activity result in OCA1B, where some melanin pigments are accumulated over time [8]. The common features between these two forms of OCA1 are nystagmus and foveal hypoplasia with reduced visual acuity. The human tyrosinase gene (TYR, 11q14–q21, MIM 606933) has 5 exons, spans about 65 kb of the genomic DNA, and encodes a 60 kDa glycoprotein - tyrosinase type I [9]. Tyrosinase catalyzes multiple steps in melanin synthesis, including the critical first and second reactions: the hydroxylation of tyrosine to L-DOPA and the oxidation of L-DOPA to DOPA-quinone. Mutations in TYR can cause complete or partial OCA depending on residual activity [10]. Chromosome 11 contains a pseudogene known as Tyrosinase-Like Gene (TYRL, 11p11.2; MIM#191270). This gene shares 98.55% sequence identity with the 3′–region of the TYR gene, including exons 4 and 5 [11]. Tomita et al (1989) reported the first pathological mutation in the TYR gene [12]. Presently, the Human Gene Mutation Database (HGMD at http://www.hgmd.org/) which is the largest general mutation database, contains approximately 320 different mutations of the TYR gene that have been documented. In this study, the TYR gene was examined in individuals who met the clinical criteria proposed for OCA1, in order to characterize the associated mutations.
Materials and Methods
Specimen Collection and Ethical Statement
Thirty Iranian OCA1 patients, including 12 females and 18 males with a mean age of 18 years, from 30 unrelated families were clinically diagnosed between February 2009 and December 2012. All of the patients had typical features of OCA1A, as summarized in Table 1. Blood samples from these 30 OCA1 patients and 100 healthy individuals as controls were obtained from the Special Medical Center, Tehran-Iran. Written informed consent, including consent to participate in the study for genetic analysis and consent to publish, was obtained from patients, parents on behalf of children, and healthy controls, and the Medical Ethics Committee of the Special Medical Center specifically approved this study (Approval No. FF-40-3224). The exclusion criterion for the control group was any history of cancer, metabolic diseases, and nuclear and mitochondrial DNA-related diseases that may affect DNA.
Table 1. Clinical features of 30 Iranian OCA1 patients.
| Patient | Gender | Age | Skin color | Hair color | Iris pigmentation | Nystagmus | Photophobia | Foveal Hypoplasia* |
| 1 | M | 21 | White | White | Hypopigmented | + | + | + |
| 2 | M | 18 | White | White | Hypopigmented | + | + | + |
| 3 | M | 29 | White | White | Hypopigmented | + | + | + |
| 4 | F | 26 | White | White | Hypopigmented | + | + | + |
| 5 | F | 22 | White | White | Hypopigmented | + | + | + |
| 6 | M | 10 | White | White | Hypopigmented | + | + | + |
| 7 | M | 17 | White | White | Hypopigmented | + | + | + |
| 8 | M | 32 | White | White | Hypopigmented | + | + | + |
| 9 | F | 13 | White | White | Hypopigmented | + | + | + |
| 10 | M | 27 | White | White | Hypopigmented | + | + | + |
| 11 | M | 6 | White | White | Hypopigmented | + | + | + |
| 12 | M | 12 | White | White | Hypopigmented | + | + | + |
| 13 | F | 19 | White | White | Hypopigmented | + | + | + |
| 14 | M | 30 | White | White | Hypopigmented | + | + | + |
| 15 | M | 4 | White | White | Hypopigmented | + | + | + |
| 16 | M | 11 | White | White | Hypopigmented | + | + | + |
| 17 | M | 23 | White | White | Hypopigmented | + | + | + |
| 18 | F | 34 | White | White | Hypopigmented | + | + | + |
| 19 | F | 10 | White | White | Hypopigmented | + | + | + |
| 20 | F | 19 | White | White | Hypopigmented | + | + | + |
| 21 | M | 26 | White | White | Hypopigmented | + | + | + |
| 22 | F | 7 | White | White | Hypopigmented | + | + | + |
| 23 | M | 35 | White | White | Hypopigmented | + | + | + |
| 24 | F | 12 | White | White | Hypopigmented | + | + | + |
| 25 | F | 18 | White | White | Hypopigmented | + | + | + |
| 26 | M | 5 | White | White | Hypopigmented | + | + | + |
| 27 | M | 28 | White | White | Hypopigmented | + | + | + |
| 28 | F | 3 | White | White | Hypopigmented | + | + | + |
| 29 | F | 11 | White | White | Hypopigmented | + | + | + |
| 30 | M | 14 | White | White | Hypopigmented | + | + | + |
*It is based on fundus exam.
DNA extraction and Polymerase Chain Reaction (PCR)
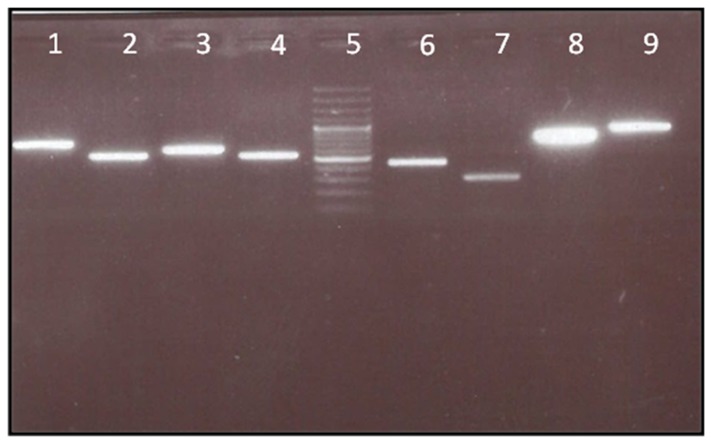
Genomic DNA was extracted from blood samples of each individual, using a QIAmapDNA micro Kit (QIAGEN#56304). The exons 1 to 3 PCR primers for amplification of the TYR gene are shown in Table 2. Briefly, PCR was performed in a 25 µl reaction volume containing 50–100 ng of genomic DNA, 2.5 µl of 10×PCR buffer, 0.1 mM of each dNTP, 1 mM of MgCl2, 0.1 µM of each primer, and 0.5 units of Taq polymerase (CinnaGen, Iran) in a thermocycler (Eppendorf, Humburg). For all amplicons, the genomic DNA was denatured at 94°C for 5 min, followed by 35 cycles of denaturation at 94°C for 1 min. The annealing temperature differed according to the Tm (°C) value of each primer set (Table 2). The extension was at 72°C for 1 min and the final extension was at 72°C for 10 min. In addition, to avoid co-amplification of the pseudogene TYRL, the Chaki PCR protocol [11] was applied for exons 4 and 5. The PCR products were examined for specificity via 1.5% agarose gel electrophoresis (Figure 1).
Table 2. PCR Primer pairs for amplification of the TYR gene in OCA1 patients.
| Gene name | Primer name | Exon | Locus (nDNA) | Primer sequence (5′ to 3′) | Amplicon size (bp) | Temperature (°C) |
| TYR | AL-1-1F | 1–1* | 88910556 | CAAACTGAAATTCAATAACATATAAGG | 678 | 59 |
| TYR | AL-1-1R | 88911233 | GTGGACAGCATTCCTTCTCC | |||
| TYR | AL-1-2F | 1–2* | 88910948 | TTCAGAGGATGAAAGCTTAAGATAAA | 521 | 59 |
| TYR | AL-1-2R | 88911468 | CGTCTCTCTGTGCAGTTTGG | |||
| TYR | AL-1-3F | 1–3* | 88911171 | CTGGCCATTTCCCTAGAGC | 605 | 60 |
| TYR | AL-1-3R | 88911775 | CCACCGCAACAAGAAGAGTC | |||
| TYR | AL-1-4F | 1–4* | 88911487 | CATCTTCGATTTGAGTGCCC | 521 | 59 |
| TYR | AL-1-4R | 88912007 | CCCTGCCTGAAGAAGTGATT | |||
| TYR | AL-2F | 2 | 88924298 | CCAACATTTCTGCCTTCTCC | 442 | 60 |
| TYR | AL-2R | 88924739 | TCAGCTAGGGTCATTGTCGAT | |||
| TYR | AL-3F | 3 | 88960909 | AGTTATAAATCAAATGGGATAATCA | 296 | 56 |
| TYR | AL-3R | 88961204 | ACATTTGATAGGCACCCTCT | |||
| TYR | AL-4F | 4 | 89017552 | CTGTTTCCAATTTAGTTTTATAC | 790 | 56 |
| TYR | AL-4R | 89018341 | TACAAAATGGCCTATGTTAAGC | |||
| TYR | AL-5F | 5 | 89028218 | TGTCTACTCCAAAGGACTGT | 924 | 55 |
| TYR | AL-5R | 89029138 | GGCACTTAGCTGGATGTGTT |
*Shows that due to the large size of exon 1, it is divided into four overlapping fragments.
Figure 1. Agarose gel electrophoresis of PCR product.
The PCR products electrophoresed on a 1.5% agarose gel. From left: lane 1: exon 1–1 (700 bp), lane 2: exon 1–2 (500 bp), lane 3: exon 1–3 (600 bp), lane 4: exon 1–4 (500 bp), Lane 5: DNA ladder (Thermo Scientific Gene Ruler 100 bp #SM0241/2/3), Lane 6: Exon 2 (450 bp), Lane 7: Exon 3 (300 bp), Lane 8 Exon 4(790 bp), Lane 9: Exon5 (920 bp).
DNA sequencing and in silico analysis of the variants
The PCR products were sequenced with the forward or reversed primers on an ABI 3700 sequencer (Kosar Company, Tehran) and compared with the wild-type TYR sequence (NM 000372.3) at the NCBI Reference Sequence Database (http://www.ncbi.nlm.nih.gov/refseq/), using the FinchTV program. Identification of the mutations at protein level was verified via the Human Gene Mutation Database (HGMD). For the novel mutations that were found in patients, further molecular tests were performed on the DNA of their parents and the 100 healthy individuals. To predict the functional effects of novel mutations, the sequence alterations were assessed by the in silico prediction algorithms SIFT [13], Polyphen-2 [14], and I-Mutant 2.0 (http://folding.biofold.org/i-mutant/i-mutant2.0.html).
Statistical analysis
Fisher's exact test using SPSS (Statistical Package for the Social Sciences, version: 13) was used to analyze the relationship between the presence of novel mutations in patients and control groups; where p-values <0.05 were regarded as statistically significant.
Result
In our study, the clinical diagnosis of 30 Iranian OCA1A patients was confirmed as through molecular screening of mutations in the TYR gene in 19 patients (Table 3). Twelve different TYR missense mutations were identified, where two of them have not been previously reported (Figure 2, Table 4).TYR mutations were homozygous in 18 of the cases. These mutations were observed in the compound heterozygous state in one patient (Patient 19). Additionally, patients 3, 9, 12, 13, 18 and 24 were heterozygous for these mutations (Table 3).
Table 3. TYR mutations and polymorphisms in 30 Iranian patients.
| Patient No. | Mutation 1 | Mutation 2 | Polymorphisms | Molecular diagnosis |
| 1 | c.265T>A (C89S) * | c.265T>A (C89S) | - | OCA1 |
| 2 | - | - | p.R402Q(Hetero) | - |
| 3 | c.606T>G (H202Q) | - | p.S192Y(Hetero) | - |
| 4 | c.1217C>T(P406L) | c.1217C>T(P406L) | - | OCA1 |
| 5 | c.1217C>T(P406L) | c.1217C>T(P406L) | - | OCA1 |
| 6 | c.1255G>A(G419R) | c.1255G>A(G419R) | - | OCA1 |
| 7 | c.606T>G (H202Q) | c.606T>G (H202Q) | - | OCA1 |
| 8 | c.896G>A(R299H) | c.896G>A(R299H) | - | OCA1 |
| 9 | c.1217C>T(P406L) | - | p.S192Y(Hetero) | - |
| 10 | c.649C>T (R217W) | c.649C>T (R217W) | - | OCA1 |
| 11 | - | - | p.S192Y(Homo) | - |
| 12 | c.1037G>A(G346E) | - | p.R402Q(Hetero) | - |
| 13 | c.98A>C(K33T) | - | p.R402Q(Hetero) | - |
| 14 | c.1255G>A(G419R) | c.1255G>A(G419R) | - | OCA1 |
| 15 | - | - | p.R402Q(Hetero) | - |
| 16 | c.996G>A(M332I) | c.996G>A(M332I) | - | OCA1 |
| 17 | c.715C>T(R239W) | c.715C>T(R239W) | - | OCA1 |
| 18 | c.606T>G (H202Q) | - | p.S192Y(Hetero) | - |
| 19 | c.539A>G (H180R) | c.1037G>A(G346E) | - | OCA1 |
| 20 | c.140G>A (G47D) | c.140G>A (G47D) | - | OCA1 |
| 21 | c.1037G>A(G346E) | c.1037G>A(G346E) | - | OCA1 |
| 22 | c.715C>T(R239W) | c.715C>T(R239W) | - | OCA1 |
| 23 | c.649C>T (R217W) | c.649C>T (R217W) | - | OCA1 |
| 24 | c.1217C>T(P406L) | - | p.S192Y(Hetero) | - |
| 25 | c.140G>A (G47D) | c.140G>A (G47D) | - | OCA1 |
| 26 | c.715C>T(R239W) | c.715C>T(R239W) | - | OCA1 |
| 27 | c.98A>C(K33T) | c.98A>C(K33T) | - | OCA1 |
| 28 | c.896G>A(R299H) | c.896G>A(R299H) | - | OCA1 |
| 29 | - | - | p.R402Q(Hetero) | - |
| 30 | - | - | p.R402Q(Hetero) | - |
*Hetero: Heterozygous; Homo: Homozygous; (-): undetected; New mutations are in bold.
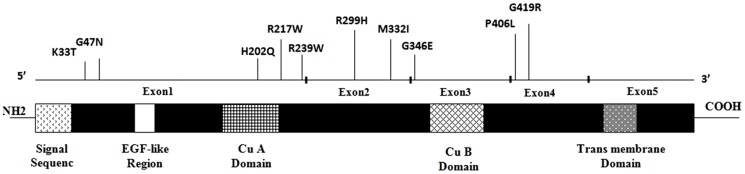
Figure 2. Reported mutations in this study are distributed on the TYR gene.
Table 4. The identified mutations in the TYR gene from OCA1 patients in our study.
| Nucleotide change | Amino acid change | Location | Frequency (%) | Status | Reference |
| c.98A>C | K33T | Exon 1 | 2(5.4) | Homo (1)*; Hetero (1) | Reported [6] |
| c.140G>A | G47D | Exon 1 | 2(5.4) | Homo (2) | Reported [42] |
| c.265T>A | C89S | Exon 1 | 1(2.7) | Homo (1) | Not reported (New) |
| c.575C>A | S192Y | Exon 1 | 5(13.5) | Homo (1); Hetero (4) | Reported [43] |
| c.539A>G | H180R | Exon 1 | 1(2.7) | Hetero (1) | Not reported (New) |
| c.606T>G | H202Q | Exon 1 | 3(8.1) | Homo (1); Hetero (2) | Reported [44] |
| c.649C>T | R217W | Exon 1 | 2(5.4) | Homo (2) | Reported [45] |
| c.715C>T | R239W | Exon 1 | 3(8.1) | Homo (3) | Reported [46] |
| c.896G>A | R299H | Exon 2 | 2(5.4) | Homo (2) | Reported [45] |
| c.996G>A | M332I | Exon 2 | 1(2.7) | Homo (1) | Reported [47] |
| c.1037G>A | G346E | Exon 3 | 3(8.1) | Homo (1); Hetero (2) | Reported [48] |
| c.1205G>A | R402Q | Exon 4 | 6(16.2) | Hetero (6) | Reported [24] |
| c.1217C>T | P406L | Exon 4 | 4(10.8) | Homo (2); Hetero (2) | Reported [49] |
| c.1255G>A | G419R | Exon 4 | 2(5.4) | Homo (2) | Reported [16] |
*The number in parenthesis in the status column shows the number of patients; New mutations are in bold.
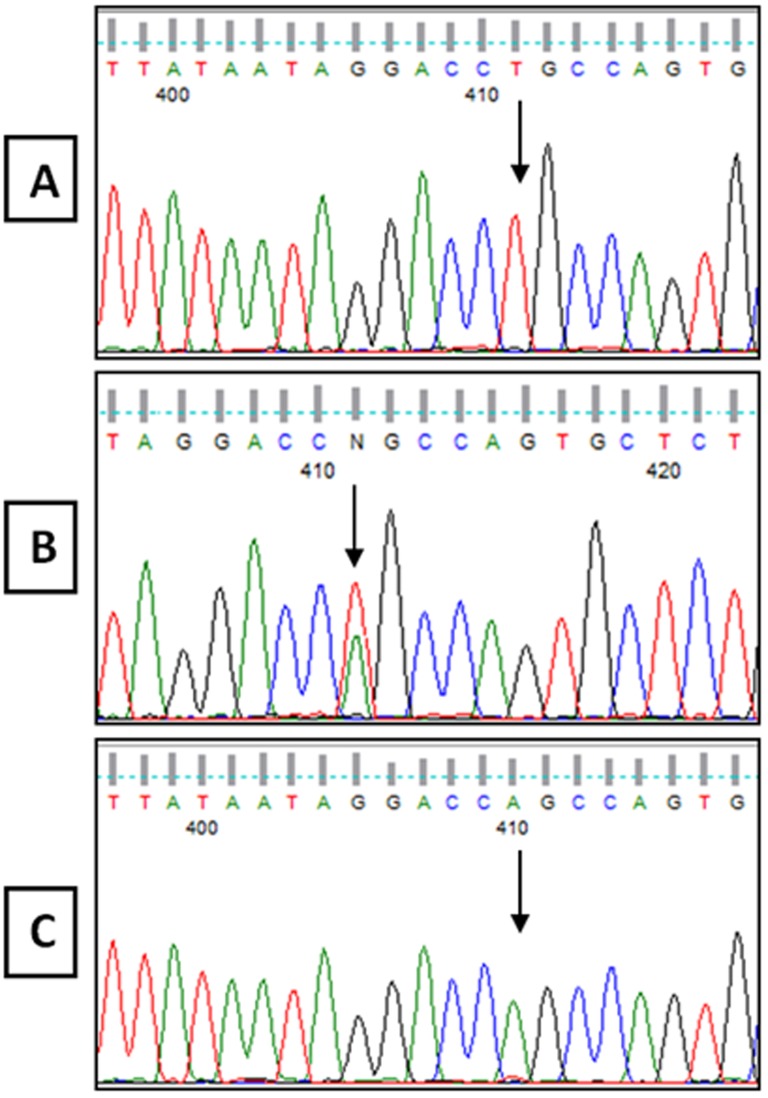
The R402Q and S192Y variants, which are common non-pathogenic polymorphisms, were detected in 6 and 5 cases, respectively (Figure 3 and Table 3). In patient 1, a new homozygous c.265T>A change in codon 89, which resulted in a cysteine to serine conversion (C89S), was identified (Figure 4). Moreover, patient 19 was compound heterozygous for the reported G346Q and new H180R (c.539A>G) mutations (Figure 5). The new p.C89S and p.H180R mutations were significantly (p<0.05) detected in the patients (Table 5), and compared to global database of TYR gene mutations (Figure 6).To predict the possible effects of these new mutations on the structure and function of tyrosinase, the mutations were analyzed using SIFT, PolyPhen and I-Mutant 2 softwares (Table 5). Here, SIFT results indicated that both C89S and H180R mutations were predicted as deleterious, with SIFT scores of −9.497 and −7.772, respectively. Using the I-Mutant server, prediction based on the sign of the free energy change value (sign of DDG) for the new C89S and H180R mutations showed that these mutations decrease the stability of the protein. Based on the PolyPhen score, both the C89S and H180R mutations were found as “Probably Damaging” to protein structure and function, with a score of 1.000, although further research is required to confirm these in silico findings.
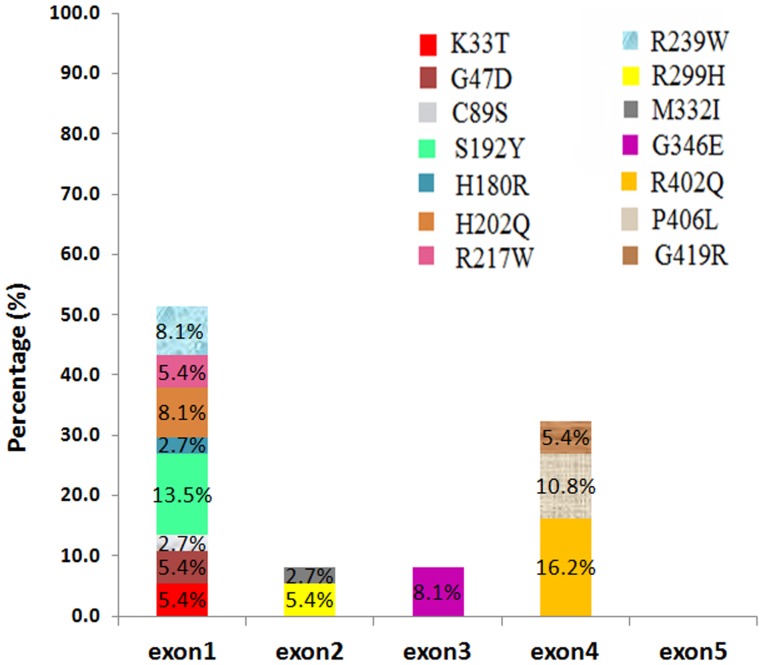
Figure 3. Distribution of mutations on the TYR gene in OCA1 patients.
Bar diagram indicates the percentage of mutations that were found in this study.
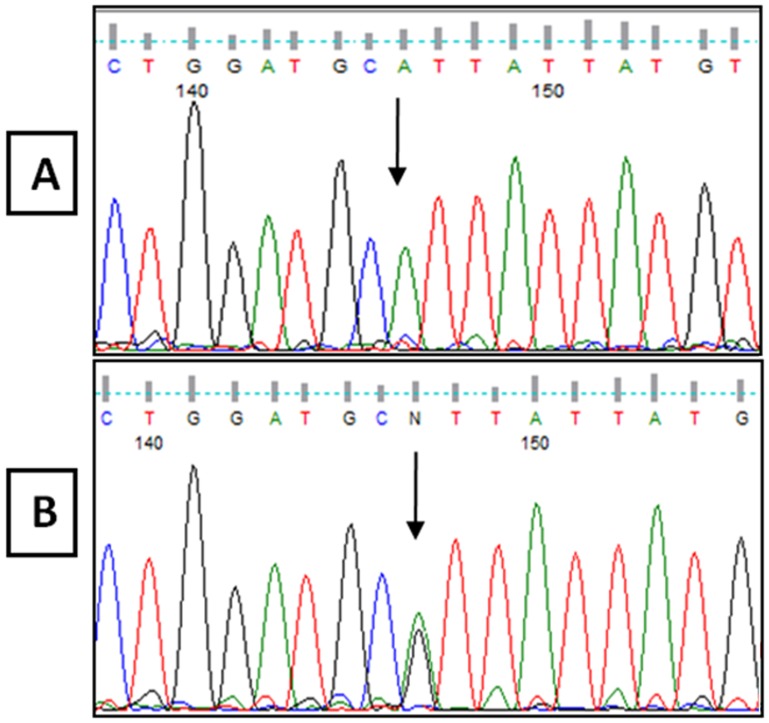
Figure 4. DNA sequencing result from Exon 1 of the TYR gene, showing c.265 T>A mutation.
A: Normal Sequence from control. B: Sequence from unaffected parents showing the c.265 T>A heterozygous mutation. C: Sequence from patient 1 with a new c.265 T>A homozygous mutation.
Figure 5. DNA sequencing result from Exon 1 of the TYR gene showing c.539 A>G mutation.
A: Normal Sequence from control and unaffected father. B: Sequence from patient 19 and an unaffected mother showing the c.539 A>G heterozygous mutation.
Table 5. Statistical and Bioinformatics Analysis of two novel pathogenic mutations.
| Novel mutation | Statistical analysis | Bioinformatic analysis | ||||
| p-value | Polyphen 2 | SIFT | I-Mutant 2.0 | |||
| Prediction | Score | Prediction | Score | Prediction (sign of DDG) | ||
| C89S | 0.048 | Probably Damaging | 1 | Deleterious | −9.497 | decrease stability |
| H180R | 0.048 | Probably Damaging | 1 | Deleterious | −7.772 | decrease stability |
Fisher's exact test using the SPSS was used for statistical analysis of novel mutations. A p-value of <0.05 is considered statistically significant. Novel mutations were analyzed by three computational methods PolyPhen 2 (benign/damaging), SIFT (tolerated/deleterious), and I-Mutant 2.0 (increase stability/decrease stability) for Bioinformatics analysis in order to predict the functional impact of novel amino acid changes.
p-value: statistically significant (p<0.05).
PolyPhen Prediction Score: benign ≤0.5; probably damaging (0.5<).
SIFT Prediction Score: deleterious (≤0.05); tolerated (≥0.05).
I-Mutant 2.0 Prediction: sign of DDG: decrease stability or increase stability.
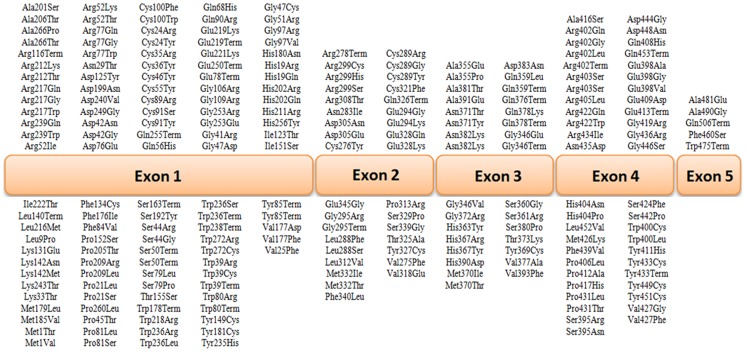
Figure 6. Database of tyrosinase (TYR) gene mutations.
[50] The mutations identified in OCA1 for each exon are described in the upper and lower half of the schema.
Discussion
Missense mutation is the most common mutational type that is correlated to OCA1 [15]. For the first time, King et al [16] recognized that most of the TYR missense mutations were located in four areas of the gene. Until now, TYR missense mutations are delineated by five key functional sites of the enzyme. Two locations are the copper-binding sites. Others are located at the 3′-end of the copper B-binding region near the amino terminus of the protein, and between the CuA and CuB domains. According to many reports, mutations in the Cu-binding site change the conformation of the a-helical regions [9] as well as the position of the histidine residues, which either could prevent proper binding of Cu to the histidine ligands or prevent TYR-Cu interaction.
In this study, the missense mutations were detected in all exons of the TYR, except for exon 5, where 51.3% and 32.4% of the mutations were found in exon 1 and exon 4, respectively, which could be considered as mutation hot spots of the TYR gene. Fukai et al (1995) reported that the R402Q mutation was associated with OCA1 incidence. At the physiological temperature (37°C), the p.R402Q tyrosinase has a reduced activity and is retained in the endoplasmic reticulum of melanocytes [17]–[20]. The contribution of the p.R402Q temperature-sensitive variant to the albino phenotype has been heavily debated in literature [10]. By itself this variant is not sufficient to cause albinism [21], [22]. It is possible that p.R402Q causes partial albinism only when paired with certain a genetic background [23], [24]. In support of this, several observers have noted that the p.R402Q variant is more common in OCA patients with one TYR mutation than in patients with two mutations [10]. However, in our study, of 6 patients (16.2%) with the heterozygous p.R402Q variant, two (Patients 12 and13) were heterozygous for one reported missense mutation in the TYR gene, G346E and K33T, while no mutation was found for the remaining 4 patients (Table 3).
The p.S192Y variant, which was reported as an SNP (rs1042602) in many populations such as Caucasians, Africans, Japanese and South Asians [25], was detected in 5 patients in our study, where 4 were heterozygous for one reported missense mutation, and one was only homozygous for the p.S192Y variant (Table 3). According to our results, the new C89S (c.265T>A) mutation was not found in the 100 control individuals. Moreover, further analysis of the parents with the normal phenotype indicated that they were heterozygous for this mutation (Figure 4). Additionally, the c.265 T>C mutation in this codon, which causes a cysteine to arginine substitution, was previously reported as a pathogenic mutation by Spritz [26]. The region spanning codons 73–107 is entirely conserved between human [27], [28] and mouse [29], [30] tyrosinases, as this region is one of the six potential N-glycosylation sites (codons 86–88), it could be vital for the function of tyrosinase [26]. Furthermore, cysteine residues in tyrosinase play an important role in the proper folding and maintenance of the tyrosinase tertiary structure in humans [31]. Alterations of these cysteine residues, such as C89S mutations in exon 1 could inactivate the protein and cause OCA1 [3]. Therefore, C89S could be considered as a pathogenic mutation.
According to our study, the new H180R (c.539A>G) mutation has not been previously reported, while in this codon (180), another pathogenic mutation (c.538C>A) that converts a His residue to an Asn residue (H180N) was previously reported. In addition, the H180R mutation is located in a sensitive domain of the enzyme, which converts one of the His residues that are bound to CU ion (metal binding site) [32]. Moreover, the father and mother of this patient were heterozygous for the G346Q and H180R mutations, respectively, and showed a normal phenotype (Figure 5). Therefore, the H180R could be considered as a pathogenic mutation.
In this study, 5 patients did not show any causative mutations in the TYR gene, which is probably due to the involvement of other OCA genes, such as the OCA2 gene, undiscovered OCA genes [33], [34], variants in the promoter or other regulatory elements which were not detected by DNA sequencing [35], synergistic or epistatic heterozygosity among known genes [36], [37], dominant mutations which are not detected as pathogenic because of ethnic/pigmentation background [23], [38], undetected splicing mutations [22], [39], undetected large deletions which are not recognized by DNA sequencing [6], [40], and undetected coding mutations because of allele dropout in sequencing [41].
In summary, we identified ten previously-reported missense mutations and two novel pathogenic mutations, c.265T>A (C89S) and c.539A>G (H180R), in Iranian patients with OCA1. However, further research is required to distinguish the subtype of OCA1 in these patients and to determine the biological role of these mutations that may affect tyrosinase enzymatic activity. In conclusion, the outcome of this study has extended the genotype spectrum of Iranian patients with pathogenic impact on oculocutaneous albinism type 1, which paves the way for a more efficient diagnosis and genetics counseling for carrier detection with this disorder in Iran.
Acknowledgments
We would like to thank all the participants for the blood donation from the Medical Genetics Department at the Special Medical Center, Tehran, Iran.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. Data are available from the Special Medical centre of Tehran Institutional Data Access / Ethics Committee for researchers who meet the criteria for access to confidential data.
Funding Statement
The authors would like to thank all the participants for the blood donation from the Medical Genetics Department at the Special Medical Center, Tehran, Iran. The authors would like to express their utmost gratitude and appreciation to University of Malaya Research Grant (RG084-13BIO), IPPP grant (PG082-2013B), BKP grant (BK020-2012) and Malaysian Ministry of Higher Education's MOHE-HIR Grants (UM.C/625/1/MOHE/MED/17 and UM.C/625/1/MOHE/MED/33) for providing partial financial supports to conduct this study. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Shin Hae P, Hyojin Ch, Yonggoo K, Myungshin K (2012) Molecular Analysis Of Korean Patients With Oculocutaneous Albinism. Jpn J Ophthalmol 56: 98–103. [DOI] [PubMed] [Google Scholar]
- 2. King RA, Hearing VJ, Creel DJ, Oetting WS (1995) Albinism In: Scriver CR, Beaudet AL, Sly WS, Valle D, Editors The Metabolic And Molecular Bases Of Inherited Disease. 3: 4353–4392. [Google Scholar]
- 3. Jing L, Kwong-Wai Ch, Leo WL Ch, Tak-Yeung Le, Pancy OS T, et al. (2010) Tyrosinase Gene (TYR) Mutations In Chinese Patients With Oculocutaneous Albinism Type 1. Clin Experiment Ophthalmol 38(1): 37–42. [DOI] [PubMed] [Google Scholar]
- 4. Grønskov K, Ek J, Brondum-Nielsen K (2007) Oculocutaneous Albinism. Orphanet J Rare Dis 2: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hutton SM, Spritz RA (2008) A Comprehensive Genetic Study of Autosomal Recessive Ocular Albinism in Caucasian Patients. Invest Ophthalmol Vis Sci 49(3): 868–872. [DOI] [PubMed] [Google Scholar]
- 6. Rooryck C, Morice-Picard F, Elçioglu NH, Lacombe D, Taieb A, et al. (2008) Molecular Diagnosis Of Oculocutaneous Albinism: New Mutations In The OCA1-4 Genes And Practical Aspects. Pigment Cell Melanoma Res 21(5): 583–587. [DOI] [PubMed] [Google Scholar]
- 7. Okoro AN (1975) Albinism in Nigeria. A clinical and social study. Br J Dermatol 92(5): 485–492. [PubMed] [Google Scholar]
- 8. Karaman A (2008) Oculocutaneous Albinism Type 1A: A Case Report. Dermatology Online Journal 14(11): 13. [PubMed] [Google Scholar]
- 9. Ray K, Chaki M, Sengupta M (2007) Tyrosinase And Ocular Diseases: Some Novel Thoughts On The Molecular Basis Of Oculocutaneous Albinism Type 1. Prog Retin Eye Res 26(4): 323–358. [DOI] [PubMed] [Google Scholar]
- 10. Simeonov DR, Wang X, Wang C, Sergeev Y, Dolinska M, et al. (2013) DNA Variations in Oculocutaneous Albinism: An Updated Mutation List and Current Outstanding Issues in Molecular Diagnostics. Hum Mutat 34(6): 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chaki M, Mukhopadhyay A, Ray K (2005) Determination Of Variants In The 3′-Region Of The Tyrosinase Gene Requires Locus Specific Amplification. Human mutation 26(1): 53–58. [DOI] [PubMed] [Google Scholar]
- 12. Tomita Y, Takeda A, Okinaga S, Tagami H, Shibahara S (1989) Human Oculocutaneous Albinism Caused By Single Base Insertion In The Tyrosinase Gene. BiochemBiophys Res Commun 164(3): 990–996. [DOI] [PubMed] [Google Scholar]
- 13. Kumar P, Henikoff S, Ng PC (2009) Predicting The Effects Of Coding Non-Synonymous Variants On Protein Function Using The SIFT Algorithm. Nat Protoc 4(7): 1073–1081. [DOI] [PubMed] [Google Scholar]
- 14. Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, et al. (2010) A Method And Server For Predicting Damaging Missense Mutations. Nat Methods 7(4): 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lin YY, Wei AH, Zhou ZY, Zhu W, He X, et al. (2011) A Novel Missense Mutation Of The TYR Gene In A Pedigree With Oculocutaneous Albinism Type 1 From China. Chin Med J 124(20): 3358–3361. [PubMed] [Google Scholar]
- 16. King RA, Mentink MM, Oetting WS (1991) Non-Random Distribution Of Missense Mutations Within The Human Tyrosine Gene In Type Ι (Tyrosinase-Related) Oculocutaneous Albinism. Mol Biol Med 8(1): 19–29. [PubMed] [Google Scholar]
- 17. Tripathi RK, Giebel LB, Strunk KM, Spritz RA (1991) A Polymorphism Of The Human Tyrosinase Gene Is Associated With Temperature-Sensitive Enzymatic Activity. GeneExpr 1(2): 103–110. [PMC free article] [PubMed] [Google Scholar]
- 18. Berson JF, Frank DW, Calvo PA, Bieler BM, Marks MS (2000) A Common Temperature-Sensitive Allelic Form Of Human Tyrosinase Is Retained In The Endoplasmic Reticulum At The Nonpermissive Temperature. J Biol Chem 275(16): 12281–12289. [DOI] [PubMed] [Google Scholar]
- 19. Halaban R, Svedine S, Cheng E, Smicun Y, Aron R, et al. (2000) Endoplasmic Reticulum Retention Is A Common Defect Associated With Tyrosinase-Negative Albinism. Proc Natl Acad Sci USA 97(11): 5889–5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Toyofuku K, Wada I, Valencia JC, Kushimoto T, Ferrans VJ, et al. (2001) Oculocutaneous Albinismtypes 1 And 3 Are ER Retention Diseases: Mutation Of Tyrosinase Or Tyrp1 Can Affect The Processing Of Both Mutant And Wild-Type Proteins. FASEB J 15(12): 2149–2161. [DOI] [PubMed] [Google Scholar]
- 21. Oetting WS, Pietsch J, Brott MJ, Savage S, Fryer JP, et al. (2009) The R402Q Tyrosinase Variant Does Not Cause Autosomal Recessive Ocular Albinism. Am J Med Genet A 149A(3): 466–469. [DOI] [PubMed] [Google Scholar]
- 22. Preising MN, Hedwig F, Miriam G, Birgit L (2011) Screening of TYR, OCA2, GPR143, and MC1R In Patients With Congenital Nystagmus, Macular Hypoplasia, And Fundus Hypopigmentation Indicating Albinism. Mol Vis 17: 939–948. [PMC free article] [PubMed] [Google Scholar]
- 23. Chiang PW, Spector E, Tsai AC (2009) Oculocutaneous Albinism Spectrum. Am J Med Genet A 149A(7): 1590–1591. [DOI] [PubMed] [Google Scholar]
- 24. Fukai K, Holmes SA, Lucchese NJ, Siu VM, Weleber RG, et al. (1995) Autosomal Recessive Ocular Albinism Associated With A Functionally Significant Tyrosinase Gene Polymorphism. Nat Genet 9(1): 92–95. [DOI] [PubMed] [Google Scholar]
- 25. Wei A, Wang Y, Long Y, Wang Y, Guo X, et al. (2010) A Comprehensive Analysis Reveals Mutational Spectra and Common Alleles in Chinese Patients with culocutaneous Albinism. J Invest Dermatol 130(3): 716–724. [DOI] [PubMed] [Google Scholar]
- 26. Spritz RA, Strunk KM, Hsieh CL, Sekhon GS, Francke U (1991) Homozygous Tyrosinase Gene Mutation In An American Black With Tyrosinase-Negative (Type IA) Oculocutaneous Albinism. Am J Hum Genet 48(2): 318–324. [PMC free article] [PubMed] [Google Scholar]
- 27. Kwon BS, Haq AK, Pomerantz SH, Halaban R (1987) Isolation And Sequence Of A Cdna Clone For Human Tyrosinase That Maps At The Mouse C-Albino Locus. Proc Natl Acad Sci USA 84(21): 7473–7477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Giebel LB, Strunk KM, Spritz RA (1991) Organization And Nucleotide Sequences Of The Human Tyrosinase Gene And A Truncated Tyrosinase-Related Segment. Genomics 9(3): 435–445. [DOI] [PubMed] [Google Scholar]
- 29. Yamamoto H, Takeuchi S, Kudo T, Makino K, Nakata A, et al. (1987) Cloning And Sequencing Of Mouse Tyrosinase cDNA. Jpn J Genet 62: 271–274. [Google Scholar]
- 30. Muller G, Ruppert S, Schmid E, Schutz G (1988) Functional Analysis Of Alternatively Spliced Tyrosinase Gene Transcripts. EMBO J 7(9): 2723–2730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia-Borron JC, Solano F (2002) Molecular Anatomy Of Tyrosinase And Its Related Proteins: Beyond The Histidine-Bound Metal Catalytic Center. Pigment Cell Res 15(3): 162–173. [DOI] [PubMed] [Google Scholar]
- 32. Opitz S, Käsmann-Kellner B, Kaufmann M, Schwinger E, Zühlke C (2004) Detection of 53 Novel DNA Variations Within the Tyrosinase Gene and Accumulation of Mutations in17 Patients with Albinism. Hum Mutat 23(6): 630–631. [DOI] [PubMed] [Google Scholar]
- 33. Lamason RL, Mohideen MA, Mest JR, Wong AC, Norton HL, et al. (2005) SLC24A5, a Putative Cation Exchanger, Affects Pigmentation in Zebrafish and Humans. Science 310(5755): 1782–1786. [DOI] [PubMed] [Google Scholar]
- 34. Wasmeier C, Romao M, Plowright L, Bennett DC, Raposo G, et al. (2006) Rab38 and Rab32 Control Post-Golgi Trafficking Of Melanogenic Enzymes. J Cell Biol 175(2): 271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Oetting WS, Fryer JP, Shriram S, King RA (2003) Oculocutaneous Albinism Type 1: The Last 100 Years. Pigment Cell Res 16(3): 307–311. [DOI] [PubMed] [Google Scholar]
- 36. Chiang PW, Fulton AB, Spector E, Hisama FM (2008a) Synergistic Interaction Of The OCA2 And OCA3 Genes In A Family. Am J Med Genet A 146A(18): 2427–2430. [DOI] [PubMed] [Google Scholar]
- 37. Zuk O, Hechter E, Sunyaev SR, Lander ES (2012) The Mystery Of Missing Heritability: Genetic Interactions Create Phantomheritability. Proc Natl Acad Sci USA 109(4): 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chiang PW, Drautz JM, Tsai AC, Spector E, Clericuzio CL (2008b) A New Hypothesis Of OCA1B. Am J Med Genet A 146A146A(22): 2968–2970. [DOI] [PubMed] [Google Scholar]
- 39. Desmet FO, Beroud C (2012) Bioinformatics And Mutations Leading To Exon Skipping. Methods Mol Biol 867: 17–35. [DOI] [PubMed] [Google Scholar]
- 40. Schnur RE, Sellinger BT, Holmes SA, Wick PA, Tatsumura YO, et al. (1996) Type I Oculocutaneous Albinism Associated With A Full-Length Deletion Of The Tyrosinase Gene. J Invest Dermatol 106(5): 1137–1140. [DOI] [PubMed] [Google Scholar]
- 41. Landsverk ML, Douglas GV, Tang S, Zhang VW, Wang GL, et al. (2012) Diagnostic Approaches To Apparent Homozygosity. Genet Med14(10): 877–882. [DOI] [PubMed] [Google Scholar]
- 42. Oetting WS, Witkop CJ Jr, Brown SA, Colomer R, Fryer JP, et al. (1993) A Frequent Tyrosinase Gene Mutation Associated With Type I-A (Tyrosinase-Negative) Oculocutaneous Albinism In Puerto Rico. Am J Hum Genet 52(1): 17–23. [PMC free article] [PubMed] [Google Scholar]
- 43. Stokowski RP, Pant PV, Dadd T, Fereday A, Hinds DA, et al. (2007) A Genomewide Association Study Of Skin Pigmentation In A South Asian Population. Am J Hum Genet 81(6): 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gargiulo A, Testa F, Rossi S, Di Iorio V, Fecarotta S, et al. (2011) Molecular And Clinical Characterization Of Albinism In A Large Cohort Of Italian Patients. Invest Ophthalmol Vis Sci 52(3): 1281–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tripathi RK, Strunk KM, Giebel LB, Weleber RG, Spritz RA (1992) Tyrosinase Gene Mutations In Type I (Tyrosinase-Deficient) Oculocutaneous Albinism Define Two Clusters Of Missense Substitutions. Am J Med Genet 43(5): 865–871. [DOI] [PubMed] [Google Scholar]
- 46. Nakamura E, Miyamura Y, Matsunaga J, Kano Y, Dakeishi-Hara M, et al. (2002) A Novel Mutation Of The Tyrosinase Gene Causing Oculocutaneous Albinism Type 1 (OCA1). J Dermatol Sci 28(2): 102–105. [DOI] [PubMed] [Google Scholar]
- 47. Grønskov K, Ek J, Sand A, Scheller R, Bygum A, et al. (2009) Birth Prevalence And Mutation Spectrum In Danish Patients With Autosomal Recessive Albinism. Invest Ophthalmol Vis Sci 50(3): 1058–1064. [DOI] [PubMed] [Google Scholar]
- 48. Oetting WS, Fryer JP, Oofuji Y, Middendorf LR, Brumbaugh JA, et al. (1994) Analysis Of Tyrosinase Gene Mutations Using Direct Automated Infrared Fluorescence DNA Sequencing Of Amplified Exons. Electrophoresis 15(2): 159–164. [DOI] [PubMed] [Google Scholar]
- 49. Giebel LB, Tripathi RK, Strunk KM, Hanifin JM, Jackson CE, et al. (1991) Tyrosinase Gene Mutations Associated With Type IB (“Yellow”) Oculocutaneous Albinism. Am J Hum Genet 48(6): 1159–1167. [PMC free article] [PubMed] [Google Scholar]
- 50. Stenson PD, Mort M, Ball EV, Shaw K, Phillips AD, et al. (2014) The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Human genetics 133(1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. Data are available from the Special Medical centre of Tehran Institutional Data Access / Ethics Committee for researchers who meet the criteria for access to confidential data.