Abstract
Different types of RNAs identified thus far represent a diverse group of macromolecules that are involved in the regulation of different biological processes. RNA is generally thought to be localized primarily in the nucleus and cytoplasm, however, some types of RNA have been detected in the extracellular milieu. These extracellular RNA (exRNA) molecules are protected from degradation and it is now widely accepted that extracellular vesicles and ribonucleoprotein particles serve as transport vehicles for exRNA among cells. The functional consequence of this transfer of genetic information probably encompasses a broad range of normal developmental and physiologic processes in many organisms. This review will focus on the role of exRNA communication in cancer. We will focus on different types of RNA species identified and characterized within tumor-derived extracellular vesicles. Further, we will describe the role of exRNAs in cancer progression, as well as their potential for use as diagnostic biomarkers and therapeutic tools for monitoring and treating cancer, respectively.
Keywords: RNA, extracellular vesicles, cancer
1. Introduction
Classical thinking dictates that endogenous RNA is found within the nucleus, where it is transcribed and regulates gene expression, and in the cytoplasm where it participates in protein translation. However, RNA outside of cells - extracellular RNA (exRNA) was identified decades ago suggesting that some RNA molecules are released from cells in a stable form resistant to degradation by ribonucleases (RNAses) [1]. What has become evident over the past few years is that different types of exRNA carried in various types of vehicles are present in the extracellular milieu. More importantly, it has been shown that these exRNA molecules, along with protein cargo, can be transferred between donor and recipient cells and influence the phenotype of the recipient cells [2–6]. This exchange of genetic information between cells with a corresponding change in the phenotype of the recipient cells has been demonstrated in human cancers in several seminal reports over the past few years [3,7–9].
Free-floating RNAs in the extracellular space are highly sensitive to degradation by RNAses found throughout the body; therefore, the ability to detect exRNAs in bodily fluids suggests that they are found in enclosed structures and protected from degradation. Several different vehicles of exRNA transport have been documented [10]. For example, exRNA has been found to associate with high-density lipoprotein (HDL) complexes, the Argonaute 2 complex, and other RNA binding proteins [11–13], as well as extracellular vesicles (EVs) [2,3].
Vesicle release is a naturally occurring process that has been observed in nearly all cell types (for review see [14,15]). EV release occurs not only in most healthy cells, but also in a number of different disease states, including human cancers [2,7,8]. In fact, EVs have been isolated from both cultured cancer cell lines and different biological fluids, including serum, plasma, ascites fluid and urine of cancer patients [3,8,9,16–19]. Most studies have shown that EVs are shed in greater numbers from cancer cells, as compared to normal cells [2,20,21].
Extracellular vesicles represent a novel vehicle for cell-cell communication which can allow transfer of cytoplasmic and membrane proteins, as well as DNA and RNA between cells (Fig. 1), and contribute to modulation of numerous biological processes [15,22–27]. Extracellular vesicles can serve as vehicles for transport of exRNA through the extracellular milieu from cancer cells to normal cells in the immediate surroundings as well as distal sites. This novel type of cell-cell communication has emerged as a means for cancer cells to both eliminate RNA and proteins that restrain their growth, and to transfer oncogenic molecules that contribute to progression of cancer and other pathogenic aspects of disease and resistance to therapies [8,9,18,28]. Since the discovery that EVs contain oncogenic molecules that are transferred to recipient cells, research has focused on elucidating the role of EVs in human cancer and the factors within EVs that contribute to disease progression [3,4,7,21,29]. Thus far, cell-to-cell communication mediated by EVs has been shown to be an important contributor in the different stages of cancer progression, such as tissue invasion, immune evasion, angiogenesis and metastasis [9,29–34].
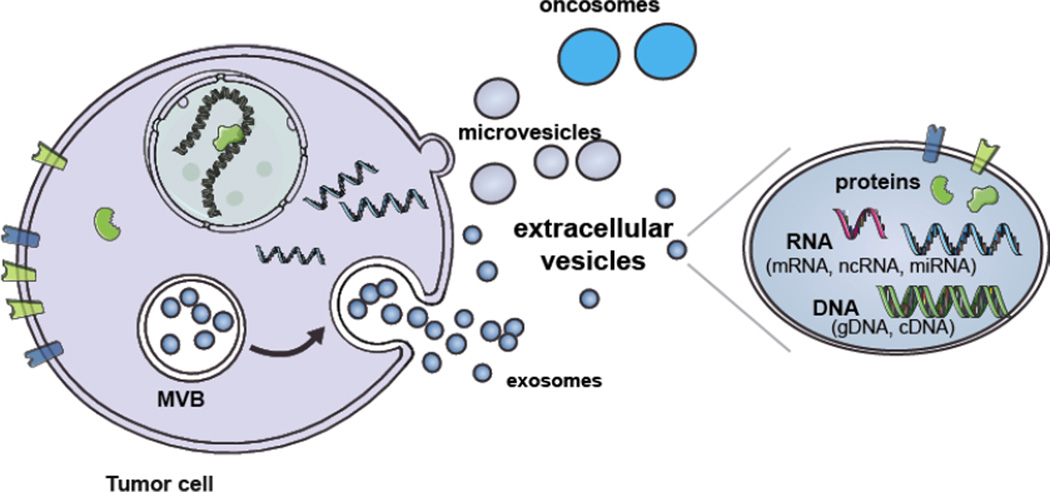
Figure 1. Biogenesis and cargo of EVs.
Extracellular vesicles are comprised of several different types of vesicles including exosomes, microvesicles and oncosomes. Exosomes are formed by the internalization of the endocytic membrane and formation of MVB inside the cell. The fusion of the MVBs with the plasma membrane results in the release of exosomes into the extracellular milieu. Microvesicles are formed by the outward budding of the plasma membrane and are directly released into the extracellular milieu. Oncosomes are a larger type of EV released by cancer cells through budding of the plasma membrane. EVs contain a variety of proteins, RNA species and types of DNA. (Modified from [62].)
In this review, we will focus on the release of the different types of exRNA molecules including mRNA, microRNA (miRNA), and other non-coding RNA (ncRNAs), as well as transposable elements and how these biomolecules contribute to cancer. Further, we will discuss the effects of exRNAs once they are taken up by recipient cells within the context of human cancers. We will also review the potential of using exRNA as biomarkers and therapeutic vehicles to treat human cancers.
2. Biogenesis and content of extracellular vesicles
Several different types of vesicles have been identified including exosomes, microvesicles, oncosomes and microparticles [7,8,35–38]. The nomenclature within the field of EVs is yet to be precisely defined, thus, in this review we will refer to all types of vesicles as EVs [36]. The different types of EVs identified to date are categorized based on their origin, size and content [37].
Exosomes are the smallest EVs and range from 30–100 nm in diameter [39]. They are believed to form by the inward budding of endocytic membranes resulting in the formation of intraluminal vesicles (ILVs), collectively termed multivesicular bodies (MVBs). Exosomes are released into the extracellular milieu upon fusion of the MVB membrane with the plasma membrane (Fig. 1) [35,40]. Microvesicles range in diameter from 100 nm to 1000 nm, are formed by the outward budding of the plasma membrane and are released directly into the extracellular milieu (Fig. 1) [36,37]. Large microvesicles (up to 5µm in diameter) derived from tumor cells have been termed oncosomes and carry oncogenic molecules that have been shown to alter the phenotype of the recipient cells in support of tumor growth [2,7,8,41].
The content of EVs is diverse and includes proteins, lipids, DNA and different types of RNA [3,21,42, 43]. Protein markers have been used to try to distinguish exosomes from microvesicles, but there is some overlap in the content of these two vesicle types. Exosome markers classically include transmembrane proteins CD63, CD81, Alix and Tsg101 [26,37]. Microvesicle protein markers are dictated in part by the proteins on the surface of the cell releasing them and Annexin V is commonly used as a marker [37,44]. The lipid content of the vesicles depends on the type of vesicle being released. For example, the lipid content of exosomes is composed of cholesterol, sphingomyelin and ceramide, while microvesicle membrane has a higher content of cholesterol [35,45].
Several different types of RNA molecules have been detected in EVs including mRNA, long non-coding RNA (lncRNA), small non-coding RNAs (sncRNAs), such as miRNA, and ribosomal RNA [4,46–49]. Some RNA molecules are enriched in EVs, compared to parental cells [e.g. 2,3,50]. RNA messages contained within EVs can be delivered to recipient cells and be translated into functional protein within the donor cell, albeit this may depend on the size and other properties of the RNAs [3,51]. Smaller miRNAs can be efficiently transferred in EVs and frequently appear to be functional in recipient cells [52–54]. The abundance of exRNA within EVs led to the concept of using EV-exRNA as biomarkers for human cancer given that tumor specific RNA, such as EGFRvIII transcript, was identified within EVs [3,48]. We will focus on specific types of RNA species detected within EVs in more detail in sections below. Since most EV preparations are obtained by differential centrifugation with the collection by ultracentrifugation yielding vesicles of all sizes, as well as protein aggregates and HDLs, preparations referred to as EVs may contain exRNA in other forms as well.
3. Proposed mechanisms of EV interaction with recipient cells
Cell-to-cell communication is mediated by a number of different mechanisms and is an important aspect of many biological processes. The role of EVs as mediators of cell-to-cell communication is being extensively investigated [23,26,55,56]. Three main mechanisms by which EVs mediate intercellular communication have been proposed [38] (Fig. 2): i) Proteins within the membrane of vesicles can serve as ligands for the receptors on the surface of recipient cells. Some of these same membrane proteins can also be cleaved by proteases generating soluble forms. The soluble form of the membrane proteins can also interact with receptors on the cell surface [57]. ii) EVs can be internalized into recipient cells via fusion of the EV membrane and cellular membrane thereby releasing EV contents into the cytoplasm. iii) EVs can be taken up into cells via endocytosis (pinocytosis and/or phagocytosis; [58]). Receptor-ligand interaction likely leads to activation of signaling pathways while internalization of EVs into recipient cells results in delivery of the EV cargo, initiating a number of different downstream events. Conceptually, it seems that direct fusion might be a more efficient means of cargo delivery as compared to endocytosis, as the latter can be associated with degradation of macromolecules.
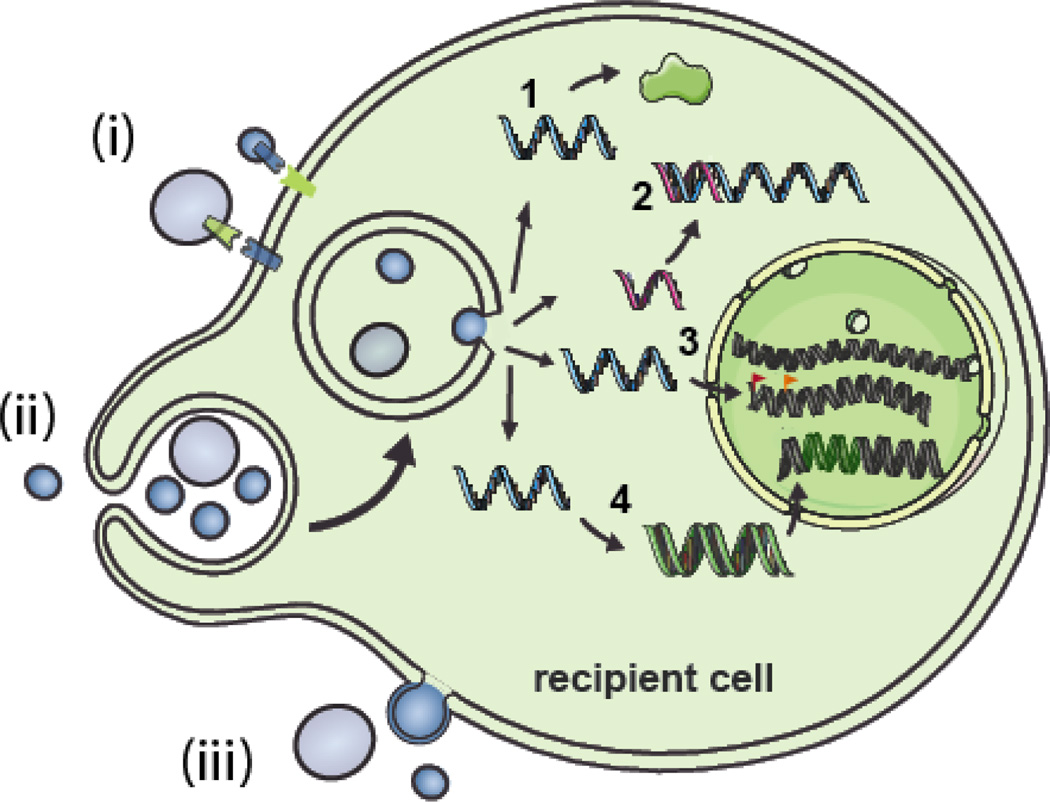
Figure 2. Interactions of EVs with cells and the potential functional roles of exRNA.
EVs can interact with recipient cells in several ways. i) The EV membrane (bearing ligands) can come in contact with receptors on the cell membrane thereby activating signaling pathways. ii) The EV membrane can fuse with the cell membrane or iii) be taken up by endocytosis/pinocytosis/phagocytosis. Depending on the type of exRNA present in the EVs these RNA molecules can have multiple effects on the phenotype of the recipient cell. 1) mRNAs can lead to translation of new proteins. 2) miRNAs can inhibit translation by binding to their mRNA targets. 3) ncRNAs can potentially alter genome methylation patterns and histone modifications by re-targeting epigenetic modifiers leading to altered gene expression patterns. 4) Repetitive elements may be integrated into the cell genome after reverse transcription. (Modified from [62].)
4. Diversity of RNA molecules in EVs
Different types of RNA have been detected in EVs purified from biological samples and conditioned media and may have a number of potential roles after cellular uptake (Fig. 2). Recent technological developments have allowed for the deep sequencing of RNA in EVs from various sources. The first studies indicate that EVs contain a diverse collection of RNA molecules, including mRNA, miRNAs, ribosomal RNA, transfer RNA, lncRNA, piwiinteracting RNA (piRNA), small nuclear RNA and small nucleolar RNA (snoRNA) [47,59]. A recent study revealed that miRNAs were the most abundant small RNA molecules in plasmaderived exRNA [49]. Cancer patients tend to have a higher yield of RNA in EVs from serum as compared with controls, most represented in the small size range (<500 nt) and of unknown cellular origin [43,60].
MRNAs
Cancer genetics is a complex puzzle characterized by genotypic changes such as mutations, deletions, insertions, amplifications and rearrangements of genes, as well as epigenetic changes in gene expression of genes that are not necessarily changed at the DNA level. In cancer, most genes have altered expression levels rather than mutations per se. The host’s response to the tumor also involves changes in gene expression in normal cells. Our group has shown that the mRNA expression pattern in serum EVs from glioblastoma patients are distinct from controls (121 genes were down-regulated at least 2-fold) and that most changes were in components of the ribosome [43]. As these genes are abundantly expressed in lymphocytes this apparent down-regulation may reflect the low white blood cell counts common in immuno-compromised glioblastoma patients [61], with white blood cells also releasing EVs into the serum [35].
The presence of mRNA sequences has been extensively described in EVs [for review see 62], albeit the size range and relative content of fragments versus full-length messages remains to be determined. Depending on the nature of the donor cell, the types of mRNAs detected and the relative enrichment in EVs can vary significantly. For example, glioblastoma cell-derived EVs are enriched in mRNAs species involved in cell proliferation, immune response, cell migration, angiogenesis and histone modification [3]. EVs from mouse mast cells (MC/9) contain mRNAs responsible for cellular development, protein synthesis, and RNA post-transcriptional modification [2]. EVs from human liver stem cells (HLSCs) and mesenchymal stem cells (MSCs) also contain a selective repertoire of mRNAs and interestingly these EVs have also been found to contain ribonuclear proteins (RNPs), such as staufen 1 (Stau1) and 2 (Stau2) that are involved in mRNA transport and stability [63] suggesting a role for RNPs in selective packaging of mRNAs into EVs. The “code(s)” for selective packaging of mRNAs in EVs has yet to be determined, but initial studies suggest the possibility of a common sequence in the 3’ UTR of mRNAs naturally enriched in EVs that can be used to increase levels of other mRNAs [64] and binding motifs in miRNAs that associate with heterogeneous nuclear ribonucleoprotein A2B1 associated with their enrichment in EVs [65].
MiRNAs
EVs also contain many non-coding RNA molecules, including miRNAs that are especially prominent in EVs derived from cancer cells and body fluids [66]. miRNAs are 21–25 nucleotides in length and can repress translation through base-pairing with the 3’UTR of mRNA molecules [for review see 67]. Processing of miRNA involves several different steps and starts with the splicing of the primary mRNA transcript into a larger precursor RNA molecule called pri-miRNA. Pri-miRNAs are cleaved in the nucleus by the enzyme Drosha, resulting in up to six different miRNA precursor molecules, pre-miRNA, per miRNA genomic locus and these pre-miRNA molecules are transported to the cytoplasm. Finally, the pre-miRNAs undergo an additional processing step by Dicer, which generates the mature, typically, 22-nucleotide miRNAs that are incorporated into the RNA-induced silencing complex (RISC). In the human genome over a 1000 putative miRNA sequences have been found. Based on conservation of miRNA binding sites in the 3’UTR of mRNA molecules it has been estimated that over 60% of human genes are targeted by one or more miRNAs, with individual miRNAs able to target hundreds of mRNAs. Due to this intricate regulatory network, miRNAs are able to influence many biological processes.
miRNAs have been detected in various body fluids, including serum, plasma, saliva, urine, breast milk and cerebrospinal fluid (CSF) [13,68]. These regulatory RNA molecules have been detected in large quantities in these body fluids despite the presence of high levels of RNAses. This indicates that these extracellular miRNAs are packaged in a manner that protects them from degradation. Indeed, miRNAs in circulation have been found inside EVs, as well as in RNA-protein and lipid complexes [13]. Vickers et al. (2011) discovered that purified HDL particles isolated from human plasma contain a number of miRNAs [11]. Interestingly, miRNAs bound to HDL particles from familial hypercholesterolemia subjects showed a different miRNA profile, compared to healthy controls supporting the use of extracellular miRNAs as biomarkers of this disease state [11]. In addition to lipoprotein complexes miRNAs are found in the circulation bound to argonaute proteins [12]. According to this study, the majority of miRNAs found in circulation are bound to argonaute proteins compared to miRNAs associated with vesicles. However, the exact percentage of vesicle-bound miRNAs vs. HDL bound and argonaute protein bound (ago-miRNA complex) miRNAs has been debated and probably differs for specific miRNAs and different biofluids. Additionally, whether ago-miRNA complexes are actively transported out of cells and taken up by recipient cells still needs to be explored. In contrast to miRNA-protein complexes, the release of miRNAs from cells within vesicles has been widely studied and it has become clear that miRNA-containing vesicles can be isolated from different biofluids, as well as conditioned medium from cells grown in culture [69]. A number of miRNAs are selectively packaged in EVs by most cells and their EV content can be further increased by elevating levels of specific miRNAs in the producer cells [70]. In 2007, Valadi et al. demonstrated the presence of miRNAs in mast cell-derived exosomes and proposed that “exosomal shuttle RNA” could mediate genetic exchange between cells [2].
Depending on the donor cells and the physiological context, the EV-mediated transfer of miRNAs can alter the phenotypic profile of the recipient cells. Importantly, this transfer of EV contents can result in changes in the levels of miRNAs in recipient cells. Examples are the transfer of miR-150 from a monocytic cell line to endothelial cells [52], transfer of miR-235 from antigen-presenting cells to T cells [71], transfer of miR-223 from macrophages to breast cancer cells [53] and the transfer of miR-126 from apoptotic endothelial cells to surviving endothelial cells [72]. While these studies show an increase of the levels of transferred miRNAs in the recipient cells they fail to discriminate whether these increased miRNA levels are the result of direct delivery via the EVs or whether they reflect transcriptional up-regulation of these miRNA due to signaling events triggered by EV uptake. An elegant study using EBV-infected B cells provided proof for direct transfer of miRNAs by showing the existence of EBV-specific miRNAs in non-infected cells [73]. These miRNAs were biologically functional as transfer of these miRNAs via isolated EVs down-regulated several target mRNAs in recipient cells.
In addition to down-regulating target mRNA molecules after EV transfer, these miRNAs could also alter the phenotypic state by binding and activating RNA-binding receptors. An interesting study showed the co-localization of EV-delivered miRNAs with Toll-like receptors (TLRs) inside endosomes in recipient cells. Furthermore, lung cancer-derived EVs induced an increase in tumor-necrosis factor (TNF) production in macrophages which was partly dependent on TLR7, suggesting that binding of miRNAs could result in functional activation of the receptor [74]. Some studies have explored the possibility of treating patients by targeting deregulated miRNAs especially in cancer stem cells [75]. miR-34 is probably the most extensively studied miRNA showing the benefits of miRNA enrichment therapy for a variety of cancer types [76].
Other ncRNAs
Many lines of evidence support the ability of EVs produced by tumor cells to influence the phenotypic state of normal proliferative cells in their environment [77]. These “heritable” changes in the recipient cells suggest epigenetic mechanisms operating on their genome, which modify their transcriptome, such as methylation of CpG islands and acetylation of histones in promoter regions [78]. The role of exRNAs in such processes is suggested by the relatively high content of regulatory RNAs in EVs. Deep sequencing of RNA from EVs derived from different cell types reveals substantial levels of many small ncRNAs, as well as lncRNAs, both of which are implicated in gene regulation [for review see 79]. These include miRNAs, as well as signal recognition particle (SRP)-RNA, vault RNA, Y-RNA and transfer RNA (tRNA) fragments in EVs from dendritic cells [59]; 7SL, Y-RNA and piRNA in EVs from a neural cell line [47]; and lncRNA, piRNA, tRNA fragments and snoRNA in EVs from plasma of glioma patients [49]. A few of these RNAs have been associated with epigenetic related mechanisms and many are up-regulated or down-regulated in cancer [80,81]. For example piRNAs (20–30 nt) are involved in maintaining genome stability and DNA methylation with aberrant expression in human tumors [82], snoRNAs (60–300 bp) may regulate gene expression by effects on other RNAs and are abnormally expressed in a number of cancers [80], and lncRNAs (>200 bp) comprise a large class of transcribed, ultra-conserved ncRNAs that mediate epigenetic modification of DNA with altered expression profiles in cancer [83]. Several studies also indicate that miR-29 and other miRNAs can inhibit mRNAs for DNA methyltransferases resulting in global hypomethylation and overall activation of the genome [84–86], a hallmark of cancer (for review see [78]). Downstream effects of EV exposure could also include histone modifications, such as acetylation, which is associated with a more active transcriptome, with the lncRNA HOTAIR causing the repressive polycomb protein, EZH2 to be retargeted across the genome, which is associated with “epigenetic switching” [87–89]. The action of DNA regulatory RNAs is an active area of research with promoter-directed RNAs acting both through sequence specific and non-specific interactions that can guide chromatin modifying proteins within the genome (for review see [80,90]). Whether sufficient quantities of these regulatory RNAs are transferred from tumor cells to stromal cells through EVs to cause epigenetic changes associated with cancer progression remains to be determined.
Repetitive elements
including transposable elements (TEs), also known as “jumping genes”, are pieces of DNA that can jump from site to site affecting the genome in a variety of ways. They can generate mutations, induce genomic instability or contribute to genomic evolution. TEs can be separated into two major classes, DNA transposons and retrotransposons: DNA transposons, which make up about 3% of TEs, can copy themselves and insert into new sites in the form of DNA. Retrotransposons move in the form of an RNA-intermediate and are reverse-transcribed into DNA, then inserted into new locations within the genome [91]. Hypomethylation of the genome is a common phenomenon in cancer and results in increased expression of TE elements [92], but it is as yet unclear to what extent higher transcription of TE elements is a driver in cancer or a by-product of cancer. Most retrotransposons were inserted into the human genome over 25 million years ago and have accumulated enough changes and mutations to render them almost entirely silent [93]. Nonetheless, a few of these sequences are still active and mobile within the genome. Most recent entries into the genome, such as the human endogenous retrovirus, HERV-K tend to have more complete sequences and be more active “jumpers”. HERV-K transcript, for example, has intriguingly been detected in some human breast cancers, but not in breast tissue from healthy controls [94] and proviruses containing mature Gag and Env proteins of retroviruses have been isolated from human melanomas, as well as lymphomas [95,96]. We have shown that certain retrotransposons, including HERV-K, HERV-H and HERV-W are highly enriched in tumor-derived EVs and can be transferred via EV-like particles to normal endothelial cells [21]. The presence and enrichment of these mobile elements in tumorderived EVs is particularly intriguing as it suggests that tumors may send out these retrotransposon “messages” via EVs to destabilize the genome of the surrounding cells. This, in turn, may make these normal cells more responsive to tumor signals.
5. The roles of extracellular RNA in cancer progression
Vesicle shedding has been observed to occur in tumor cells for a number of different human cancer types [3,9,41,62,97,98]. In fact, EVs have been isolated from both cultured tumor cells and biological samples, such as plasma, urine and ascites fluid of cancer patients, as well as controls [3,8, 9,16–19]. Current evidence supports an influence of tumor EVs on many aspects of cancer progression, such as invasiveness, angiogenesis, immune evasion, metastasis and coagulation [30,31,99,100]. However, the numbers of studies that have directly shown the effect of EV-mediated exRNAs on recipient cells with respect to cancer progression are limited.
A specific example of the role of miRNA in cancer is illustrated by the ability of EV-mediated transfer of miR-223 from activated macrophages to breast cancer cells with an associated increase in their invasiveness [53]. Using DiscovArray it was shown that miR-223 was expressed in the activated macrophages, but not in two breast cancer cell lines. Further, miR-223 was released from activated macrophages within EVs and transferred to the breast cancer cells. The recipient breast cancer cells internalized the EVs and this transfer was associated with increased invasiveness of the breast cancer cells, as measured by a trans-well invasion assay. EVs purified from unactivated macrophages did not significantly stimulate invasiveness of the breast cancer cells. Importantly, the invasiveness of the tumor cells was dependent on miR-223 within the EVs, as incubation of breast cancer cells with EVs purified from activated macrophages followed by the treatment of the recipient cells with an miR-223 antisense oligonucleotides (ASOs) reduced the invasiveness of the breast cancer cells. These effects were not observed using EVs from non-activated macrophages. The authors did not probe for upregulation of the premiR for 223 in the recipient cells to rule out a secondary effect. Thus, EV-mediated transfer of miR-223 released by activated macrophages, directly or indirectly, promotes tumor invasiveness of breast cancer cells, albeit other factors in these EVs may also contribute to this cancer supportive phenotype.
Angiogenesis is an important process in the development of human cancers as a constant and expanding blood supply is required for tumor growth. In 2012, Zhuang et al. showed that miRNA increase in endothelial cells mediated by EVs stimulated tumor angiogenesis [101]. Specifically, using a tumor-endothelial cell co-culture system, the authors identified several highly elevated miRNAs in endothelial cells co-incubated with tumor cells, but not with endothelial cells grown alone, as previously shown by Würdinger et al. [102]. Zhang et al. went on to show that the effect was due specifically to factors released into the conditioned media by tumor cells. Further, the observed increase in the miRNAs in the endothelial cells could be attributed to the uptake of tumor EVs, as knock-down of Drosha and inhibition of miR-9 and miR-183 in tumor cells led to a reduction in the levels of these miRNAs in recipient endothelial cells. In subsequent co-culture assays focusing on the effects of miR-9, they showed that knockdown of miR-9 in the tumor cells led to a reduced rate of endothelial cell migration. One of the caveats of the study is that the authors did not directly show that miR-9 in tumor-derived EVs delivered to endothelial cells was responsible for induction of angiogenesis. However, the finding that delivery of miRNAs via EVs results in changes to the miRNA profile of endothelial cells has important implications for angiogenesis formation and tumor progression.
These studies represent major steps forward in elucidating contributions of specific RNA molecules transferred within EVs to cancer progression. Future studies are needed to establish the role of other exRNAs in progression of tumors.
6. exRNA as biomarkers for cancer
The presence of exRNAs from cancer cells in biofluids provides the potential for their use as biomarkers to provide a “snapshot” of the macromolecular composition of tumor cells. EVs have been found in all biofluids, including serum, plasma [3], CSF [103], urine [104], breast milk and saliva [105]. Cancer diagnosis, as well as treatment decisions, stratification for clinical trials, longitudinal profiling of dynamic genetic changes in tumors, outcomes of clinical trials, and follow-up for populations at risk, all have the potential to be informed by exRNA assays on biofluids, thereby, bypassing the need for tumor biopsy in many cases. exRNA-based assays could also provide an important companion diagnostic to determine specific pathways mutated or altered in individual cancers. As cancer molecular diagnostics becomes increasingly sophisticated, knowledge about specific aberrant pathways on a patient-by-patient basis could open the opportunity for specific drug targeting for individualized care [106]. exRNAs are ideal biomarkers because they have the potential to be highly sensitive, predictive, robust, translatable and most importantly minimally invasive.
The use of exRNAs in biofluids as biomarkers of cancer is being explored using a number of different biofluids and types of cancer with analysis of levels and mRNA mutations, as well as levels of miRNAs and other non-coding RNAs (see Table 1 for examples). Biomarker detection in EVs can be challenging because tumor-derived EVs are not very abundant relative to the bulk of circulating EVs which are derived from normal host cells [107]. Examples of promising EV-based mRNA markers for peripheral cancers include detection of mRNA for a fusion protein, TMPRSS2-ERG in urine EVs in prostate cancer [108], elevated VEGF, IL-6, RANTES mRNAs in EVs from blood in gastric cancer [109] and LISCH7 mRNA in EVs from plasma in colon cancer [110]. Several papers describe distinctive mRNA transcriptomes in saliva from patients with breast cancer [111]; pancreatic cancer [112] and ovarian cancer [113], as compared to controls. mRNA encoding for a tRNA synthetase unique splice variant has also been shown to be released in EVs from prostate cancer cells in culture [114], but this has yet to be evaluated in urine from these patients. In addition, microarray analysis of mRNAs in serum EVs from glioblastoma patients and controls can correctly separate these two groups by unsupervised clustering analysis, although in this case the critical differences in mRNA levels seem to reflect the response of the patient cells to the tumor rather than being mRNAs in tumor-derived EVs [43]; albeit abnormally high levels of the tenascin C mRNA characteristic of glioblastoma tumors was found in serum EVs from these patients [43].
Table 1.
Extracellular vesicle RNA as a source for cancer biomarkers
| Cancer | miRNA | Biofluid | Reference |
|---|---|---|---|
| Esophageal squamous cell cancer (ESCC) |
miR-21 | Serum | [145] |
| Lung cancer | miR levels | Plasma/serum | [46] |
| Gliomas | miRNA, LINE, SINE | Conditioned medium | [50] |
| Cancer | mRNA | Biofluid | Reference |
| Ewing’s sarcoma | Microarray | Conditioned medium | [146] |
| Prostate cancer | Survivin | Plasma/serum | [147] |
| Pancreatic cancer | Apbb1ip, Aspn, BCO31781, Daf2, Foxp1, Gng2, Incenp |
Saliva | [148] |
| Glioblastoma | EGFRvIII, miR-21 | Serum | [3] |
| Medulloblastoma | cMyc amplification | Mouse serum | [21] |
| Gliomas | IDH1 | CSF | [116] |
| Glioblastoma | Ribosomal RNA | Serum | [43] |
| Prostate cancer | PCA-3 and TMPRSS2-ERG |
Urine | [108] |
| Gastric cancer | VEGF, IL-6, RANTES |
Blood | [109] |
| Colon cancer | LISCH7 | Plasma | [110] |
In brain tumors, the constitutively active form of EGFR, the EGFRvIII variant, present in about 30% of glioblastomas [115], was detected in serum-derived EVs from patients whose tumors were confirmed to contain this mutant/variant [3]. More recently a point mutation in the isocitrate dehydrogenase 1 (IDH1) gene was detected in CSF EVs from glioma patients [116]. This mutation occurs in 80% of low-grade gliomas and 20% of secondary glioblastomas and correlates with longer survival and better response to temozolomide treatment [117]. Large deletions and rearrangements, such as EGFRvIII, are relatively easy to detect using real-time PCR because the background genes are different enough so they can be excluded in the PCR assay. Small genetic changes, such as the point mutation in the IDH1 gene can be extremely challenging and require more sensitive assays. BEAMing (Bead, Emulsion, Amplification, Magnetics) PCR, and more recently digital PCR instrumentation allow amplification of very rare events, even when there is a prominent wild-type background signal [118]. In BEAMing assays, molecules of interest are diluted into thousands (or millions) of individual water compartments and re-suspended in oil such that each PCR reaction occurs individually, thereby reducing interference from the background signal. This platform was used to develop an assay for the mutant IDH1 with detection sensitivity estimated at 1 mutant per 10,000 normal copies of the mRNA in CSF [116]. Next generation sequencing platforms, especially direct targeted sequencing, should increase the sensitivity of the exRNA-based biomarker detection even further.
Great emphasis is currently being placed on levels of miRNAs in biofluids as a source of biomarkers for cancer (for review see [119]). For example, studies have reported abnormal distinctive miRNA profiles in serum from patients with ovarian cancer [48], lung cancer [46] and esophageal squamous cell carcinoma [120], as well as in saliva from patients with oral cancer [121]. Other cancers, including prostate [108,122], liver [123] and colorectal cancer [124] also contain unique profiles of miRNAs within their released EVs. We and others have detected abnormally high levels of miR21 and other specific miRNAs in serum and CSF from brain tumor patients [3,125,126]. To facilitate use of miRNAs as tumor biomarkers, extensive efforts have been undertaken to collate EV miRNA associated with different types of cancer into public databases [127], with a companion databases for EV mRNA and protein [128]. ncRNAs, including lncRNA, siRNA, piRNA, snRNA and others have also been detected at high levels in EVs in cancer patients, thus supporting a multi-variant platform for cancer biomarker discovery [21,50,59,62] with a peak of small (<200 nt) RNAs distinctively found in serum EVs from glioblastoma patients, as compared to controls [59]. Collectively there is great promise in the use of exRNAs in biofluids as biomarkers for cancer.
7. exRNA as therapeutics for cancer
In considering the potential therapeutic impact of exRNA for cancer, it is important to bear in mind the large amount of information EVs normally contain in the form of protein, lipid, DNA and RNA, and how these constituents vary depending on the cells from which the EVs are derived. In general, EVs from tumor cells are considered part of the pathogenesis of cancer, with a substantial component within these tumor EVs being exRNA (for review see [107]).
One potential therapeutic strategy would be to reduce the release of EVs from tumor cells, for example using short interfering RNAs (siRNAs) which down-regulate Rab proteins involved in the MVB release process [129] or to remove EVs from circulation by dialysis [130]. EVs from normal cells on the other hand, such as immune cells, are seen as active components in fighting cancer with an important aspect being their RNA content, as found in EVs from immune cells (see below). In this case, strategies would seek to selectively augment this cancer-targeting component, such as enriching EVs from appropriate cell types in patients.
Given the backdrop of the complex cargo in EVs, one has to consider that any therapeutic RNA incorporated into EVs will need to function in the background of a host of other RNA species and components, so the cell of origin used to generate EVs will be critical. When properly selected, these therapeutic EVs can contain inherent, as well as engineered RNAs. Several methods have been used to incorporate therapeutic RNA into EVs, as well as to retarget EVs to specific tissues (Fig. 3). 1) In one of the first attempts, EVs from autologous dendritic cells in culture were targeted by transduction with a membrane ligand. EVs were then harvested from transduced cells and loaded with siRNA by electroporation followed by intravenous administration into mice [131]. Although carried out to produce a knock-down of neurodegenerative mRNAs in mice, this study offers a paradigm for targeting EVs for uptake by specific tissues and delivering therapeutic RNA via protective and non-immunogenic EVs, with obvious applications to cancer. 2) Another approach has been to transfect a DNA expression construct into cells to achieve high expression levels of a therapeutic mRNA/protein in the EV donor cells in culture, such that substantial amounts of these therapeutic agents are incorporated into the EVs. This was used to deliver a prodrug activating system (cytosine deaminase-uracil phosphoribosyltransferase) to schwannoma cells by direct injection of loaded EVs into the tumor, with subsequent systemic treatment with the prodrug (5-fluorocytosine) resulting in regression of the tumor in a mouse nerve model [132]. Again, this provides a paradigm for delivery of therapeutic RNAs, such as short hairpins (shRNAs), albeit strategies to load specific RNAs into EVs still need to be elucidated, with a potential EV-targeting “zipcode” being one option [64]. Introduction of a targeting peptide ligand for epidermal growth factor receptor (EGFR), which is highly expressed on many tumor types, into the membrane of EVs has also been used to deliver the tumor suppressor miRNA let-7a to inhibit growth of breast cancer in a murine model [133]. 3) Other studies also implicate EV-mediated transfer of endogenous miRNAs in cancer, such as miR-143 in EVs from normal prostate epithelial cells which inhibits proliferation of prostate cancer cells [134] and functional transfer of miR-335 from T cell EVs to antigen-presenting cells through the immune synapse [71], which might be harnessed to promote immune rejection of tumor cells. 4) Parallels between the incorporation of RNA into EVs and production of RNA viruses in enveloped virions, e.g. retroviruses [135] can provide important insights into EV cargo loading. In a sense many virus vectors in use, such as lentivirus, are de facto members of the EV class and can deliver therapeutic genes to many cell types, for example, stable delivery of tumor antigen expression cassettes to dendritic cells for cancer immunotherapy [136], and viral proteins can be used to promote fusion of EVs with the recipient cell [e.g. 137]. Notably, expression of human endogenous retrovirus sequences is up-regulated in many cancers [e.g. 21] with retroviral-like particles being released by some cancer cells [96] (see HERV section above). Although this is generally seen as a potential oncogenic mechanism, components of these endogenous human sequences may be engineered to serve as efficient therapeutic RNA delivery vehicles.
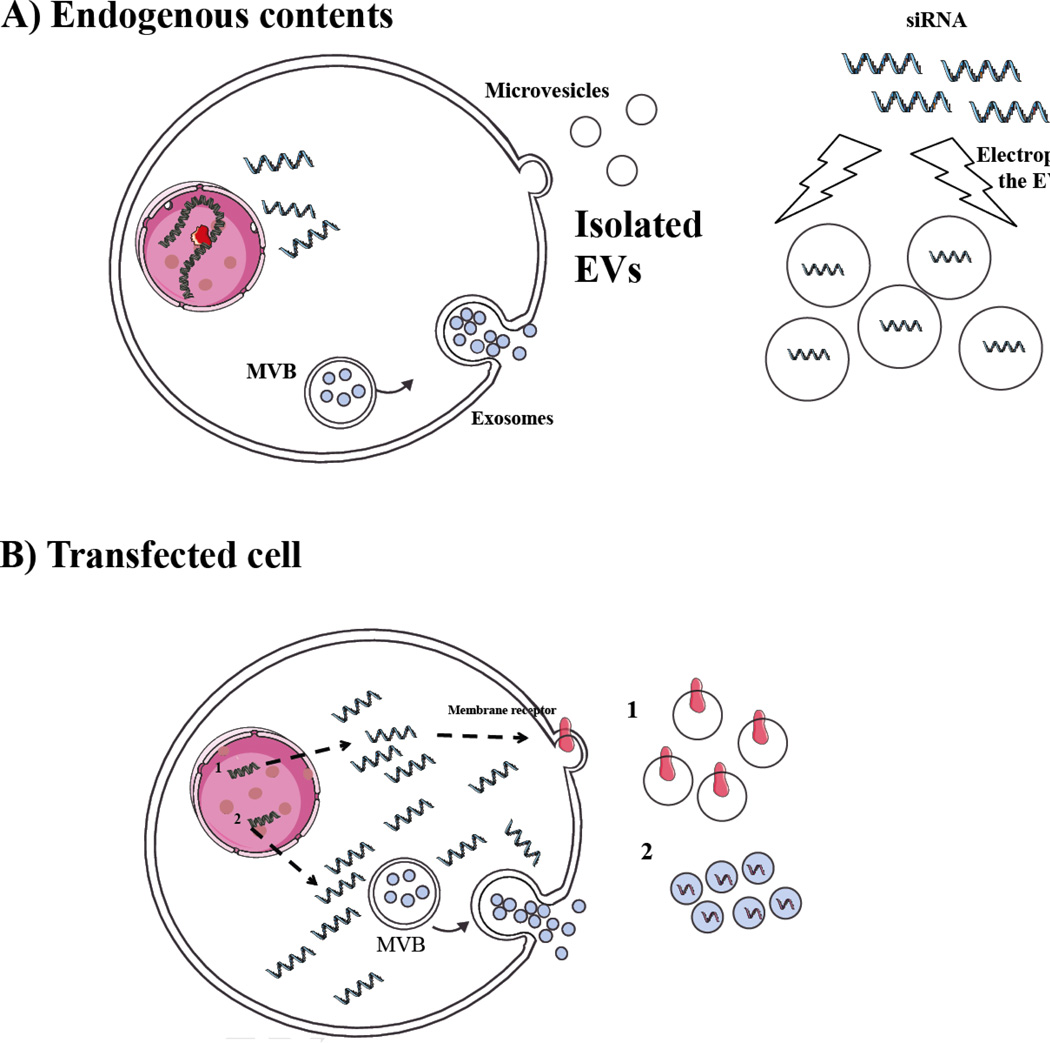
Figure 3. Generation of therapeutic Evs.
A) EVs can be isolated from cultured cells derived from a patient, e.g. dendritic cells, hematopoietic cells and fibroblasts, and then loaded with a therapeutic RNA, e.g. siRNA or miRNA ex vivo, thus providing a non-immunogenic vehicle for RNA delivery. The endogenous contents of these EVs will vary depending on the cell type from which they are derived and in some cases these contents may also promote therapy. B) Cells can also be transfected in culture with an expression cassette for an mRNA encoding a targeting ligand (1) or therapeutic protein for incorporation into the EVs. Other RNAs, e.g. shRNA, miRNA or mRNAs can be expressed at high levels in the donor cell, possibly including EV incorporation signals, and used to deliver therapeutic RNAs.
One can envision future approaches which promote release of vesicles from the plasma membrane, such as by expression of oligomeric proteins [138], and elucidation of RNA sequence – protein binding combinations in the EVs that can load specifically engineered RNAs into the vesicles. Such RNAs could include mRNAs encoding therapeutic proteins, as above, as well as miRNA or other regulatory ncRNAs alone or in combination. (For a broader review of the therapeutic potential of EVs for cancer one is referred to [139–143]).
8. Considerations for future research
A recent review highlights the importance of RNA as extracellular signaling molecules [90]. Release of RNAs within EVs or other stabilizing vehicles for transport of functional RNAs into the extracellular milieu, and delivery to recipient cells is well documented in other recent literature reports [2–4]. It is evident that the cargo contained within these exRNA vehicles, contributes to different biological processes that can influence the phenotype of recipient cells [3,8,15,22–26, 28]. Studies have also shown that specific RNA molecules are enriched in EVs suggesting a special role for these messages or regulatory molecules in delivering information to recipient cells [3,50]. One of the limitations of the current studies is the focus on the presence of specific RNA molecules rather than the entire RNA transcriptome contained within EVs, as well as other proteins and lipids which probably act in concert. This is particularly critical given that the cargo of the EVs is dictated by the cell of origin and should be evaluated on a cell-to-cell basis [37]. Further, several recent reports identifying RNA within EVs using next generation sequencing methods highlight the differences and diversity of exRNAs in different samples, with EVs having a high content of small non-coding RNAs [49,59,144]. Establishing the profile of different RNA molecules released within EVs, specifically with respect to EVs derived from cancer cells as compared to normal cells, will shed light on the particular RNA molecules that may play a role in the different biological processes important for cancer development, progression and metastasis. Using deep sequencing and next generation sequencing methods will help to move forward in better understanding of the composition of RNA within EVs and other extracellular carriers [49].
Equally important, it is necessary to establish the role that specific RNA molecules released via EVs play with regard to disease progression in the context of different types of human cancer. Although many studies have shown that specific exRNA molecules are released and taken up by other cells, a major goal of the future research should focus on determining the potential functions of these identified exRNAs in the recipient cells.
Acknowledgements
We thank Ms. Suzanne McDavitt for skilled editorial assistance and Dr. Emanuele Cocucci for insights into EV trafficking. Funding was provided by NIH/NCI CA069246 and U19 CA179563 and Voices Against Brain Tumor Foundation (XOB) by the NIH Common Fund, through the Office of Strategic Coordination/Office of the NIH Director. Funding was provided by the Richard Floor Biorepository Fund (XOB and LB). LB is supported by the FMD Fellowship (ECOR MGH). KEvdV is supported by the Dutch Scientific Organisation (NWO-Rubicon). Funding was provided by NIH 2T32AG000222-21 (JSR).
Abbreviations
- ASO
antisense oligonucleotides
- CSF
cerebrospinal fluid
- EGFR
epidermal growth factor receptor
- ESCC
esophageal squamous cell cancer
- EV
extracellular vesicle
- exRNA
extracellular RNA
- HDL
high-density lipoprotein
- HLSC
human liver stem cell
- IDH1
isocitrate dehydrogenase 1
- ILVs
intraluminal vesicles
- lncRNA
long non-coding RNA
- miRNA
microRNA
- MSC
mesenchymal stem cells
- MVB
multivesicular body
- ncRNA
non-coding RNA
- piRNA
piwi-interacting RNA
- RNAse
ribonuclease
- RNP
ribonuclear protein
- shRNA
short hairpin RNA
- siRNA
short interfering RNA
- sncRNA
small non-coding RNA
- snoRNA
small nucleolar RNA
- SRP
signal recognition particle
- Stau1
staufen 1
- TE
transposable element
- TLR
Toll-like receptor
- TNF
tumor-necrosis factor
- tRNA
transfer RNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest disclosure
The authors declare that there are no conflicts of interest.
Contributor Information
Jasmina S. Redzic, Email: jasmina.redzic@ucdenver.edu.
Leonora Balaj, Email: Balaj.Leonora@mgh.harvard.edu.
Kristan E. van der Vos, Email: kristanvandervos@hotmail.com.
Xandra O. Breakefield, Email: breakefield@hms.harvard.edu.
References
- 1.Kolodny GM. Evidence for transfer of macromolecular RNA between mammalian cells in culture. Exp Cell Res. 1971;65:313–324. doi: 10.1016/0014-4827(71)90007-3. [DOI] [PubMed] [Google Scholar]
- 2.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 3.Skog J, Würdinger T, van Rijn S, Meijer D, Gainche L, Curry WTJ, et al. Glioblastoma microvesicles transport RNA and protein that promote tumor growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10:1470–1476. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Urbanowicz B, Branski P, et al. Tumour-derived microvesicles carry several surface determinants and mRNA of tumour cells and transfer some of these determinants to monocytes. Cancer Immunol Immunother. 2006;55:808–818. doi: 10.1007/s00262-005-0075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Panagopoulos K, Cross-Knorr S, Dillard C, Pantazatos D, Del Tatto M, Mills D, et al. Reversal of chemosensitivity and induction of cell malignancy of a non-malignant prostate cancer cell line upon extracellular vesicle exposure. Mol Cancer. 2013;12:118. doi: 10.1186/1476-4598-12-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renzulli JF, Del Tatto M, Dooner G, Aliotta J, Goldstein L, Dooner M, et al. Microvesicle induction of prostate specific gene expression in normal human bone marrow cells. J Urol. 2010;184:2165–2171. doi: 10.1016/j.juro.2010.06.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rak J. Microparticles in cancer. Semin Thromb Hemost. 2010;36:888–906. doi: 10.1055/s-0030-1267043. [DOI] [PubMed] [Google Scholar]
- 8.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10:619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 9.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012;18:883–891. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Etheridge A, Gomes CP, Pereira RW, Galas D, Wang K. The complexity, function and applications of RNA in circulation. Front Genet. 2013;4:115. doi: 10.3389/fgene.2013.00115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen X, Liang H, Zhang J, Zen K, Zhang CY. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Camussi G, Deregibus MC, Bruno S, Cantaluppi V, Biancone L. Exosomes/microvesicles as a mechanism of cell-to-cell communication. Kidney Int. 2010;78:838–848. doi: 10.1038/ki.2010.278. [DOI] [PubMed] [Google Scholar]
- 15.Castellana D, Toti F, Freyssinet JM. Membrane microvesicles: macromessengers in cancer disease and progression. Thromb Res. 2010;125:S84–S88. doi: 10.1016/S0049-3848(10)70021-9. [DOI] [PubMed] [Google Scholar]
- 16.Logozzi M, De Milito A, Lugini L, Borghi M, Calabrò L, Spada M, et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS One. 2009;4:e5219. doi: 10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen C, Skog J, Hsu C-H, Lessard R, Breakefield XO, Toner M, et al. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip. 2010;5:505–511. doi: 10.1039/b916199f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keller S, König AK, Marmé F, Runz S, Wolterink S, Koensgen D, et al. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009;278:73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 19.Pisitkun T, Shen RF, Knepper MA. Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A. 2004;101:13368–13373. doi: 10.1073/pnas.0403453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Inal JM, Ansa-Addo EA, Stratton D, Kholia S, Antwi-Baffour SS, Jorfi S, et al. Microvesicles in health and disease. Arch Immunol Ther Exp. 2012;60:107–121. doi: 10.1007/s00005-012-0165-2. [DOI] [PubMed] [Google Scholar]
- 21.Balaj L, Lessard R, Dai L, Cho Y-J, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011;2:180. doi: 10.1038/ncomms1180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipazzi P, Burdek M, Villa A, Rivoltini L, Huber VFilipazzi P, Burdek M, Villa A, Rivoltini L, Huber V. Recent advances on the role of tumor exosomes in immunosuppression and disease progression. Semin Cancer Biol. 2012;22:342–349. doi: 10.1016/j.semcancer.2012.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nat Rev Immunol. 2009;9:581–593. doi: 10.1038/nri2567. [DOI] [PubMed] [Google Scholar]
- 24.Hulsmans M, Holvoet P. MicroRNA-containing microvesicles regulating inflammation in association with atherosclerotic disease. Cardiovasc Res. 2013;100:7–18. doi: 10.1093/cvr/cvt161. [DOI] [PubMed] [Google Scholar]
- 25.Meckes DGJ, Shair KH, Marquitz AR, Kung CP, Edwards RH, Raab-Traub N. Human tumor virus utilizes exosomes for intercellular communication. Proc Natl Acad Sci U S A. 2010;107:20370–20375. doi: 10.1073/pnas.1014194107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simons M, Raposo G. Exosomes--vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 27.Bellingham SA, Guo BB, Coleman BM, Hill AF. Exosomes: vehicles for the transfer of toxic proteins associated with neurodegenerative diseases. Front Physiol. 2012;3:124. doi: 10.3389/fphys.2012.00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Baj-Krzyworzeka M, Szatanek R, Weglarczyk K, Baran J, Zembala M. Tumour-derived microvesicles modulate biological activity of human monocytes. Immunol Lett. 2007;113:176–82. doi: 10.1016/j.imlet.2007.07.014. [DOI] [PubMed] [Google Scholar]
- 29.Al-Nedawi K, Meehan B, Kerbel RS, Allison AC, Rak J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc Natl Acad Sci U S A. 2009;106:3794–3799. doi: 10.1073/pnas.0804543106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kahlert C, Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med (Berl) 2013;91:431–437. doi: 10.1007/s00109-013-1020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pap E, Pallinger E, Falus A. The role of membrane vesicles in tumorigenesis. Crit Rev Oncol Hematol. 2011;79:213–223. doi: 10.1016/j.critrevonc.2010.07.015. [DOI] [PubMed] [Google Scholar]
- 32.Antonyak MA, Li B, Boroughs LK, Johnson JL, Druso JE, Bryant KL, et al. Cancer cell-derived microvesicles induce transformation by transferring tissue transglutaminase and fibronectin to recipient cells. Proc Natl Acad Sci U S A. 2011;108:4852–4857. doi: 10.1073/pnas.1017667108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valenti R, Huber V, Iero M, Filipazzi P, Parmiani G, Rivoltini L. Tumor-released microvesicles as vehicles of immunosuppression. Cancer Res. 2007;67:2912–2915. doi: 10.1158/0008-5472.CAN-07-0520. [DOI] [PubMed] [Google Scholar]
- 34.Iero M, Valenti R, Huber V, Filipazzi P, Parmiani G, Fais S, et al. Tumour-released exosomes and their implications in cancer immunity. Cell Death Diff. 2008;15:80–88. doi: 10.1038/sj.cdd.4402237. [DOI] [PubMed] [Google Scholar]
- 35.Cocucci E, Racchetti G, Meldolesi J. Shedding microvesicles: artefacts no more. Trends Cell Biol. 2009;19:43–51. doi: 10.1016/j.tcb.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Gould SJ, Raposo G. As we wait: coping with an imperfect nomenclature for extracellular vesicles. J Extracell Ves. 2013;2:20389. doi: 10.3402/jev.v2i0.20389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.György B, Szabó TG, Pásztói M, Pál Z, Misják P, Aradi B, et al. Membrane vesicles, current state-of-the-art: emerging role of extracellular vesicles. Cell Mol Life Sci. 2011;68:2667–2688. doi: 10.1007/s00018-011-0689-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mathivanan S, Ji H, Simpson RJ. Exosomes: extracellular organelles important in intercellular communication. J Proteomics. 2010;73:1907–1920. doi: 10.1016/j.jprot.2010.06.006. [DOI] [PubMed] [Google Scholar]
- 39.Théry C, Zitvogel L, Amigorena S. Exosomes: composition, biogenesis and function. Nat Rev Immunol. 2002;2:569–579. doi: 10.1038/nri855. [DOI] [PubMed] [Google Scholar]
- 40.Keller S, Sanderson MP, Stoeck A, Altevogt P. Exosomes: from biogenesis and secretion to biological function. Immunol Lett. 2006;107:102–108. doi: 10.1016/j.imlet.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 41.Di Vizio D, Morello M, Dudley AC, Schow PW, Adam RM, Morley S, et al. Large oncosomes in human prostate cancer tissues and in the circulation of mice with metastatic disease. Am J Pathol. 2012;181:1573–1584. doi: 10.1016/j.ajpath.2012.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathivanan S, Fahner CJ, Reid GE, Simpson RJ. ExoCarta 2012: database of exosomal proteins, RNA and lipids. Nucleic Acids Res. 2012;40:D1241–D1244. doi: 10.1093/nar/gkr828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Noerholm M, Balaj L, Limperg T, Salehi A, Zhu LD, Hochberg FH, et al. RNA expression patterns in serum microvesicles from patients with glioblastoma multiforme and controls. BMC Cancer. 2012;12:22. doi: 10.1186/1471-2407-12-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lee Y, El Andaloussi S, Wood MJ. Exosomes and microvesicles: extracellular vesicles for genetic information transfer and gene therapy. Hum Mol Genet. 2012;21:R125–R134. doi: 10.1093/hmg/dds317. [DOI] [PubMed] [Google Scholar]
- 45.Raposo G, Stoorvogel W. Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol. 2013;200:373–383. doi: 10.1083/jcb.201211138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabinowits G, Gerçel-Taylor C, Day JM, Taylor DD, Kloecker GH. Exosomal microRNA: a diagnostic marker for lung cancer. Clin Lung Cancer. 2009;10:42–46. doi: 10.3816/CLC.2009.n.006. [DOI] [PubMed] [Google Scholar]
- 47.Bellingham SA, Coleman BM, Hill AF. Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res. 2012;40:10937–10949. doi: 10.1093/nar/gks832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 49.Huang X, Yuan T, Tschannen M, Sun Z, Jacob H, Du M, et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genomics. 2013;14:319. doi: 10.1186/1471-2164-14-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Li CC, Eaton SA, Young PE, Lee M, Shuttleworth R, Humphreys DT, et al. Glioma microvesicles carry selectively packaged coding and non-coding RNAs which alter gene expression in recipient cells. RNA Biol. 2013;10:1333–1344. doi: 10.4161/rna.25281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006;20:847–856. doi: 10.1038/sj.leu.2404132. [DOI] [PubMed] [Google Scholar]
- 52.Zhang Y, Liu D, Chen X, Li J, Li L, Bian Z, et al. Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell. 2010;39:133–144. doi: 10.1016/j.molcel.2010.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Yang M, Chen J, Su F, Yu B, Su F, Lin L, et al. Microvesicles secreted by macrophages shuttle invasion-potentiating microRNAs into breast cancer cells. Mol Cancer. 2011;10:117. doi: 10.1186/1476-4598-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. Methods Mol Biol. 2013;1024:41–51. doi: 10.1007/978-1-62703-453-1_4. [DOI] [PubMed] [Google Scholar]
- 55.Lopez-Verrilli MA, Court FA. Exosomes: mediators of communication in eukaryotes. Biol Res. 2013;46:5–11. doi: 10.4067/S0716-97602013000100001. [DOI] [PubMed] [Google Scholar]
- 56.Muralidharan-Chari V, Clancy JW, Sedgwick A, D’Souza-Schorey C. Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci. 2010;123:1603–1611. doi: 10.1242/jcs.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Kilsdonk JW, van Kempen LC, van Muijen GN, Ruiter DJ, Swart GW. Soluble adhesion molecules in human cancers: sources and fates. Eur J Cell Biol. 2010;89:415–427. doi: 10.1016/j.ejcb.2009.11.026. [DOI] [PubMed] [Google Scholar]
- 58.Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, et al. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem. 2013;288:17713–17724. doi: 10.1074/jbc.M112.445403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nolte-'t Hoen EN, Buermans HP, Waasdorp M, Stoorvogel W, Wauben MH, 't Hoen PA. Deep sequencing of RNA from immune cell-derived vesicles uncovers the selective incorporation of small non-coding RNA biotypes with potential regulatory functions. Nucleic Acids Res. 2012;40:9272–9285. doi: 10.1093/nar/gks658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Taylor DD, Gercel-Taylor C. The origin, function, and diagnostic potential of RNA within extracellular vesicles present in human biological fluids. Front Genet. 2013;4:142. doi: 10.3389/fgene.2013.00142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grossman SA, Ye X, Lesser G, Sloan A, Carraway H, Desideri S, et al. Immunosuppression in patients with high-grade gliomas treated with radiation and temozolomide. Clin Cancer Res. 2011;17:5473–5480. doi: 10.1158/1078-0432.CCR-11-0774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van der Vos KE, Balaj L, Skog J, Breakefield XO. Brain tumor microvesicles: Insights into intercellular communication in the nervous system. Cell Mol Neurobiol. 2011;31:949–959. doi: 10.1007/s10571-011-9697-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Collino F, Deregibus MC, Bruno S, Sterpone L, Aghemo G, Viltono L, et al. Microvesicles derived from adult human bone marrow and tissue specific mesenchymal stem cells shuttle selected pattern of miRNAs. PLoS One. 2010;5:e11803. doi: 10.1371/journal.pone.0011803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bolukbasi MF, Mizrak A, Ozdener BG, Madlener S, Ströbel T, Skog J, et al. miR-1289 and “zipcode”-like sequence enrich mRNAs in microvesicles. Mol Ther Nucleic Acids. 2012;1:1:e10. doi: 10.1038/mtna.2011.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Villarroya-Beltri C, Gutiérrez-Vázquez C, Sánchez-Madrid F, Mittelbrunn M. Analysis of microRNA and protein transfer by exosomes during an immune synapse. Methods Mol Biol. 2013;1024:41–51. doi: 10.1007/978-1-62703-453-1_4. [DOI] [PubMed] [Google Scholar]
- 66.Hannafon BN, Ding WQ. Intercellular Communication by Exosome-Derived microRNAs in Cancer. Int J Mol Sci. 2013;14:14240–14269. doi: 10.3390/ijms140714240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liang H, Gong F, Zhang S, Zhang CY, Zen K, Chen X. The origin, function, and diagnostic potential of extracellular microRNAs in human body fluids. Wiley Interdiscip Rev RNA. 2014;5:285–300. doi: 10.1002/wrna.1208. [DOI] [PubMed] [Google Scholar]
- 69.Simpson RJ, Kalra H, Mathivanan S. ExoCarta as a resource for exosomal research. J Extracell Ves. 2012;1 doi: 10.3402/jev.v1i0.18374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guduric-Fuchs J, O’Connor A, Camp B, O’Neill CL, Medina RJ, Simpson DA. Selective extracellular vesicle-mediated export of an overlapping set of microRNAs from multiple cell types. BMC Genomics. 2012;13:357. doi: 10.1186/1471-2164-13-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mittelbrunn M, Gutiérrez-Vázquez C, Villarroya-Beltri C, González S, Sánchez-Cabo F, González MÁ, et al. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat Commun. 2011;2:282. doi: 10.1038/ncomms1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zernecke A, Bidzhekov K, Noels H, Shagdarsuren E, Gan L, Denecke B, et al. Delivery of microRNA-126 by apoptotic bodies induces CXCL12-dependent vascular protection. Sci Signal. 2009;2:ra81. doi: 10.1126/scisignal.2000610. [DOI] [PubMed] [Google Scholar]
- 73.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans E, Lindengey JL, et al. Functional delivery of viral miRNAs via exosomes. Proc Natl Acad Sci U S A. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fabbri M, Paone A, Calore F, Galli R, Gaudio E, Santhanam R, et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc Natl Acad Sci U S A. 2012;109:E2110–E2116. doi: 10.1073/pnas.1209414109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mizoguchi M, Guan Y, Yoshimoto K, Hata N, Amano T, Nakamizo A, et al. Clinical implications of microRNAs in human glioblastoma. Front Oncol. 2013;3:19. doi: 10.3389/fonc.2013.00019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bader AG. miR-34 - a microRNA replacement therapy is headed to the clinic. Front Genet. 2012;3:120. doi: 10.3389/fgene.2012.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Deregibus MC, Cantaluppi V, Calogero R, Lo Iacono M, Tetta C, Biancone L, et al. Endothelial progenitor cell derived microvesicles activate an angiogenic program in endothelial cells by a horizontal transfer of mRNA. Blood. 2007;110:2440–2448. doi: 10.1182/blood-2007-03-078709. [DOI] [PubMed] [Google Scholar]
- 78.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Patil VS, Zhou R, Rana TM. Gene regulation by non-coding RNAs. Crit Rev Biochem Mol Biol. 2014;49:16–32. doi: 10.3109/10409238.2013.844092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 81.Hu S, Wu J, Chen L, Shan G. Signals from noncoding RNAs: unconventional roles for conventional pol III transcripts. Int J Biochem Cell Biol. 2012;44:1847–1851. doi: 10.1016/j.biocel.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 82.Cheng J, Guo JM, Xiao BX, Miao Y, Jiang Z, Zhou H, et al. piRNA, the new non-coding RNA, is aberrantly expressed in human cancer cells. Clin Chim Acta. 2011;412:1621–1625. doi: 10.1016/j.cca.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 83.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 84.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc Natl Acad Sci U S A. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, et al. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Suzuki H, Maruyama R, Yamamoto E, Kai M. DNA methylation and microRNA dysregulation in cancer. Mol Oncol. 2012;6:567–578. doi: 10.1016/j.molonc.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gal-Yam EN, Egger G, Iniguez L, Holster H, Einarsson S, Zhang X, et al. Frequent switching of Polycomb repressive marks and DNA hypermethylation in the PC3 prostate cancer cell line. Proc Natl Acad Sci U S A. 2008;105:12979–12984. doi: 10.1073/pnas.0806437105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fraga MF, Esteller M. Towards the human cancer epigenome: a first draft of histone modifications. Cell Cycle. 2005;4:1377–1381. doi: 10.4161/cc.4.10.2113. [DOI] [PubMed] [Google Scholar]
- 89.Friedman JM, Liang G, Liu CC, Wolff EM, Tsai YC, Ye W, et al. The putative tumor suppressor microRNA-101 modulates the cancer epigenome by repressing the polycomb group protein EZH2. Cancer Res. 2009;69:2623–2629. doi: 10.1158/0008-5472.CAN-08-3114. [DOI] [PubMed] [Google Scholar]
- 90.Dinger ME, Mercer TR, Mattick JS. RNAs as extracellular signaling molecules. J Mol Endocrinol. 2008;40:151–159. doi: 10.1677/JME-07-0160. [DOI] [PubMed] [Google Scholar]
- 91.Goodier JL, Kazazian HHJ. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell. 2008;135:23–35. doi: 10.1016/j.cell.2008.09.022. [DOI] [PubMed] [Google Scholar]
- 92.Daskalos A, Nikolaidis G, Xinarianos G, Savvari P, Cassidy A, Zakopoulou R, et al. Hypomethylation of retrotransposable elements correlates with genomic instability in non-small cell lung cancer. Int J Cancer. 2009;124:81–87. doi: 10.1002/ijc.23849. [DOI] [PubMed] [Google Scholar]
- 93.Ruprecht K, Mayer J, Sauter M, Roemer K, Mueller-Lantzsch N. Endogenous retroviruses and cancer. Cell Mol Life Sci. 2008;65:3366–3382. doi: 10.1007/s00018-008-8496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 95.Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- 96.Contreras-Galindo R, Kaplan MH, Leissner P, Verjat T, Ferlenghi I, Bagnoli F, et al. Human endogenous retrovirus K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Baran J, Baj-Krzyworzeka M, Weglarczyk K, Szatanek R, Zembala M, Barbasz J, et al. Circulating tumour-derived microvesicles in plasma of gastric cancer patients. Cancer Immunol Immunother. 2010;59:841–850. doi: 10.1007/s00262-009-0808-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Silva J, Garcia V, Rodriguez M, Compte M, Cisneros E, Veguillas P, et al. Analysis of exosome release and its prognostic value in human colorectal cancer. Genes Chromosomes Cancer. 2012;51:409–418. doi: 10.1002/gcc.21926. [DOI] [PubMed] [Google Scholar]
- 99.Millimaggi D, Mari M, D’Ascenzo S, Carosa E, Jannini EA, Zucker S, et al. Tumor vesicle-associated CD147 modulates the angiogenic capability of endothelial cells. Neoplasia. 2007;9:349–357. doi: 10.1593/neo.07133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Delves GH, Stewart AB, Cooper AJ, Lwaleed BA. Prostasomes, angiogenesis, and tissue factor. Semin Thromb Hemost. 2007;33:75–79. doi: 10.1055/s-2006-958465. [DOI] [PubMed] [Google Scholar]
- 101.Zhuang G, Wu X, Jiang Z, Kasman I, Yao J, Guan Y, et al. Tumour-secreted miR-9 promotes endothelial cell migration and angiogenesis by activating the JAK-STAT pathway. EMBO J. 2012;31:3513–3523. doi: 10.1038/emboj.2012.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Würdinger T, Tannous BA, Saydam O, Skog J, Grau S, Soutschek J, et al. miR-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14:382–393. doi: 10.1016/j.ccr.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Street JM, Barran PE, Mackay CL, Weidt S, Balmforth C, Walsh TS, et al. Identification and proteomic profiling of exosomes in human cerebrospinal fluid. J Transl Med. 2012;10:15. doi: 10.1186/1479-5876-10-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Miranda KC, Bond DT, McKee M, Skog J, Păunescu TG, Da Silva N, et al. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010;78:191–199. doi: 10.1038/ki.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011;9:9. doi: 10.1186/1479-5876-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yen KE, Bittinger MA, Su SM, Fantin VR. Cancer-associated IDH mutations: biomarker and therapeutic opportunities. Oncogene. 2010;29:6409–6417. doi: 10.1038/onc.2010.444. [DOI] [PubMed] [Google Scholar]
- 107.Rak J. Extracellular vesicles - biomarkers and effectors of the cellular interactome in cancer. Front Pharmacol. 2013;4:21. doi: 10.3389/fphar.2013.00021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nilsson J, Skog J, Nordstrand A, Baranov V, Mincheva-Nilsson L, Breakefield XO, et al. Prostate cancer-derived urine exosomes: A novel approach to biomarkers for prostate cancer. Brit J Cancer. 2009;100:1603–1607. doi: 10.1038/sj.bjc.6605058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kim HK, Song KS, Park YS, Kang YH, Lee YJ, Lee KR, et al. Elevated levels of circulating platelet microparticles, VEGF, IL-6 and RANTES in patients with gastric cancer: possible role of a metastasis predictor. Eur J Cancer. 2003;39:184–191. doi: 10.1016/s0959-8049(02)00596-8. [DOI] [PubMed] [Google Scholar]
- 110.García JM, García V, Peña C, Domínguez G, Silva J, Diaz R, et al. Extracellular plasma RNA from colon cancer patients is confined in a vesicle-like structure and is mRNA-enriched. RNA. 2008;14:1424–1432. doi: 10.1261/rna.755908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Zhang L, Xiao H, Karlan S, Zhou H, Gross J, Elashoff D, et al. Discovery and preclinical validation of salivary transcriptomic and proteomic biomarkers for the non-invasive detection of breast cancer. PLoS One. 2010;5:e15573. doi: 10.1371/journal.pone.0015573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang L, Farrell JJ, Zhou H, Elashoff D, Akin D, Park NH, et al. Salivary transcriptomic biomarkers for detection of resectable pancreatic cancer. Gastroenterology. 2010;138:949–957. doi: 10.1053/j.gastro.2009.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee YH, Kim JH, Zhou H, Kim BW, Wong DT. Salivary transcriptomic biomarkers for detection of ovarian cancer: for serous papillary adenocarcinoma. J Mol Med. 2012;90:427–434. doi: 10.1007/s00109-011-0829-0. [DOI] [PubMed] [Google Scholar]
- 114.Wang F, Xu Z, Zhou J, Lo WS, Lau CF, Nangle LA, et al. Regulated capture by exosomes of mRNAs for cytoplasmic tRNA synthetases. J Biol Chem. 2013;288:29223–29228. doi: 10.1074/jbc.C113.490599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Johnson H, Del Rosario AM, Bryson BD, Schroeder MA, Sarkaria JN, White FM. Molecular characterization of EGFR and EGFRvIII signaling networks in human glioblastoma tumor xenografts. Mol Cell Proteomics. 2012;11:1724–1740. doi: 10.1074/mcp.M112.019984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chen WW, Balaj L, Liau LM, Samuels M, Kotsopoulos S, Maguire CA, et al. BEAMing and digital droplet of qRT-PCR analysis of mutant IDH1 mRNA in tumor extracellular vesicles. Mol Ther Nucl Acids. 2013;2:e109. doi: 10.1038/mtna.2013.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Borodovsky A, Seltzer MJ, Riggins GJ. Altered cancer cell metabolism in gliomas with mutant IDH1 or IDH2. Curr Opin Oncol. 2012;24:83–89. doi: 10.1097/CCO.0b013e32834d816a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Diehl F, Li M, He Y, Kinzler KW, Vogelstein B, Dressman D. BEAMing: single-molecule PCR on microparticles in water-in-oil emulsions. Nat Methods. 2006;3:551–559. doi: 10.1038/nmeth898. [DOI] [PubMed] [Google Scholar]
- 119.Wittmann J, Jäck HM. Serum microRNAs as powerful cancer biomarkers. Biochim Biophys Acta. 2010;1806:200–207. doi: 10.1016/j.bbcan.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 120.Takeshita N, Hoshino I, Mori M, Akutsu Y, Hanari N, Yoneyama Y, et al. Serum microRNA expression profile: miR-1246 as a novel diagnostic and prognostic biomarker for oesophageal squamous cell carcinoma. Br J Cancer. 2013:108. doi: 10.1038/bjc.2013.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yoshizawa JM, Wong DT. Salivary microRNAs and oral cancer detection. Methods Mol Biol. 2013;936:313–324. doi: 10.1007/978-1-62703-083-0_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bryant RJ, Pawlowski T, Catto JW, Marsden G, Vessella RL, Rhees B, et al. Changes in circulating microRNA levels associated with prostate cancer. Br J Cancer. 2012;106:768–774. doi: 10.1038/bjc.2011.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Gailhouste L, Gomez-Santos L, Ochiya T. Potential applications of miRNAs as diagnostic and prognostic markers in liver cancer. Front Biosci. 2013;18:199–223. doi: 10.2741/4096. [DOI] [PubMed] [Google Scholar]
- 124.Chiba M, Kimura M, Asari S. Exosomes secreted from human colorectal cancer cell lines contain mRNAs, microRNAs and natural antisense RNAs, that can transfer into the human hepatoma HepG2 and lung cancer A549 cell lines. Oncol Rep. 2012;28:1551–1558. doi: 10.3892/or.2012.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Akers JC, Ramakrishnan V, Kim R, Skog J, Nakano I, Pingle S, et al. miR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): A Platform for Glioblastoma Biomarker Development. PLoS One. miR-21 in the Extracellular Vesicles (EVs) of Cerebrospinal Fluid (CSF): A Platform for Glioblastoma Biomarker Development. 2013;8:e78115. doi: 10.1371/journal.pone.0078115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Teplyuk NM, Mollenhauer B, Gabriely G, Giese A, Kim E, Smolsky M, et al. MicroRNAs in cerebrospinal fluid identify glioblastoma and metastatic brain cancers and reflect disease activity. Neuro Oncol. 2012;14:689–700. doi: 10.1093/neuonc/nos074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Russo F, Di Bella S, Nigita G, Macca V, Laganà A, Giugno R, et al. miRandola: extracellular circulating microRNAs database. PLoS One. 2012;7:e47786. doi: 10.1371/journal.pone.0047786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mathivanan S, Simpson RJ. ExoCarta: A compendium of exosomal proteins and RNA. Proteomics. 2009;9:4997–5000. doi: 10.1002/pmic.200900351. [DOI] [PubMed] [Google Scholar]
- 129.Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, et al. Rab27a supports exosome-dependent and -independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res. 2012;72:4920–4930. doi: 10.1158/0008-5472.CAN-12-0925. [DOI] [PubMed] [Google Scholar]
- 130.Marleau AM, Chen CS, Joyce JA, Tullis RH. Exosome removal as a therapeutic adjuvant in cancer. J Transl Med. 2012;10:134. doi: 10.1186/1479-5876-10-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- 132.Mizrak A, Bolukbasi MF, Ozdener GB, Brenner GJ, Madlener S, Erkan EP, et al. Genetically engineered microvesicles carrying suicide mRNA/protein inhibit schwannoma tumor growth. Mol Ther. 2013;21:101–108. doi: 10.1038/mt.2012.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ohno S, Takanashi M, Sudo K, Ueda S, Ishikawa A, Matsuyama N, et al. Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells. Mol Ther. 2013;21:185–191. doi: 10.1038/mt.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Kosaka N, Iguchi H, Yoshioka Y, Hagiwara K, Takeshita F, Ochiya T. Competitive interactions of cancer cells and normal cells via secretory microRNAs. J Biol Chem. 2012;287:1397–1405. doi: 10.1074/jbc.M111.288662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gould SJ, Booth AM, Hildreth JE. The Trojan exosome hypothesis. Proc Natl Acad Sci U S A. 2003;100:10592–10597. doi: 10.1073/pnas.1831413100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Breckpot K, Emeagi PU, Thielemans K. Lentiviral vectors for anti-tumor immunotherapy. Curr Gene Ther. 2008;8:438–448. doi: 10.2174/156652308786848058. [DOI] [PubMed] [Google Scholar]
- 137.Temchura VV, Tenbusch M, Nchinda G, Nabi G, Tippler B, Zelenyuk M, et al. Enhancement of immunostimulatory properties of exosomal vaccines by incorporation of fusion-competent G protein of vesicular stomatitis virus. Vaccine. 2008;26:3662–3672. doi: 10.1016/j.vaccine.2008.04.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Fang Y, Wu N, Gan X, Yan W, Morrell JC, Gould SJ. Higher-order oligomerization targets plasma membrane proteins and HIV gag to exosomes. PLoS Biol. 2007;5:e158. doi: 10.1371/journal.pbio.0050158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.El Andaloussi S, Mäger I, Breakefield XO, Wood MJ. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat Rev Drug Discov. 2013;12:347–357. doi: 10.1038/nrd3978. [DOI] [PubMed] [Google Scholar]
- 140.Kosaka N, Takeshita F, Yoshioka Y, Hagiwara K, Katsuda T, Ono M, et al. Exosomal tumor-suppressive microRNAs as novel cancer therapy: “exocure” is another choice for cancer treatment. Adv Drug Deliv Rev. 2013;65:376–382. doi: 10.1016/j.addr.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 141.Seow Y, Wood MJ. Biological Gene Delivery Vehicles: Beyond Viral Vectors. Mol Ther. 2009;17:767–777. doi: 10.1038/mt.2009.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mobergslien A, Sioud M. Exosome-derived miRNAs and cellular miRNAs activate innate immunity. J Innate Immum. 2014;6:105–110. doi: 10.1159/000351460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Koppers-Lalic D, Hogenboom MM, Middeldorp JM, Pegtel DM. Virus-modified exosomes for targeted RNA delivery; a new approach in nanomedicine. Adv Drug Deliv Rev. 2013;65:348–356. doi: 10.1016/j.addr.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Batagov AO, Kurochkin IV. Exosomes secreted by human cells transport largely mRNA fragments that are enriched in the 3’-untranslated regions. Biol Direct. 2013;8:12. doi: 10.1186/1745-6150-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Tanaka Y, Kamohara H, Kinoshita K, Kurashige J, Ishimoto T, Iwatsuki M, et al. Clinical impact of serum exosomal microRNA-21 as a clinical biomarker in human esophageal squamous cell carcinoma. Cancer. 2013;119:1159–1167. doi: 10.1002/cncr.27895. [DOI] [PubMed] [Google Scholar]
- 146.Miller IV, Raposo G, Welsch U, Prazeres da Costa O, Thiel U, Lebar M, et al. First identification of Ewing’s sarcoma-derived extracellular vesicles and exploration of their biological and potential diagnostic implications. Biol Cell. 2013;105:289–303. doi: 10.1111/boc.201200086. [DOI] [PubMed] [Google Scholar]
- 147.Khan S, Jutzy JM, Valenzuela MM, Turay D, Aspe JR, Ashok A, et al. Plasma-derived exosomal survivin, a plausible biomarker for early detection of prostate cancer. PLoS One. 2012;7:e46737. doi: 10.1371/journal.pone.0046737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Lau C, Kim Y, Chia D, Spielmann N, Eibl G, Elashoff D, et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J Biol Chem. 2013;288:26888–26897. doi: 10.1074/jbc.M113.452458. [DOI] [PMC free article] [PubMed] [Google Scholar]