Abstract
An innovative drug delivery technology is urgently needed to satisfy unmet medical needs in treating various brain disorders. As a fundamental carrier for plasmid DNA or nucleic acids, we developed a liposomal nanoparticle (multifunctional envelope-type nano device [MEND]) containing a proton-ionizable amino lipid (YSK-MEND). Here we report on the impact of apolipoprotein E (ApoE) modification on the function of YSK-MEND in terms of targeting brain cells. The cellular uptake and function of YSK-MEND encapsulating short interference RNA or plasmid DNA were significantly improved as a result of ApoE modification in mouse neuron-derived cell lines (Neuro-2a and CAD). Intracerebroventricular administration of ApoE-modified YSK-MEND (ApoE/YSK-MEND) encapsulating plasmid DNA also resulted in higher transgene expression in comparison with YSK-MEND that was not modified with ApoE. Moreover, observation of fluorescence-labeled ApoE/YSK-MEND and expression of mCherry (fluorescence protein) derived from plasmid DNA indicated that this carrier might be useful for delivering and conferring transgene expression in neural stem cells and/or neural progenitor cells. Thus, this system may be a useful tool for the treatment of neurodegenerative disease.
Keywords: brain, DNA, oligonucleotide, delivery, nanoparticles
Introduction
A technology for curing brain disorders, such as Alzheimer’s disease and Parkinson’s disease, constitutes an unmet medical need. Gene therapy or treatment with functional nucleic acid, ie, short interference RNA (siRNA), is an attractive method for meeting these needs. To realize these therapies, a nanosized carrier that is capable of delivering plasmid DNA and siRNA to brain parenchymal cells is essential. Topical administration into the brain is one of the rational approaches. Recently, a Phase I study of gene therapy for curing Parkinson’s disease using the adeno-associated virus encoding L-amino acid decarboxylase is currently under way.1 While the adeno-associated virus-based carrier is less immunogenic in comparison with other viral vectors,2 an effective nonviral carrier would still be desirable for use as a safe gene carrier.
Previously, we developed a liposomal nanoparticle known as a multifunctional envelope-type nano device (MEND) for delivering plasmid DNA or siRNA to liver,3,4 cancer,5,6 lung,7 and adipose tissue.8 Further, to improve the in vivo kinetic and intracellular fate of MEND, neutral MEND (YSK-MEND) was prepared using a pH-sensitive cationic lipid with tertiary amines (YSK05) as a key component of the envelope structure.9 Several previous studies revealed that the neutral lipid particles could accumulate effectively in the liver with the aid of serum apolipoprotein E (ApoE).10,11 ApoE is a member of the soluble apolipoprotein family that plays a key role in the cell-to-cell delivery of lipoproteins via the low-density lipoprotein (LDL) receptor family or heparin sulfate proteoglycan.12 Our recent findings demonstrated that preliminary incubation of YSK-MEND with recombinant ApoE stimulated cellular uptake of these particles in a cell line derived from mouse brain endothelial cells.13 In that study, ApoE was spontaneously absorbed onto the surface of the MEND by simple incubation, and was then recognized, presumably via lipoprotein-related receptors.
Meanwhile, the physiological function of ApoE in the central nervous system is currently being clarified, especially in relation to the mechanism of Alzheimer’s disease and lipid metabolism.14,15 In the brain, ApoE is produced by glial cells, and then functions as a ligand for cellular uptake of the cholesterol-rich high-density-like lipoproteins, that target the LDL receptor family, including LDLR and LDLR-related protein (LRP)1, which is expressed on neuronal cells, glial cells, and astrocytes.14 Therefore, ApoE/YSK-MEND was expected to accelerate its cellular uptake into brain parenchymal cells when administered in the brain. In this study, our first goal was to evaluate the in vitro function of ApoE/YSK-MEND in neuronal cells as a pDNA or siRNA carrier in vitro. The second goal was to evaluate the in vivo function of ApoE as a ligand for brain parenchymal cells via intracerebroventricular injection using plasmid DNA as the cargo.
Materials and methods
Chemicals
1,1′-dioctadecyl-3,3′-tetramethylindocarbocyamine perchlorate (DiD) was purchased from Invitrogen (Carlsbad, CA, USA). Hoechst 33342 was purchased from Dojindo Laboratories (Kumamoto, Japan). Recombinant human ApoE3 was purchased from BioVision Incorporated (Milpitas, CA, USA). 1-(monomethoxy polyethyleneglycol 2000)-2,3-dimyristoylglycerol (PEG-DMG) was purchased from NOF Corporation (Kanagawa, Japan). Cholesterol was purchased from Avanti Polar Lipids (Alabaster, AL, USA). For preparation of plasmid DNA encoding luciferase that is completely free from unmethylated CpG motifs [pCpGfree-Luc(0)], the multiple cloning site of pCpGfree-mcs (InvivoGen, San Diego, CA, USA) was preliminarily replaced with a new one (5′-Bgl II-Pvu II-Nco I-Sca I-Xba I-Nhe I-3′) by ligating the hybridized oligonucleotide fragments to Bgl II/Nhe I digestion of pCpGfree-mcs (pCpGfree-NEWmcs). Thereafter, a fragment encoding CpG-free luciferase plus one CpG motif just below the stop codon; Luc (+1) was obtained by Nco I/Nhe I digestion of pORF-Luc; Sh-CpG (Invivogen), and then ligated to the Nco I/Nhe I digested site of pCpGfree-NEWmcs [pCpGfree-Luc (+1)]. Finally, the last remaining CpG motif was removed by inserting a CpG motif-free nucleotide fragment into a Dra III/Nhe I digested site of pCpGfree-Luc (+1), where we refer to it as a pCpGfree-Luc (0).
Animals
ICR mice were purchased from Japan SLC Inc (Shizuoka, Japan). The experimental protocols were reviewed and approved by the Hokkaido University Animal Care Committee in accordance with the Guidelines for the Care and Use of Laboratory Animals.
Cell culture
Neuro-2a cells derived from mouse neuroblastoma cells were maintained in Dulbecco’s Modified Eagle’s Medium-high glucose (Sigma-Aldrich, St Louis, MO, USA) supplemented with 10% fetal bovine serum, NaHCO3 (3.7 g/L), antibiotic-antimycotic (Gibco, Thermo Fisher Scientific, Waltham, MA, USA), and L-glutamine (Gibco; final concentration 0.2 mM). CAD cells were maintained in Dulbecco’s Modified Eagle’s Medium/F-12 (Sigma-Aldrich) supplemented with 10% fetal bovine serum, penicillin (10,000 units/mL), and streptomycin (10 mg/mL). The cells were cultured under an atmosphere of 5% CO2/air at 37°C.
ApoE/YSK-MEND formation
YSK-MEND composed of YSK05, cholesterol (70/30) plus 3 mol% of PEG-DMG was prepared by the tertiary butanol (t-BuOH) dilution procedure as described previously.9 Briefly, 10 mM of total lipids were dissolved in a 90% t-BuOH solution. siRNA or plasmid DNA was dissolved in 20 mM citrate buffer (pH 4.0) and added dropwise into the lipid solution under vigorous mixing to avoid a low local concentration of t-BuOH. Subsequently, the solution was quickly diluted with citrate buffer to a final concentration of <20% t-BuOH. The t-BuOH was removed by ultrafiltration, followed by replacement with phosphate-buffered saline (pH 7.4). Finally, the solution was again ultrafiltrated to concentrate the YSK-MEND. Recombinant ApoE was preliminarily dissolved in 5 mM phosphate buffer/0.5 mM dithiothreitol and stocked as 1 mg/mL ApoE solution. For in vitro study, ApoE/YSK-MEND was prepared by adding ApoE stock solution to YSK-MEND at the indicated concentration, and incubated at 4°C for 6 hours. For instance, to prepare the 1 μg/mL ApoE/YSK-MEND, 50 μL of YSK-MEND (0.1 mM of lipid concentration) was added to 2.5 μL of ApoE stock solution (1 mg/mL) and then incubated. For in vivo use, 192 μL of YSK-MEND (0.2 ng/μL of plasmid DNA concentration) was added to 4.8 μL of ApoE stock solution (1 mg/mL) and 43.2 μL of phosphate-buffered saline. The final concentration of ApoE was adjusted to 20 μg/mL. The average diameter and zeta potential of the ApoE/YSK-MEND were determined using a Zetasizer Nano ZS ZEN3600 (Malvern Instruments, Malvern, UK). DiD-labeled ApoE/YSK-MEND was prepared by adding DiD (0.5 mol% of total lipid) in the lipid composition.
Quantification of cellular uptake of ApoE/YSK-MEND by flow cytometry
The cellular uptake of the ApoE/YSK-MEND was assessed by flow cytometry. Neuro-2a cells were seeded at a density of 1×105 cells per well in six-well plates (Corning Inc, New York, NY, USA) in 2 mL of the culture medium at 37°C under an atmosphere with 5% CO2. After 24 hours, the cells were incubated with DiD-labeled ApoE/YSK-MEND at a concentration of 1 μM of total lipid in culture medium for 1 hour. The cells were washed with phosphate-buffered saline and then trypsinized in 1 mL of phosphate-buffered saline containing 0.5% bovine serum albumin and 2 mM ethylenediamine tetraacetic acid (MACS buffer). After filtration through nylon mesh to remove cell aggregates and dust, the cells were analyzed using a FACS Calibur™ flow cytometer (Becton Dickinson, Franklin Lakes, NJ, USA). Cellular uptake of DiD-labeled MENDs was expressed as the mean fluorescence intensity, calculated using CellQuest software (Becton Dickinson).
Quantification of gene silencing effect of ApoE/YSK-MEND by quantitative realtime (RT)-PCR
Neuro-2a cells were seeded at a density of 1×105 cells per well in six-well plates in 2 mL of culture medium at 37°C. After 24 hours, ApoE/YSK-MEND encapsulating anti-β-site amyloid precursor protein cleaving enzyme 1 (BACE1) siRNA was transfected into these cells in 1 mL of culture medium at an siRNA concentration of 150 nM. The medium was replaced with fresh culture medium after 1 hour. Cells were harvested 24 hours after transfection. Total RNA was isolated from cells by TRIzol reagent (Invitrogen) and reverse transcribed using a high capacity RNA-to-cDNA kit (Applied Biosystems, Waltham, MA, USA) according to the manufacturer’s protocol. A quantitative polymerase chain reaction (PCR) analysis was performed using SYBR Green Master Mix (Applied Biosystems) and an Mx3500P real-time quantitative PCR system (Agilent Technologies, Santa Clara, CA, USA). All reactions were performed using appropriate primer sets for BACE1 and β-actin (Hokkaido System Science Co Ltd, Hokkaido, Japan). The sequences of the siRNA and primers are summarized in Table 1. Amplification was carried out by primary denaturing at 95°C (10 minutes), 40 cycles of denaturation at 95°C (15 seconds), and annealing at 60°C (1 minute). The β-actin endogenous gene was used for relative gene quantification. Specificity was preliminarily verified by melting curb analysis. The sequences of siRNA used in the present study are listed in Table 1.
Table 1.
Sequences of siRNA and primers used in the present study
| siRNA | Sense | Antisense |
| BACE1 | GAACCUAUGCGAUGCGAAUDTDT | AUUCGCAUCGCAUAGGUUCDTDT |
| Primers | Forward | Reverse |
| BACE1 | AGATCTCAAGATGGACTGCAAGGA | TCTTGGGCAAGCGAAGGTTG |
| β-actin | AGAGGGAAATCGTGCGTGAC | CAATAGTGATGACCTGGCCGT |
Abbreviations: BACE1, β-site amyloid precursor protein cleaving enzyme 1; siRNA, small interfering RNA.
In vitro transfection studies
For transfection, 8×104 Neuro-2a or CAD cells were cultured in a 24-well dish for 1 day. The cells were then washed with phosphate-buffered saline, and YSK-MEND and ApoE/YSK-MEND containing 0.2 μg of plasmid DNA [pCpGfree-Luc (0)] were applied to the medium, and then was incubated for 24 hours. The cells were collected by centrifugation (2,000 g, 4°C) for 2 minutes, and then solubilized with 75 μL of reporter lysis buffer (Promega, Madison, WI, USA). Luciferase activity in a 40 μL aliquot of cell lysate was then measured using a luminometer (Luminescencer-PSN; ATTO Corporation, Tokyo, Japan). The amount of protein in the cell lysate was determined using a bicinchoninic acid protein assay kit (Pierce, Rockford, IL, USA). Luciferase activity is expressed as relative light units per mg of protein.
Intracerebroventricular injection
ICR mice were anesthetized with isoflurane before and throughout treatment. ApoE/YSK-MEND was injected at 1 mm lateral and 0 mm caudal to bregma, and 3 mm deep from the dura using animal testing injection needles (two-step injection needle for tuberculin, Natsume Seisakusho Co., Ltd.) and a 50 μL Hamilton syringe (Hamilton Company, Reno, NV, USA). The ApoE/YSK-MEND (1.6 μg of total plasmid DNA) was injected slowly over 20 seconds. The syringe was kept in place for 1 minute after injection to prevent backflow.
Luciferase assay
ICR mice (aged 10–11 weeks, male) were administered ApoE/YSK-MEND encapsulating plasmid DNA encoding luciferase [pCpGfree-Luc (0)] by intracerebroventricular injection at a plasmid DNA dose of 1.6 μg per mouse. After 24 hours, the brains were harvested, separated into four sections, and frozen in liquid nitrogen. Each brain quarter was homogenized using Micro Smash MS-100R (Tomy Digital Microbiology Co Ltd, Tokyo, Japan) in 800 μL of lysis buffer (100 mM Tris-HCl, 2 mM ethylenediamine tetraacetic acid, 0.1% Triton X-100, pH 7.8), and the lysate was then centrifuged at 12,000 g for 10 minutes at 4°C. The four sections of supernatants individually prepared from each brain were mixed, and luciferase activity in a 20 μL aliquot of supernatant was then measured using a luminometer (Luminescencer-PSN). The amount of protein in the supernatant was determined using a bicinchoninic acid protein kit (Pierce). Luciferase activity is expressed as relative light units per mg of protein.
Analysis of gene expression and distribution of ApoE/YSK-MEND after injection
ICR mice (aged 10–11 weeks, male) were treated with DiD-labeled ApoE/YSK-MEND encapsulating pmCherry-N1 (Clontech Laboratories Inc, Palo Alto, CA, USA), an mCherry-encoding plasmid DNA at a plasmid DNA dose of 1.6 μg per mouse by intracerebroventricular injection. At 24 hours post-injection, the mice were sacrificed and perfused intracardially with 4% paraformaldehyde in 0.1 M phosphate buffer. In order to observe the distribution of ApoE/YSK-MEND in brain tissue, the brain was excised and 100 μm thick coronal and horizontal brain sections were prepared using a DSK-100 Microslicer™ (Dosaka EM Co Ltd, Irvine, CA, USA). For immunohistochemistry, excised brain tissue was soaked in 4% paraformaldehyde overnight and then cryoprotected in 20% sucrose in phosphate buffer for 1 day. Subsequently, the brain was frozen in powdered dry ice. Next, 40 μm thick coronal tissue sections were cut on a cryostat (CM1900; Leica Microsystems, Wetzlar, Germany). All slices were placed into 0.05% sodium azide in phosphate buffer and stored at 4°C until use.
Immunohistochemistry
The sections were incubated with 0.3% Triton-X/phosphate-buffered saline at room temperature for 10 minutes, and blocked in 5% goat serum/Triton-X/phosphate-buffered saline (blocking buffer) for 1 hour. Primary antibodies against NeuN, GFAP, Iba1, and Sox2 were applied to tissue sections in blocking buffer at 4°C overnight. Information on the antigen, host species, source, and specificity is summarized in Table 2. The slices were rinsed three times in Triton-X/phosphate-buffered saline, followed by incubation with Alexa Fluor 488-labeled species-specific secondary antibodies (at a dilution of 1:500 in blocking buffer) at room temperature for 1 minute. Sections were rinsed three times in phosphate-buffered saline. Thereafter, the nuclei were stained with 40 μM of Hoechst 33,342. Sections were then coverslipped with Vectashield® hard set mounting medium (Vector Laboratories Inc, Burlingame, CA, USA). The stained samples were observed by confocal laser scanning microscopy (Nikon A1; Nikon Co Ltd, Tokyo, Japan).
Table 2.
Primary antibodies used in the present study
| Molecules | Host | Clonality, isotype | Dilution ratio | Catalog number (supplier) |
|---|---|---|---|---|
| NeuN | Mouse | Monoclonal, IgG1 | 1:1,000 | MaB337 Chemicon International Inc., Temecula, CA, USA |
| GFAP | Mouse | Monoclonal, IgG1 | 1:600 | MAB360 Chemicon International Inc. |
| Iba1 | Rabbit | Polyclonal, IgG | 1:1,000 | 019-19741 Wako Pure Chemicals, Osaka, Japan |
| Sox2 | Rabbit | Polyclonal, IgG | 1:1,000 | Ab97959 Abcam, Cambridge, MA, USA |
Abbreviation: Ig, immunoglobulin.
Results and discussion
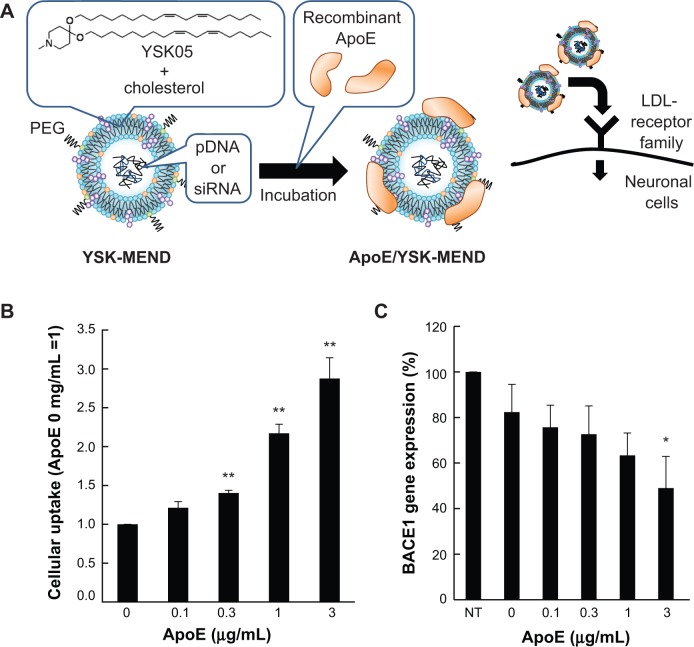
To validate the activity of ApoE as a cell targeting ligand in vitro, cellular uptake of YSK-MEND and ApoE/YSK-MEND encapsulating siRNA was first compared using a mouse neuron-derived cell line (Neuro-2a). To prepare ApoE/YSK-MEND, YSK-MEND was incubated with recombinant human ApoE3 at indicated concentrations for 6 hours (Figure 1A). The physicochemical properties of the ApoE/YSK-MEND encapsulating siRNA are shown in Table 3. The zeta potential tended to decrease depending on the ApoE modifications, while their sizes remained fairly constant. To evaluate the cellular uptake of MENDs, the particles were labeled with DiD, a fluorescent marker. Cellular uptake was analyzed by flow cytometry. As shown in Figure 1B, cellular uptake was increased as a function of modified ApoE concentration. This result indicates that ApoE may well be useful as a ligand for targeting neuronal cells.
Figure 1.
Effect of ApoE modification on cellular uptake and gene knockdown efficacy of YSK-MEND in vitro.
Notes: (A) Schematic diagram illustrating the probable structure of ApoE/YSK-MEND, and a scheme for cellular uptake. (B) Neuro-2a cells were incubated with DiD-labeled ApoE/YSK-MEND encapsulating anti-BACE1 siRNA at a concentration of 1 μM of total lipids of MEND for 1 hour. Cellular uptake of ApoE/YSK-MEND was analyzed by flow cytometry. The data are represented as the mean ± standard deviation (n=3). Statistical analyses were performed by one-way analysis of variance, followed by Dunnett’s test (**P<0.01) relative to ApoE-unmodified YSK-MEND. (C) Neuro-2a cells were treated with ApoE/YSK-MEND at a dose of 150 nM of total siRNA for 1 hour in the presence of serum. The gene silencing effect was then measured at 24 hours after transfection. The data are represented as the mean ± standard deviation (n=3). Statistical analyses were performed by one-way analysis of variance, followed by Dunnett’s test (*P<0.05, **P<0.01) relative to no treatment.
Abbreviations: DiD, 1,1′-dioctadecyl-3,3′-tetramethylindocarbocyamine perchlorate; LDL, low-density lipoprotein; ApoE, apolipoprotein E; BACE1, β-site amyloid precursor protein cleaving enzyme 1; pDNA, plasmid DNA; siRNA, small interfering RNA; PEG, polyethylene glycol; MEND, multifunctional envelope-type nano device; NT, no treatment.
Table 3.
Physicochemical properties of siRNA-encapsulating YSK-MEND with or without ApoE modification
| ApoE (μg/mL) | 0 | 0.1 | 0.3 | 1 | 3 |
|---|---|---|---|---|---|
| Size (nm) | 103±22 | 104±19 | 105±19 | 105±17 | 105±19 |
| Zeta potential (mV) | −1.12±0.94 | −4.49±5.84 | −2.24±0.85 | −3.01±0.65 | −4.44±1.11 |
Note: Data are expressed as the mean ± standard deviation (n=3).
Abbreviations: ApoE, apolipoprotein E. siRNA, short interference RNA; YSK-MEND, multifunctional envelope-type nano device prepared with YSK05.
To evaluate the potential of ApoE/YSK-MEND as a carrier for nucleic acid delivery, Neuro-2a cells were incubated with ApoE/YSK-MEND encapsulating siRNA, targeting BACE1, since knockdown of this gene is considered to be one of the strategies for curing Alzheimer’s disease.16 In parallel with enhanced cellular uptake (Figure 1B), the gene silencing effects increased with increasing ApoE concentration (Figure 1C).
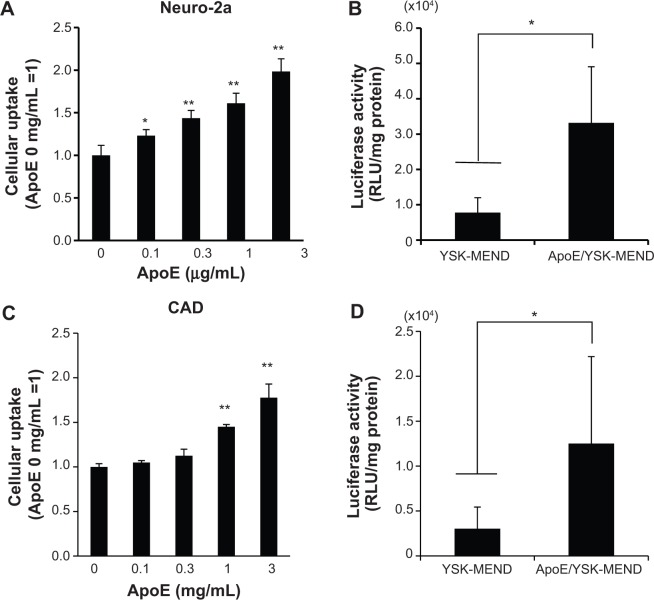
To demonstrate the in vivo benefit of using ApoE as a ligand, YSK-MEND or ApoE/YSK-MEND was then administered to the mouse brain by intracerebroventricular injection. However, a recent study revealed that gene knockdown was achieved only in a narrow area within 1 mm of the site of injection when siRNA-encapsulating particles were administered.17 Thus, in the in vivo studies, plasmid DNA encoding luciferase or mCherry instead of siRNA was used as a cargo to investigate the impact of ApoE modification on the function of YSK-MEND, since transgene expression can be quantitatively evaluated using whole brain tissue or visualized even when only a portion of the cells exhibited transgene expression. In this case, the plasmid DNA encoding luciferase was first encapsulated in YSK-MEND. For ApoE modification for in vivo use, YSK-MEND (6 mmol of total lipid in 240 μL of phosphate-buffered saline) was incubated with recombinant ApoE3 at a concentration of 20 μg/mL overnight at 4°C. Prior to the in vivo study, the function of plasmid DNA-encapsulating ApoE/YSK-MEND was validated in vitro. In this study, another type of neuron-derived cell line (CAD), along with Neuro-2a, was used to address the generality of these phenomena. As shown in Figure 2A and C, cellular uptake of YSK-MEND was increased with increasing levels of ApoE modification in both Neuro-2a and CAD cells. Consistent with these data, uptake of ApoE/YSK-MEND (3 μg/mL ApoE) was significantly higher than that of YSK-MEND (Figure 2B and D). Thus, the ApoE-dependent uptake machinery is available regardless of the type of cargo.
Figure 2.
Effect of ApoE-modification on cellular uptake and gene transfection activity of YSK-MEND in vitro.
Notes: Neuro-2a (A) and CAD (C) cells were incubated with DiD-labeled YSK-MEND or ApoE/YSK-MEND. Cellular uptake of ApoE/YSK-MEND was analyzed by flow cytometry. The data are represented as the mean ± standard deviation (n=3). Statistical analyses were performed by one-way analysis of variance, followed by Dunnett’s test (*P<0.05, **P<0.01) relative to ApoE-unmodified YSK-MEND. Neuro-2a (B) or CAD (D) cells were treated with YSK-MEND or ApoE/YSK-MEND (modified with 3 μg/mL of ApoE) encapsulating pCpGfree-Luc (0) for 1 hour in the presence of serum. Gene transfection activity was then measured at 24 hours after transfection. The data are represented as the mean ± standard deviation (n=3). Statistical analyses were performed by one-way analysis of variance, followed by the Student’s t-test (*P<0.05).
Abbreviations: DiD, 1,1′-dioctadecyl-3,3′-tetramethylindocarbocyamine perchlorate; ApoE, apolipoprotein E; MEND, multifunctional envelope-type nano device; RLU, relative light units; YSK-MEND, multifunctional envelope-type nano device prepared with YSK05.
In vivo luciferase gene expression in whole brain was evaluated after 24 hours post-intracerebroventricular administration. Gene expression of YSK-MEND was significantly enhanced by modification with ApoE (Figure 3). Because of the existence of the blood–brain barrier, the protein concentration in cerebrospinal fluid is kept lower than that in serum. On the other hand, ApoE is abundantly supplied by astrocytes.18 Ulrich et al reported that ApoE was also found in interstitial fluid at a concentration of 2 μg/mL.19 Thus, it is plausible that neutral liposomal particles, including YSK-MEND, might be inherently recognized by endogenous ApoE, and thereby gradually accumulated in brain cells. In fact, Rungta et al reported that neutralized lipid nanoparticles formed using an ionizable lipid successfully delivered siRNA to neurons when injected intracranially.17 Uno et al also reported that α-tocopherol-conjugated siRNA accumulated efficiently in neuronal cells after incorporation into high-density lipoprotein after intracerebroventricular injection.16 Even in this situation, premodification of ApoE significantly enhanced gene expression in the brain. Thus, binding of ApoE to YSK-MEND is a rate-limiting process for cellular uptake.
Figure 3.
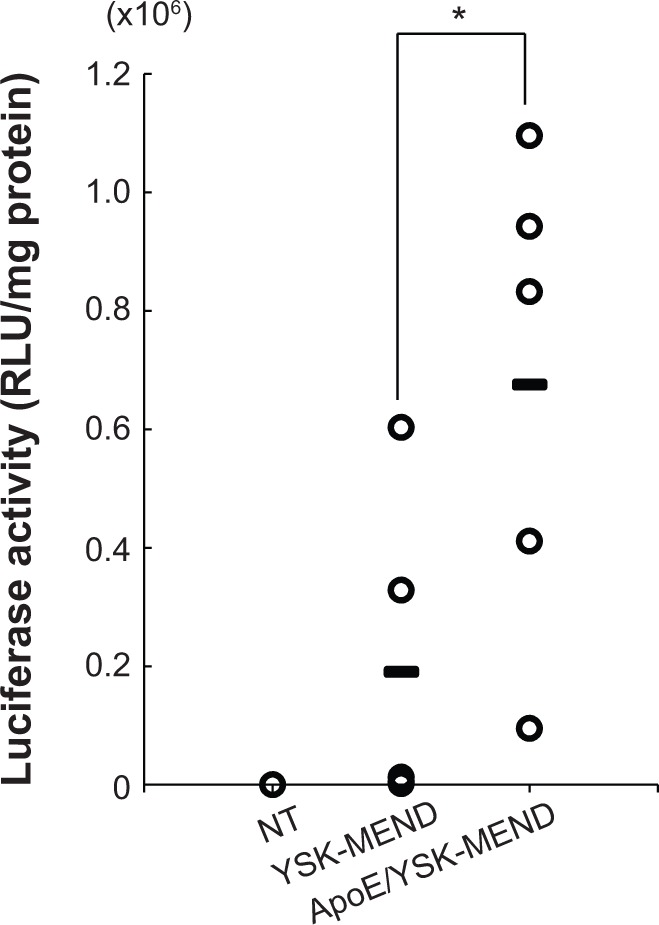
Comparison of transgene expression of YSK-MEND and ApoE/YSK-MEND encapsulating plasmid DNA encoding luciferase after intracerebroventricular injection. ICR mice (aged 10–11 weeks, male) were administered ApoE/YSK-MEND at a dose of 1.6 μg plasmid DNA/mouse by intracerebroventricular injection. After 24 hours, the mice were sacrificed and the luciferase activity was evaluated. Luciferase activity is expressed as relative light units per mg of protein. An open circle symbol and bars represent raw data for each mouse and the mean value, respectively (n=4). Statistical analyses were performed using the unpaired t-test and the Mann–Whitney test. (*P<0.05).
Abbreviations: ApoE, apolipoprotein E; MEND, multifunctional envelope-type nano device; NT, no treatment; RLU, relative light units; YSK-MEND, multifunctional envelope-type nano device prepared with YSK05.
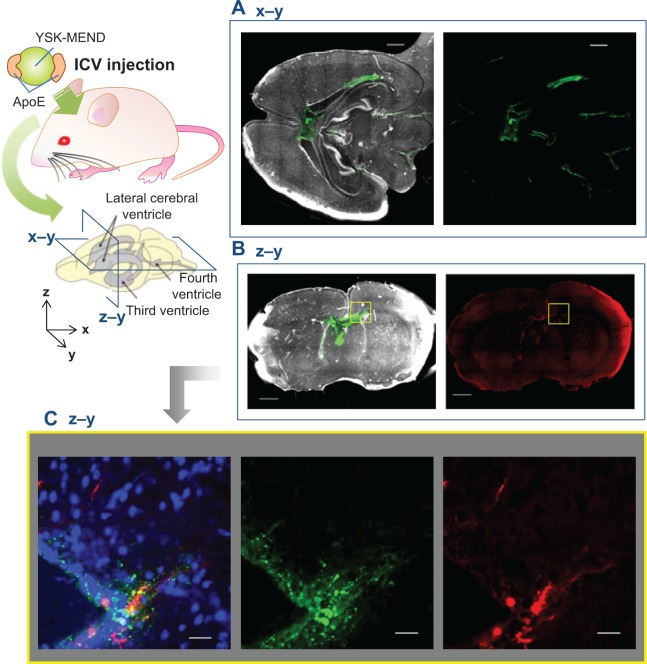
In order to identify the distribution of ApoE/YSK-MEND and the cells that are transfected, plasmid DNA encoding mCherry was encapsulated in DiD-labeled ApoE/YSK-MEND. Localization of ApoE/YSK-MEND and expression of mCherry are shown in Figure 4. Figure 4A and B represent images in brain sections that were sliced along the coronal and horizontal planes, respectively. Figure 4C represents the magnified image from the rectangular area in the horizontal plane (Figure 4B). While ApoE/YSK-MEND was mainly observed in the paraventricular region, close to the site of administration, a fraction of the particles was detected on the surface of the cerebral cortex and cerebellum (Figure 4A). Therefore, ApoE/YSK-MEND could spread in the cerebrospinal fluid and become internalized in cells that are in direct contact with cerebrospinal fluid. The fluorescence derived from mCherry was detected in a part of the cells that take up ApoE/YSK-MEND, especially in the subventricular zone, just below the ependymal cell layer (Figure 4B and C).
Figure 4.
Distribution of ApoE/YSK-MEND and mCherry expression in the brain after ICV administration of ApoE/YSK-MEND. ICR mice (10–11 weeks, male) were administered DiD-labeled ApoE/YSK-MEND encapsulating plasmid DNA encoding mCherry at a dose of 1.6 μg plasmid DNA per mouse by intracerebroventricular injection. The frozen section was prepared 24 hours after administration and observed by confocal laser scanning microscopy. Nuclei were stained by Hoechst 33,342 (λex/λem 352 nm/461 nm; (A) and (B), white; (C), blue). DiD (λex/λem 644 nm/665 nm) was indicated by green. mCherry (λex/λem 587 nm/610 nm) was pseudocolored in red. (A) Coronal section, (B) horizontal section, and (C) magnified image of the region of interest surrounded by yellow rectangle: merged cells. Scale bars 1,000 μm (A, B) and 25 μm (C).
Abbreviations: DiD, 1,1′-dioctadecyl-3,3′-tetramethylindocarbocyamine perchlorate; ICV, intracerebroventricular; ApoE, apolipoprotein E; MEND, multifunctional envelope-type nano device; ex, excitation; em, emission; YSK-MEND, multifunctional envelope-type nano device prepared with YSK05.
The receptor for ApoE/YSK-MEND in the central nervous system remains to be elucidated. Deletion of LDLR or LRP1 in mice increases the concentration of ApoE in the central nervous system, so these two receptors play a key role in the metabolism and/or clearance of ApoE. Thus, it is most plausible that LDLR20 or LRP121 contributed to cellular uptake of ApoE/YSK-MEND. The ApoE-dependent increase in cellular uptake and function of YSK-MEND in Neuro-2a in vitro also supports our hypothesis, since LDLR and/or LRP are also expressed in this cell line.22
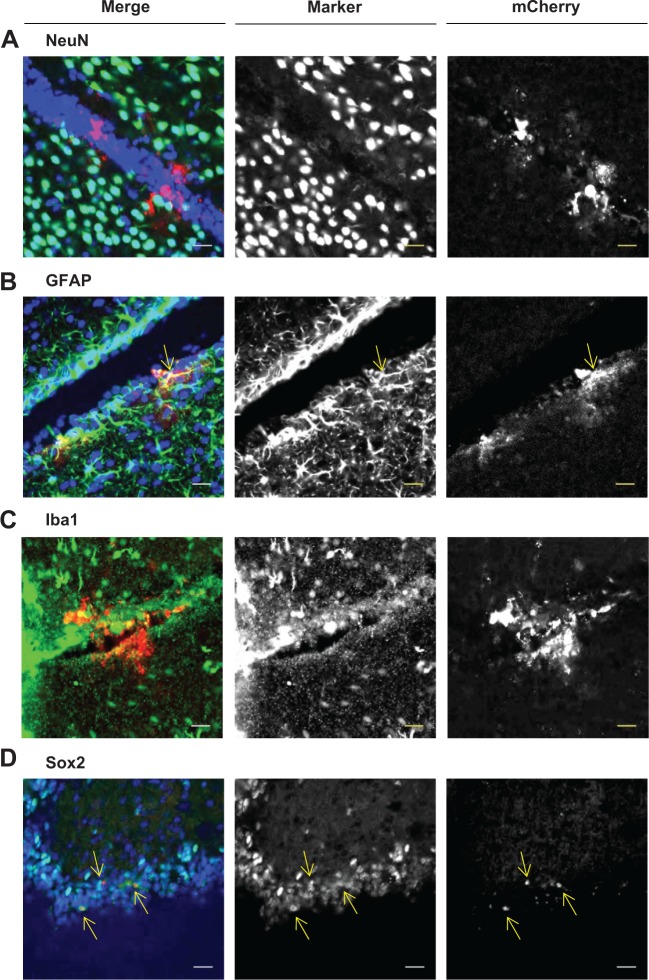
To identify the type of cells transfected, we stained the brain sections with antibodies for typical brain cell markers. Expression of mCherry was found to be colocalized with Sox2- and/or GFAP-positive cells (Figures 5B and D), while colocalization was poorly observed in cells stained with NeuN and Iba1, markers for neuronal cells and microglia, respectively (Figure 5A and C). It has been reported that neural stem cells and neural progenitor cells are found in the subventricular zone.23 Sox2 and GFAP are expressed at relatively high levels in these cells, and become negative after differentiation. Therefore, it is possible that mCherry is expressed in neural stem cells and neural progenitor cells.
Figure 5.
Identification of mCherry-expressing cells by immunostaining. ICR mice (aged 10–11 weeks, male) were administered with DiD-labeled ApoE/YSK-MEND at a dose of 1.6 μg pDNA/mouse by intracerebroventricular injection. The frozen section was prepared 24 hours after administration and stained with antibodies which were typical cell markers for brain cells. (A) NeuN, (B) GFAP, (C) Iba1, and (D) Sox2, left panels: merged (blue; nuclei, green; immunostaining, red; mCherry), middle panels: immunostaining, right panels: mCherry. Yellow arrows indicated the mCherry-/Sox2-positive cells. Scale bars represent 20 μm.
Abbreviations: DiD, 1,1′-dioctadecyl-3,3′-tetramethylindocarbocyamine perchlorate; ApoE, apolipoprotein E; MEND, multifunctional envelope-type nanodevice.
Rungta et al and Uno et al reported that their ApoE-associated or high-density lipoprotein-associated carriers functioned in neural cells.16,17 However, in our system, no gene expression was detected in neuronal cells. This discrepancy can be explained by differences in the cargo. In the previous studies, siRNA was delivered to the cytoplasm.24 However, in this study, we delivered the necessary plasmid DNA to the nucleus.25,26 The nuclear entry of plasmid DNA was severely limited by the double-layered nuclear envelope structure. Thus, non-dividing neuronal cells express hardly any transgenes, even when a particle can enter the cells. Unlike mature neurons, neural stem cells and neural progenitor cells actively undergo cell division, even in the adult brain.18 Since the nuclear transfer of plasmid DNA and the resulting transgene expression was achieved in the mitosis phase when the nuclear membrane structure is temporarily diminished,27,28 dividing neural stem cells and/or neural progenitor cells represent attractive targets for gene delivery.
Conclusion
In conclusion, we demonstrated that ApoE/YSK-MEND may be useful as a nucleic acid delivery carrier when introduced by means of intracerebroventricular injection. This nanotechnology is one of the potential solutions to unmet medical needs regarding development of a cure for neurodegenerative disease.
Acknowledgments
This work was supported by the Funding Program for Next Generation World-Leading Researchers (NEXT program; LR001), and in part by a grant for Industrial Technology Research (09A06019a) from the New Energy and Industrial Technology Development Organization. HA is also supported by the Uehara Memorial Foundation and the Asahi Glass Foundation. The authors would also like to thank Dr MS Feather for his helpful advice in writing the English manuscript.
Author contributions
Mina Tamaru, Taichi Nakatani, Yusuke Sato and Hiroto Hatakeyama acquired data. Kazuaki Kajimoto contributed to the analysis and interpretation of the results. Hidetaka Akita and Hideyoshi Harashima supervised and designed the experiments. Mina Tamaru and Hidetaka Akita wrote the manuscript with critical input from the coauthors. All authors contributed to discussion of the results.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Muramatsu S, Fujimoto K, Kato S, et al. A phase I study of aromatic L-amino acid decarboxylase gene therapy for Parkinson’s disease. Mol Ther. 2010;18:1731–1735. doi: 10.1038/mt.2010.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCaffrey AP, Fawcett P, Nakai H, et al. The host response to adenovirus, helper-dependent adenovirus, and adeno-associated virus in mouse liver. Mol Ther. 2008;16:931–941. doi: 10.1038/mt.2008.37. [DOI] [PubMed] [Google Scholar]
- 3.Khalil IA, Hayashi Y, Mizuno R, et al. Octaarginine- and pH sensitive fusogenic peptide-modified nanoparticles for liver gene delivery. J Control Release. 2011;156:374–380. doi: 10.1016/j.jconrel.2011.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Yamauchi J, Hayashi Y, Kajimoto K, et al. Comparison between a multifunctional envelope-type nano device and lipoplex for delivery to the liver. Biol Pharm Bull. 2010;33:926–929. doi: 10.1248/bpb.33.926. [DOI] [PubMed] [Google Scholar]
- 5.Hatakeyama H, Ito E, Akita H, et al. A pH-sensitive fusogenic peptide facilitates endosomal escape and greatly enhances the gene silencing of siRNA-containing nanoparticles in vitro and in vivo. J Control Release. 2009;139:127–132. doi: 10.1016/j.jconrel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 6.Sakurai Y, Hatakeyama H, Akita H, et al. Efficient short interference RNA delivery to tumor cells using a combination of octaarginine, GALA and tumor-specific, cleavable polyethylene glycol system. Biol Pharm Bull. 2009;32:928–932. doi: 10.1248/bpb.32.928. [DOI] [PubMed] [Google Scholar]
- 7.Kusumoto K, Akita H, Ishitsuka T, et al. Lipid envelope-type nanoparticle incorporating a multifunctional peptide for systemic siRNA delivery to the pulmonary endothelium. ACS Nano. 2013;7:7534–7541. doi: 10.1021/nn401317t. [DOI] [PubMed] [Google Scholar]
- 8.Hossen MN, Kajimoto K, Akita H, et al. Ligand-based targeted delivery of a peptide modified nanocarrier to endothelial cells in adipose tissue. J Control Release. 2010;147:261–268. doi: 10.1016/j.jconrel.2010.07.100. [DOI] [PubMed] [Google Scholar]
- 9.Sato Y, Hatakeyama H, Sakurai Y, et al. A pH-sensitive cationic lipid facilitates the delivery of liposomal siRNA and gene silencing activity in vitro and in vivo. J Control Release. 2012;163:267–276. doi: 10.1016/j.jconrel.2012.09.009. [DOI] [PubMed] [Google Scholar]
- 10.Akinc A, Querbes W, De S, et al. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan X, Kuipers F, Havekes LM, et al. The role of apolipoprotein E in the elimination of liposomes from blood by hepatocytes in the mouse. Biochem Biophys Res Commun. 2005;328:57–62. doi: 10.1016/j.bbrc.2004.12.137. [DOI] [PubMed] [Google Scholar]
- 12.Hatters DM, Peters-Libeu CA, Weisgraber KH. Apolipoprotein E structure: insights into function. Trends Biochem Sci. 2006;31:445–454. doi: 10.1016/j.tibs.2006.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Tamaru M, Akita H, Kajimoto K, et al. An apolipoprotein E modified liposomal nanoparticle: ligand dependent efficiency as a siRNA delivery carrier for mouse-derived brain endothelial cells. Int J Pharm. 2014;465:77–82. doi: 10.1016/j.ijpharm.2014.02.016. [DOI] [PubMed] [Google Scholar]
- 14.Fan QW, Iosbe I, Asou H, et al. Expression and regulation of apolipoprotein E receptors in the cells of the central nervous system in culture: a review. J Am Aging Assoc. 2001;24:1–10. doi: 10.1007/s11357-001-0001-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayashi H. Lipid metabolism and glial lipoproteins in the central nervous system. Biol Pharm Bull. 2011;34:453–461. doi: 10.1248/bpb.34.453. [DOI] [PubMed] [Google Scholar]
- 16.Uno Y, Piao W, Miyata K, et al. High-density lipoprotein facilitates in vivo delivery of alpha-tocopherol-conjugated short-interfering RNA to the brain. Hum Gene Ther. 2011;22:711–719. doi: 10.1089/hum.2010.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rungta RL, Choi HB, Lin PJ, et al. Lipid nanoparticle delivery of siRNA to silence neuronal gene expression in the brain. Mol Ther Nucleic Acids. 2013;2:e136. doi: 10.1038/mtna.2013.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Curtis MA, Faull RL, Eriksson PS. The effect of neurodegenerative diseases on the subventricular zone. Nat Rev Neurosci. 2007;8:712–723. doi: 10.1038/nrn2216. [DOI] [PubMed] [Google Scholar]
- 19.Ulrich JD, Burchett JM, Restivo JL, et al. In vivo measurement of apolipoprotein E from the brain interstitial fluid using microdialysis. Mol Neurodegener. 2013;8:13. doi: 10.1186/1750-1326-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fryer JD, Demattos RB, McCormick LM, et al. The low density lipoprotein receptor regulates the level of central nervous system human and murine apolipoprotein E but does not modify amyloid plaque pathology in PDAPP mice. J Biol Chem. 2005;280:25754–25759. doi: 10.1074/jbc.M502143200. [DOI] [PubMed] [Google Scholar]
- 21.Liu Q, Zerbinatti CV, Zhang J, et al. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron. 2007;56:66–78. doi: 10.1016/j.neuron.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeMattos RB, Rudel LL, Williams DL. Biochemical analysis of cell-derived apoE3 particles active in stimulating neurite outgrowth. J Lipid Res. 2001;42:976–987. [PubMed] [Google Scholar]
- 23.Zhao C, Deng W, Gage FH. Mechanisms and functional implications of adult neurogenesis. Cell. 2008;132:645–660. doi: 10.1016/j.cell.2008.01.033. [DOI] [PubMed] [Google Scholar]
- 24.Elbashir SM, Harborth J, Lendeckel W, et al. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 25.Brunner S, Sauer T, Carotta S, et al. Cell cycle dependence of gene transfer by lipoplex, polyplex and recombinant adenovirus. Gene Ther. 2000;7:401–407. doi: 10.1038/sj.gt.3301102. [DOI] [PubMed] [Google Scholar]
- 26.Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–434. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James MB, Giorgio TD. Nuclear-associated plasmid, but not cell-associated plasmid, is correlated with transgene expression in cultured mammalian cells. Mol Ther. 2000;1:339–346. doi: 10.1006/mthe.2000.0054. [DOI] [PubMed] [Google Scholar]
- 28.Tseng WC, Haselton FR, Giorgio TD. Mitosis enhances transgene expression of plasmid delivered by cationic liposomes. Biochim Biophys Acta. 1999;1445:53–64. doi: 10.1016/s0167-4781(99)00039-1. [DOI] [PubMed] [Google Scholar]