Summary
Local Ca2+ transients such as puffs and sparks form the building blocks of cellular Ca2+ signaling in numerous cell types. They have traditionally been studied by line scan confocal microscopy, but advances in TIRF microscopy together with improved electron-multiplied CCD (EMCCD) camera snow enable rapid (>500 frames s−1) imaging of subcellular Ca2+ signals with high spatial resolution in two dimensions. This approach yields vastly more information (ca. 1GB per minute) than line scan imaging, rendering visual identification and analysis of local events imaged both laborious and subject to user bias. Here we describe a routine to rapidly automate identification and analysis of local Ca2+ events. This features an intuitive graphical user-interfaces and runs under Matlab and the open-source Python software. The underlying algorithm features spatial and temporal noise filtering to reliably detect even small events in the presence of noisy and fluctuating baselines; localizes sites of Ca2+ release with sub-pixel resolution; facilitates user review and editing of data; and outputs time-sequences of fluorescence ratio signals for identified event sites along with Excel-compatible tables listing amplitudes and kinetics of events.
Keywords: Calcium, Imaging, Total internal reflection microscopy, Algorithm, Automation, Fluorescence
Introduction
Ca2+ ions serve ubiquitous roles in intracellular signaling pathways across numerous cell types (1). Much of the versatility and specificity of Ca2+ signals derives from the fact that they can be highly localized in both space and time; a consequence of the rapid release of Ca2+ ions into the cytosol through channels in the plasma and endoplasmic reticulum (ER) membranes together with the restricted diffusion of Ca2+ in the face of stationary cytosolic buffers (2). The study of these local Ca2+ signals first became possible owing to the development of highly sensitive fluorescent indicator dyes (3). Following the initial discovery of local Ca2+ puffs originating through inositol trisphosphate receptors (IP3Rs) in the ER membrane (4–6), numerous other local signals have been identified, notably including sparks arising from ryanodine receptors in the SR of muscle (7, 8).
The imaging, detection and analysis of local Ca2+ signals present several difficulties. The fluorescence signals provided by even the best indicators are faint and have poor signal-to-noise ratio; signals are often superimposed on a non-stationary background level; and their durations are brief (typically a few tens of ms or shorter). Moreover, the optimal resolution of the underlying channel kinetics requires monitoring Ca2+-dependent fluorescence from a very small volume immediately adjacent to the Ca2+ release channels (9), further exacerbating these problems. For many years, the favored technique for recording local Ca2+ signals was line scan confocal microscopy (5, 8,10). In this approach, fluorescence is monitored from a focused laser spot that is rapidly scanned along a single line in the cell to generate a line scan image, visualized with spatial information in one dimension and time in the other. Advantages of this technique include the relatively small dimensions (ca. 300 × 300 × 800 nm; x-y-z) of the confocal spot; the ability to use highly sensitive and low noise photomultipliers to detect emitted fluorescence; and fast time resolution because relatively few pixels are sampled along the single line.
Analysis of line scan images is hampered, however, by the high degree of photon (shot) noise normally present as a consequence of the low number of photons collected per pixel at the high scan rates required. Detection of local Ca2+ events by visual inspection is thus subjective and prone to bias, as well as being laborious. Several automated detection algorithms have thus been developed, employing varying strategies to detect, localize and measure events from line scan images (11–13). Nevertheless, confocal line scan imaging suffers inherent limitations, most importantly the restriction of spatial information to a single line and the resulting difficulty in accounting for signals that arise from local Ca2+ release at sites distant from the scan line.
More recently, technological advances in total internal reflection fluorescence (TIRF) microscopy and improved capabilities of electron-multiplied CCD (EMCCD) cameras have enabled rapid imaging of Ca2+ signals in two spatial dimensions (14, 15). The evanescent field created by TIRF microscopy is used to excite the fluorescence of cytosolic Ca2+ indicator within a region only ~100 nm deep adjacent to the cell membrane, providing an optical ‘section’ appreciably narrower than is possible by confocal microscopy. Moreover, this sheet is continuously illuminated, as opposed to the sequential scanning of a confocal spot, so that large areas of a cell can be simultaneously imaged by a camera. Currently available CCD cameras enable imaging with 128 × 128 pixel spatial resolution at frame rates of >500 frames s−1. This approach yields vastly more information than line scan imaging, but the amount of data generated (ca. 1 GB per minute) render visual identification and analysis impracticable, and places the onus on development of automated computer algorithms.
Here, we describe a routine we developed, running under both MatLab and the open-source software environment Python, to automate processing of TIRF imaging data of local signals (Ca2+ puffs) originating from IP3Rs in cultured mammalian cells (15, 16). Although several algorithms are available to automate identification and analysis of Ca2+ signals from 3-dimensional (x, y, time) image stacks (12, 17–21), these were developed and optimized for Ca2+ signals emanating from RyR’s (Ca2+ sparks). In contrast to the relatively large and stereotyped fluorescence spark signals, puffs are more heterogeneous and of smaller average amplitude. We evaluated two of the more recent published algorithms (18, 20), and found them inadequate for our purposes in terms of their ability to detect smaller events and lack of kinetic analysis. Moreover, unlike previous algorithms, our algorithm is site-oriented, grouping together data from discrete individual puff sites which display heterogeneous behaviors. The algorithm features an intuitive user interface to facilitate visual inspection and editing of the fluorescence signals. Spatial and temporal noise filtering enables reliable detection of even single-channel signals in the presence of noisy and fluctuating baselines, and data are output as Excel-compatible files including localizations of sites of Ca2+ release with sub-pixel resolution, and time-sequences of fluorescence ratio for all identified event sites, and tables listing amplitudes and kinetics of all events.
Materials and Methods
Cell Culture
Human SH-SY5Y neuroblastoma cells were cultured as previously described (22) in a mixture (1:1) of Ham’s F12 medium and Eagle’s minimal essential medium, supplemented with 10% (v/v) fetal bovine serum (FBS) and 1% nonessential amino acids. Cells were incubated at 37 °C in a humidified incubator with a 95% air and 5% CO2 atmosphere, pass aged every 7 days, and used for up to 20 passages. A few days prior to imaging, cells were harvested in phosphate-buffered saline (PBS) without Ca2+ or Mg2+ and sub-cultured in Petri dishes with glass cover slips as the base (MatTek) at a seeding density of 3×104 cells/ml.
Loading of cell permeant esters
Cells were loaded a few hours before use by incubation with HEPES-buffered saline (HBS: in mM; NaCl 135, KCl 5, MgCl2 1.2, CaCl2 2.5, HEPES 5, glucose 10) containing 1 μM ci-IP3-(propionyloxymethyl) ester (ci-IP3/PM) (SiChem, Bremen, Germany) and 5 μM Cal520AM (AAT Bioquest, Sunnyvale, CA) at room temperature for 60 min. Following this cells were loaded for 1 hr with 5 μM EGTA acetoxymethyl ester (Anaspec, Freemont, CA). For ratiometric experiments, cell were first loaded with Cal520AM, ci-IP3 and EGTA as described above and were subsequently incubated for 45 min with 100 nm calcein red-orange, AM (Invitrogen, Carlsbad, CA).
Total Internal Reflection Microscopy
Imaging of changes in [Ca2+]i was accomplished using a custom-built TIRF microscope system (15) based around an Olympus IX 70 microscope equipped with an Olympus X60 TIRFM objective (NA 1.45). Fluorescence of cytosolic Cal520 was excited within the ~100 nm evanescent field formed by total internal reflection of a 488 nm laser beam incident through the microscope objective at the cover glass/aqueous interface. Images of emitted fluorescence (λ>510 nm) were captured at a resolution of 128 × 128 pixels (1 pixel = 0.33 μm) at a rate of 420 frames s−1 by a Cascade 128 electron multiplied CCD camera (Roper Scientific). Photorelease of i-IP3 from a caged precursor was evoked by flashes of UV (350–400 nm) light derived from a fiber-optic arc lamp source introduced through a UV reflecting dichroic mirror in the upper side-port of the microscope. The UV light was adjusted to uniformly irradiate a region slightly larger than the imaging frame, and any given imaging field was exposed to only a single flash.
Ratiometric imaging in cells loaded with both Cal520 and calcein red-orange utilized a DV2 dual-view image splitter (Photometrics, Tucson, AZ) coupled to an Evolve 512 EMCCD (Photometrics) operated at a resolution of 512 × 512 pixels and a frame rate of 30 ms. A 565 DCXR dichroic filter and 520/20 nm band pass and 635 nm long pass filters were used to spectrally separate signals from the two dyes, which were imaged onto two halves of the camera chip and were subsequently aligned and used to generate a ratio image using MetaMorph software (Molecular Devices, Sunnyvale, CA). Control experiments with cells loaded with only one or other of the dyes showed no detectable bleed-through between the two spectral channels.
Algorithm
The MatLab algorithm was developed using MatLab version R2012b with the following additional toolboxes installed; Signal Processing, Image Processing, Mapping and Optimization toolbox. The Python algorithm was developed using Python 2.7.6 with the following additional toolboxes installed; Scipy, Numpy, and Matplotlib. In this study we used a PC running Windows 7 (64 bit) with a 3.4 GHz, quad core PC and 8 Gb RAM. 16 bit multi-plane TIFF image stacks (128 × 128 pixels) were acquired at 5 ms frame rate for 20 seconds (~4000 frames) using MetaMorph.
Routines running under MatLab or Python, together with sample experimental data and detailed user instructions are freely available by e-mailing the corresponding author (ismith@uci.edu).
Generation of synthetic data
To evaluate the performance of the algorithm, we generated synthetic data by creating an ‘idealized’ puff sequence, and adding this to baseline experimental data (‘noise’) at different known locations after scaling to different amplitudes. The idealized puff record was generated by selecting 7 large amplitude events (ΔF/F0>1) from raw experimental data, cropping out 20 × 20 pixel regions around the centroid of each event and forming an averaged x, y, t sequence after aligning the individual image stacks to synchronize their times of peak fluorescence. The resulting, essentially noise-free puff was normalized in amplitude to ΔF/F0=1, and after scaling by various factors, idealized puffs were added at known locations and times to 1200 frames of experimentally recorded resting Cal520 fluorescence.
Results
Manual processing of calcium puff image data
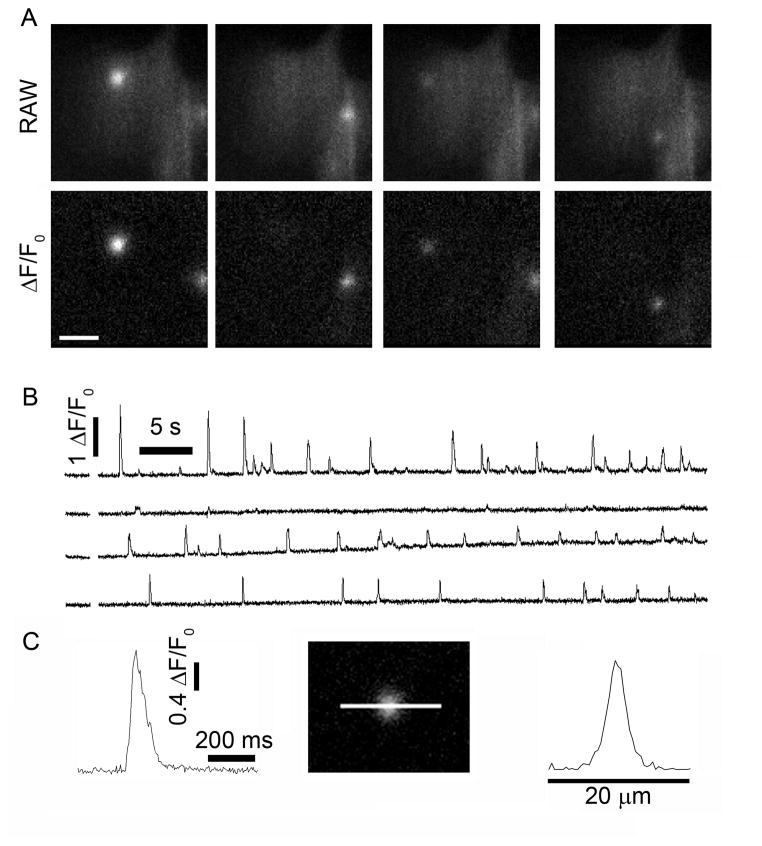
Figure 1 illustrates representative examples of Ca2+ puff activity at localized sites in human neuroblastoma SH-SY5Y cells, recorded as previously described (15, 16, 22). Cells were loaded with the Ca2+ indicator Cal520, a caged precursor (ci-IP3) of the slowly-metabolized IP3 analog iIP3, and the slow Ca2+ buffer EGTA by incubation with membrane-permeant esters of these compounds. Cells were essentially quiescent before photorelease of ci-IP3, while puffs were elicited shortly after a photolysis flash and subsequently continued for tens of seconds (Fig 1A & B; see also left panel Video 1).
Fig. 1.
Local puff activity recorded in SH-SY5Y cells following photo-release of ci-IP3. (A) Representative image frames taken from a video sequence of raw fluorescence, illustrating the appearance of puffs at different locations within a cell. Puffs appear as bright white ‘localized’ fluorescence signals over a darker gray background of resting cytosolic Cal520 fluorescence. Puff activity becomes easier to visually identify when it is expressed as a ΔF/F0. (B) Traces illustrate puffs evoked at 4 different sites within a cell following photo-release of ci-IP3. All traces show fluorescence ratio changes (ΔF/Fo) averaged within a 1 × 1 μm region of interest centered on puff sites. (C) Selected example of a calcium puff shown on an expanded time scale. Image shows a calcium puff in an SH-SY5Y cell. The traces shows measurement of calcium dependent fluorescence along the line in the image passing through the center of the puff.
We had previously (15, 23) processed and analyzed images of such local Ca2+ signals using MetaMorph 7.7.0 (Molecular Dynamics). After subtraction of the camera black offset level, image sequences were processed by dividing each frame by an average of ~100 frames captured before the photolysis flash, so that fluorescence represents a ratio (ΔF/F0) of the increase in fluorescence (ΔF) at each pixel relative to the mean resting fluorescence (F0) prior to stimulation. The resulting image stack was then further processed by frame-by-frame subtraction of heavily smoothed (16 × 16 pixel low-pass filter) images, so as to correct for slow drift in basal fluorescence and fluctuations in laser power (See right panel Video 1). Fluorescence traces like those in Fig. 1B were then derived by visually identifying sites of localized activity, and manually positioning regions of interest (typically 3×3 pixels; 1.2×1.2μm) centered on these Ca2+ release sites. Use of a camera operating at a frame rate of 420 frames s−1 provides sufficient temporal resolution to resolve the kinetics of individual puffs and to determine their spatial profile (Fig. 1C).
Nevertheless, this procedure suffers several drawbacks. Although the subtraction of a heavily smoothed image is effective in correcting for baseline changes due to rising global [Ca2+], it introduces negative-going artifacts from adjacent puff sites and attenuates the puff magnitude to a variable extent depending upon the spatial spread of the event and the selected width of the low-pass filter. Moreover, visual identification of active sites is laborious and subjective. In particular, sites showing low amplitude and/or infrequent events are likely to be overlooked.
An automated algorithm for detection and analysis of local Ca2+ events
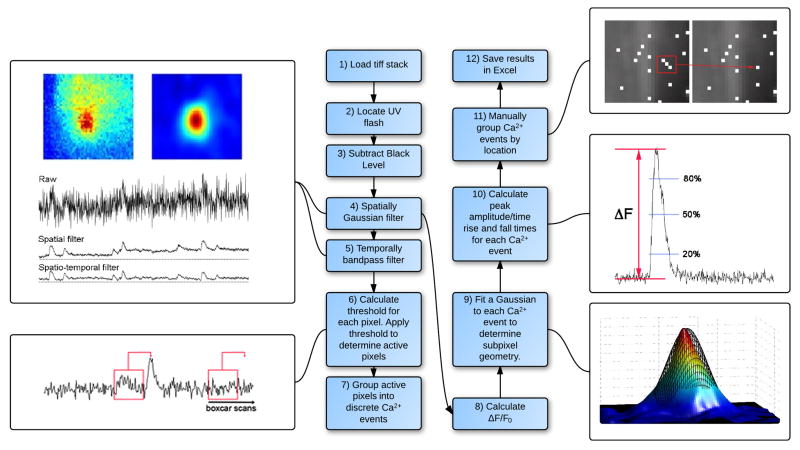
For these reasons, we developed an algorithm to enable rapid automated, unbiased detection of local Ca2+ events imaged with fast EMCCD cameras. The working of the algorithm is summarized by the flow chart in Figure 2, and further details are presented in the Supplementary Information. For clarity, we separately describe here the functioning of the algorithm in terms of event detection and event analysis, but all functions are in fact performed seamlessly.
Fig. 2.
Flow chart of event detection and analysis. Image stacks in Tiff format are loaded into MatLab. UV flash artifacts (if present) are identified and removed from the image stack. The camera black level is subtracted from all image frames and the stack subsequently undergoes spatio-temporal filtering to reduce noise. Events are detected by analyzing each pixel to determine if they exceed a ‘slope-threshold’ criterion with adjacent active pixels being grouped together. Once identification of active sites is accomplished the spatially Gaussian filtered image stack is further processed to render data as a ΔF/Fo. Sub-pixel locations of active sites are then determined along with multiple analysis criteria. Results are subsequently exported into Excell.
Event detection
The algorithm accepts image data stored as the widely used multi-plane TIFF format. Many of our experiments employ photorelease of caged compounds to initiate Ca2+ signals, and the resulting flash artifact is optionally removed by the algorithm based on detection of the maximal positive and negative temporal derivatives of averaged fluorescence throughout the frame; i.e. the beginning and end of the flash artifact [step 2]. The user is then prompted to select an area of the image without cells, and the average value throughout this region during the pre-stimulation frames is subtracted, pixel-by pixel, from all frames within the image stack to correct for the black level offset of the camera and stray light [step 3]. The image stack is spatially smoothed with a Gaussian function of user-specified width (typically 3 pixels; about 1μm)[step 4]. A temporally filtered copy of this stack is then created using two non-causal 1st order Butterworth filters [step 5]; this copy is used only for initial detection of local Ca2+ events and not for analysis. A high frequency cutoff is employed to reduce high-frequency photon shot noise while minimizing attenuation of rapid local Ca2+ signals, and low frequencies are attenuated to compensate for baseline drift over several seconds. Utilizing the spatio-temporal filtered image stack, the baseline standard deviation is calculated for every pixel from image frames captured before stimulation, and is used to generate a threshold for that pixel by multiplying by a user-specified value (for our data typically in the range of 7–10 times the baseline standard deviation). The filtered image stack copy is scanned, pixel by pixel, by a boxcar window that calculates the difference in intensity at any given time from that of a local minimum during a user-specified preceding interval (6–20 frames). Pixels exceeding the threshold value are assigned a value of TRUE in a Boolean x, y, t matrix corresponding to the image stack [step 6]. After completing the matrix the algorithm then groups together pixels separated in space and time by less than user-specified values (here, ‘true’ pixels occurring in the same frame within 10 pixels of one another (~3.3 μm)), and considers them a discrete event [step 7]. Events comprised of fewer than a user-specified number of pixels (in x, y, t; typically 10) are considered to be artifacts, and are rejected for analysis.
Event analysis
The Boolean matrix is used to create three-dimensional (x, y, t) boxes encompassing the pixels of each unique event, and the dimensions of the boxes are further enlarged (padded) by adding a user-specified value to the x, y dimensions (typically 40). A box thus encompasses a spatial region extending spatially beyond the spread of its associated local Ca2+ signal, and temporally beginning at its onset and ending at its termination. The coordinates of the boxes are mapped onto a ratio (F/F0) image stack, formed from the original spatially-filtered image stack created in step 4 by dividing the value of every pixel in the stack by the mean value of that pixel prior to stimulation (frames 5 through the onset of the photolysis flash)[step 8]. A mean spatial image of each event is then created by averaging across all frames within the time window. The image is normalized relative to the intensity of the brightest pixel, and the event is fitted by a two-dimensional Gaussian function with free parameters (determined with sub-pixel resolution) of x and y centroid positions; x and y standard deviations; and angle of the long axis of the resulting elliptical function [step 9]. Fluorescence ratio (F/F0) profiles are then generated from the spatially-filtered for the pixel corresponding to the centroid location of each event, and can optionally be viewed by the user for verification and possible re-analysis after selecting different parameters. The algorithm automatically calculates the peak fluorescence ratio (ΔF) and kinetic parameters (rise time and times to fall to 20%, 50% and 80% of peak) of every event [step 10].
Linking sites
Events whose centroids are localized to within one pixel of one another are considered as arising from the same site. However, it is frequently the case that repeated events show localizations within just a few pixels (< 1 μm) of one another, and may thus be considered to be generated by the same underlying cluster of channels. The user is thus given the option to link together adjacent sites; either by specifying a radius within which adjacent sites will automatically be linked and considered as a single site, or by manually drawing around individual pixel locations to be linked [step 11].
Data export
Data are exported in the Microsoft Excel .xls file format. A single Excel file is generated for each image stack, and comprises three tabs containing a record of user-defined input parameters; fluorescence traces (F/F0) traces for all identified sites; and parameters of all identified sites and events (Table 1). The latter tab includes the following parameters for each site; the sequential event number for each identified site; the mean subpixel x, y localization of each site; the number of events at each location; and the maximum event amplitude observed at that site. Further data are output characterizing the properties of all events originating at a site: the subpixel x, y localization and x, y widths (standard deviation) of each event; peak event amplitude (ΔF/F0); time of peak amplitude; event rise and fall times, reported at 20, 50 and 80% of peak amplitude [step 12].
Table 1.
Output data from algorithm.
|
User-defined constants contains all input parameters used to detect and analyze image stack
Puff data contains all the information as detailed in Table 1
Group traces contains F/F0 traces of all sites in each stack
User interface and interaction
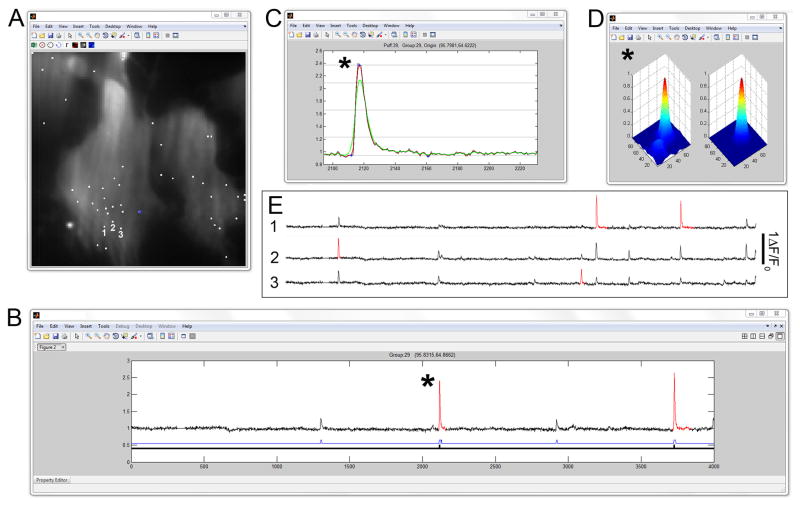
Following loading of the TIFF stack, specification of various parameters (user-defined spatio-temporal filters, threshold detection level and selection of the background level to be subtracted) the algorithm proceeds to run. The main interactive window generated at the end of this process is shown in Figure 3. Fig. 3A shows a monochrome image of resting fluorescence from cells being analyzed. Superimposed on this are event locations (white squares), determined as the centroid of the two-dimensional Gaussian fitted to events. The user can scroll through the different sites to review traces showing background-subtracted, Gaussian smoothed fluorescence ratio changes (F/F0) at each of these sites (Fig. 3B). Red highlighted events indicate those identified by the algorithm as arising at that particular site, and not from ‘bleed-through’ from activity localized to adjacent sites. Clicking on a red event then opens two further interactive windows showing the temporal evolution of the event on an expanded timescale (Fig. 3C), and a spatial profile of the event averaged over its time course together, with the spatial profile of the fitted Gaussian function (Fig. 3D). Any events that are judged to be artifactual can be deleted by right-clicking. The lower blue trace (Fig 3B) marks all instances when the fluorescence signal at the selected site exceeded the detection threshold. Clicking on a marker for events not highlighted in red brings up a new display showing fluorescence at the site where that event originated.
Fig. 3.
User interface. (A) Screenshot of the window showing a monochrome image of resting Cal520 fluorescence on which are superimposed the locations of all sites where activity was localized (white squares). The user can cycle between sites using the cursor key or by manually clicking on a square. (B) Window showing the fluorescence ratio trace (F/F0) at the selected site. Red-highlighted events are identified as having arisen at that site. The lower blue bar marks all events which exceeded the threshold for identification at that site. Those events not highlighted in red have been localized by the algorithm to other adjacent but distinct sites; clicking a marker on the blue bar takes the user to that event site. Clicking on a red event with the mouse opens up two further interactive windows, showing the temporal evolution of the highlighted event on an expanded timescale (C) and spatial profiles of the event averaged over its time course together with the spatial profile of the fitted Gaussian (D). (E) Traces illustrating how the algorithm assigns events to their sites of origin. The traces show fluorescence signals recorded at the three numbered sites in A. See text for further explanation.
Fig. 3E further illustrates the method to associate fluorescence signals exclusively to their site of origin. Traces show fluorescence from the three adjacent sites (1,2,3) numbered in panel A. The first event in the traces is highlighted red only in the record from site 2, indicating that the Gaussian fitting localized it to that site. Although large fluorescence signals are present at the same time in the other two traces, they are not considered as arising at sites 1 or 2, and are not associated by the algorithm with those sites in the final data output. Subsequent events in the traces illustrate further examples where puffs were localized to site 3 and site 1, and are thus not highlighted or considered for analysis in the record from site 2.
A final consideration before exporting the data is to consider sites identified as having arisen from closely adjacent pixels as being a single site. This may be accomplished by selecting the
 button in Panel A and inputting a radius value in pixels within which all identified sites are grouped and considered as a single site located at the centroid of the original localizations. Alternatively, the
button in Panel A and inputting a radius value in pixels within which all identified sites are grouped and considered as a single site located at the centroid of the original localizations. Alternatively, the
 button allows an outline to be manually drawn around the sites of interest. Data are then exported into Excel by selecting the
button allows an outline to be manually drawn around the sites of interest. Data are then exported into Excel by selecting the
 button.
button.
Evaluation of the algorithm using synthetic data
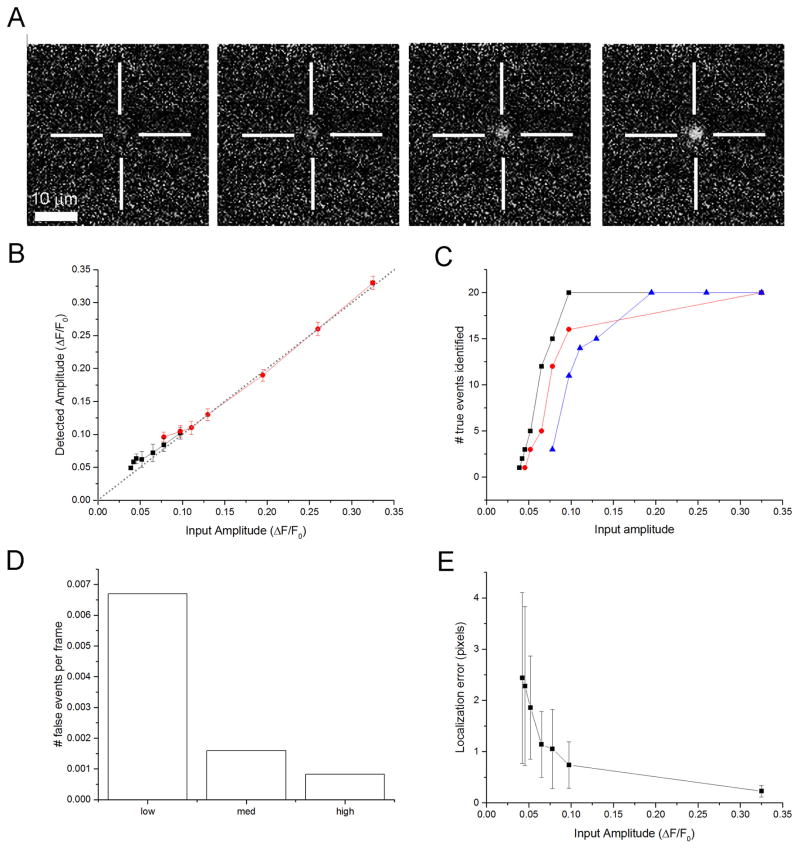
In order to validate and characterize the performance of the algorithm we synthesized ‘ideal’ puffs scaled to varying amplitudes and embedded these at known locations and times in an experimental record of baseline ‘noise’ fluorescence from quiescent cells (Fig 4A). The resulting image stacks were then processed by the algorithm, using different detection thresholds (low, medium and high; respectively 7,9 and 10-times the standard deviation of baseline noise).
Fig. 4.
Evaluation of the algorithm using synthetic puff data. (A) Representative image frames taken from simulated puffs at their peak fluorescence. Each frame is obtained from ΔF/F0 video sequences and shows amplitudes (from left to right of 0.05, 0.1, 0.2 and 0.3 respectively). (B) The reported puff amplitude closely matches the input puff amplitude. The graph plots mean amplitudes of puffs detected at the locations and times where scaled, idealized puffs were embedded in the data stack as a function of the true peak amplitude of the embedded puffs. Black squares and red circles show amplitudes of events detected using, respectively, low and high threshold thresholds. Symbols overlap for input amplitudes >0.1 ΔF/Fo. Data are means + SD from measurements on 20 detected puffs. The dotted grey line indicates a one-to-one relationship. (C) The proportion of puffs detected decreases with decreasing amplitude. Synthetic data were generated embedding 20 idealized puffs of varying scaled amplitudes, and the graph shows the numbers of those events successfully identified by the algorithm employing low (black squares), medium (red circles) and high (black triangles) detection thresholds. (D) Identification of false positives as a function of detection threshold. Bars indicate the number of events falsely identified in a record of resting fluorescence without added events, expressed as number of false identifications per image frame. (E) Localization error of the centroid position of puffs reduces with increasing puff amplitude. Data points plot the mean difference (in pixels) from the known centroid of embedded puffs to the centroid as located by the algorithm. Error bars are means ± SD, N= 20 events.
Figure 4B plots the mean amplitude of the embedded events reported by the algorithm as a function of their true amplitude. Using the high detection threshold, events with amplitudes greater than about 0.1 ΔF/Fo were reliably detected, and over this range there was a close to one-to-one relationship between input and reported amplitudes (R > 0.99). Smaller events (down to ΔF/F0 0.04) could be detected using the low threshold value. With both thresholds, the amplitudes of small events close to the detection limit were slightly overestimated, presumably because those events whose amplitude was diminished by underlying noise fluctuations were not detected at different threshold levels as a function of true amplitude of these events.
Figure 4C shows the proportions of embedded events that were detected as a function of true event amplitude, using different threshold settings. Appreciable numbers of ‘true’ events with amplitudes less than about ΔF/Fo 0.15 were missed using the high threshold, whereas using the low threshold events as small as ΔF/Fo 0.08 could still be reliably detected. The trade-off for using a low detection threshold, however, is an increasing likelihood of false positives; i.e. detection of apparent events in the baseline noise record without any embedded ‘true’ events (Figure 4D). As expected, the lowest threshold setting produced the highest false positive rate, about 0.007 ‘events’ per frame, corresponding roughly to one event per cell every 3 s, with a mean amplitude of about 0.036 ± 0.001 ΔF/Fo. To place these event amplitudes in context, in our recordings from SHSY-5Y cells the fluorescence signals arising from opening of individual IP3R channels have a mean amplitude of about 0.11 ΔF/Fo. Following strong photorelease of IP3 events large puffs are typically evoked at a rate of about 2 per cell per second.
Figure 4E shows how the error in localizing events improves with increasing event amplitude. Ideal puffs of varying amplitudes were embedded in the baseline fluorescence record, centered at specific pixel locations, and the graph plots the mean localization error (difference between the known centroid position and the centroid position as determined by the algorithm) as a function of true event amplitude. For events with amplitudes comparable to single IP3R channel openings (ΔF/Fo~ 0.1) the localization error was around 1 pixel (333 nm on our imaging system), reducing to ~0.2 pixel (70 nm) for larger puffs (ΔF/Fo>0.3).
Analysis of experimental data
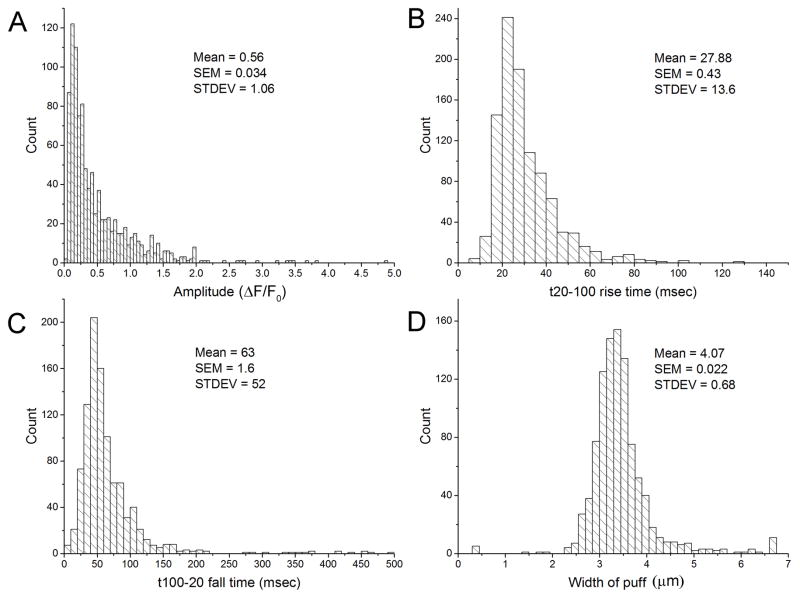
Fig. 5 shows representative output data from the algorithm following analysis of 18 image stacks (each 20 seconds long; 4000 frames) encompassing 674 event locations before grouping (977 events) from SHSY-5Y cells loaded with Cal520 that were stimulated by photorelease of IP3. The panels show distributions of peak event amplitudes (A), event kinetics (rise times, (B); fall times, (C) and spatial width of the Gaussian profile fitted to the average time course of the event (D). The entire algorithm including processing, detection and analysis took ~ 35 s to run each stack on a 3.4 GHz, quad core PC with 8Gb RAM, excluding time for user interaction. By comparison, using the same computer to process a single image stack as ΔF/F0 as described for Fig 1 and then manually identify event locations took about 30 minutes.
Fig. 5.
Representative data provided by the algorithm from analysis of many experimental imaging record similar to that shown in Fig 1A. A total of 977 events were detected following photorelease of IP3 (3640 frames), whereas no events were detected prior to the photolysis flash (316 Frames). (A) Distribution of experimental event amplitudes. (B,C) Event kinetics. Panels show respective distributions of event rise times (from 20% to 100% peak amplitude) and fall times (time to fall to 20% from peak). (D) Distribution of spatial extent of events (mean standard deviation of the Gaussian functions fitted to each event.
The algorithm described here was optimized for TIRF imaging of Ca2+ events in cells co-loaded with EGTA, which localizes fluorescence signals and produces a flatter baseline by suppressing Ca2+ waves and global increases. However, as illustrated in Fig. 6A, it remains effective in detecting and analyzing local events from records obtained by conventional wide-field fluorescence imaging in cells not loaded with EGTA, even when these are superimposed upon fluctuating baselines.
Fig. 6.
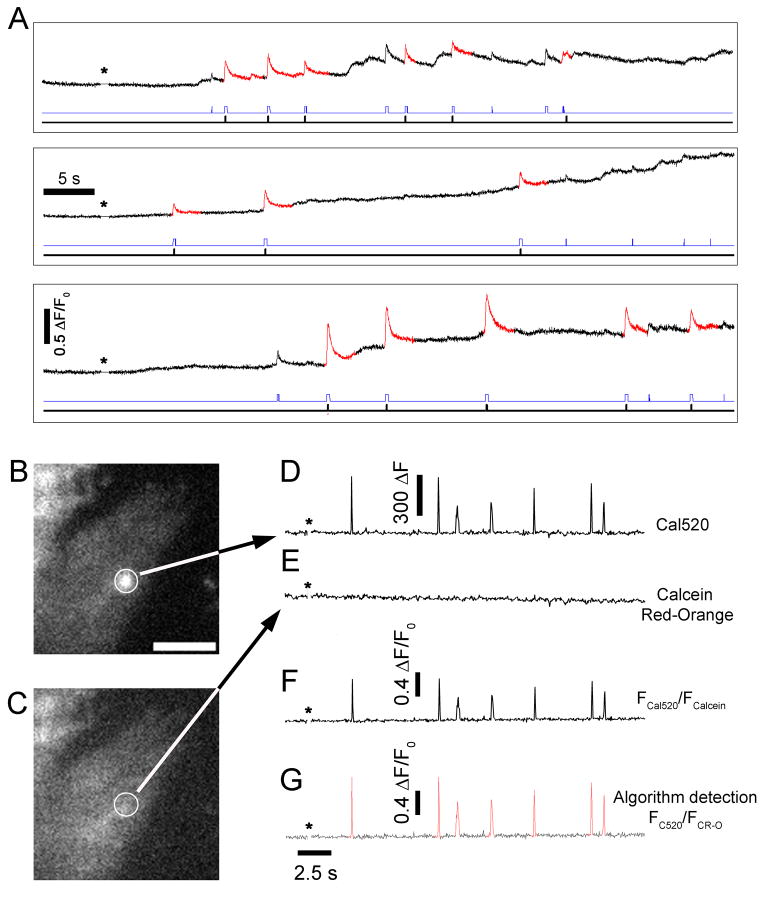
Widefield imaging of local Ca2+ events in cells without EGTA loading that showed showed fluctuating baselines. (A) Representative fluorescence traces (ΔF/F0) from identified sites in cells loaded only with Cal520 and ci-IP3. Red highlighted sections of these traces shows puffs identified as originating from that particular site (also indicated by inflections on the black bar underneath each trace). Inflections in the blue bar indicate that the fluorescence at the site reached the threshold for detection but originated from a nearby site. Asterisks indicate time of UV flash.
(B–G) Ratiometric TIRFM recording and analysis of local puff activity in SH-SY5Y cells dual loaded with the green fluorescent Ca2+ sensitive indicator Cal520 and the Ca2+ insensitive indicator calcein red-orange. (B) Representative image frame taken from a video sequence of raw Cal520 fluorescence captured during a puff (circled). Scale bar = 10 μm. (C) Corresponding image frame of simultaneously recorded calcein red-orange fluorescence, with the location of the puff site circled. (D,E) Fluorescence recordings in the green and red channels from the circled region of interest. A photolysis flash was delivered when indicated by the asterisks. Fluorescence is indicated in arbitrary units. (F) Ratiometric trace derived by dividing the traces in C and D. (G) A ratiometric image sequence was obtained by frame-by-frame division of the background subtracted Cal520 sequence by that of the background subtracted calcein red-orange sequence. This ratiometric image was processed by the algorithm, and the trace shows activity detected at the same site as in B–F. Calibration bar indicates fractional change in FCal520/Fcalcein ratio relative to the mean ratio at that site before photorelease of IP3.
Single wavelength Ca2+ sensitive indicators, such as the fluo dyes and Cal520, do not provide true ratiometric signals. To further extend the utility of the algorithm, we explored its use for processing ratiometric Ca2+ measurements obtained in cells loaded with both Ca2+-sensitive (Cal520) and Ca2+-insensitive (calcein red-orange) fluorescent dyes (24). Figure 6B illustrates a single frame of Cal520 fluorescence capturing a local Ca2+ event (circled), with the corresponding intensity profile over time shown to the right (Fig. 6D). As expected, simultaneous recordings of calcein red-orange fluorescence showed no change (Figs. 6C,E). Ratiometric image stacks were generated by diving the background subtracted Cal520 image stack by the background subtracted calcein red-orange stack (e.g. trace in Fig. 6F). The ratiometric image stack was then processed and analyzed by the algorithm in the identical way as for single-wavelength image data (Fig. 6G).
Discussion
We describe an algorithm, implemented on both MatLab and the open-source software Python, that enables fast processing, analysis and rapid user evaluation of local cellular Ca2+ transients. This was developed for the specific purpose of analyzing IP3-mediated Ca2+ puffs in cultured mammalian cell lines. However, numerous other examples of analogous ‘elementary’ local Ca2+ signals have been discovered in a wide variety of cell types (25), and we anticipate that the algorithm will be more generally applicable to such events, as well as to ‘fundamental’ signals arising from single Ca2+-permeable channels (14, 23). Because the activity of individual channels and clusters of small numbers of channels is stochastic in nature (26), very large data sets (ideally comprising thousands of events) are required for analysis. For example, we were able to reveal correlations between amplitudes and intervals of successive puffs emerge only by statistical analysis of hundreds of events collected laboriously by manual localization and measurement (27). Automated, high-throughput and objective measurements of Ca2+ signals that both display non-Gaussian parameter distributions and are subject to stochastic variability will thus greatly facilitate our understanding of the mechanisms underlying their activity.
A unique feature of our algorithm, not shared by previous programs for analysis of local Ca2+ signals (19, 21), is that it is site-oriented. Ca2+ puffs arise at sparsely distributed, discrete, locations throughout a cell, which are comprised from varying numbers of tightly clustered IP3Rs. A given puff site remains almost stationary over tens of minutes, and can give rise to numerous, repeated events (15, 28,29). In light of the wide heterogeneity between different sites, it is thus important to derive site-specific statistics, because this important information is lost and contributes to greater variability in overall population measures. The algorithm includes powerful features for identifying the location of origin of events and assigning them automatically or with user intervention to specific sites; and for each site outputs data including a trace of fluorescence activity over time, and mean statistics of event frequency, amplitude and kinetics.
In order to facilitate the dissemination and use of our algorithm we have implemented it to run under both MatLab and the open-source software Python, and designed intuitive graphical user interfaces that greatly facilitate data analysis.
Supplementary Material
Video 1. High-speed TIRF imaging of local Ca2+ events in SHSY-5Y cells following photo-release of i-IP3 in cells co-loaded with EGTA and Cal520. UV flash occurs at about 1 second and is seen as a bright white uniform increase in fluorescence across the field of view. Left panel shows raw fluorescence recordings from cells with local Ca2+ signals appearing as bright white localized increases in fluorescence intensity on a background of resting Cal520 fluorescence. The right panel shows the same data processed in Metamorph as a ΔF/F0 (see methods for details). Time stamp reads in s:ms.
Acknowledgments
This work was supported by National Institutes of Health grants GM 100201 to I.F.S, and GM 048071 and GM 065830 to I.P.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berridge MJ, Lipp P, Bootman MD. The versatility and universality of calcium signalling. Nat Rev Mol Cell Biol. 2000;1:11–21. doi: 10.1038/35036035. [DOI] [PubMed] [Google Scholar]
- 2.Schwaller B. Cytosolic Ca2+ buffers. Cold Spring Harbor perspectives in biology. 2010;2:a004051. doi: 10.1101/cshperspect.a004051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Grynkiewicz G, Poenie M, Tsien RY. A new generation of calcium indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 4.Parker I, Yao Y. Regenerative release of calcium from functionally discrete subcellular stores by inositol trisphosphate. Proc Biol Sci. 1991;246:269–274. doi: 10.1098/rspb.1991.0154. [DOI] [PubMed] [Google Scholar]
- 5.Yao Y, Choi J, Parker I. Quantal puffs of intracellular calcium evoked by inositol trisphosphate in Xenopus oocytes. J Physiol. 1995;482:533–553. doi: 10.1113/jphysiol.1995.sp020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao Y, Parker I. Ca2+ influx modulation of temporal and spatial patterns of inositol trisphosphate-mediated Ca2+ liberation in Xenopus oocytes. J Physiol. 1994;476:17–28. [PMC free article] [PubMed] [Google Scholar]
- 7.Cheng H, Lederer WJ. Calcium sparks. Physiological reviews. 2008;88:1491–1545. doi: 10.1152/physrev.00030.2007. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H, Lederer WJ, Cannell MB. Calcium sparks: elementary events underlying excitation-contraction coupling in heart muscle. Science. 1993;262:740–744. doi: 10.1126/science.8235594. [DOI] [PubMed] [Google Scholar]
- 9.Shuai J, Parker I. Optical single-channel recording by imaging calcium flux through individual ion channels: theoretical considerations and limits to resolution. Cell Calcium. 2005;37:283–299. doi: 10.1016/j.ceca.2004.10.008. [DOI] [PubMed] [Google Scholar]
- 10.Bootman M, Niggli E, Berridge M, Lipp P. Imaging the hierarchical calcium signalling system in HeLa cells. J Physiol. 1997;499:307–314. doi: 10.1113/jphysiol.1997.sp021928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng H, Song LS, Shirokova N, Gonzalez A, Lakatta EG, Rios E, Stern MD. Amplitude distribution of calcium sparks in confocal images: theory and studies with an automatic detection method. Biophys J. 1999;76:606–617. doi: 10.1016/S0006-3495(99)77229-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong CH, Soeller C, Cannell MB. Increasing sensitivity of Ca2+ spark detection in noisy images by application of a matched-filter object detection algorithm. Biophys J. 2008;95:6016–6024. doi: 10.1529/biophysj.108.135251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wegner Fv, Both M, Fink RH. Automated detection of elementary calcium release events using the a trous wavelet transform. Biophys J. 2006;90:2151–2163. doi: 10.1529/biophysj.105.069930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Demuro A, Parker I. Imaging the activity and localization of single voltage-gated Ca(2+) channels by total internal reflection fluorescence microscopy. Biophys J. 2004;86:3250–3259. doi: 10.1016/S0006-3495(04)74373-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smith IF, Parker I. Imaging the quantal substructure of single inositol trisphosphate receptor channel activity during calcium puffs in intact mammalian cells. Proc Natl Acad Sci U S A. 2009;106:6404–6409. doi: 10.1073/pnas.0810799106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith IF, Wiltgen SM, Shuai J, Parker I. Calcium puffs originate from preestablished stable clusters of inositol trisphosphate receptors. Sci Signal. 2009;2:ra77. doi: 10.1126/scisignal.2000466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banyasz T, Chen-Izu Y, Balke CW, Izu LT. A new approach to the detection and statistical classification of Ca2+ sparks. Biophys J. 2007;92:4458–4465. doi: 10.1529/biophysj.106.103069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bray MA, Geisse NA, Parker KK. Multidimensional detection and analysis of Ca2+ sparks in cardiac myocytes. Biophys J. 2007;92:4433–4443. doi: 10.1529/biophysj.106.089359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francis M, Qian X, Charbel C, Ledoux J, Parker JC, Taylor MS. Automated region of interest analysis of dynamic Ca(2)+ signals in image sequences. Am J Physiol Cell Physiol. 2012;303:C236–243. doi: 10.1152/ajpcell.00016.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shkryl VM, Blatter LA, Rios E. Properties of Ca2+ sparks revealed by four-dimensional confocal imaging of cardiac muscle. J Gen Physiol. 2012;139:189–207. doi: 10.1085/jgp.201110709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steele EM, Steele DS. Automated detection and analysis of Ca(2+) sparks in x-y image stacks using a thresholding algorithm implemented within the open-source image analysis platform Image J. Biophys J. 2014;106:566–576. doi: 10.1016/j.bpj.2013.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith IF, Wiltgen SM, Parker I. Localization of puff sites adjacent to the plasma membrane: Functional and spatial characterization of calcium signaling in SH-SY5Y cells utilizing membrane-permeant caged inositol trisphosphate. Cell Calcium. 2009;45:65–76. doi: 10.1016/j.ceca.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Demuro A, Parker I. “Optical patch-clamping”: single-channel recording by imaging calcium flux through individual muscle acetylcholine receptor channels. J Gen Physiol. 2005;126:179–192. doi: 10.1085/jgp.200509331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paredes RM, Etzler JC, Watts LT, Zheng W, Lechleiter JD. Chemical calcium indicators. Methods. 2008;46:143–151. doi: 10.1016/j.ymeth.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niggli E, Shirokova N. A guide to sparkology: the taxonomy of elementary cellular Ca2+ signaling events. Cell Calcium. 2007;42:379–387. doi: 10.1016/j.ceca.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Thurley K, I, Smith F, Tovey SC, Taylor CW, Parker I, Falcke M. Timescales of IP(3)-evoked Ca(2+) spikes emerge from Ca(2+) puffs only at the cellular level. Biophys J. 2011;101:2638–2644. doi: 10.1016/j.bpj.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dickinson GD, Swaminathan D, Parker I. The probability of triggering calcium puffs is linearly related to the number of inositol trisphosphate receptors in a cluster. Biophys J. 2012;102:1826–1836. doi: 10.1016/j.bpj.2012.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marchant JS, Parker I. Role of elementary calcium puffs in generating repetitive calcium oscillations. Embo J. 2001;20:65–76. doi: 10.1093/emboj/20.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thomas D, Lipp P, Berridge MJ, Bootman MD. Hormone-evoked elementary calcium signals are not stereotypic, but reflect activation of different size channel clusters and variable recruitment of channels within a cluster. J Biol Chem. 1998;273:27130–27136. doi: 10.1074/jbc.273.42.27130. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video 1. High-speed TIRF imaging of local Ca2+ events in SHSY-5Y cells following photo-release of i-IP3 in cells co-loaded with EGTA and Cal520. UV flash occurs at about 1 second and is seen as a bright white uniform increase in fluorescence across the field of view. Left panel shows raw fluorescence recordings from cells with local Ca2+ signals appearing as bright white localized increases in fluorescence intensity on a background of resting Cal520 fluorescence. The right panel shows the same data processed in Metamorph as a ΔF/F0 (see methods for details). Time stamp reads in s:ms.