Abstract
Background
Antibiotic resistance in Helicobacter pylori contributes to failure in eradicating the infection and is most often due to point and missense mutations in a few key genes.
Methods
The antibiotic susceptibility profiles of H. pylori isolates from 46 Pakistani patients were determined by Etest. Resistance and pathogenicity genes were amplified, and sequences were analyzed to determine the presence of mutations.
Results
A high percentage of isolates (73.9%) were resistant to metronidazole (MTZ), with considerable resistance to clarithromycin (CLR; 47.8%) and amoxicillin (AML; 54.3%) also observed. Relatively few isolates were resistant to tetracycline (TET; 4.3%) or to ciprofloxacin (CIP; 13%). However, most isolates (n = 43) exhibited resistance to one or more antibiotics. MTZ-resistant isolates contained missense mutations in oxygen-independent NADPH nitroreductase (RdxA; 8 mutations found) and NADH flavin oxidoreductase (FrxA; 4 mutations found). In the 23S rRNA gene, responsible for CLR resistance, a new point mutation (A2181G) and 4 previously reported mutations were identified. Pathogenicity genes cagA, dupA, and vacA s1a/m1 were detected frequently in isolates which were also found to be resistant to MTZ, CLR, and AML. A high percentage of CagA and VacA seropositivity was also observed in these patients. Phylogenetic analysis of partial sequences showed uniform distribution of the 3′ region of cagA throughout the tree.
Conclusions
We have identified H. pylori isolates in Pakistan which harbor pathogenicity genes and worrying antibiotic resistance profiles as a result of having acquired multiple point and missense mutations. H. pylori eradication regimens should therefore be reevaluated in this setting.
Keywords: 23S rRNA gene, antibiotic resistance, cagA, clarithromycin resistance, Helicobacter pylori infection, metronidazole resistance
Helicobacter pylori is a common pathogen infecting approximately 50% of the world’s population and is the causative infectious agent in the development of diseases including gastritis, peptic ulcer, and gastric cancer [1]. Emerging resistance of H. pylori strains to several classes of commonly used, widely available antibiotics is the major factor contributing toward the failure of eradication therapy. Wide variations in antibiotic resistance patterns have been described according to differing geographic regions [2,3]. Antibiotic resistance has been attributed to key mutations in a relatively small number of nucleotide and amino acid sequences. Amino acid substitutions in the sequences of oxygen-independent NADPH nitroreductase (RdxA) and NADH flavin oxidoreductase (FrxA) have been reported to be associated with metronidazole (MTZ) resistance [4,5]. Point mutations in 23S ribosomal RNA (23S rRNA) and amino acid changes in penicillin binding protein 1 (Pbp1) have been shown to cause clarithromycin (CLR) and amoxicillin (AML) resistance, respectively [6–9].
Several virulence factors are also known to influence the pathogenicity of H. pylori. These include the presence of the cytotoxin-associated gene pathogenicity island (cag PAI) [10,11], the induced by contact with epithelium gene (iceA) [12], the blood group antigen-binding adhesin (babA) [13], the duodenal ulcer-promoting gene A (dupA) [14], and vacuolating cytotoxin (vacA) [12]. Geographic variations in the distributions of these pathogenicity genes have also been described. This is also likely to affect the outcome of infection, particularly whether the more serious consequences such as gastric cancer result [15].
Polymorphism in CagA occurs as a result of the presence of variable number of repeat regions at the 3′ region of the cagA gene [16–18]. These repeat regions represent combinations of the EPIYA motif (Glu-Pro-Ile-Tyr-Ala). EPIYA-A and EPIYA-B motifs occur widely in all CagA proteins, whereas EPIYA-C and EPIYA-D motifs allow classification of strains as “Western” or “East Asian” types [19]. The “East Asian” variant is regarded as being more harmful than the “Western” type [20]. Sequence analysis of the 3′ region of the cagA gene from H. pylori strains which have been cultured from patients with gastroduodenal diseases has been performed in many countries, but no such sequence analysis has previously been reported from Pakistan, although one key study has shown that clinical strains of H. pylori from Pakistan positive for the cagA promoter region to be significantly associated with gastric inflammation, ulceration, and carcinoma [21].
In Pakistan, H. pylori infection is highly prevalent and there is also indiscriminate consumption of commonly used antibiotics, which can be purchased over the counter without prescription from a healthcare professional. Therefore, we designed a study to investigate in this population; [1] the resistance patterns to commonly used antibiotics of H. pylori cultured from patients undergoing diagnostic endoscopy for investigation of upper gastrointestinal symptoms, [2] the gene mutations associated with this antibiotic resistance, and [3] the frequency and associations of H. pylori pathogenicity genes in the same cohort.
Materials and Methods
Patients
A total of 93 adult patients (with symptoms of acid reflux, abdominal pain, dyspepsia, heartburn, vomiting, or bloating) attending for endoscopy at the Gastrointestinal Endoscopy Department, Military Hospital, Rawalpindi, were enrolled in the study from July 2011 to March 2012. Seventy-one patients were male (mean age 45.8 ± 16.4; range 20–80 years) and 22/93 were female (mean age 49.1 ± 15.1; range 19–78 years). Informed written consent was obtained from each patient, and the study was approved by the Board of Advance Studies and Research, Quaid-i-Azam University, Islamabad. Patients were confirmed to have not taken any antibiotics or gastric acid inhibitors for at least 4 weeks prior to the time of their enrollment into the study. However, in view of the widespread over the counter use of antimicrobial agents in Pakistan, it was not possible to determine accurately to what extent patients had previously taken antibiotics to treat infections with bacteria including H. pylori.
Assessment of Helicobacter pylori Infection Status
Endoscopic biopsy tissue specimens were taken from the gastric antrum within 3 cm of the pylorus. The presence of H. pylori within these gastric antral mucosal biopsies was evaluated by routine histopathology. Briefly, each tissue specimen was formalin-fixed, paraffin-embedded, sectioned and stained with hematoxylin/eosin and Giemsa. Helicobacter pylori load, the degree of neutrophil and mononuclear cell infiltration, and the presence of atypia, atrophy, and intestinal metaplasia were each scored on the basis of the updated Sydney System (0: none, 1: mild, 2: moderate, 3: marked) [22], by an experienced pathologist who was “blinded” to the specimens and their H. pylori infection status. Helicobacter pylori infection was further confirmed by rapid urease test using Christensen urea agar slope (CM0071; Oxoid, Basingstoke, UK) and/or bacterial culture. Helicobacter pylori status was considered positive when histopathology showed positive results, and this was also confirmed in the majority of cases by rapid urease test and/or culture.
Isolation and Culture of Helicobacter pylori
Gastric antral biopsies were placed in 20% glucose solution [23], then chopped, and inoculated on Columbia blood agar (CM0331B; Oxoid) supplemented with Dent’s H. pylori selective medium (SR0147E; Oxoid) and 7% laked horse blood (SR0048; Oxoid). Plates were incubated at 37 °C for up to 7 days in a moist microaerophilic atmosphere of 10% CO2, 5% O2, and 85% N2 established using a Campylobacter gas generating kit (BR0056A, Oxoid). After 3 days of initial incubation, plates were evaluated for growth on a daily basis. Where no growth of H. pylori was observed after incubation for 7 days, plates were recorded as negative for H. pylori culture. Following positive growth, individual small rounded, translucent colony-forming units (CFU) were selected and subcultured twice to ensure pure cultures. Isolates were identified as H. pylori on the basis of morphology following Gram stain, positive findings of oxidase, catalase and urease tests as per [24], and by PCR for 16S rRNA (see Table 1).
Table 1.
Scores of histopathologic grades in Helicobacter pylori-positive patients (n = 57)
| Grade | ||||
|---|---|---|---|---|
| Histopathology | None (n) | Mild (n) | Moderate (n) | Marked (n) |
| H. pylori load | 0 | 20 | 25 | 12 |
| Neutrophil infiltration | 9 | 15 | 22 | 11 |
| Mononuclear cell infiltration | 1 | 7 | 39 | 10 |
| Atrophy | 16 | 27 | 9 | 5 |
| Atypia | 23 | 22 | 7 | 5 |
| Intestinal metaplasia | 48 | 7 | 2 | 0 |
n, number patients.
In Vitro Antibiotic Susceptibility Tests
Susceptibility testing to the antibiotics MTZ, CLR, AML, TET, and CIP was performed using commercial Epsilometer test (Etest®) for minimum inhibitory concentration (MIC) (bioM_erieux SA, Marcy-ÍEtoile, France). Briefly, Columbia blood agar supplemented with 7% vol/vol laked horse blood (SR0048, Oxoid) was inoculated with a bacterial cell suspension calibrated at 3.0 McFarland and allowed to surface dry before application of Etest® strips as per the manufacturer’s instructions. Following 3-day incubation at 37 °C under microaerophilic conditions, Etest® strips were read and resistance to an antibiotic determined using breakpoint concentrations as follows: MTZ (>8 µg/mL), CLR (>1 µg/mL), AML (>1 µg/mL), TET (>1 µg/mL), and CIP (>1 µg/mL). Breakpoint concentrations were defined on the basis of previously approved clinical laboratory standards [6,25–27]. Helicobacter pylori strain 26695 was included as an antibiotic susceptibility testing quality control.
Genomic DNA Extraction
Total genomic DNA was extracted from all H. pylori isolates by ethanol precipitation and proteinase K treatment, using a modification of a method previously described [28]. Briefly, a single bacterial colony was selected, resuspended in 20 µL of 1% sodium dodecyl sulfate, 40 µL proteinase K (100 µg/mL), 80 µL of proteinase K buffer (4M NaCl, 0.5M EDTA; pH 7.5), and incubated at 55 °C for 1 hour. Subsequently, 100 µL 6M NaCl was added, vortexed for 1 minute, and centrifuged at 16,863 × g for 1 minute, at 4 °C. Nucleic acids were then precipitated by adding absolute ethanol and harvested by centrifugation (16,863 × g at 4 °C for 1 minute). Resultant DNA pellets were each washed with 70% vol/vol ethanol and then resuspended in 100 µL 10 mM Tris, 1 mM EDTA buffer; pH 7.5. Samples were stored at −20 °C prior to PCR and DNA sequence analysis.
PCR Amplification of Helicobacter pylori Virulence Determinant Genes
PCR was performed on purified genomic DNA from all H. pylori to examine for the presence of 16S rRNA and markers of pathogenicity including, the cag PAI (consisting of cagA, cagE, cagM), the 3′ region of cagA, iceA1, iceA2, babA, dupA (jhp0917 and jhp0918), and allelic variants of vacA (s1a, s1b, s1c, s2, m1, m2). Oligonucleotide primers for 16S rRNA were synthesized by Alpha DNA (Montreal, QC, Canada), primers for the 3′ region of cagA were supplied by Eurogentec (Hampshire, UK), and primers for all other pathogenicity genes were from Gene Link (Hawthorne, NY, USA). Primer sequences, PCR cycling conditions, and expected amplicon sizes are shown in Supporting Information File S1. Each PCR consisted of 5× FIREPol master mix (Solis BioDyne, Tartu, Estonia) and 5 µL of genomic DNA in a final reaction volume of 25 µL. Thermal cycling was performed using either a T1 Thermalcycler (Biometra GmbH, Goettingen, Germany) or a Multi-Gene OptiMax (Labnet, Edison, NJ, USA). Aliquots (10 µL) of PCR were subjected to electrophoresis on 1% wt/vol agarose gels in Tris-acetate buffer, with ethidium bromide staining for detection of amplicons visualized on a Transilluminator T12 (Biometra GmbH). Presence of dupA was defined as positive with PCR amplification for both jhp0917 and jhp0918.
PCR Amplification and Sequencing of Helicobacter pylori Genes Associated with Antibiotic Resistance
PCR was performed on purified genomic DNA from all H. pylori (resistant and sensitive) strains to obtain amplicons of genes rdxA, frxA, 23S rRNA, and pbp1 to support DNA and amino acid sequence identification of point and missense mutations conferring resistance to MTZ (RdxA, FrxA), CLR (23S rRNA), and AML (Pbp1). Oligonucleotide primers for rdxA and 23S rRNA were synthesized by Gene Link, and the others were supplied by Eurogentec. Primer sequences, PCR cycling conditions, and expected amplicon sizes are shown in Supporting Information File S1. Each PCR mixture for the 3′ region of cagA and for the antibiotic resistance genes consisted of 2× DreamTaq Green PCR master mix (Thermo Scientific, Madison, WI, USA), 5 µL of genomic DNA. Aliquots (10 µL) from the PCR were subjected to electrophoresis on 1% wt/vol agarose gels in Tris-acetate buffer, with GelRed staining for detection of amplicons, visualized on a GelDoc™ XR System (Bio-Rad Laboratories, Hertfordshire, UK).
Prior to DNA sequencing, PCR-generated amplicons of the 3′ region of cagA and genes associated with antibiotic resistance (rdxA, frxA, 23S rRNA, and pbp1) were each purified using a QIAquick PCR purification kit (Qiagen GmbH, Hilden, Germany). DNA was quantified using a NanoDrop Lite Spectrophotometer (Thermo Scientific) followed by sequencing by two commercial services, Eurofins MWG Operon (London, UK) and GATC Biotech (Köln, Germany). Point and missense mutations were identified in resistant and sensitive H. pylori isolates and compared to available sequences of H. pylori reference strain 26695. Sequence comparisons were performed using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) and MEGA software v5.1. Nucleotide sequences were deposited in GenBank (http://www.ncbi.nlm.nih.gov/genbank/) using BankIt.
CagA EPIYA Motifs and Phylogenetic Analysis
CagA EPIYA segments were characterized according to motif pattern as described previously [19]. Partial sequences of the 3′ region of the cagA gene of H. pylori reference strains 26695, J99, G27, SS1, and other deposited sequences retrieved from the National Center for Biotechnology Information Web site (http://www.ncbi.nlm.nih.gov/) were compared with our sequencing data.
CagA and VacA Seropositivity
Venous blood (5 mL) was collected from each patient to evaluate for the presence of specific serum antibodies against CagA and VacA. CagA and VacA seropositivity was determined using a Helico Blot 2.1 Western blot assay kit (MP Biomedicals SAS, Cedex, France) containing H. pylori CagA and VacA antigens with molecular weights of 116 kDa and 89 kDa, respectively. Test strips were incubated with sera for 1 hour at 25 °C and then incubated with an alkaline phosphatase-conjugated goat anti-human immunoglobulin G antibody for a further 1 hour. Strips were then developed with 5-bromo-4-chloro-2-indolylphosphate and nitroblue tetrazolium for 15 minutes. Helico Blot kit reactive and nonreactive control sera were assayed along with patients’ samples for each test run performed. Results were interpreted according to the manufacturer’s instructions.
Statistical Analysis
Statistical analysis was performed using SPSS v20.0 software (SPSS Inc, Chicago, IL, USA). Chi-square (χ2) was used to assess the relationships between pathogenicity genes and clinical findings, histopathologic grades, and antibiotic resistance. Differences were considered significant when p ≤ .05.
Results
Demographics of Helicobacter pylori-Colonized Patients
The frequency of H. pylori infection observed in this patient cohort was 57/93 (61.3%) as assessed by gastric histopathology. For males, 41/71 were positive for H. pylori [57.7%], and in females, 16/22 [72.7%] were found to be positive. From the 57 H. pylori-positive patients, a total of 46 H. pylori isolates (49.5%) were cultured from gastric biopsy tissue. In all cases, positive culture was confirmed by 16S rRNA PCR. In those patients in whom H. pylori was identified by histopathology, 26/46 (56.5%) patients aged between 19 and 44 years were infected, and 31/47 (66.0%) patients aged above 45 years were infected. Frequent symptoms in patients with H. pylori colonization were abdominal pain (45/57; 78.9%), heartburn (43/57; 75.4%), acid reflux (31/57; 54.4%), vomiting (29/57; 50.9%), and bloating (29/57; 50.9%). Regarding clinical disease states, 45/75 (60.0%) of patients with gastritis, 8/12 (66.7%) of those with peptic ulcer disease (PUD), and 4/6 (66.7%) of those with gastric cancer (GC) were found to be H. pylori positive. The histopathologic findings in H. pylori-colonized patients are described in Table 1. To note, we were not able to culture H. pylori organisms from all patients in whom this bacterium was detected by histopathology. A variety of reasons including density and localization of infection, strain type, and technical difficulties in a developing world setting may have contributed to this, as previously described [29,30].
Antibiotic Susceptibilities of Helicobacter pylori Clinical Isolates
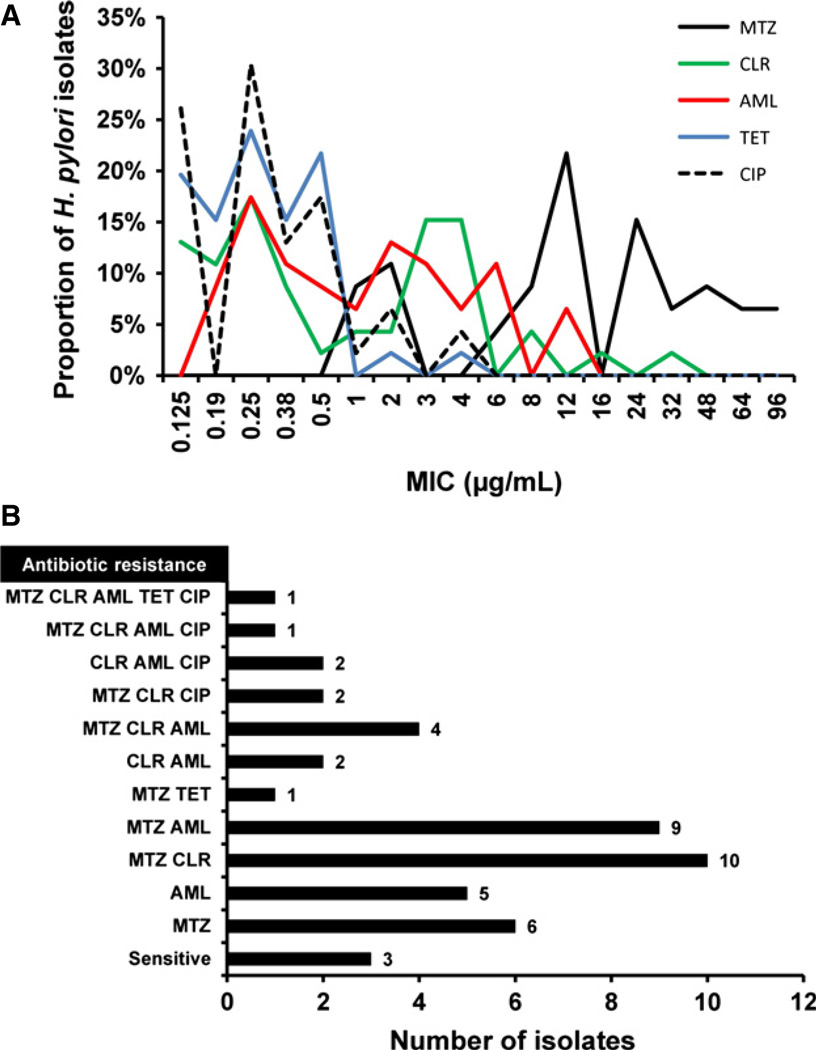
Of the 46 H. pylori isolates cultured, 34/46 (73.9%) displayed resistance to MTZ, with MICs ranging from 8 to 96 µg/mL, while only 12/46 were seen to be sensitive to this nitroimidazole antibiotic (MIC range 1–6 µg/mL). Over half the H. pylori isolates (25/46; 54.3%) were also found to be resistant to AML (MIC 1–12 µg/mL), with the rest being sensitive to this β-lactam antibiotic (MIC 0.19–0.5 µg/mL). Similarly, clinical isolates also displayed resistance to the macrolide antibiotic CLR in 22/46 (47.8%) cases, with MICs ranging from 1 to 32 µg/mL, while the remaining 24 isolates were all sensitive to CLR (MIC 0.125–0.50 µg/mL). The majority of isolates (44/46; 95.7%) were found to highly sensitive to TET (MIC 0.125–0.5 µg/mL), with only 2 isolates showing resistance (MIC 2–4 µg/mL). Likewise, isolates were also sensitive to CIP (40/46; 87.0%) with MICs ranging from 0.125 to 0.5 µg/mL. Resistance to CIP was observed in only 13.0% (6/46) of H. pylori isolates, with MIC ranging from 1 to 4 µg/mL (see Fig. 1A).
Figure 1.
Antibiotic susceptibilities of Helicobacter pylori clinical isolates. (A) Susceptibility testing to amoxicillin (AML), ciprofloxacin (CIP), clarithromycin (CLR), metronidazole (MTZ), and tetracycline (TET) among 46 H. pylori isolates cultured from gastric antral biopsy tissue. MICs were determined by Etest. (B) Distribution of multidrug resistant clinical isolates (i.e., resistance to two or more antibiotics), isolates showing resistance to MTZ or AML alone and those isolates sensitive to all five antibiotics. Resistance was determined using breakpoint concentrations as follows: MTZ (>8 µg/mL), CLR (>1 µg/mL), AML (>1 µg/mL), TET (>1 µg/mL), and CIP (>1 µg/mL).
Resistance against a single antibiotic was only found for a small number of H. pylori clinical isolates; MTZ (n = 6) and AML (n = 5). More frequently the isolates were resistant to more than one antibiotic, with dual antibiotic resistance being observed for MTZ & CLR (n = 10), MTZ & AML (n = 9), CLR & AML (n = 2), and MTZ & TET (n = 1). Multidrug resistance including triple resistance was seen to MTZ, CLR, & AML (n = 4), MTZ, CLR, & CIP (n = 2), and CLR, AML, & CIP (n = 2), and quadruple resistance to MTZ, CLR, AML, & CIP was found in one H. pylori isolate. Only one H. pylori isolate showed resistance against all five antibiotics tested and 3 clinical isolates were found to be sensitive to all five antibiotics studied (see Fig. 1B).
Nucleotide (and Amino Acid) Substitutions Identified in Antibiotic Resistant Helicobacter pylori
PCR-amplified DNA sequences of both resistant and sensitive clinical isolates were examined for nucleotide substitutions as compared to H. pylori reference strain 26695. We were able to sequence amplicons for rdx and frxA from 16 MTZ-resistant H. pylori isolates (with MICs of 12–96 µg/mL) and also amplicons from isolates resistant to CLR (n = 10; MICs of 4–32 µg/mL) and AML (n = 6; MICs of 2–12 µg/mL). For all sensitive H. pylori isolates (sensitive to MTZ, CLR, and AML), no nucleotide substitutions were found, with DNA amplicon sequences obtained all being identical to the reference strain.
MTZ resistance
Nucleotide sequence information for rdxA (Accession no. KC733771-90) and frxA (KC831769-70) was submitted to GenBank. Amino acid substitutions were identified in RdxA in the 16 H. pylori isolates that were resistant to MTZ (MICs 12–96 µg/mL) at positions R16C/H (n = 4/16 isolates), C19Y (n = 1), T31E (n = 8), R90K (n = 5), H97T (n = 8), S108A (n = 2), A118T/S (n = 11), and G189C (n = 1).
Likewise, four known amino acid substitutions were identified in FrxA in two H. pylori isolates which were resistant to MTZ (for isolate SUK55, at F72S, and G73S; for isolate SA18, F72S, G73S, N111H, and A153V), with MICs of 12 and 24 µg/mL, respectively. Interestingly, both of these isolates also possessed amino acid substitutions in RdxA (isolate SUK55, T31E, and H97T; isolate SA15, T31E, and A118T). All other MTZ-resistant H. pylori isolates which possessed amino acid substitutions in RdxA had no identified substitutions in FrxA. Amino acid substitutions such as R16C, C19Y, S108A, and G189C in RdxA and N111H and A153V in FrxA were connected with high MICs (from 24 to 96 µg/mL) in MTZ-resistant H. pylori isolates.
CLR resistance
A previously unreported adenine to guanine (A>G) nucleotide substitution was identified at position 2181A>G in the 23S rRNA gene sequence of two H. pylori clinical isolates found to be resistant to CLR following antibiotic susceptibility testing (with MICs 4 and 16 µg/mL). In addition, four previously reported nucleotide substitutions were identified in 9 other isolates with MICs ranging from 4 to 32 µg/mL, 2116A>G (n = 3/10 CLR-resistant isolates), 2142A>G (n = 1), 2143A>G (n = 3), and 2182T>C (n = 2) [6,7,31]. Variation at position 2116A>G was associated high MICs to CLR (16 and 32 µg/mL) as compared to other observed nucleotide substitutions in the 23S rRNA gene. Nucleotide sequence information for 23S rRNA was submitted to GenBank (KC556778 and KC733766-70).
AML resistance
Helicobacter pylori isolates (n = 6, with MICs ranging from 2 to 12 µg/mL) were found to have previously reported amino acid substitutions in Pbp1 at positions D535N (n = 5 of 6 isolates), S543R (n = 3), and T556S (n = 1) [8,32,33]. Nucleotide sequence information for pbp1 was submitted to GenBank (KC763636-41).
Frequency of Pathogenicity Genes in Clinical Isolates
Of the 46 H. pylori clinical isolates, 37 (80.4%) were positive for cagA, with other genes within the cag PAI, namely cagE and cagM, being detected with frequencies of 34.8% (16/46) and 54.3% (25/46), respectively. The frequency of iceA1 among the isolates was 41.3% (19/46), whereas iceA2 was not found in any isolate. Thirtythree of 46 isolates (71.7%) possessed dupA, and 18/46 (39.1%) babA. Notably, all H. pylori clinical isolates expressed vacA, with 42/46 possessing the allelic variant s1a (91.3%) and 27/46 the variant m1 (58.7%). Other vacA allelic variants found in clinical isolates included s2 (4/46, 8.7%) and m2 (19/46, 41.3%). Variants vacA s1b and s1c were not observed in any isolates tested. The three main vacA genotypes were therefore categorized as vacA s1a/m1 (27/46 isolates, 58.7%), vacA s1a/m2 (15/46, 32.6%), and vacA s2/m2 (4/46, 8.7%).
Presence of the cagA gene was predominant among H. pylori isolates that had vacA alleles: s1a (89.2%, 33/37) and m2 (51.4%, 19/37), whereas cagA-negative isolates harbored the vacA alleles s1a (100%, 9/9) and m1 (100%, 9/9). When vacA genotypes were compared with presence of the cagA gene, there was a significant association between presence of the cagA gene and vacA s1a/m1 48.7% (18/37), vacA s1a/m2 40.5% (15/37), and vacA s2/m2 10.8% (4/37) genotypes (all p < .05, χ2 test).
Association of Pathogenicity Genes with Key Clinical Findings
The pathogenicity genes cagA, cagM, dupA, and vacA genotype s1a/m1 were detected more frequently in H. pylori-colonized patients who had gastritis. In patients with PUD, H. pylori isolates had higher frequencies of cagA, cagM, iceA1, dupA, and vacA genotype s1a/m1. H. pylori containing cagA, babA, dupA, and vacA genotype s1a/m1 were also more frequently observed in GC patients. However, none of these differences were statistically significant. For detailed information on these genotypes, see Supplementary Information File S2.
Single (vacA genotype s1a/m1)- and double (vacA, cag PAI)-positive genotypes were identified only in a few H. pylori isolates (n = 2 and n = 4, respectively, with these specific isolates all cultured from gastritis patients). Seven H. pylori isolates displayed a triple-positive genotype (vacA, cag PAI, and dupA), two of which were isolated from patients with PUD, one from patient with GC, and four from patients with gastritis. The most frequent quadruple-positive genotype was vacA, cag PAI, dupA, and iceA1, which was observed in six of H. pylori isolates. One of the isolates which had a quadruple-positive genotype was cultured from the stomach of a PUD patient, whereas all other isolates containing quadruple-positive genotype were isolated from gastritis patients. Commonly occurring quintuplepositive genotype (vacA, cag PAI, dupA, iceA1, and babA) was found in six isolates. Two H. pylori isolates which had quintuple-positive genotype were associated with PUD, and 4 H. pylori isolates were cultured from patients suffering from gastritis.
Association of Pathogenicity Genes with Histopathologic Grade
There were high frequencies of most of the H. pylori pathogenicity genes with the more marked histopathologic grades of H. pylori load, neutrophil infiltration, and mononuclear cell infiltration as compared to the scores of atypia, atrophy, and intestinal metaplasia. However, significant differences were only found between babA- and dupA-positive strains and scores of H. pylori load (Supplementary Information File S3). Among H. pylori-colonized patients with moderate mononuclear cell infiltration, vacA s1a/m1 was frequently observed (66.7%), while those with a marked score possessed vacA s1a/m2, and those with a mild score were vacA s2/m2 positive. The overall difference was found to be statistically significant (p = .032). However, no significant associations between vacA genotypes and H. pylori load, neutrophil infiltration, atypia, atrophy, and intestinal metaplasia were observed (Supplementary Information File S4).
Association of Antibiotic Resistance and Pathogenicity Genes in Clinical Isolates
High frequencies of cagA, cagM, and dupA genes were found in MTZ-resistant H. pylori isolates, and similar observations (with the exception of cagM) were made in cases of CLR and AML resistance. There were only two isolates that showed resistance against TET and both these isolates also had dupA and iceA1. The majority of CIP-resistant isolates harbored dupA and iceA1, whereas CIP-resistant isolates showed an even distribution of H. pylori-containing vacA genotypes s1a/m1 and s1a/m2. Most H. pylori isolates that showed resistance against all antibiotics had the vacA genotype s1a/m1 (Table 2). Differences between antibiotic resistance and pathogenicity genes were found to be nonsignificant.
Table 2.
Percentage distribution of key pathogenicity genes in Helicobacter pylori isolates demonstrating antibiotic resistance
| Gene/allelic varianta |
MTZ resistance (n = 34) |
CLR resistance (n = 22) |
AML resistance (n = 25) |
TET resistance (n = 2) |
CIP resistance (n = 6) |
|---|---|---|---|---|---|
| cagA+ | 82.4 (28) | 77.3 (17) | 76.0 (19) | 0 (0) | 50.0 (3) |
| cagE+ | 35.3 (12) | 22.7 (5) | 32.0 (8) | 0 (0) | 16.7 (1) |
| cagM+ | 58.8 (20) | 45.5 (10) | 48.0 (12) | 0 (0) | 33.3 (2) |
| babA+ | 44.1 (15) | 45.5 (10) | 40.0 (10) | 50.0 (1) | 33.3 (2) |
| dupA+ | 70.6 (24) | 59.1 (13) | 80.0 (20) | 100 (2) | 83.3 (5) |
| iceA1+ | 38.2 (13) | 40.9 (9) | 52.0 (13) | 100 (2) | 83.3 (5) |
| vacA s1a/m1 | 52.9 (18) | 59.1 (13) | 56.0 (14) | 100 (2) | 50.0 (3) |
| vacA s1a/m2 | 38.2 (13) | 31.8 (7) | 36.0 (9) | 0 (0) | 50.0 (3) |
| vacA s2/m2 | 8.8 (3) | 9.1 (2) | 8.0 (2) | 0 (0) | 0 (0) |
AML, amoxicillin; CIP, ciprofloxacin; CLR, clarithromycin; MTZ, metronidazole; TET, tetracycline.
Presence or absence of pathogenicity genes is based on PCR assay.
CagA EPIYA Motifs and Phylogenetic Analysis
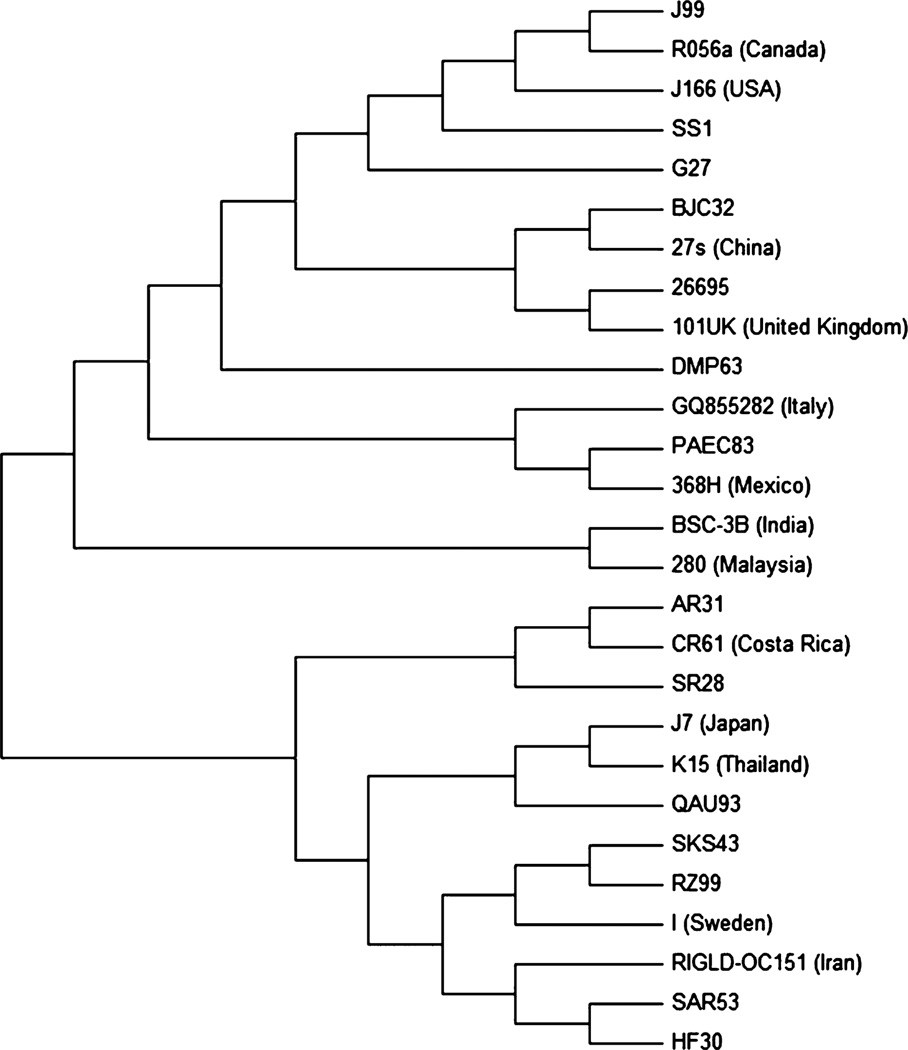
We attempted to sequence the 3′ end of cagA in all H. pylori isolates, but only obtained sequencing data in 10 cases. Several of these sequences contained multiple motifs, but only two types of motifs were found, namely EPIYA (22 occurrences in total) and EPIYT (seven occurrences in total). EPIYA-ABC was detected in most of the sequences, and this was classified as a Western type of cagA, while EPIYA-ABD was only found in the sequence of a single H. pylori isolate (QAU93). To study variations in sequences of cagA gene in H. pylori of different countries, un-rooted phylogenetic tree was drawn. Partial sequences of the cagA gene were close to the sequences published from other developed and developing nations, that is, QAU93 was genetically similar to J7 (Japan) and K15 (Thailand), SAR30 and HF30 were similar to RIGLD-OC151 (Iran), and SKS43 and RZ99 were similar to I (Sweden). The sequences of H. pylori isolates SR28, AR31, BJC32, and PAEC83 were similar to CR61 (Costa Rica), 27s (China), and 368H (Mexico), whereas DMP63 was similar to 26695 and 101UK (United Kingdom); see Fig. 2 for un-rooted phylogenetic tree. DNA sequence information for the 3′ region of cagA (Accession no. KC626079-88) was submitted to GenBank.
Figure 2.
Phylogenetic affiliation of 3′ region of cagA gene sequences of Helicobacter pylori isolates. An un-rooted neighbor-joining consensus tree was constructed with CagA EPIYA segments from H. pylori reference strains, sequences deposited for different H. pylori strains, and sequences of H. pylori isolates of the present study. Gene sequence of I (Sweden) was from gastric biopsy, while all other were sequences of H. pylori strains deposited by authors from developed and developing countries. CagA EPIYA sequences of this study were similar to the sequences published from these countries. MEGA software version 5.1 was used to generate and view the phylogenetic tree by bootstrapping at 1000 bootstrap method.
CagA and VacA Seropositivity
All patients infected with a cagA-positive H. pylori strain were seropositive, suggesting that cagA was fully transcribed and translated into CagA protein. Antibodies to vacuolating cytotoxin VacA were found in 71.7% of H. pylori-infected patients. In patients infected with type s1a/m1 and s1a/m2 H. pylori strains, antibodies to vacuolating cytotoxin were observed in 70.4% and 80.0%, respectively. Only 50% of patients infected with type s2/m2 had antibodies against VacA. There were no significant associations between histopathologic findings and CagA or VacA seropositivity.
Discussion
This study was designed to determine the molecular basis of antibiotic resistance and pathogenicity in H. pylori bacteria isolated from gastric biopsies from symptomatic patients in Pakistan, a developing country where the frequency of H. pylori infection is high and emerging antibiotic resistance is a threatening crisis.
Classical triple therapy eradication regimes for H. pylori eradication have recently shown disappointing efficacy mainly due to the emergence of resistant strains of this bacterium to MTZ and CLR [34,35]. We also report high MTZ resistance, considerable resistance to CLR and AML but little resistance against TET and CIP in the Northern region of Pakistan. Three previous studies investigating H. pylori resistance have been published from the Southern part of Pakistan (approximately 1400 km from the Northern region). These showed wide variations in the frequencies of antibiotic resistance to MTZ (89.0% and 48% of H. pylori isolates with resistance to MTZ not being analyzed in the third study), CLR resistance in 36.0%, 64.0%, and 33.0% of isolates, and AML resistance in 37.0%, 98.0%, and 2.0% of isolates, respectively [36–38]. Geographic variations in the antibiotic resistance of H. pylori are related to the consumption of antibiotics in different communities [39]. In Pakistan, there is unregulated and extensive use of antibiotics specially MTZ, CLR, and AML to treat various infections including respiratory and intestinal disorders. This is likely to have contributed to the considerable resistance of H. pylori isolates toward these antibiotics. We therefore suggest that health authorities in Pakistan should pay serious attention to regulating the availability to these frequently consumed antibiotics.
The mechanism of antibiotic resistance in H. pylori is associated with point mutations and amino acid substitutions in a key resistance-specific gene (23S rRNA) and proteins (RdxA, FrxA, and PBP1). Resistance to MTZ in H. pylori isolates in the present cohort was high, and we detected previously reported amino acid substitutions at various sites (R16C/H, C19Y, T31E, R90K, H97T, S108A, A118T, and G189C) in RdxA [9,25,40]. T31E, R90K, H97T, and A118T were the most common amino acid substitutions found in RdxA associated with MTZ resistance. However, it is very difficult when multiple mutations are present to determine which are significant particularly given that structural analysis of RdxA has shown that some of the amino acid changes noted may not in fact be responsible for the drug resistance [41]. Substitutions in both the MTZ resistance-associated proteins RdxA and FrxA were only identified in two of our MTZ-resistant H. pylori isolates. This suggests that most MTZ resistance in this setting resulted from inactivation of rdxA, which is in agreement with previous reports [4,42]. Jeong and colleagues concluded that development of MTZ resistance requires inactivation of rdxA alone or both rdxA and frxA depending on genotype. Only rarely does MTZ resistance occur as a result of inactivation of frxA alone, and in this case, it may be due to a difference in the regulation of nitroreductase gene expression [42]. High resistance against MTZ in H. pylori isolates supports a previous study which highlights that the extensive use of MTZ against other pathogens in persons positive with chronic H. pylori infection may stimulate the increased frequency of mutation in H. pylori, thus inducing the emergence of resistance against MTZ and other antibiotics [43].
Globally, the most frequently reported nucleotide substitutions within the 23S rRNA gene responsible for CLR resistance in H. pylori clinical isolates are 2142A>G and 2143A>G (along with 2116A>G and 2182T>C) [6,7,31]. Our findings of nucleotide substitutions in 23S rRNA for CLR resistance are in accordance with these previous studies. Of significant interest, we identified a new nucleotide substitution (2181A>G) in the 23S rRNA gene of two clinical H. pylori isolates that were identified as being resistant to CLR by antibiotic susceptibility analysis only. Given that nucleotide substitutions in other antibiotic resistance-associated genes, such as rdxA associated with MTZ resistance, may not in fact be responsible for the drug resistance, clearly further experiments (mutagenic analysis by transformation) are required to substantiate any direct association of this novel nucleotide variation in the 23S rRNA gene to CLR resistance. It is also worth noting that despite the high CLR resistance rates seen in H. pylori isolates from Pakistan, a recent study has reported a low incidence of recurrence of H. pylori infection in patients [38].
Amino acid substitutions were found at three positions (D535N, S543R, and T556S) in AML in H. pylori isolates which were resistant to this antibiotic. These substitutions have all been reported in previous studies [8,32,33]. In the current study, the most common amino acid substitution found in Pbp1 that is also responsible for AML resistance was D535N.
We also report a high frequency of the cagA gene (80.4%) in these Pakistani H. pylori isolates. This percentage was higher than in our recent previous report from the same geographic area. This reported 61.9% prevalence of the cagA gene in H. pylori detected in gastric biopsy specimens, but these were collected from children [28]. Other studies from the same region of Pakistan have shown 24.2% [44] and 56% [45] cagA prevalence in adult dyspeptic patients. The high frequency of cagA-positive H. pylori strains in our study is similar to reports from neighboring South and East Asian countries including India (78.4%) [46], China (82.3%) [47], Malaysia (94.0%) [48], and Japan (96.3%) [49]. Lower frequencies of cagA-positive strains have been reported in H. pylori isolates from Iran (67.0%) and Afghanistan (60.0%) [15].
The frequency of cagE in our H. pylori isolates was also similar to previous reports from South Asia (Iran, 44.0%) [15] and East Asia (Malaysia, 59.0%) [50]. cagE and cagM genes were highly prevalent with frequencies of 96.0% and 97.0%, respectively, in H. pylori isolates cultured in Taiwan (East Asia) [51], whereas we found comparatively lower frequencies of cagE and cagM genes in our Pakistani clinical H. pylori isolates.
In East Asia, iceA1 is found commonly [52], whereas iceA2 is more frequent in H. pylori strains from the USA [53]. In the present study, the considerable frequency of the more virulent allele iceA1 in H. pylori isolates suggests that the predominant allelic form of iceA in Pakistan, like East Asia, is iceA1. The frequency of babA-positive H. pylori isolates in the current investigation was also similar to recent studies from South Asia (31.4%, India and 40.6%, Iran) [46,54], but lower than reports published from East Asia [55–57]. In the present study, the dupA gene was highly prevalent in clinical H. pylori isolates, and this frequency was higher than in other reports from South and East Asian countries [58–60].
The vacA s1a/m1 genotype was predominant among H. pylori isolates in this Pakistani cohort as compared to vacA s1a/m2, whereas the vacA s2/m2 genotype was found infrequently. This again contrasts with our previous study, in which the vacA s1a/m2 genotype was predominantly detected in H. pylori cultured from children from the same region [28]. In addition, the vacA s1b/m2 genotype has been reported among adult patients with dyspepsia in another study from the same region [44]. In previous studies from Karachi (Southern region of Pakistan), vacA s1a/m1 was found to be the predominant genotype [45,61]. It has been observed in this investigation and previous reports from Pakistan that different vacA genotypes were present among H. pylori strains. Frequently observed vacA genotypes in H. pylori from neighboring South and East Asian countries were s1/m2 (Iran), s1/m1 (Afghanistan), [15], s1a/m2 (India and China) [45,62], and s1c/m1b (Japan) [63].
These pathogenicity genes were therefore frequently found in these Pakistani H. pylori clinical isolates in accordance with previous studies from South and East Asia [46,54–57, 64]. However, the presence of these pathogenicity genes was not significantly associated with the severity of disease. cagA-positive H. pylori strains have previously been associated with more severe gastroduodenal diseases [12]. Similarly in this study, cagA-positive H. pylori strains were associated with more marked scores among histopathologic grades including H. pylori load, neutrophil infiltration, and mononuclear cell infiltration. Earlier reports suggested that H. pylori strains with vacA s1/m1 were likely to be more virulent than those with s1/m2 [65–67]. This is in agreement with our findings where the vacA s1a/m1 genotype was found more frequently in patients with high histopathologic scores as compared to other vacA genotypes.
In a recent report, MTZ resistance was found frequently in cagA-negative H. pylori strains, and it was suggested that absence of the cagA gene was the reason for acquisition of MTZ resistance [68]. However, our report of a relationship between MTZ resistance and cagA positivity is in agreement with another study in which cagA genotype (cagA 2a) was found more frequently in MTZ-resistant H. pylori strains [69]. In addition, high frequencies of the cagA and dupA genes as compared to other pathogenicity genes were found in MTZ-, CLR-, and AML-resistant H. pylori isolates. The most frequent vacA genotype in those H. pylori isolates which were resistant to all antibiotics was the more toxin-producing genotype vacA s1a/m1. The high frequencies of these pathogenicity genes in resistant H. pylori strains highlight the potential threat and crisis in treatment of H. pylori infection.
In this study, the Western type of cagA was found more commonly in H. pylori isolates than the East Asian type of cagA, but our data are not sufficient to provide a conclusive prevalence. To analyze whether any evolutionary connection or mutational variation exists in H. pylori strains from Pakistan and other countries, an un-rooted phylogenetic analysis of partial gene sequences of Western and East Asian type of cagA was performed. A partial sequence of the cagA gene of H. pylori strain QAU93, which had East Asian type cagA, was genetically similar to H. pylori strains J7 (Japan) and K15 (Thailand), both of which also contained the East Asian type of cagA. Sequences of all other H. pylori isolates which possessed the Western type cagA were close to H. pylori strains isolated in East Asian, South Asian, and Western countries. This suggests that there is a uniform distribution of partial cagA gene sequences of Pakistani H. pylori isolates throughout the phylogenetic tree.
We observed that anti-CagA and anti-VacA antibodies were frequently detected in this cohort, in agreement with other studies [70,71]. This high frequency may be due to the different immunogenetic properties of vacuolating cytotoxin and CagA proteins. All the cagA-positive H. pylori strains in our population expressed CagA protein (100%) against which antibodies were produced in the sera of the corresponding H. pylori-colonized patients. Discrepancy has been reported between the results of Western blot and PCR assays by another group, in which they showed that Western blot analysis detected serum antibodies against CagA, while PCR did not reveal the presence of the cagA gene in some cases [72]. This is in contrast with our findings where we found that all patients infected with cagA-positive strains produced anti-CagA antibodies. This suggests that the cagA gene was translated into CagA protein and recognized by the host immune response.
The high frequency of anti-VacA antibodies (73.9%) in patients’ sera also suggested that the vacA gene was highly expressed into VacA protein. H. pylori harboring vacA s2/m2 were isolated from patients (n = 2) who had antibodies against VacA protein in their sera. This shows the either mixed but patchy infection of H. pylori or viable but nonculturable H. pylori containing other vacA genotypes instead of vacA s2/m2 which could not be isolated from same patients. In the current analysis, our results of PCR and Western blot assay are in agreement with CagA and VacA status evaluation.
In conclusion, our findings show that the H. pylori strains which colonized Pakistani symptomatic patients had considerable resistance against commonly used antibiotics and that in some cases this was due to acquisition of novel gene variations. The isolates were also likely to be highly pathogenic and virulent due to the presence of several pathogenicity genes. This study therefore emphasizes the importance of reevaluation of treatment regimens for H. pylori in Pakistan, as the population of this country is currently at high risk of developing severe gastric pathologies.
Supplementary Material
Acknowledgments
We thank Aiza Saadia of the Department of Histopathology, Army Medical College, Rawalpindi, Pakistan, for the histopathologic examinations. The work described here was supported by Higher Education Commission (Pakistan) under the Indigenous PhD Fellowship Program (5000 Fellowships, 117-3838-BM7-017) and the International Research Support Initiative Programme (22BMS03).
Footnotes
Disclosures
Competing interests: The authors have no competing interests.
Supporting Information
Additional Supporting Information may be found in the online version of this article:
Table S1 PCR primer pairs and amplification conditions for H. pylori pathogenicity genes.
Table S2 PCR primers pairs and amplification conditions for H. pylori genes associated with antibiotic resistance.
Table S3 Association of pathogenicity genes of H. pylori with key clinical findings.
Table S4 Frequency and association of pathogenicity genes with histopathological grade.
Table S5 Frequency and association of vacA genotypes with histopathological grades.
References
- 1.Blaser MJ, Atherton JC. Helicobacter pylori persistence: biology and disease. J Clin Invest. 2004;11:321–333. doi: 10.1172/JCI20925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adamek RJ, Suerbaum S, Pfaffenbach B, Opferkuch W. Primary and acquired Helicobacter pylori resistance to clarithromycin, metronidazole and amoxicillin–influence on treatment outcome. Am J Gastroenterol. 1998;93:386–389. doi: 10.1111/j.1572-0241.1998.00386.x. [DOI] [PubMed] [Google Scholar]
- 3.Glupczynski Y, Megraud F, Lopez-Brea M, Andersen LP. European multicentre survey of in vitro antimicrobial resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 2001;20:820–823. doi: 10.1007/s100960100611. [DOI] [PubMed] [Google Scholar]
- 4.Jeong JY, Mukhopadhyay AK, Dailidiene D, et al. Sequential inactivation of rdxA (HP0954) and frxA (HP0642) nitroreductase genes causes moderate and high-level metronidazole resistance in Helicobacter pylori. J Bacteriol. 2000;182:5082–5090. doi: 10.1128/jb.182.18.5082-5090.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goodwin A, Kersulyte D, Sisson G, Veldhuyzen van Zanten SJ, Berg DE, Hoffman PS. Metronidazole resistance in Helicobacter pylori is due to null mutations in a gene (rdxA) that encodes an oxygen-insensitive NADPH nitroreductase. Mol Microbiol. 1998;28:383–393. doi: 10.1046/j.1365-2958.1998.00806.x. [DOI] [PubMed] [Google Scholar]
- 6.Ribeiro ML, Vitiello L, Miranda MC, Benvengo YH, Godoy AP, Mendonca S, Pederazzoli J., Jr Mutations in the 23S rRNA gene are associated with clarithromycin resistance in Helicobacter pylori isolates in Brazil. Ann Clin Microbiol Antimicrob. 2003;2:11. doi: 10.1186/1476-0711-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Versalovic J, Shortridge D, Kibler K, Griffy MV, Beyer J, Flamm RK, Tanaka SK, Graham DY, Go MF. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerrits MM, Godoy AP, Kuipers EJ, Ribeiro ML, Stoof J, Mendonca S, van Vliet AH, Pedrazzoli J, Jr, Kusters JG. Multiple mutations in or adjacent to the conserved penicillin-binding protein motifs of the penicillin-binding protein 1A confer amoxicillin resistance to Helicobacter pylori. Helicobacter. 2006;11:181–187. doi: 10.1111/j.1523-5378.2006.00398.x. [DOI] [PubMed] [Google Scholar]
- 9.Paul R, Postius S, Melchers K, Schafer KP. Mutations of the Helicobacter pylori genes rdxA and pbp1 cause resistance against metronidazole and amoxicillin. Antimicrob Agents Chemother. 2001;45:962–965. doi: 10.1128/AAC.45.3.962-965.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mattar R, Marques SB, Monteiro Mdo S, Dos SA, Iriya K, Carrilho FJ. Helicobacter pylori cag pathogenicity island genes: clinical relevance for peptic ulcer disease development in Brazil. J Med Microbiol. 2007;56:9–14. doi: 10.1099/jmm.0.46824-0. [DOI] [PubMed] [Google Scholar]
- 11.Tomasini ML, Zanussi S, Sozzi M, Tedeschi R, Basaglia G, De Paoli P. Heterogeneity of cag genotypes in Helicobacter pylori isolates from human biopsy specimens. J Clin Microbiol. 2003;41:976–980. doi: 10.1128/JCM.41.3.976-980.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Doorn LJ, Figueiredo C, Sanna R, Plaisier A, Schneeberger P, de Boer W, Quint W. Clinical relevance of the cagA, vacA, and iceA status of Helicobacter pylori. Gastroenterology. 1998;115:58–66. doi: 10.1016/s0016-5085(98)70365-8. [DOI] [PubMed] [Google Scholar]
- 13.Sheu BS, Sheu SM, Yang HB, Huang AH, Wu JJ. Host gastric Lewis expression determines the bacterial density of Helicobacter pylori in babA2 genopositive infection. Gut. 2003;52:927–932. doi: 10.1136/gut.52.7.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lu H, Hsu PI, Graham DY, Yamaoka Y. Duodenal ulcer promoting gene of Helicobacter pylori. Gastroenterology. 2005;128:833–848. doi: 10.1053/j.gastro.2005.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, Nakhjavani FA, Mirsalehian A, Zali MR. Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, Iran. J Gastroenterol Hepatol. 2009;24:1380–1386. doi: 10.1111/j.1440-1746.2009.05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamaoka Y, Kodama T, Kashima K, Graham DY, Sepulveda AR. Variants of the 3’ region of the cagA gene in Helicobacter pylori isolates from patients with different H. pylori-associated diseases. J Clin Microbiol. 1998;36:2258–2263. doi: 10.1128/jcm.36.8.2258-2263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yamaoka Y, El-Zimaity HM, Gutierrez O, Figura N, Kim JG, Kodama T, Kashima K, Graham DY. Relationship between the cagA 3’ repeat region of Helicobacter pylori, gastric histology, and susceptibility to low pH. Gastroenterology. 1999;117:342–349. doi: 10.1053/gast.1999.0029900342. [DOI] [PubMed] [Google Scholar]
- 18.Shiota S, Matsunari O, Watada M, Yamaoka Y. Serum Helicobacter pylori CagA antibody as a biomarker for gastric cancer in east-Asian countries. Future Microbiol. 2010;5:1885–1893. doi: 10.2217/fmb.10.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xia Y, Yamaoka Y, Zhu Q, Matha I, Gao X. A comprehensive sequence and disease correlation analyses for the C-terminal region of CagA protein of Helicobacter pylori. PLoS ONE. 2009;4:e7736. doi: 10.1371/journal.pone.0007736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Higashi H, Tsutsumi R, Fujita A, Yamazaki S, Asaka M, Azuma T, Hatakeyama M. Biological activity of the Helicobacter pylori virulence factor CagA is determined by variation in the tyrosine phosphorylation sites. Proc Natl Acad Sci USA. 2002;99:14428–14433. doi: 10.1073/pnas.222375399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yakoob J, Jafri W, Abbas Z, Abid S, Khan R, Jafri N, Ahmad Z. Low prevalence of the intact cag pathogenicity island in clinical isolates of Helicobacter pylori in Karachi, Pakistan. Br J Biomed Sci. 2009;66:137–142. doi: 10.1080/09674845.2009.11730260. [DOI] [PubMed] [Google Scholar]
- 22.Dixon MF, Genta RM, Yardley JH, Correa P. Classification and grading of gastritis. The updated Sydney System. International Workshop on the Histopathology of Gastritis, Houston 1994. Am J Surg Pathol. 1996;20:1161–1181. doi: 10.1097/00000478-199610000-00001. [DOI] [PubMed] [Google Scholar]
- 23.Christopher LC, Harry LTM. Helicobacter pylori Protocols (Methods in Molecular Medicine) 2nd edn. Totowa, NJ: Humana Press; 1997. [Google Scholar]
- 24.Enroth H, Nyren O, Engstrand L. One stomach–one strain: does Helicobacter pylori strain variation influence disease outcome? Dig Dis Sci. 1999;44:102–107. doi: 10.1023/a:1026658301825. [DOI] [PubMed] [Google Scholar]
- 25.Marais A, Bilardi C, Cantet F, Mendz GL, Megraud F. Characterization of the genes rdxA and frxA involved in metronidazole resistance in Helicobacter pylori. Res Microbiol. 2003;154:137–144. doi: 10.1016/S0923-2508(03)00030-5. [DOI] [PubMed] [Google Scholar]
- 26.Nishizawa T, Suzuki H, Tsugawa H, Muraoka H, Matsuzaki J, Hirata K, Ikeda F, Takahashi M, Hibi T. Enhancement of amoxicillin resistance after unsuccessful Helicobacter pylori eradication. Antimicrob Agents Chemother. 2011;55:3012–3014. doi: 10.1128/AAC.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Toledo H, Lopez-Solis R. Tetracycline resistance in Chilean clinical isolates of Helicobacter pylori. J Antimicrob Chemother. 2010;65:470–473. doi: 10.1093/jac/dkp457. [DOI] [PubMed] [Google Scholar]
- 28.Rasheed F, Ahmad T, Ali M, Ali S, Ahmed S, Bilal R. High frequency of cagA and vacA s1a/m2 genotype among Helicobacter pylori infected gastric biopsies of Pakistani children. Mal J Microbiol. 2011;7:167–170. [Google Scholar]
- 29.Logan RP, Polson RJ, Misiewicz JJ, Rao G, Karim NQ, Newell D, Johnson P, Wadsworth J, Walker MM, Baron JH. Simplified single sample 13Carbon urea breath test for Helicobacter pylori: comparison with histology, culture, and ELISA serology. Gut. 1991;32:1461–1464. doi: 10.1136/gut.32.12.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barthel JS, Everett ED. Diagnosis of Campylobacter pylori infections: the “gold standard” and the alternatives. Rev Infect Dis. 1990;12:S107–S114. doi: 10.1093/clinids/12.supplement_1.s107. [DOI] [PubMed] [Google Scholar]
- 31.Ahmad N, Zakaria WR, Abdullah SA, Mohamed R. Characterization of clarithromycin resistance in Malaysian isolates of Helicobacter pylori. World J Gastroenterol. 2009;15:3161–3165. doi: 10.3748/wjg.15.3161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwon DH, Dore MP, Kim JJ, Kato M, Lee M, Wu JY, Graham DY. High-level beta-lactam resistance associated with acquired multidrug resistance in Helicobacter pylori. Antimicrob Agents Chemother. 2003;47:2169–2178. doi: 10.1128/AAC.47.7.2169-2178.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tseng YS, Wu DC, Chang CY, Kuo CH, Yang YC, Jan CM, Su YC, Kuo FC. Amoxicillin resistance with beta-lactamase production in Helicobacter pylori. Eur J Clin Invest. 2009;39:807–812. doi: 10.1111/j.1365-2362.2009.02166.x. [DOI] [PubMed] [Google Scholar]
- 34.O’Connor A, Gisbert JP, McNamara D, O’Morain C. Treatment of Helicobacter pylori infection 2010. Helicobacter. 2010;15:46–52. doi: 10.1111/j.1523-5378.2010.00774.x. [DOI] [PubMed] [Google Scholar]
- 35.Kim JS, Ji JS, Lee BI, Choi H, Kim JH. Sequential therapy or triple therapy for Helicobacter pylori infection in Asians: systematic review and meta-analysis. Clin Res Hepatol Gastroenterol. 2014;38:118–125. doi: 10.1016/j.clinre.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Khan A, Farooqui A, Manzoor H, Akhtar SS, Quraishy MS, Kazmi SU. Antibiotic resistance and cagA gene correlation: a looming crisis of Helicobacter pylori. World J Gastroenterol. 2012;18:2245–2252. doi: 10.3748/wjg.v18.i18.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yakoob J, Abid S, Abbas Z, Jafri SN. Antibiotic susceptibility patterns of Helicobacter pylori and triple therapy in a high-prevalence area. Br J Biomed Sci. 2010;67:197–201. doi: 10.1080/09674845.2010.11730319. [DOI] [PubMed] [Google Scholar]
- 38.Yakoob J, Abid S, Jafri W, Abbas Z, Mumtaz K, Hamid S, Ahmed R. Low rate of recurrence of Helicobacter pylori infection in spite of high clarithromycin resistance in Pakistan. BMC Gastroenterol. 2013;13:33. doi: 10.1186/1471-230X-13-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Graham DY. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272–1277. doi: 10.1016/s0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 40.Mendz GL, Megraud F. Is the molecular basis of metronidazole resistance in microaerophilic organisms understood? Trends Microbiol. 2002;10:370–375. doi: 10.1016/s0966-842x(02)02405-8. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Júlvez M, Rojas AL, Olekhnovich I, Espinosa AV, Hoffman PS, Sancho J. Structure of RdxA - an oxygen-insensitive nitroreductase essential for metronidazole activation in Helicobacter pylori. FEBS J. 2012;279:4306–4317. doi: 10.1111/febs.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jeong JY, Mukhopadhyay AK, Akada JK, Dailidiene D, Hoffman PS, Berg DE. Roles of FrxA and RdxA nitroreductases of Helicobacter pylori in susceptibility and resistance to metronidazole. J Bacteriol. 2001;183:5155–5162. doi: 10.1128/JB.183.17.5155-5162.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sisson G, Jeong JY, Goodwin A, Bryden L, Rossler N, Lim-Morrison S, Raudonikiene A, Berg DE, Hoffman PS. Metronidazole activation is mutagenic and causes DNA fragmentation in Helicobacter pylori and in Escherichia coli containing a cloned H. pylori RdxA(+) (Nitroreductase) gene. J Bacteriol. 2000;182:5091–5096. doi: 10.1128/jb.182.18.5091-5096.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ahmad T, Sohail K, Rizwan M, Mukhtar M, Bilal R, Khanum A. Prevalence of Helicobacter pylori pathogenicity-associated cagA and vacA genotypes among Pakistani dyspeptic patients. FEMS Immunol Med Microbiol. 2009;55:34–38. doi: 10.1111/j.1574-695X.2008.00492.x. [DOI] [PubMed] [Google Scholar]
- 45.Yakoob J, Abid S, Abbas Z, Jafri W, Ahmad Z, Ahmed R, Islam M. Distribution of Helicobacter pylori virulence markers in patients with gastroduodenal diseases in Pakistan. BMC Gastroenterol. 2009;9:87. doi: 10.1186/1471-230X-9-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxena A, Shukla S, Prasad KN, Ghoshal UC. Virulence attributes of Helicobacter pylori isolates & their association with gastroduodenal disease. Indian J Med Res. 2011;133:514–520. [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Ou Z, Wang F, Guo Y, Zhang R, Zhang J, Li P, Xu W, He Y. Distinctiveness of the cagA genotype in children and adults with peptic symptoms in South China. Helicobacter. 2009;14:248–255. doi: 10.1111/j.1523-5378.2009.00690.x. [DOI] [PubMed] [Google Scholar]
- 48.Ramelah M, Aminuddin A, Alfizah H, Isa MR, Jasmi AY, Tan HJ, Rahman AJ, Rizal AM, Mazlam MZ. cagA gene variants in Malaysian Helicobacter pylori strains isolated from patients of different ethnic groups. FEMS Immunol Med Microbiol. 2005;44:239–242. doi: 10.1016/j.femsim.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 49.Yamaoka Y, Kodama T, Kita M, Imanishi J, Kashima K, Graham DY. Relationship of vacA genotypes of Helicobacter pylori to cagA status, cytotoxin production, and clinical outcome. Helicobacter. 1998;3:241–253. doi: 10.1046/j.1523-5378.1998.08056.x. [DOI] [PubMed] [Google Scholar]
- 50.Tan HJ, Rizal AM, Rosmadi MY, Goh KL. Distribution of Helicobacter pylori cagA, cagE and vacA in different ethnic groups in Kuala Lumpur, Malaysia. J Gastroenterol Hepatol. 2005;20:589–594. doi: 10.1111/j.1440-1746.2005.03783.x. [DOI] [PubMed] [Google Scholar]
- 51.Lai CH, Perng CL, Lan KH, Lin HJ. Association of IS605 and cag-PAI of Helicobacter pylori isolated from patients with gastrointestinal diseases in Taiwan. Gastroenterol Res Pract. 2013;2013:356217. doi: 10.1155/2013/356217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamaoka Y, Kodama Y, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome: studies in four different countries. J Clin Microbiol. 1999;37:2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Podzorski RP, Podzorski DS, Wuerth A, Tolia V. Analysis of the vacA, cagA, cagE, iceA, and babA2 genes in Helicobacter pylori from sixty-one pediatric patients from the Midwestern United States. Diagn Microbiol Infect Dis. 2003;46:83–88. doi: 10.1016/s0732-8893(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 54.Talebi TB, Taghvaei T, Mohabbati Mobarez A, Vaira G, Vaira D. High correlation of babA(2)-positive strains of Helicobacter pylori with the presence of gastric cancer. Intern Emerg Med. 2011;8:497–501. doi: 10.1007/s11739-011-0631-6. [DOI] [PubMed] [Google Scholar]
- 55.Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B, Tor-Udom S, Vilaichone RK. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30–36. doi: 10.1016/j.ijid.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 56.Kim SY, Woo CW, Lee YM, Son BR, Kim JW, Chae HB, Youn SJ, Park SM. Genotyping cagA, vacA subtype, iceA1, and babA of Helicobacter pylori isolates from Korean patients, and their association with gastroduodenal diseases. J Korean Med Sci. 2001;16:579–584. doi: 10.3346/jkms.2001.16.5.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mizushima T, Sugiyama T, Komatsu Y, Ishizuka J, Kato M, Asaka M. Clinical relevance of the babA2 genotype of Helicobacter pylori in Japanese clinical isolates. J Clin Microbiol. 2001;39:2463–2465. doi: 10.1128/JCM.39.7.2463-2465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Douraghi M, Mohammadi M, Oghalaie A, Abdirad A, Mohagheghi MA, Hosseini ME, Zeraati H, Ghasemi A, Esmaieli M, Mohajerani N. dupA as a risk determinant in Helicobacter pylori infection. J Med Microbiol. 2008;57:554–562. doi: 10.1099/jmm.0.47776-0. [DOI] [PubMed] [Google Scholar]
- 59.Arachchi HSJ, Kalra V, Lal B, et al. Prevalence of duodenal ulcer-promoting gene (dupA) of Helicobacter pylori in patients with duodenal ulcer in North Indian population. Helicobacter. 2007;12:591–597. doi: 10.1111/j.1523-5378.2007.00557.x. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Z, Zheng Q, Chen X, Xiao S, Liu W, Lu H. The Helicobacter pylori duodenal ulcer promoting gene, dupA in China. BMC Gastroenterol. 2008;8:49. doi: 10.1186/1471-230X-8-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanif M, Zaidi P, Rasool A, Hameed A, Ahmed L. Cytotoxin genes of Helicobacter pylori in gastroduodenal disease patients of Karachi. As Pac J Mol Biol Biotechnol. 2010;18:333–340. [Google Scholar]
- 62.Chen XJ, Yan J, Shen YF. Dominant cagA/vacA genotypes and coinfection frequency of H. pylori in peptic ulcer or chronic gastritis patients in Zhejiang Province and correlations among different genotypes, coinfection and severity of the diseases. Chin Med J (Engl) 2005;20:460–467. [PubMed] [Google Scholar]
- 63.Yamazaki S, Yamakawa A, Okuda T, et al. Distinct diversity of vacA, cagA, and cagE genes of Helicobacter pylori associated with peptic ulcer in Japan. J Clin Microbiol. 2005;43:3906–3916. doi: 10.1128/JCM.43.8.3906-3916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Atherton JCCP, Peek RM, Jr, Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. J Biol Chem. 1995;270:17771–17777. doi: 10.1074/jbc.270.30.17771. [DOI] [PubMed] [Google Scholar]
- 65.Atherton JC, Peek RM, Jr, Tham KT, Cover TL, Blaser MJ. Clinical and pathological importance of heterogeneity in vacA, the vacuolating cytotoxin gene of Helicobacter pylori. Gastroenterology. 1997;112:92–99. doi: 10.1016/s0016-5085(97)70223-3. [DOI] [PubMed] [Google Scholar]
- 66.Moss SF, Malfertheiner P. Helicobacter and gastric malignancies. Helicobacter. 2007;12:23–30. doi: 10.1111/j.1523-5378.2007.00539.x. [DOI] [PubMed] [Google Scholar]
- 67.Alfizah H, Ramelah M, Rizal AM, Anwar AS, Isa MR. Association of Malaysian Helicobacter pylori virulence polymorphisms with severity of gastritis and patients’ ethnicity. Helicobacter. 2012;17:340–349. doi: 10.1111/j.1523-5378.2012.00956.x. [DOI] [PubMed] [Google Scholar]
- 68.Taneike I, Nami A, O’Connor A, Fitzgerald N, Murphy P, Qasim A, O’Connor H, O’Morain C. Analysis of drug resistance and virulence-factor genotype of Irish Helicobacter pylori strains: is there any relationship between resistance to metronidazole and cagA status? Aliment Pharmacol Ther. 2009;30:784–790. doi: 10.1111/j.1365-2036.2009.04095.x. [DOI] [PubMed] [Google Scholar]
- 69.Vilaichone RK, Mahacahai V, Tumwasorn S, Kachintorn U. cagA genotype and metronidazole resistant strain of Helicobacter pylori in functional dyspepsia in Thailand. J Gastroenterol Hepatol. 2011;26:46–48. doi: 10.1111/j.1440-1746.2011.06652.x. [DOI] [PubMed] [Google Scholar]
- 70.Cover TL. The vacuolating cytotoxin of Helicobacter pylori. Mol Microbiol. 1996;20:241–246. doi: 10.1111/j.1365-2958.1996.tb02612.x. [DOI] [PubMed] [Google Scholar]
- 71.Basso D, Navaglia F, Brigato L, Piva MG, Toma A, Greco E, Galeotti F, Roveroni G, Corsini A, Plebani M. Analysis of Helicobacter pylori vacA and cagA genotypes and serum antibody profile in benign and malignant gastroduodenal diseases. Gut. 1998;43:182–186. doi: 10.1136/gut.43.2.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Paoluzi OA, Rossi P, Montesano C, Bernardi S, Carnieri E, Marchione OP, Nardi F, Iacopini F, Pica R, Paoluzi P. Discrepancy between polymerase chain reaction assay and Western blot analysis in the assessment of CagA status in dyspeptic patients. Helicobacter. 2001;6:130–135. doi: 10.1046/j.1523-5378.2001.00019.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.