Abstract
Easily screening markers for early detection of chronic heart failure (CHF) are lacking. We identified twenty differently expressed proteins including orosomucoid 1(ORM1) in urine between patients with CHF and normal controls by proteomic methods. Bioinformatics analyses suggested ORM1 could be used for further analysis. After verification by western blotting, the urinary levels of ORM1 were quantified with enzyme-linked immunosorbent assay (ELISA) by correcting for creatinine expression. The ORM1-Cr was significantly elevated in CHF patients than normal controls (6498.83±4300.21 versus 2102.26±1069.24 ng/mg). Furthermore, a Spearman analysis indicated that the urinary ORM1 levels had a high positive correlation with the classification of CHF, and the multivariate analysis suggested that the urinary ORM1 content was associated with the plasma amino-terminal pro- brain natriuretic peptide (NT-proBNP) (OR: 2.106, 95% CI: 1.213–3.524, P = 0.002) and the New York Heart Association (NYHA) classification (OR: 3.019, 95% CI: 1.329–4.721, P<0.001). In addition, receiving operating curve (ROC) analyses suggested that an optimum cut-off value of 2484.98 ng/mg with 90.91% sensitivity and 85.48% specificity, respectively, could be used for the diagnosis of CHF. To sum up, our findings indicate that ORM1 could be a potential novel urinary biomarker for the early detection of CHF.
Introduction
Chronic heart failure (CHF), a clinical syndrome of left ventricular systolic and/or diastolic dysfunction, remains a major threat to public health [1]. It has been estimated that there are currently over 5 million CHF patients, and more than 550,000 new patients diagnosed yearly in the USA [2]. With the ageing of the global population, the CHF populations are increasing. Recent studies have indicated that approximately 20% of people in the world will suffer CHF at some point through their lifetime [3]. Many CHF patients do not receive accurate diagnoses or optimum pharmacological treatments, therefore, resulting in substantial mortality and morbidity.
Although great progress has been made in diagnostic intensity and treatment, the long-term prognosis for CHF is still poor. In addition, the rising incidence of CHF is a cause for concern, and there is currently no gold standard for its diagnosis [4]. Early diagnosis and treatment are extremely important in this disease. Therefore, the development of a reliable, non-invasive biomarker is of considerable clinical importance, aiming at increasing the early detection rate of heart failure and/or predicting the progression of the disease in time.
Proteomic approaches, which could simultaneously analyze thousands of proteins and peptides, can be used to search for novel biomarkers for the early detection of the disease, their development and prognostic assessment, and the selection and monitoring of proper treatment modalities [5]. Proteomic patterns in body fluids present new opportunities for identification of novel, highly-sensitive specific markers for early detection of disease [6]. Obviously, urine samples are derived from minimally invasive procedures in clinical analysis, and recent advances in urinary proteomics have generated interest into new potential biomarkers for various systemic diseases such as various cancers and cardiovascular diseases [7], [8]. However, to date, easily applicable screening biomarkers for the early detection of CHF based on urine samples are lacking.
In our study, we applied two-dimensional differential gel electrophoresis (2D-DIGE) proteomic technologies to identify differentially expressed proteins in urinary protein extracts of CHF patients and healthy donors. Furthermore, we suggest that orosomucoid 1 (ORM1) is a potential novel urinary biomarker for the early detection of CHF.
Material and Methods
a) Urinary samples collection and preparation
All subjects in this study were recruited from the Chinese Han population at Nanfang Hospital (Guangzhou, China) from May 2011 to August 2012. The study protocol was approved by the Nanfang Hospital Ethics Committee. All subjects were informed about the purpose of the study and gave their written consent. In all, 197 patients with CHF were confirmed according to the Framingham Heart Study (FHS) diagnostic criteria of CHF [9]. The diagnosis of CHF was based on a complete clinical evaluation, laboratory testing, echocardiography, chest X-ray, and coronary angiography, coupled with plasma amino-terminal pro-brain natriuretic peptide (NT-proBNP) content ≥1000 pg/ml. The diagnosis was conducted independently by two experienced clinicians in Nanfang Hospital, Southern Medical University. Cardiac function for each CHF patient was divided into class II, class III, and class IV according to the Cardiac Function Standard of the New York Heart Association (NYHA). The main exclusion criteria were as follows: unconscious patients; other systemic diseases such as cancer, lung disease, liver cirrhosis, bleeding disorders, severe acute infectious and metabolic disorders. 83 healthy volunteers with no evidence of disease were used as control.
The second voided clean-catch urine samples form subjects were collected in the early morning. Each urine sample (20 ml) was directly collected into a sterile plastic tube and then immediately centrifuged at 2500×g for 10 min at 4°C to remove cell debris and particulate matter. The supernatant was stored at −80°C for further analysis. Repeated freeze-thaw cycles were avoided. All the clinical data of those subjects were collected as well.
b) Proteomic analysis
Equal volume urine specimens from 15 CHF patients and 15 controls were pooled respectively for 2D-DIGE analysis. Of these CHF patients, there were five cases in each NYHA class (II, III and IV). For processing, samples were first thawed on ice with the addition of protease inhibitors, 1 mmol/L phenylmethylsulfonyl fluoride, 5 mmol/L phenanthroline, and 5 mmol/L benzamidine (Sigma, St. Louis, Missouri, USA), and then centrifuged using Centricon Plus-20, 10,000 MWCO devices (Millipore, Bedford, MA, USA). Following extraction, other interfering components in the concentrated urine were removed using a 2D Clean-Up Kit (GE Healthcare, Piscataway, New Jersey, USA) according to the manufacturer's instructions. Protein concentration was measured with the 2D Quant Kit (GE Healtcare, Uppsala, Sweden).
Protein extracts obtained from the pooled urine samples were labeled with Cy2, Cy3 and Cy5 dyes (CyDye DIGE Fluor minimal dyes, GE Healthcare) according to the Ettan two-dimensional difference gel electrophoresis (DIGE) protocol. Briefly, 50 µg of urine protein samples from CHF patients and control group were minimally labeled with 400 pmol of Cy5 and Cy3 fluorescent dyes respectively. An internal pool was labeled with Cy2 and used to assess the reproducibility and statistical inferences. It was generated by combining equal amounts of extracts from all CHF and healthy control samples included in the study. All the labeled reactions were incubated for 30 min on ice protected from light, and then stopped by adding 1 ml of 10 mM lysine for 10 min [10]. Following the labeling reaction, all the three labeled samples were mixed and resolved in one gel.
An immobilized pH gradient (IPG) strip (24 cm, pH 3–10, nonlinear, GE Healthcare) was used for isoelectric focusing. For rehydration, samples were brought to 450 µl with rehydration buffer and 1 ml of mineral oil was added to avoid evaporation. The IPG strips were passively rehydrated overnight at room temperature. Isoelectric focusing was performed according to the protocol provided by the manufacturer (GE Healthcare) as follows: 1 h at 500 V, 1 h at 1000 V, 1 h at 5000 V, 1 h at 8000 V, then maintaining at 8000 V until a total of 60 kVh. After focusing, the strips were prepared for the second dimension gels by incubation for 15 min in equilibrium buffers I and II, respectively. Standard continuous SDS-PAGE electrophoresis for the second dimension (12%) was carried out using an Ettan DALT twelve system (GE Healthcare) until the dye front reached the bottom of the gels. Following SDS-PAGE, image scans were performed immediately using a Typhoon 9410 scanner (GE Healthcare) at the excitation emission of 488/520 nm (Cy2), 532/580 nm (Cy3) and 633/670 nm (Cy5), respectively. In addition, another strip with 1000 mg of proteins loaded was performed in parallel as a preparative gel for spots picking as marked in 2D-DIGE. The gel was stained with Coomassie Brilliant Blue [11]. Identical samples were run in three times.
After scanning, the gel images were analyzed using the DeCyder 5.01 software (GE Healthcare). Its differential in-gel analysis (DIA) module was used for pairwise comparisons of each sample with the internal standard within each gel by calculating the normalized spot volumes. The spot volumes were calculated by the background intensities combined with the borders of the spots. Spot volumes were normalized by dividing each Cy3 or Cy5 spot volume with the corresponding internal standard (Cy2). The average abundance changes for each spot across the different spot maps were calculated with the DeCyder biological variation analysis (BVA) module. The spots whose ratios of Cy5/Cy2 and Cy3/Cy2 changed by 1.5-fold or greater were regarded as differently up- or down- regulated expressed spots and were considered for further protein identification.
As described in our previous studies [12], [13], the protein spots were identified by matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF/TOF-MS). Shortly, the differentially expressed protein spots were excised from the Coomassie-stained gels with an Ettan Spot Picker (GE Healthcare), and then subjected to trypsin digestion, peptide extraction and desalting. The peptide mixtures were analyzed using an ABI Voyager DE-STR mass spectrometer (ABI 4700 Proteomic Analyzer, Applied Biosystems, Foster City, CA, USA). A trypsin- fragment peak was served as an internal standard for mass calibration. A list of the corrected mass peaks was the peptide mass fingerprinting (PMF).
Protein identification using peptide mass fingerprinting was performed using the MASCOT search engine (http://www.matrixscience.com/, Matrix Science Ltd, London, UK) against the MSDB protein database. The search was defined as the Homo sapiens subsets of the sequences in the Swiss–Prot and NCBI nonredundant protein sequence databases. The following search parameters were used during the searche: tolerance of one missed trypsin cleavages, the errors in peptide mass were within 25 ppm for both the precursor mass tolerance and the fragments mass. Carbamidomethylation of cysteine and oxidation of methionine as fixed and variable modifications, respectively, were taken into account for database searching. The proteins matched more than four peptides and with a MASCOT score higher than 63 were considered statistically significant (P<0.05). The identification results were filtered by peakErazor software (Lighthouse Data, Odense, Denmark). All matched sequences were manually validated [12], [13].
c) Bioinformatic analysis
The interaction network of differentially expressed proteins was performed automatically by STRING (Search Tool for the Retrieval of Interacting Genes/Proteins; version 8.3; http://string.embl.de/) with following analysis parameters[14]: species — Homo sapiens, confidence level — 0.400, active prediction methods — all. The PubGene Analysis tool (http://www.pubgene.org/) was used to search for literature bio-association analysis of these proteins [15]. The bio-associations had the following categories in Pubgene: Process, Function and Component. The key bio-processes related to the identified proteins were retrieved for further analysis.
d) Western blotting analysis
For western blot analyses, the samples involved six CHF patients (n = 2 in each class II, III and IV of NYHA), and six healthy controls. A total of 30 µg prepared urine proteins were separated by 12% SDS-PAGE. The gels were then transferred onto Polyvinylidene Xuoride (PVDF) (Millipore) membranes. The membranes were blocked in a solution of TBS containing 5% nonfat milk powder and 0.1% Tween-20 for 1 h at room temperature and then incubated overnight at 4°C with the monoclonal antibody against the human ORM1 protein (diluted 1∶500; Abcam, UK). After three 10-min washes in TBS-T, the membranes were incubated with horseradish peroxidase horseradish (HRP) conjugate of goat anti-rabbit IgG (Bioworld Technology, Louis Park, MN, USA) at a 1∶5000 dilution at room temperature for 1 h. The proteins were detected using an enhanced chemiluminescence (ECL, Pierce, Rockford, IL, USA) detection system. Relative intensities were documented and analyzed by densitometry.
e) Enzyme-linked immunosorbent assay (ELISA) analysis
The concentration of ORM1 protein in urine samples was measured with a commercially available ELISA kit (R&D Systems, Minneapolis, USA) according to the manufacturer's instructions. The assay has a minimum detectable dose of ORM1 at 0.538 ng/ml, exhibiting linearity between 3.12 and 200 ng/ml. The standard curve was created using the suppliers' lyophilized human ORM1. The levels of urinary ORM1 were corrected by urinary creatinine (Cr) concentrations to avoid the influence of urine volume. Thereby, the results were expressed as ORM1 -to-Cr ratio (ORM1/Cr nanograms per miligrams of creatinine). Urine creatinine (Cr) levels were measured at the Department of Clinical Laboratory of Nanfang Hospital (Guangzhou, China).
f) Statistical methods
All data were collected and used for statistical analyses with SPSS software 13.0 (SPSS, Chicago, IL, USA). Values were presented as mean ± standard deviation (SD). The difference among two groups or three groups was compared with Student's t test or one-way ANOVA. A Spearman analysis was performed to explore the relationship between urinary ORM1-Cr and the clinical characteristics of subjects where appropriate. Univariate (nonparametric rank sum test) and multivariate (logistic regression) analyses were conducted to evaluate the relationship between urinary ORM1 expression and clinical parameters of CHF, including age, sex, diabetes, hypertension, coronary heart disease, cardiomyopathy, renal dysfunction, left ventricular ejection fraction (LVEF), NT-proBNP and NYHA classification. Receiving operating curve (ROC) analyses were used to define the most optimal diagnostic cutoff as well as the diagnostic performance given by the area under the curve (AUC), estimating the sensitivity versus its false-positive rate at optimal cutoffs. The best statistical cut-off value of ORM1-Cr was defined, which means the point at which the sum of sensitivity and specificity is more than other points. The results were considered statistically significant at P-value <0.05.
Results
a) Clinical characteristics of subjects
Table 1 has list the clinical characteristics of all subjects in our study. There was no statistical difference in clinical characteristics (e.g. age, sex, serum creatinine and urinary creatinine). The urine samples form 15 CHF patients and 15 controls were randomly selected for 2D-DIGE analysis. Among the 15 CHF patients, there were five cases in each class II, III and IV of NYHA. Furthermore, there were 176 CHF patients and 62 healthy volunteers recruited for ELISA quantitative analysis. Of these 176 subjects with CHF, the CHF cases belonging to class II, III and IV numbered 34, 87 and 55 respectively.
Table 1. Clinical data for subjects in the study.
| Samples for 2D-DIGE | Samples for ELISA | |||
| Controls | CHF | Controls | CHF | |
| Number | 15 | 15 | 62 | 176 |
| Gender (Male/Female) | 8/7 | 6/9 | 30/32 | 99/78 |
| Age (years) | 58.67±14.89 | 61.73±13.82 | 59.82±13.17 | 62.62±14.59 |
| Urine creatinine (mg/dl) | 97.91±46.03 | 113.29±63.08 | 97.91±46.03 | 113.29±63.08 |
| Serum creatinine (umol/L) | 99.72±46.57 | 115.47±68.94 | 100.63±49.29 | 118.45±70.57 |
| NT-proBNP (pg/ml) | / | 6852.12±7816.69 | / | 7703.96±8695.85 |
b) 2D-DIGE analysis and twenty differently expressed proteins were identified by MALDI-FOF/TOF-MS
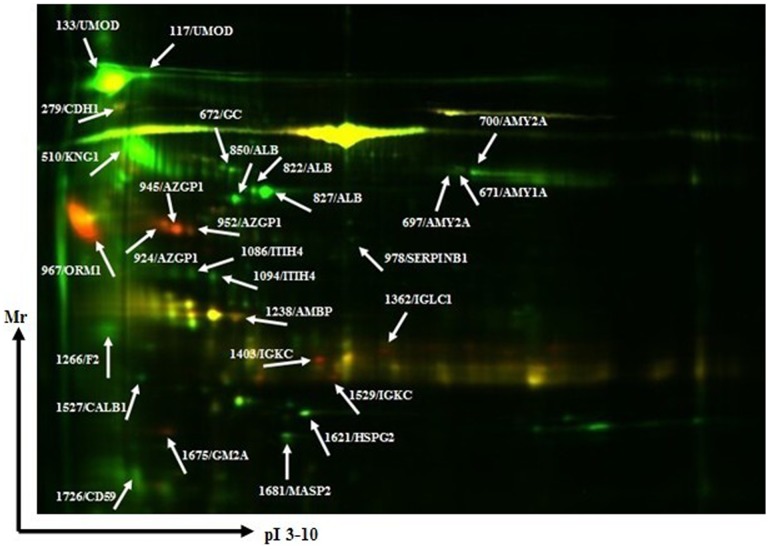
Figure 1 has shown a representative 2D-DIGE image from proteomic profiling of urine samples from patients with CHF and healthy controls. After quantitative and statistical analysis, the 28 differential protein spots with volumes changed by 1.5-fold or more were selected for further identification by MALDI-FOF/TOF-MS. These 28 differently expressed protein spots corresponded to twenty different protein accession numbers (Fig. 1). Seven proteins were significantly up-regulated in CHF and the other thirteen were down- regulated. The up-regulated proteins were Cadherin-1 (CDH1), Zinc-alpha-2-glycoprotein (AZGP1), Alpha-1-acid glycoprotein 1 (ORM1), Protein AMBP (AMBP), Ig kappa chain C region (IGKC), Ig lambda-1 chain C regions (IGLC1),Ganglioside GM2 activator (GM2A). The down-regulated proteins involved Uromodulin (UMOD), Kininogen-1 (KNG1), Alpha-amylase 1 (AMY1A), Vitamin D-binding protein (GC), Pancreatic alpha-amylase (AMY2A), Serum albumin (ALB), Leukocyte elastase inhibitor (SERPINB1), Inter-alpha-trypsin inhibitor heavy chain H4 (ITIH4), Prothrombin (F2), Calbindin (CALB1), Basement membrane-specific heparan sulfate proteoglycan core protein (HSPG2), Mannan-binding lectin serine protease 2 (MASP2), CD59 glycoprotein (CD59). Information about these protein spots concerning access numbers, gene names, molecular weight, PI, total ion scores, fold changes of spots volumes and overall trends is presented in Table 2.
Figure 1. Distribution of all 28 differentially expressed protein spots in a representative two- dimensional fluorescent differential gel electrophoresis (2D-DIGE) image from proteomic profiling of urine samples from patients with CHF and healthy controls.
Further identified protein spots are indicated by spot ID. Protein spots, protein name and spot ID are indicated (arrows).
Table 2. Identification of differentially expressed proteins in urine from chronic heart failure and normal control.
| Master number | Accession No. | Gene Name | Protein molecular weight | Protein PI | Pep. Counta | Total ion score | Fold changesb | Overall trendc |
| 117 | P07911 | UMOD | 72451.4 | 5.05 | 9 | 518 | −12.77±3.46 | down |
| 133 | P07911 | UMOD | 72451.4 | 5.05 | 9 | 323 | −10.37±3.18 | down |
| 279 | P12830 | CDH1 | 97852 | 4.58 | 3 | 212 | 3.15±0.29 | up |
| 510 | P01042 | KNG1 | 72995.6 | 6.34 | 7 | 263 | −2.54 ±0.33 | down |
| 671 | P04745 | AMY1A | 58415.2 | 6.47 | 7 | 213 | −3.05±2.19 | down |
| 672 | P02774 | GC | 54525.6 | 5.4 | 3 | 134 | −3.33±1.26 | down |
| 697 | P04746 | AMY2A | 58354.3 | 6.6 | 8 | 106 | −2.45±1.06 | down |
| 700 | P04746 | AMY2A | 58354.3 | 6.6 | 8 | 215 | −2.33±0.52 | down |
| 822 | P02768 | ALB | 71317.2 | 5.92 | 8 | 208 | −4.33±0.51 | down |
| 827 | P02768 | ALB | 71317.2 | 5.92 | 9 | 229 | −6.31±1.21 | down |
| 850 | P02768 | ALB | 71317.2 | 5.92 | 7 | 163 | −6.24±0.89 | down |
| 924 | P25311 | AZGP1 | 34079 | 5.57 | 5 | 265 | 7.31±1.54 | up |
| 945 | P25311 | AZGP1 | 34079 | 5.57 | 4 | 245 | 7.55±1.89 | up |
| 952 | P25311 | AZGP1 | 34079 | 5.57 | 6 | 349 | 7.46±2.27 | up |
| 967 | P02763 | ORM1 | 23724.8 | 4.93 | 3 | 302 | 14.81±3.49 | up |
| 978 | P30740 | SERPINB1 | 42828.7 | 5.9 | 5 | 233 | −3.96±1.12 | down |
| 1086 | Q14624 | ITIH4 | 103521.1 | 6.51 | 4 | 189 | −3.96±1.23 | down |
| 1094 | Q14624 | ITIH4 | 103521.1 | 6.51 | 5 | 212 | −2.89±0.63 | down |
| 1238 | P02760 | AMBP | 39886.3 | 5.95 | 4 | 298 | 5.63±2.03 | up |
| 1266 | P00734 | F2 | 71474.7 | 5.64 | 3 | 167 | −4.55±1.96 | down |
| 1362 | P01842 | IGLC1 | 11400.6 | 6.92 | 1 | 32 | −11.72±3.69 | up |
| 1403 | P01834 | IGKC | 11772.7 | 5.58 | 1 | 94 | 17.63±5.67 | up |
| 1527 | P05937 | CALB1 | 30291.1 | 4.7 | 3 | 77 | −4.09±0.86 | down |
| 1529 | P01834 | IGKC | 11772.7 | 5.58 | 1 | 50 | 4.43±1.49 | up |
| 1621 | P98160 | HSPG2 | 479220.5 | 6.06 | 6 | 253 | −2.89±1.34 | down |
| 1675 | P17900 | GM2A | 21280.9 | 5.17 | 2 | 152 | 6.31±3.63 | up |
| 1681 | O00187 | MASP2 | 77224.2 | 5.47 | 7 | 351 | −3.15±1.74 | down |
| 1726 | P13987 | CD59 | 14795 | 6.02 | 1 | 59 | 2.99±1.36 | down |
Calculated by amino acid count.
Fold changes of spot intensities represented as mean ± SD.
up: up-regulated in the heart failure group. down: down-regulated in the heart failure group.
c) ORM1 was indicated for further investigation based on bioinformatics analysis
Pubgene analysis of the twenty identified proteins was performed to search for the associated bio-processes. As a result, this analysis predicted 1,600 different bio-processes to be associated with identified proteins. The top 15 closely related bio-processes included growth, induction, signal transduction, pathogenesis, secretion, catabolic process, digestion, localization, translation, coagulation, death, RNA splicing, apoptosis, phosphorylation and complement activation. Noticeably, ORM1 was predicted to play important roles in many of these 15 bio-processes such as growth, induction, signal transduction, pathogenesis, catabolic process, coagulation and phosphorylation (Table 3). Furthermore, we observed tight connections between ORM1 and other identified proteins such as ALB, AMBP and ITIH4 from the Network based on co-occurrence in articles, which was consistent with the result by STRING 9.0 program. Those findings suggested that ORM1 was a key function partner with ALB, AMBP and ITIH4 (Fig. 2B), which play vital roles in the development of acute-phase reaction and inflammation indicated by Pubgene (not shown).
Table 3. Top 15 of total 1600 bio-process for the twenty identified proteins in Pubgene.
| Description | Associated Terms | Article (p-value) |
| Growth | ALB, AMBP, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC ITIH4 KNG1, MASP2, ORM1, SERPINB1, UMOD | 7.96 |
| Induction | ALB, AMBP, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, ORM1, SERPINB1, UMOD | 3.97 |
| Signal transduction | ALB, AMBP, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, ORM1, SERPINB1, UMOD | 3.93 |
| Pathogenesis | ALB, AMBP, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, ORM1, SERPINB1, UMOD | 3.76 |
| Secretion | ALB, AMBP, AMY1A, AMY2A, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, ITIH4, KNG1, SERPINB1, UMOD | 3.15 |
| Catabolic process | ALB, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, ORM1, SERPINB1, UMOD | 3.1 |
| Digestion | ALB, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, SERPINB1, UMOD | 2.49 |
| Localization | ALB, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, SERPINB1, UMOD | 2.44 |
| Translation | ALB, AMBP, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, SERPINB1, UMOD | 1.95 |
| Coagulation | ALB, AMY1A, AMY2A, CD59, CDH1, F2, GC, HSPG2, ITIH4, KNG1, MASP2, ORM1, SERPINB1, UMOD | 1.53 |
| Death | ALB, AMY1A, AMY2A, AZGP1, CALB1, CD59, CDH1, F2, GC, GM2A, HSPG2, IGKC, ITIH4, KNG1, MASP2, SERPINB1, UMOD | 1.33 |
| RNA splicing | ALB, CDH1, IGKC, IGLC1 | 1.1 |
| Apoptosis | ALB, AMBP, AMY1A, AMY2A, CALB1, CD59, CDH1, F2, GC, HSPG2, ITIH4, KNG1, SERPINB1, UMOD | 1.04 |
| phosphorylation | ALB, AMY1A, AMY2A, CALB1, CD59, CDH1, F2, GC, HSPG2, IGKC, ITIH4, KNG1, ORM1, UMOD | 1 |
| complement activation | ALB, CD59, CDH1, ITIH4, KNG1, MASP2 | 0.982 |
P = term/total (number of records of terms according to a specific function divided by their total number of records in the MEDLINE).
Figure 2. Bioinformatic analysis of bio-association network by Pubgene and String programs.
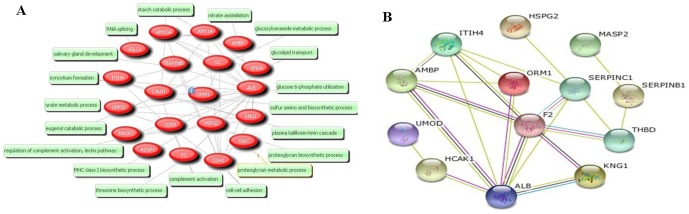
(A) Bio-process associated with the twenty identified proteins was indicated by Pubgene. (B)The ORM1 protein function partner network was indicated by String analysis.
d) Verification of higher urinary ORM1 levels in patients with CHF
To further verify our proteomic findings, we used western blotting analysis to study the expression of urinary ORM1 in individual samples from patients with CHF and control groups. The results clearly demonstrated that urinary ORM1 was significantly up-regulated in CHF cases in comparison to controls. These variations in urinary ORM1 expression were consistent with our DIGE results (Fig. 3).
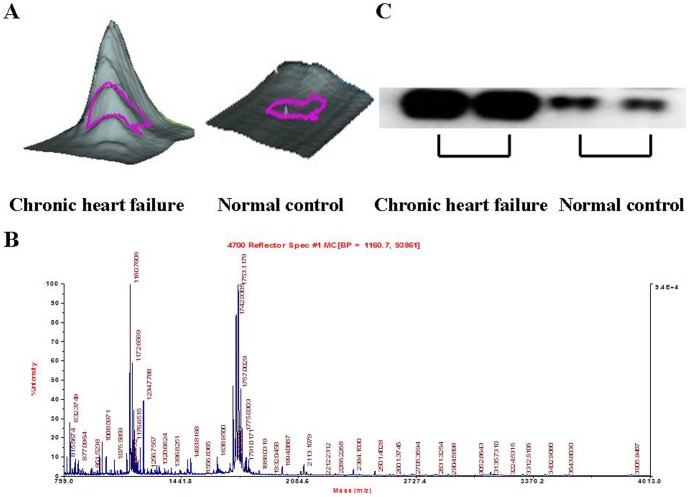
Figure 3. Verification of ORM1 protein expression in urine samples form patients with CHF and normal controls.
(A) A 3-D view of ORM1 protein was to show the higher expression of ORM1 protein in urine of patients with CHF compared with normal controls. (B) Verification of ORM1 expression in individual urine samples of patients with CHF and normal controls by western blotting. (C) MS of in-gel trypsin digests of the protein and analysis of the depicted peptide spectrum resulted in the identification of ORM1.
e) Quantification of urinary ORM1 by ELISA
ELISA was used to quantify urinary ORM1 levels in 176 patients with CHF and 62 healthy controls. After normalization by creatinine level, the urinary ORM1 was markedly elevated in patients with CHF compared to controls (6498.83±4300.21 versus 2102.26±1069.24 ng/mg, P<0.0001) (Fig. 4A). When the CHF patients were stratified by NYHA classification, the urinary ORM1 concentrations were 3086.24±1474.91, 6284.97±4088.02 and 8946.71±4298.05 ng/mg for CHF patients with class II, III and IV, respectively. The expression level of urinary ORM1 was positively associated with the class of CHF classification (P<0.0001). The correlation indicated by the Spearman test is 0.499, suggesting that the level of ORM1 was markedly elevated in urine as heart failure worsened. In addition, the relationship between urinary ORM1 levels and the clinical characteristics of patients with CHF is presented in Table 4. There was no significant association between the urinary ORM1 expression and other clinical features of CHF such as age, sex, diabetes, hypertension, coronary heart disease, cardiomyopathy, renal dysfunction and left ventricular ejection fraction (LVEF) except for NT-proBNP and the classification of CHF. Furthermore, the multivariate analysis suggested that the urinary ORM1 content was associated with the NT-proBNP (OR: 2.106, 95% CI: 1.213–3.524, P = 0.002) and the NYHA classification of CHF (OR: 3.019, 95% CI: 1.329–4.721, P<0.001) (Table 5).
Figure 4. Urinary ORM1 as a potential biomarker for CHF.
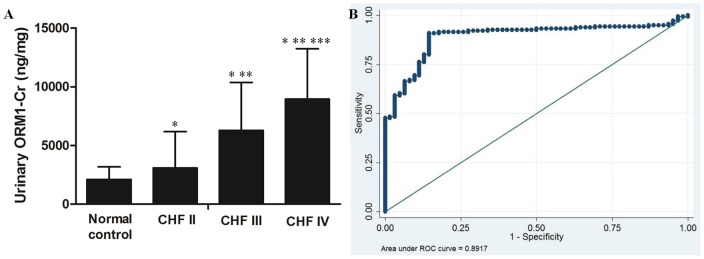
(A) ELISA was used to quantify urinary ORM1 levels in 176 patients with CHF and 62 healthy controls. The expression level of ORM1 in different NYHA functional classification of CHF. *P <0.05 compared with control group; **P <0.05 compared with CHF I; ***P <0.05 compared with CHF III. (B) ROC curve of urinary ORM1 as a detection marker for CHF was based on a series of 238 urine samples. The optimal cut off was 2484.98 ng/mg, and the area under the ROC curve (AUC) for diagnosis of CHF was 0.892 (95% confidence interval (CI) 0.848–0.936).
Table 4. Relationship between urinary ORM1/Cr level and chronic heart failure.
| Clinical features | Number | Urinary ORM1 levels (ng/mg) | P |
| age≥65 years | 83 | 6608.92±4207.72 | 0.750 |
| age<65 years | 93 | 6400.57±4401.53 | |
| Male | 108 | 6254.27±4292.88 | 0.343 |
| Female | 68 | 6887.25±4314.96 | |
| With diabetes | 28 | 6284.09±4181.24 | 0.664 |
| Without diabetes | 148 | 6437.29±4333.47 | |
| With hypertension | 68 | 5894.90±3791.18 | 0.140 |
| Without hypertension | 108 | 6879.08±4567.93 | |
| With coronary heart disease | 62 | 6885.65±4416.60 | 0.380 |
| Without coronary heart disease | 114 | 6288.45±4240.41 | |
| With cardiomyopathy | 42 | 6033.04±3915.83 | 0.423 |
| Without cardiomyopathy | 134 | 6644.82±4417.51 | |
| With renal dysfunction | 31 | 7503.97±4472.75 | 0.152 |
| Without renal dysfunction | 145 | 6283.94±4247.46 | |
| LVEF<50% | 136 | 6922.04±4432.13 | 0.153 |
| LVEF≥50% | 40 | 5990.78±4106.26 | |
| NT-proBNP≥7704* pg/ml | 62 | 8344.42±4719.76 | <0.001 |
| NT-proBNP<7704 pg/ml | 114 | 5495.09±3705.04 | |
| NYHA II | 34 | 3086.24±1474.91 | <0.001 |
| NYHA III | 87 | 6284.97±4088.02 | |
| NYHA IV | 55 | 8946.71±4298.05 |
* The mean value of NT-proBNP in CHF patients.
Table 5. The multivariate analysis of urinary ORM1/Cr level and clinical parameters.
| Parameter | p-value(uni) | p-value(multi) | 95% CI | OR |
| Age | 0.815 | 0.637 | 0.563–1.426 | 1.012 |
| Sex | 0.567 | 0.346 | 0.852–1.639 | 1.156 |
| Diabetes | 0.342 | 0.266 | 0.899–1.963 | 1.818 |
| Hypertension | 0.459 | 0.501 | 0.603–2.507 | 1.962 |
| Coronary heart disease | 0.147 | 0.193 | 0.786–3.722 | 2.321 |
| Cardiomyopathy | 0.861 | 0.632 | 0.479–1.892 | 1.523 |
| Renal dysfunction | 0.201 | 0.153 | 0.463–1.557 | 0.678 |
| LVEF | 0.747 | 0.518 | 0.902–1.010 | 0.998 |
| NT-proBNP | <0.001 | 0.002 | 1.213–3.524 | 2.106 |
| NYHA classification | <0.001 | <0.001 | 1.329–4.721 | 3.019 |
f) Evaluation of urinary ORM1 as a potential biomarker for CHF
After quantitative measurement of ORM1 in 238 urine samples by ELISA, we applied ROC curves to assess the potential utility of urinary ORM1 in diagnosing and monitoring CHF. ROC analyses rendered an optimum cut-off value of 2484.98 ng/mg corresponding to 90.91% sensitivity and 85.48% specificity. The area under the ROC curve (AUC) of ORM1-Cr for diagnosis of CHF was 0.892 (95% confidence interval (CI) 0.848–0.936).
Discussion
The exact incidence and prevalence of CHF remain probably underestimated due to the difficulties in accurate diagnosis [16]. Easily applicable screening markers for the early detection of the clinical course of CHF are currently lacking. A number of published studies have used proteomic methods to identify novel biomarkers for a variety of diseases. Notably, urinary proteomic biomarker models have recently been developed and shown potential for the accurate identification of several different disorders including ischaemic heart disease [17], endometriosis [18], diabetic nephropathy [19] and some cancers [20], [21].
In our present study, we focused on the urinary proteome, because urine is easy to collect relatively large quantities using non-invasive procedures compared with other body fluids. Moreover, the clinical importance of some urinary proteins has been demonstrated not only in urogenital diseases, but also in other systemic diseases [22]–[24]. Here, we conducted a 2D-DIGE analysis, which is an accurate quantitative comparison proteomic method, to compare the different protein expression profiles in the urine proteomes between CHF patients and healthy controls. Twenty differentially expressed proteins were identified as candidate biomarkers correlated with CHF. We further identified ORM1 as a novel CHF-related biomarker, which was significantly up-regulated in CHF patients, by bioinformatic analysis, western blotting and ELISA analyses.
Our bioinformatic analyses indicated that ORM1 played important roles in many bio-processes such as growth, induction, signal transduction, pathogenesis, catabolic process, coagulation and phosphorylation. ORM1 was also suggested to be a key function partner with ALB, AMBP and ITIH4 (Fig. 2B). Furthermore, inflammation was demonstrated to be associated with the pathophysiology of CHF [25]. ORM1 is a prominent component of the temporary proteinuria, which could be associated with exercise and acute inflammation [26]. To date, a study regarding the association between urinary ORM1 and CHF has not been reported. Thus, ORM1 was selected for further investigation in our study.
ORM1 (Orosomucoid-1, α1-acid glycoprotein 1), a single-chain polypeptide with a high carbohydrate moiety (42%) and a strong negative charge (isoelectric point, 2.7), functions as a transport protein in the bloodstream [27], [28]. It binds various ligands in the interior of its beta-barrel domain, and also binds synthetic drugs and influences their distribution and availability in the body. Human liver cells are normally the site of ORM1 production, but it can also be produced in endothelial cells and some tumor cells [29]. Some studies have demonstrated that ORM1 could be also synthesized by lymphocytes, macrophages, monocytes and granulocytes [26], [30]. ORM1, a member of immunocalins family, has a regulatory dampening effect on the inflammatory cascade, thereby protecting against tissue damage from excessive inflammation [28], [31]. Some studies have demonstrated that the concentrations of ORM1 in plasma and urine are usually much lower than albumin, but increased in urine to concentrations equal to or higher than albumin in conditions of acute inflammation and tissue repair [32]–[34].
To the best of our knowledge, this study is the first time that ORM1 has been identified in urine from patients with CHF by 2D-DIGE proteomic means. Our urinary proteome findings showed that the expression of ORM1 was significantly increased 14.81-fold in the CHF group compared with healthy controls. Meanwhile, the expression of elevated ORM1 in urine from CHF patients was further confirmed by western blotting. After quantitative ELISA analysis of urinary ORM1 in 238 individual samples, we observed that the levels of urinary ORM1 were elevated in CHF patients than controls, with significant difference found. Furthermore, the Spearman analysis showed that urinary ORM1 had a high positive correlation with the NYHA functional classification of CHF, indicating that the increased level of urinary ORM1 was associated with the CHF worsened, and the multivariate analysis indicated that the urinary ORM1 expression correlated with the plasma NT-proBNP (P = 0.002) and NYHA classification (P<0.001). In addition, ROC curves suggested at an optimum cut-off value of 2484.98 ng/mg with 90.91% sensitivity and 85.48% specificity respectively could be used for the diagnosis of CHF. Thus, all those results demonstrate the urinary ORM1 protein has potential value for the early diagnosis of CHF.
However, the pathophysiologic mechanisms responsible for those findings are unclear. One possible explanation could be that the increased ORM1 may be correlated with the inflammatory activation in patients with CHF. Previous studies have provided solid evidence to support a pivotal role of inflammation in the underlying pathophysiology of CHF through both animal and human research [35], [36]. Cytokines are mainly produced by activated monocytes and macrophages, and these inflammatory proteins have formed the basis of the CHF inflammatory paradigm. Notably, monocyte activation, which plays a core role in the inflammatory pathophysiology of CHF, results in the subsequent release of inflammatory cytokines, their migration to the myocardium, adhesion to the endothelial wall, and other complex processes of the immune system [36], [37]. It has been demonstrated that C-reactive protein levels are increased in patients with CHF and could activate monocytes and then stimulate their production of inflammatory cytokines in a dose-dependent manner. Moreover, this cytokine production is significantly enhanced in those patients with ongoing myocardial damage than in those without [37], [38]. Thereby, we suppose that the levels of urinary ORM1, similar to the C-reactive protein, may be closely related to inflammatory condition of CHF. Further studies are needed to confirm the reasons leading to the exaggerated excretion of urinary ORM1 in CHF patients.
Most studies have investigated the levels of ORM in serum. Only recently have some studies begun to look at changes in ORM levels in the urine of patients with certain diseases. Christiansen et al. [39] reported that levels of ORM were found to be significantly elevated in the urine of patients with type 2 diabetes, even in normoalbuminuric patients. Furthermore, they suggested that the increased excretion of urinary ORM could be an independent, powerful predictor of cardiovascular mortality in patients with type 2 diabetes, as determined through five years of follow-up [30]. Their explanation was that the elevated urinary ORM is associated with cardiovascular risk factors such as inflammation, impaired left ventricular function and endothelial dysfunction [40]. Exaggerated excretion of urinary ORM was also found to predict preeclampsia in pregnant women with pregestational type 1 diabetes, which was probably caused by an increased local renal inflammatory response [41]. The enhanced urinary ORM concentrations were also detected in hypertensive subjects, along with other proteins such as albumin, a-1-antitrypsin, transferrin and retinol binding proteins [42]. In addition, Irmak S et al. [43] identified that ORM is significantly increased in urine samples from bladder cancer patients, particularly in those with invasive tumor stages.
Several limitations of the current study, especially about the specificity of this potential biomarker, are needed to be considered when contemplating the potential value of urinary ORM1 in early diagnosis of CHF. First, all the participants were recruited from Chinese Han populations in the same hospital. As a result, our sample must be regarded as a homogeneous cohort that may not represent the overall CHF population. Second, although we did not detect an association between urinary ORM1 levels and other clinical characteristics of CHF such as age, sex, diabetes, hypertension, coronary heart disease, cardiomyopathy, renal dysfunction and LVEF, we could not exclude the possibility that other unknown factors may affect the urinary ORM1 expression. Another point that needs to be addressed is the control group. Inflammation could lead to an elevated level of ORM, and healthy controls made up the only control group in our study, probably resulting in an overestimation of the specificity of ORM1. Therefore, our preliminary findings require further exploration through larger-scale studies with adequate control groups, in particular controls that include subjects with inflammation.
Above all, this study firstly reports the identification and validation of the significantly up-regulated protein of urinary ORM1 in CHF. The elevated urinary ORM1 levels were detected in patients with CHF than healthy controls. Moreover, urinary ORM1 expression had a high positive correlation with the NYHA functional classification of CHF. Thus, our findings suggest that urinary ORM1 could be a potential biomarker for detecting CHF. Further research is warranted to investigate the possible role of urinary ORM1 in the pathogenesis of CHF.
Funding Statement
The authors have no support or funding to report.
References
- 1. Remme WJ, Swedberg K (2001) The Task Force for the Diagnosis and Treatment of Chronic Heart Failure of the European Society of Cardiology. Guidelines for the diagnosis and treatment of chronic heart failure. Eur Heart J 22: 1527–1560. [DOI] [PubMed] [Google Scholar]
- 2. Lloyd-Jones D, Adams RJ, Brown TM, Carnethon M, Dai S, et al. (2010) Executive summary: heart disease and stroke statistics: 2010 update: a report from the American Heart Association. Circulation 121: 948–954. [DOI] [PubMed] [Google Scholar]
- 3. Lloyd-Jones DM, Larson MG, Leip EP, Beiser A, D'Agostino RB, et al. (2002) Lifetime risk for developing chronic heart failure: the Framingham Heart Study. Circulation 106: 3068–3072. [DOI] [PubMed] [Google Scholar]
- 4. Schocken DD (2008) American Society for Preventive Cardiology. Prev Cardiol 11: 127–128. [DOI] [PubMed] [Google Scholar]
- 5. Dawson J, Walters M, Delles C, Mischak H, Mullen W (2012) Urinary proteomics to support diagnosis of stroke. PLoS One 7: e35879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kaiser T, Wittke S, Just I, Krebs R, Bartel S, et al. (2004) Capillary electrophoresis coupled to mass spectrometer for automated and robust polypeptide determination in body fluids for clinical use. Electrophoresis 25: 2044–2055. [DOI] [PubMed] [Google Scholar]
- 7. Ye B, Skates S, Mok SC, Horick NK, Rosenberg HF, et al. (2006) Proteomic-based discovery and characterization of glycosylated eosinophil-derived neurotoxin and COOH-terminal osteopontin fragments for ovarian cancer in urine. Clin Cancer Res 12: 432–441. [DOI] [PubMed] [Google Scholar]
- 8. Zimmerli LU, Schiffer E, Zurbig P, Good DM, Kellmann M, et al. (2008) Urinary proteomic biomarkers in coronary artery disease. Mol Cell Proteomics 7: 290–298. [DOI] [PubMed] [Google Scholar]
- 9. Hunt SA, Abraham WT, Chin MH, Feldman AM, Francis GS, et al. (2005) ACC/AHA 2005 guideline update for the diagnosis and management of chronic heart failure in the adult. A report of the American College of Cardiology/American Heart Association Task Force on practical guidelines. Circulation 112: e154–e235. [DOI] [PubMed] [Google Scholar]
- 10. Jiang H, Guan G, Zhang R, Liu G, Cheng J, et al. (2009) Identification of urinary soluble E-cadherin as a novel biomarker for diabetic nephropathy. Diabetes Metab Res Rev 25: 232–41. [DOI] [PubMed] [Google Scholar]
- 11. Zhao L, Wang H, Liu C, Liu Y, Wang X, et al. (2010) Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut 59: 1226–35. [DOI] [PubMed] [Google Scholar]
- 12. Li J, Guo W, Li F, He J, Yu Q, et al. (2012) HnRNPL as a key factor in spermatogenesis: Lesson from functional proteomic studies of azoospermia patients with sertoli cell only syndrome. J Proteomics 75: 2879–91. [DOI] [PubMed] [Google Scholar]
- 13. Li F, Chen DN, He CW, Zhou Y, Olkkonen VM, et al. (2012) Identification of urinary Gc-globulin as a novel biomarker for bladder cancer by two-dimensional fluorescent differential gel electrophoresis (2D-DIGE). J Proteomics 77: 225–36. [DOI] [PubMed] [Google Scholar]
- 14. Jenssen TK, Laegreid A, Komorowski J, Hovig E (2001) A literature network of human genes for high-throughput analysis of gene expression. Nat Genet 28: 21–8. [DOI] [PubMed] [Google Scholar]
- 15. Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, et al. (2009) STRING 8 — a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res 37: D412–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guha K, McDonagh T (2013) Heart failure epidemiology: European perspective. Curr Cardiol Rev 9: 123–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Delles C, Schiffer E, von Zur Muhlen C, Peter K, Rossing P, et al. (2010) Urinary proteomic diagnosis of coronary artery disease: identification and clinical validation in 623 individuals. Journal of Hypertension 28: 2316–22. [DOI] [PubMed] [Google Scholar]
- 18. Cho S, Choi YS, Yim SY, Yang HI, Jeon YE, et al. (2012) Urinary vitamin D-binding protein is elevated in patients with endometriosis. Hum Reprod 27: 515–22. [DOI] [PubMed] [Google Scholar]
- 19. Snell-Bergeon JK, Maahs DM, Ogden LG, Kinney GL, Hokanson JE, et al. (2009) Evaluation of urinary biomarkers for coronary artery disease, diabetes, and diabetic kidney disease. Diabetes Technol Ther 11: 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ye B, Skates S, Mok SC, Horick NK, Rosenberg HF, et al. (2006) Proteomic-based discovery and characterization of glycosylated eosinophil-derived neurotoxin and COOH-terminal osteopontin fragments for ovarian cancer in urine. Clin Cancer Res 12: 432–441. [DOI] [PubMed] [Google Scholar]
- 21. Jou YJ, Lin CD, Lai CH, Chen CH, Kao JY, et al. (2010) Proteomic identification of salivary transferring as a biomarker for early detection of oral cancer. Anal Chim Acta 681: 41–48. [DOI] [PubMed] [Google Scholar]
- 22. Smith ER, Zurakowski D, Saad A, Scott RM, Moses MA (2008) Urinary biomarkers predict brain tumor presence and response to therapy. Clin Cancer Res 14: 2378–2386. [DOI] [PubMed] [Google Scholar]
- 23. Cho SH, Oh YJ, Nam A, Kim HY, Park JH, et al. (2007) Evaluation of serum and urinary angiogenic factors in patients with endometriosis. Am J Reprod Immunol 58: 497–504. [DOI] [PubMed] [Google Scholar]
- 24. Buhimschi CS, Norwitz ER, Funai E, Richman S, Guller S, et al. (2005) Urinary angiogenic factors cluster hypertensive disorders and identify women with severe preeclampsia. Am J Obstet Gynecol 192: 734–741. [DOI] [PubMed] [Google Scholar]
- 25. Bozkurt B, Mann DL, Deswal A (2010) Biomarkers of inflammation in heart failure. Heart Failure Review 15: 331–341. [DOI] [PubMed] [Google Scholar]
- 26. Andersson LC, Gahmberg CG (1979) Surface glycoproteins of resting and activated human T lymphocytes. Mol Cell Biochem 27: 117–131. [DOI] [PubMed] [Google Scholar]
- 27. Ito S, Tsuda A, Momotsu T, Igarashi K, Kasahara S, et al. (1989) Urinary orosomucoid excretion rate in patients with non-insulin-dependent diabetes mellitus. Acta Endocrinol (Copenh) 120: 584–590. [DOI] [PubMed] [Google Scholar]
- 28. Christiansen MS, Hommel E, Friberg L, Mølvig J, Magid E, et al. (2010) Increased urinary orosomucoid excretion is not related to impaired renal function in patients with type 2 diabetes. J Diabetes Complications 24: 28–36. [DOI] [PubMed] [Google Scholar]
- 29. Sorensson J, Matejka GL, Ohlson M, Haraldsson B (1999) Human endothelial cells produce orosomucoid, an important component of the capillary barrier. Am J Physiol 276: H530–534. [DOI] [PubMed] [Google Scholar]
- 30. Gahmberg CG, Andersson LC (1978) Leukocyte surface origin of human alpha1- acid glycoprotein (orosomucoid). J Exp Med 148: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christiansen MS, Hommel E, Magid E, Feldt-Rasmussen B (2005) Orosomucoid in urine is a powerful predictor of cardiovascular mortality in normoalbuminuric patients with type 2 diabetes at five years of follow-up. Diabetologia 48: 386–393. [DOI] [PubMed] [Google Scholar]
- 32. Kuller LH, Tracy RP, Shaten J, Meilahn EN (1996) Relation of C-reactive protein and coronary heart disease in the MRFIT nested case-control study. Multiple Risk Factor Intervention Trial. Am J Epidemiol 144: 537–547. [DOI] [PubMed] [Google Scholar]
- 33. Fournier T, Medjoubi N, Porquet D (2000) Alpha-1-acid glycoprotein. Biochim Biophys Acta 1482: 157–171. [DOI] [PubMed] [Google Scholar]
- 34. Logdberg L, Wester L (2000) Immunocalins: a lipocalin subfamily that modulates immune and inflammatory responses. Biochim Biophys Acta 1482: 284–297. [DOI] [PubMed] [Google Scholar]
- 35. Maekawa Y, Anzai T, Yoshikawa T, Asakura Y, Takahashi T, et al. (2002) Prognostic significance of peripheral monocytosis after reperfused acute myocardial infarction: a possible role for left ventricular remodeling. J Am Coll Cardiol 39: 241–246. [DOI] [PubMed] [Google Scholar]
- 36. Wrigley BJ, Lip GY, Shantsila E (2011) The role of monocytes and inflammation in the pathophysiology of heart failure. Eur J Heart Fail 13: 1161–71. [DOI] [PubMed] [Google Scholar]
- 37. Yin WH, Chen JW, Jen HL, Chiang MC, Huang WP, et al. (2004) Independent prognostic value of elevated high-sensitivity C-reactive protein in chronic heart failure. Am Heart J 147: 931–938. [DOI] [PubMed] [Google Scholar]
- 38. Nakagomi A, Seino Y, Endoh Y, Kusama Y, Atarashi H, et al. (2010) Upregulation of monocyte proinflammatory cytokine production by C-reactive protein is significantly related to ongoing myocardial damage and future cardiac events in patients with chronic heart failure. J Card Fail 16: 562–571. [DOI] [PubMed] [Google Scholar]
- 39. Christiansen MS, Hommel E, Magid E, Feldt-Rasmussen B (2002) Orosomucoid in urine predicts cardiovascular and over-all mortality in patients with Type II diabetes. Diabetologia 45: 115–120. [DOI] [PubMed] [Google Scholar]
- 40. Christiansen MS, Iversen K, Larsen CT, Goetze JP, Hommel E, et al. (2009) Increased urinary orosomucoid excretion: a proposed marker for inflammation and endothelial dysfunction in patients with type 2 diabetes. Scand J Clin Lab Invest 69: 272–81. [DOI] [PubMed] [Google Scholar]
- 41. Christiansen MS, Hesse D, Ekbom P, Hesse U, Damm P, et al. (2010) Increased urinary orosomucoid excretion predicts preeclampsia in pregnant women with pregestational type 1 diabetes. Diabetes Res Clin Pract 9: 16–21. [DOI] [PubMed] [Google Scholar]
- 42. Lisowska-Myjak B, Krych A, Kołodziejczyk A, Pachecka J, Gaciong Z (2011) Urinary proteins, N-acetyl-β-D-glucosaminidase activity and estimated glomerular filtration rate in hypertensive patients with normoalbuminuria and microalbuminuria. Nephrology (Carlton) 16: 403–9. [DOI] [PubMed] [Google Scholar]
- 43. Irmak S, Tilki D, Heukeshoven J, Oliveira-Ferrer L, Friedrich M, et al. (2005) Stage-dependent increase of orosomucoid and zinc-alpha2-glycoprotein in urinary bladder cancer. Proteomics 5: 4296–304. [DOI] [PubMed] [Google Scholar]