Abstract
The last several years have witnessed an explosion in the understanding and use of novel, versatile trans-acting elements. TALEs, CRISPR/Cas, and sRNAs can be easily fashioned to bind any specific sequence of DNA (TALEs, CRISPR/Cas) or RNA (sRNAs) because of the simple rules governing their interactions with nucleic acids. This unique property enables these tools to repress the expression of genes at the transcriptional or post-transcriptional levels, respectively, without prior manipulation of cis-acting and/or chromosomal target DNA sequences. These tools are now being harnessed by synthetic biologists, particularly those in the eukaryotic community, for genome-wide regulation, editing, or epigenetic studies. Here we discuss the exciting opportunities for using TALEs, CRISPR/Cas, and sRNAs as synthetic trans-acting regulators in prokaryotes.
Introduction
The core challenge of any synthetic biology project is designing the correct regulatory elements to optimize expression of a system's critical RNA and/or protein components. Many tools, both computational and physical, have been developed to predict the impact of cis-acting regulatory elements, such as promoters and ribosome binding sites, on gene expression [1,2]. However, when these cis elements are applied to alter native gene expression, the regulation of the evolved system may be compromised. In other situations, the tools needed to modify an organism's genome do not exist and therefore methods of manipulating native gene expression in cis are not possible. Problems such as manipulating native metabolic pathways, studying orphan gene clusters in novel bacteria, and optimizing the expression of genes for multiple conditions could be solved with the development of trans-acting regulatory tools that conditionally manipulate expression of a target gene without altering its native regulation. There are many examples of natural trans-acting regulators that act at the transcriptional (e.g. DNA-binding repressors), post-transcriptional (e.g. RNA stability), and translational (e.g. small interfering RNAs) levels to both induce and repress gene expression. Here, we will discuss the potential synthetic utilization of three trans-acting regulatory tools as repressors of gene expression in Escherichia coli.
Well-characterized trans-acting repressors, such as LacI, TetR, and cI, have been used to great effect to regulate recombinant protein production as well as serve as fundamental elements in complex genetic circuits [1]. The affinity of these proteins for their unique cognate DNA sequences (i.e. operators) has made them reliable trans-acting regulators; however, this same repressor-sequence relationship fundamentally limits their flexibility. The recent development of transcription activator-like effectors (TALEs), the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas system, and synthetic small RNAs (sRNAs) as novel customizable regulatory tools has garnered the attention of the biotechnology community [3-5]. The widespread interest in, and potential of, these systems stems from the opportunity to tailor each of these tools to specifically interact with any desired target DNA (TALEs, CRISPR) or RNA (sRNAs, CRISPR) sequence, rather than a single operator. This powerful feature of TALEs and CRISPR/Cas has been harnessed in conjunction with fusions to activation/repression domains and DNA nucleases to evoke changes in gene expression and perform rapid genome editing in eukaryotic species such as yeast [6], Caenorhabditis elegans [7], plants [8,9], zebra-fish [10-12], mice [13,14], and human cells [15-19].
These tools are enabling a wide range of synthetic biology and metabolic engineering applications that are just beginning to be realized (Fig. 1); for an in-depth discussion of each of these regulators the reader is directed to recent excellent reviews elsewhere [20-22]. Here we provide the reader with a basic understanding of the mechanisms of these three tools, a comparative analysis of their potential utility, and a discussion of their prospective roles in prokaryotic biotechnology (summarized in Table 1).
Figure 1.
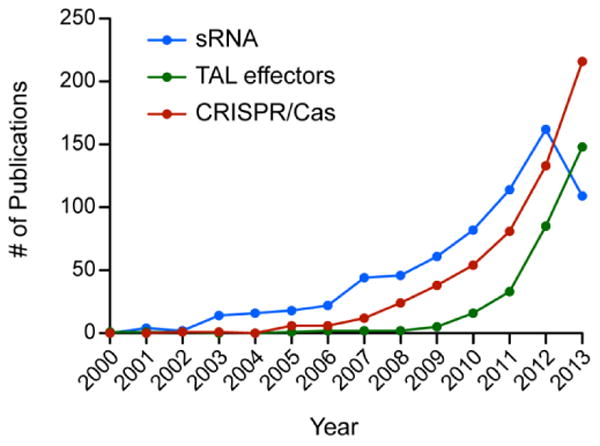
Plot highlighting the increasing popularity of sRNAs, TALEs and CRISPR/Cas in the published primary literature over the last 13 years. Web of Science was used to quantify the number of publications for each year using the following search criteria: (i) sRNA, TS=(sRNA) OR TI=(sRNA), (ii) TALEs, TS=(TAL effector) OR TS=(Transcription activator-like effector) OR TI=(TAL effector) OR TI=(Transcription activator-like effector), and (iii) CRISPR/Cas, TS=(CRISPR) OR TS=(clustered regularly interspaced short palindromic repeats) OR TI=(CRISPR) OR TI=(clustered regularly interspaced short. For each search TS = Topic and TI = Title.
Table 1.
Summary of the relevant characteristics of each trans-regulatory repressor discussed in this review.
| Mode of Action | Molecules Involved | Implementation | Design Variables | Specificity | |
|---|---|---|---|---|---|
|
| |||||
| TALEs |
|
|
|
|
|
|
| |||||
| CRISPR/Cas |
|
|
|
|
|
|
| |||||
| sRNAs |
|
|
|
|
|
Mechanism, Design, and Use of TALEs
Mode of Action
Species of the plant pathogen genus Xanthomonas inject proteins known as transcription activator-like effectors (TALEs) into plant cells to elicit changes in gene expression that contribute to host infection [23]. TALEs are organized into three sections: (i) an N-terminal domain containing a type III secretion signal, (ii) a C-terminal domain containing a nuclear localization signal and an acidic activation domain, and (iii) a central repeat domain that determines DNA-binding specificity [24]. In an archetypal TALE, the DNA binding domain consists of between 15.5 to 19.5 repeat regions. Each repeat typically contains 34 amino acids except for the last repeat, which is only 20 amino acids in length (i.e. the 0.5-mer). The 34 amino acids in each repeat are highly conserved except for those at the 12th and 13th positions, which are referred to as the repeat variable diresidues (RVDs) [22]. The amino acid identity of the RVDs is responsible for DNA nucleotide recognition, enabling the design of TALEs to target unique DNA sequences [25,26]. Crystal structures of TALEs bound to their target sequences show that the protein wraps around the major groove of the DNA, permitting the RVDs to make base-specific contacts with the sense strand [27-29]. While TALEs have primarily been used in eukaryotic applications [15,16], a TALE designed to bind the lacO1 operator strongly repressed expression of a plasmid-based fluorescent reporter gene in E. coli, presumably by blocking transcription initiation in a manner akin to LacI [30,31]. This initial demonstration motivates further use of TALEs as trans-acting transcriptional repressors in bacteria (Fig. 2A, B).
Figure 2.
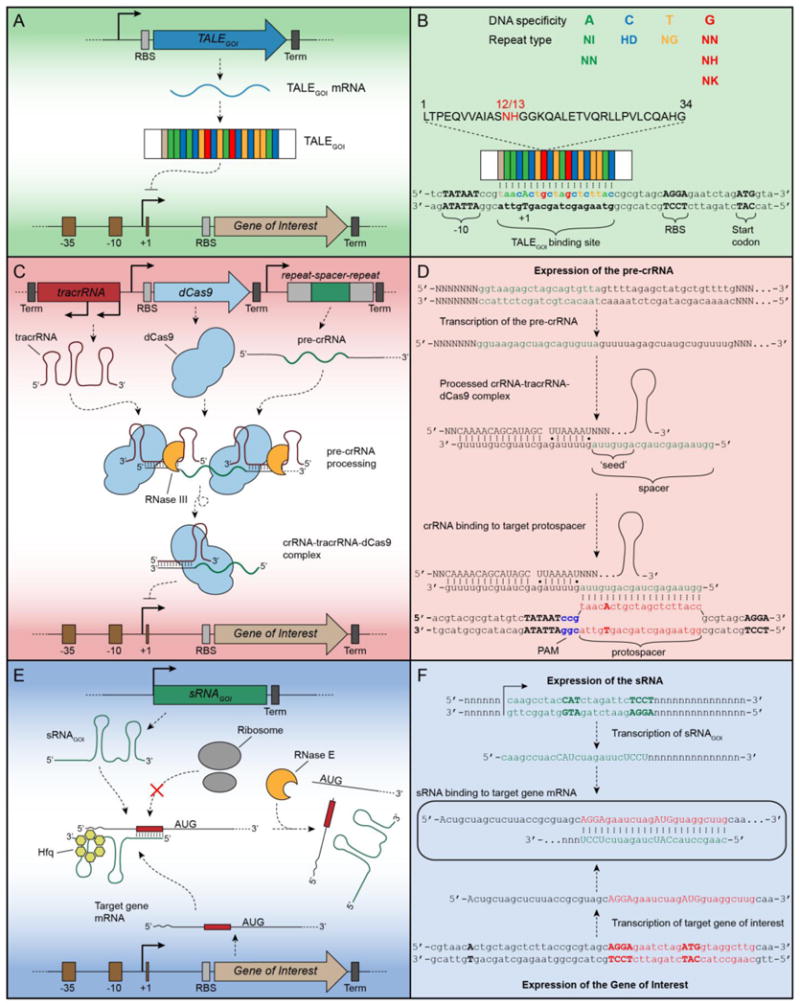
Cartoon schematic and binding mechanism of TALE, CRISPR/Cas, and sRNA to inhibit bacterial gene expression. Panels A, C, and E depict the expression of each trans-regulatory tool and their respective mode of action to repress the expression of a gene of interest. Panels B, D, and F detail the binding interactions of each trans-regulator at base-pair resolution. (A, B) This hypothetical 17.5-mer TALE contains a repeat domain designed to bind a 19-bp DNA sequence located several base pairs 3′ of a canonical -10 hexamer. TALE binding to this region occludes RNA polymerase association with the promoter, thereby repressing transcription initiation. The repeat variable diresidues (RVDs) at the 12th and 13th positions within each 34 amino acid repeat are specific for a particular DNA base. (C, D) In the type II CRISPR/dCas9 system, a pre-crRNA is processed by tracrRNA, RNase III, and dCas9 complex. The mature crRNA-tracrRNA partners with dCas9 to bind a promoter proximal region and inhibit transcription initiation. Specifically, the 20-bp of the 5′-end of the crRNA known as the spacer region (shown in green in D) are responsible for recognizing and binding to a complementary sequence in the DNA target known as the protospacer (shown in red in D). The dCas9 protein requires the presence of an NGG protospacer adjacent motif (PAM), shown here in blue as CGG at the 3′ end of the protospacer. (E, F) An engineered sRNA is expressed and interacts with its cognate mRNA target in an Hfq-dependent process. To prevent ribosome binding and translation, the most 5′ 24-bp of the sRNA (shown in green in F) are designed to complementarily bind the translation initiation region of the target mRNA (shown in red in F). The interacting RNAs are often degraded by RNase E and associated degradosome proteins.
Implementation and Efficacy
Like many native repressors, a TALE designed to inhibit transcription initiation should target a region proximal to a gene's promoter in order to occlude RNA polymerase binding [32]. A TALE-based repression strategy is implemented by expressing, in trans, a TALE that encodes the proper series of RVDs for binding a targeted DNA sequence. Native TALE binding sites generally begin with a 5′ thymine, although recently TALEs have been evolved to bypass this requirement [29,33]. Once a DNA target is identified, an RVD for each target base is selected according to the following code: C = HD, T = NG, A = NI, G = NN, NH, NK [25,26]. When selecting an RVD, other factors to consider are: (i) RVD composition affects TALE-DNA binding affinity [34], (ii) the NN RVD also interacts with adenine, however the alternative RVDs NK and NH enable the specific targeting of G [35-37], (iii) the number of 34 amino acid repeats affects DNA binding [25], and (iv) there is a potential N-terminally biased polarity to TALE binding [34]. The native type III secretion and nuclear localization signals and activation domain are unnecessary when using a TALE for prokaryotic gene repression and as such the N- and C- termini can be significantly reduced in size [16,30,38]. The largest challenge to using TALEs is the need to construct a new DNA-binding domain, roughly 1.8 kb for a 17.5-mer, for each DNA target sequence. The highly conserved nature of each 34 amino acid repeat complicates the use of traditional molecular biology approaches. Fortunately, the biotechnology community has developed methods to enable rapid de novo TALE construction, including Golden Gate cloning [39], ligation-independent cloning [40], and solid-phase synthesis strategies [41].
For TALEs to be useful as trans-regulators, they must specifically bind their target sequence to preclude unwanted off-target effects. Two or more evenly spaced RVD-DNA base mismatches have been shown to reduce DNA binding in eukaryotes (quantified as the degree of gene activation by the TALE) while three mismatches completely abolish TALE-mediated gene activation [42]. Little to no off-target TALE activity was observed in global gene expression studies of eukaryotes expressing TALEs [6,15,23]. Analogous ChIP- and RNA-seq studies need to be performed to investigate TALE infidelity in prokaryotes. However, it is reasonable to assume that the smaller size of bacterial genomes (e.g. E. coli genome = 4.6 × 106 bp vs. human genome = 3.3 × 109 bp) precludes significant off-target effects.
Mechanism, Design, and Use of CRISPR/Cas
Mode of Action
The CRISPR/Cas system, like TALEs, is natively involved in a host-pathogen interaction. When a bacterial host is invaded by phage or plasmid DNA, small bacterial RNAs recognize and bind the foreign nucleic acid, targeting it for degradation by CRISPR associated (Cas) proteins [43]. The CRISPR/Cas of Streptococcus pyogenes is most commonly used in biotechnology applications [4]. Natively, CRISPR/Cas-mediated DNA degradation occurs in three steps: (i) transcription of a CRISPR repeat-spacer array, where each spacer encodes a unique CRISPR RNA (crRNA) sequence complementary to a foreign DNA, (ii) association of this pre-crRNA with trans-activating crRNAs (tracrRNAs) for processing by RNase III into mature crRNAs, and (iii) targeting and degradation of invading DNA by a complex of crRNA, tracrRNA, and Cas9 [20,44,45]. The spacer region of the crRNA recognizes a complementary sequence of the foreign DNA, known as a protospacer, leading to specific binding of the RNA-Cas9 complex and subsequent dsDNA cleavage [20,44]. The simplicity of this RNA-guided endonuclease system has led to widespread use of the S. pyogenes type II CRISPR/Cas9 system for eukaryotic genome-editing [14,18]. Inactivation of Cas9 nuclease activity (a.k.a. dCas9) converts it into an RNA-guided DNA-binding protein, which has proven useful for modulating eukaryotic gene expression [19,46,47]. In two recent independent studies crRNA-dCas9 was shown to repress reporter gene expression in E. coli by binding to its promoter region, analogous to the mechanism described above for TALE-mediated gene repression in prokaryotes [48,49] (Fig. 2C, D).
Implementation and Efficacy
Implementation of the native type II CRISPR/Cas9 system involves multiple moving pieces including the crRNA, tracrRNA, dCas9 (˜160 kDa), and RNase III. The tracrRNA is expressed as a separate transcript from the pre-crRNA spacer such that the hybrid-complex of the two RNAs becomes a substrate for RNase III processing [20,44]. The system complexity can be reduced by fusing the 3′ end of the crRNA to the 5′ end of the tracrRNA to form a chimeric molecule, termed a small guide RNA (sgRNA) [18,48,50,51]. In this implementation, only two components, the sgRNA and dCas9, are needed.
sgRNAs or tracrRNA/crRNA complexes can be used to guide dCas9 to sequences in the vicinity of the bacterial promoter to inhibit transcription initiation [48,49]. It has also been shown that sgRNAs can inhibit transcription elongation by targeting regions of the open-reading frame as well as non-coding DNA regions. Small guide RNAs targeted to the non-template DNA strand (i.e. the coding strand) of an open-reading frame were most effective in reducing gene expression, while inhibition of transcription initiation appears to be independent of the DNA strand targeted [48,49]. It is currently not clear which type of RNA guide is most efficient in repressing transcriptional events.
Unlike TALEs, the number of sites that can be targeted by CRISPR/Cas9 is constrained by the need for a protospacer adjacent motif (PAM), which is NGG for the S. pyogenes system [50,52] (Fig. 2D). Truncation studies of CRISPR/Cas9 spacers indicate that a spacer length of 20 bp provides optimal repression, however only 12 bp of complementarity between the 3′ end of the spacer sequence and its target protospacer are minimally required for gene repression [48,49]. This is consistent with data showing that mutations in the 7 bp at the 3′ end of spacer, the so-called “seed” region, eliminate binding while multiple mutations outside of this seed region in either the spacer or the protospacer are tolerated [50,53].
The small size of this seed region suggests that it might be difficult to achieve gene specific repression for some CRISPR/Cas9 applications. Indeed, using Cas9 for genome editing in higher organisms with genomes orders of magnitude larger than most prokaryotes has been shown to result in off-target effects [54]. In E. coli, however, it appears that the seed region is adequate to prevent spurious repression, at least when using an sgRNA. A set of RNA-seq experiments demonstrated that sgRNAs targeted to the chromosomal loci lacI and lacZ, respectively, affect only the intended genes [48].As E. coli's genome is approximately 4.6 Mbp in length, we might expect it to contain about 580,000 PAM sequences. Given this and a minimum requirement of 12 contiguous complementary bases for repression, there is an ˜3% chance that an off-target site would exist for any selected protospacer. However, there has been at least one report of an off-target effect when using CRISPR/Cas9 in prokaryotes; Marraffini and co-workers [49] could not clone a particular spacer sequence, presumably because its off-target site was an essential gene.
Mechanism, Design, and Use of Small RNAs
Mode of Action
Small, trans-acting RNAs are frequently used by bacteria to post-transcriptionally repress gene expression. Often sRNAs are expressed under cell-taxing conditions (e.g. iron limitation, oxidative and temperature stress), where they bind to complementary mRNAs to prevent translation of encoded genes [55]. Repression by sRNAs generally requires Hfq, a homomultimeric RNA chaperone that stabilizes the binding of sRNA to mRNA [56]. In many cases, the sRNA prevents translation from its cognate mRNA by blocking recruitment of the ribosome to the ribosome-binding site (RBS) and/or access to a gene's start codon. The pairing of sRNA-mRNA is in some cases sufficient to repress gene expression; however, this pairing is also commonly accompanied by the degradation of the mRNA by RNase E and its associated proteins, collectively known as the degradosome in E. coli (Fig. 3E, F) [21,56,57].
Figure 3.
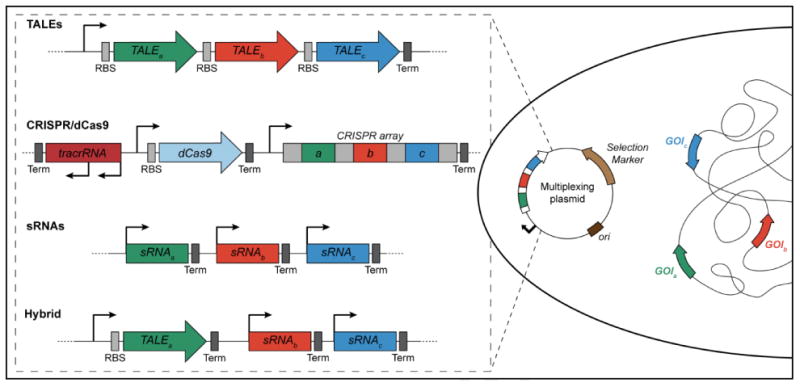
Using multiple TALEs, CRISPR/Cas, and sRNAs to simultaneously target and repress the expression of multiple chromosomal targets. In the illustration, three different TALEs, each designed to target a different gene (genes of interest A, B, and C, respectively) are expressed in trans from a single operon. A similar outcome is achieved using the type II CRISPR/Cas system, where three unique spacers are integrated together into a single repeat-spacer-repeat CRISPR array. To simultaneously express three sRNAs, each gene encoding an individual sRNA is expressed from its own promoter. These tools can also be combined together to repress the same gene at both the transcriptional and post-transcriptional level or, as shown in the hybrid scenario, selectively and simultaneously repress multiple genes with different trans-regulators.
Since small, non-coding RNA regulatory systems are well characterized in many bacterial species, it is perhaps not surprising that it has been applied as a trans-regulatory tool for synthetic biology. Examples of sRNA applications include: (i) identifying essential genes in Staphylococcus aureus [58], (ii) knocking down recA expression in E. coli [59], (iii) improving succinate production in E. coli [60], and (iv) down-regulating genes associated with acetone biosynthesis in Clostridium acetobutylicum [61]. More recently, Lee and colleagues [62] used sRNAs to simultaneously target 130 genes in E. coli, including those that encode for transporters, transcription factors, and central metabolic enzymes, in order to substantially improve tyrosine and cadaverine production.
Implementation and Efficacy
A single designer RNA is all that needs to be introduced for effective post-transcriptional repression of a target gene, perhaps making sRNAs simpler to implement than either TALEs or CRISPR/Cas9, especially considering sRNAs are not generally processed prior to use [55]. Several native sRNA scaffolds have been successfully implemented as synthetic regulators [63,64]. In one study, the MicC scaffold was demonstrated as optimal for gene repression in E. coli [62]. Using this system, a gene of interest can be repressed simply by re-designing the ˜24 bp target-binding sequence of the MicC scaffold [65]. While this scaffold functions with native E. coli Hfq and RNase E proteins, new scaffolds may be required when using trans-acting sRNAs in other bacterial species. Alternatively, E. coli Hfq and RNase E could be heterologously expressed to enable sRNA function, but cross-talk between imported and native systems would need to be managed. High levels of synthetic sRNAs may sequester Hfq and interfere with its native uses, as Hfq binds one mRNA and sRNA at a time [66]. Therefore, the levels of sRNA and Hfq expression will need to be optimized to maintain a stable strain. Also, unlike TALEs and CRISPR/Cas, sRNAs and their cognate mRNAs may be rapidly turned over, which may require increased sRNA expression, relative to the other trans-regulators, to maintain intended levels of repression [56].
Both native and synthetic small regulatory RNAs have been shown to effectively repress gene expression when targeted to a region spanning the ribosome binding site and translation start site. Targeting this translation initiation region (TIR) with synthetic sRNAs expressed from a strong promoter results in the greatest levels of gene repression (> 90%), though sRNAs targeted to the middle of the encoded open-reading frame are also capable of greatly reducing target gene expression [62]. The target binding sites of native sRNAs vary in length and usually span more than 30 bp in total. Base pairing between the sRNA and its cognate mRNA is imperfect, consisting of multiple 8-9 nucleotide stretches of complementarity [55]. In contrast, synthetic sRNAs are usually shorter, on the order of 20-25 bp in length, and are designed to perfectly complement their target mRNA. The binding energy of an sRNA:mRNA pair correlates with gene repression, where data suggests that a maximum binding energy of -20 kcal/mol (i.e. a minimum length of 24 bp) is sufficient to achieve a high level of repression [62,65]. This relationship may also enable a user to tune gene expression levels by altering the length and complementarity to a target site or by selecting alternate sites in the TIR. However, extending the sRNA length to 30 bp and beyond in order to achieve lower binding energies (and thus hypothetically greater repression) has been shown to come at the cost of target specificity [62]. Again, global transcriptomic and proteomic studies are needed to further probe the off-target effects of trans-expressed synthetic sRNAs.
Opportunities
The ability to tailor TALEs, CRISPR/Cas, and sRNAs to bind specific DNA sequences of interest affords researchers the opportunity for unprecedented control of gene expression in both native and synthetic prokaryotic systems. Researchers have only begun to harness the most exciting applications of these tools, including multiplexing, fine-tuning of gene expression, and targeted gene activation.
Multiplexing
All three systems can be used for multiplexing (i.e. targeting multiple genes simultaneously for activation, repression or genome-editing Fig 3), though the inherent characteristics of each suggest that all three are not equally amenable for this application. Though more cumbersome to assemble, TALEs have been used for multiplexing in human cells to synergistically and dynamically regulate gene expression [67]. While multiple TALEs could be expressed from a single operon in prokaryotes, their large size places a limit on the number that can be cloned into a single bacterial expression vector.
While small in size, the RNA-based methods present their own challenges for implementation in multiplexing studies. sRNAs must either be transcribed from separate promoters [62] or potentially processed from a larger transcript to generate individual RNAs. The P. aeruginosa endoRNase Csy4 could be used to liberate multiple sRNAs from a polycistronic transcript by recognizing and cleaving 28-nt repetitive sequences inserted between separate encoded sRNAs, though the intrinsic sRNA scaffold terminator may impede this possibility [68]. However, simultaneous expression of four or more copies of an sRNA without a target binding sequence (i.e. an sRNA unable to bind mRNA), increases metabolic burden [62].
The type II CRISPR/Cas9 system is perhaps the best equipped for multiplexing [14,18,19,46,69]. Targeting multiple genes simply requires the redesign of a native CRISPR array to encode multiple, unique spacers. Following transcription, this array would then be processed to yield the desired crRNAs. Small guide RNAs have also proven popular for multiplexing in eukaryotes as only the sgRNA and Cas9 need to be coexpressed, eliminating the need for the tracrRNA [14,46]. However, their use in bacteria and eukaryotes poses a similar problem to that described above for sRNAs, that is, each sgRNA must be transcribed from its own promoter in order to produce the functional RNAs. Separate sgRNAs have been used in E. coli to simultaneously down regulate two different reporter genes and have been shown to exert a synergistic effect on gene repression when targeted to the same open reading frame [48].
Tuning Gene Expression
Many biotechnology applications require fine control of gene expression to achieve optimal results. Unfortunately, most tools for fine control require cis-modifications to the gene expression cassette. In principle, regulation of a target gene by any of these three systems may be modulated by adjusting the promoters and/or RBS signals driving the expression of the system's components (e.g. the TALE, the dCas9/crRNA-tracrRNA/sgRNA, sRNA/Hfq/RNAse E). By adjusting the expression levels of these system components and/or their affinity for their target nucleic acid, it should be possible to achieve the goal of predictable a priori tuning of target gene expression levels using these trans-regulators. Experiments required to quantitatively demonstrate this fine degree of control are beginning to emerge [70].
Gene Activation
While TALEs and CRISPR/Cas have primarily been used as repressors in prokaryotes, they can also be engineered to activate gene expression. The CRISPR/Cas system can activate expression of a reporter gene by fusing RpoZ (the omega subunit of RNA polymerase) to dCas9 in an E. coli strain devoid of its native rpoZ gene [49]; in principle, this same approach could be replicated to make bacterial TALE activators. Alternative fusions to convert TALEs and dCas9 to prokaryotic activator proteins might further expand the repertoire of trans-activators and ultimately permit the simultaneous activation and repression of a desired set of genes independent of strain background. Gene activation can also be achieved by targeting sRNAs to mRNA sequences that, in the absence of the sRNA, block translation initiation in cis. This strategy requires the target mRNA to possess this rare regulatory element [55]. Lastly, TALEs have recently been converted into ligand-inducible activators of human cell gene expression [71], raising the possibility of converting TALEs into site-specific, ligand responsive transcription factors for the inducible control of any native gene without chromosomal modification.
Conclusions
TALEs, CRISPR/dCas9, and sRNAs collectively offer the potential for an unprecedented level of control of native and heterologous gene expression. The popularity of these tools is apparent (Fig. 1), and the next step is to transition them from proof-of-concept tools into everyday molecular biology and biotechnology workhorses. Which system(s) will emerge as the best tool for a given application remains an unanswered question, but several predictions can be made from current knowledge. First, sRNAs are the most amenable for initial high throughput target validation studies. Second, TALEs are the more robust tool for gene-silencing efforts. Last, CRISPR/Cas is the most facile all-purpose tool and the most promising option for multiplexing. In the coming years, these predictions will be tested in head-to-head experiments that compare the ability of each regulator to repress, tune, and activate bacterial gene expression while minimizing the impact on cellular resources and physiology. Further study of binding relationships between regulator and target will enable design algorithms that maximize fidelity and efficiency. Regardless of the experimental outcomes, the future is bright for these tools to play a major role as the next generation of trans-regulatory elements for both engineering and molecular biology applications.
Highlights.
trans-acting repressors enable synthetic regulation of target genes.
The DNA binding domain of a TALE can be targeted to promoters to inhibit transcription.
Non-cleaving Cas9 can be guided to target DNA by small RNAs to repress transcription.
Synthetic sRNAs can inhibit translation and decrease message stability by binding mRNAs.
Acknowledgments
This work was funded by the National Science Foundation (CBET-1149678). M.F.C. is supported by NHGRI HG002760. M.C.P. is supported by NIH T32 GM08349.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Khalil AS, Collins JJ. Synthetic biology: applications come of age. Nat Rev Genet. 2010;11:367–379. doi: 10.1038/nrg2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lanza AM, Crook NC, Alper HS. Innovation at the intersection of synthetic and systems biology. Current Opinion in Biotechnology. 2012;23:712–717. doi: 10.1016/j.copbio.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 3.Doyle EL, Stoddard BL, Voytas DF, Bogdanove AJ. TAL effectors: highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends in Cell Biology. 2013 doi: 10.1016/j.tcb.2013.1004.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat Meth. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rusk N. Engineers meet small RNA. Nature Methods. 2007;4:986–987. [Google Scholar]
- 6.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Research. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood AJ, Lo TW, Zeitler B, Pickle CS, Ralston EJ, Lee AH, Amora R, Miller JC, Leung E, Meng X, et al. Targeted genome editing across species using ZFNs and TALENs. Science. 2011;333:307. doi: 10.1126/science.1207773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc Natl Acad Sci USA. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morbitzer R, Römer P, Boch J, Lahaye T. Regulation of selected genome loci using de novo-engineered transcription activator-like effector (TALE)-type transcription factors. Proc Natl Acad Sci USA. 2010;107:21617–21622. doi: 10.1073/pnas.1013133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang P, Xiao A, Zhou MG, Zhu ZY, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nature Biotechnology. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- 11.Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JRJ. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnology. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- 13.Malina A, Mills JR, Cencic R, Yan Y, Fraser J, Schippers LM, Paquet M, Dostie J, Pelletier J. Repurposing CRISPR/Cas9 for in situ functional assays. Genes Dev. 2013;27:2602–2614. doi: 10.1101/gad.227132.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, Cost GJ, Zhang L, Santiago Y, Miller JC, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nat Biotechnol. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16*.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. This paper describes a truncated TALE scaffold that has proven useful for biotechnology applications and demonstrates its utility for genome editing and gene activation in human cells. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Moore R, Guinn M, Bleris L. Transcription activator-like effector hybrids for conditional control and rewiring of chromosomal transgene expression. Sci Rep. 2012;2:897. doi: 10.1038/srep00897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, Yang L, Church GM. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013 doi: 10.1038/nbt.2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–266. doi: 10.1146/annurev-biochem-072911-172315. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman S. The small RNA regulators of Escherichia coli: Roles and mechanisms. Annual Review of Microbiology. 2004;58:303–328. doi: 10.1146/annurev.micro.58.030603.123841. [DOI] [PubMed] [Google Scholar]
- 22.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 23.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: finding plant genes for disease and defense. Current Opinion in Plant Biology. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 24.Mak ANS, Bradley P, Bogdanove AJ, Stoddard BL. TAL effectors: function, structure, engineering and applications. Curr Opin Struct Biol. 2013;23:93–99. doi: 10.1016/j.sbi.2012.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, Lahaye T, Nickstadt A, Bonas U. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 26.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 27.Stella S, Molina R, Yefimenko I, Prieto J, Silva G, Bertonati C, Juillerat A, Duchateau P, Montoya G. Structure of the AvrBs3-DNA complex provides new insights into the initial thymine-recognition mechanism. Acta Crystallographica Section D-Biological Crystallography. 2013;69:1707–1716. doi: 10.1107/S0907444913016429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28*.Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, Shi Y, Yan N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335:720–723. doi: 10.1126/science.1215670. Presents a high-quality crystal structure of an engineered TALE, helping to elucidate the RVD-DNA interactions responsible for TALE DNA-binding specificity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Politz MC, Copeland MF, Pfleger BF. Artificial repressors for controlling gene expression in bacteria. Chem Commun (Camb) 2013;49:4325–4327. doi: 10.1039/c2cc37107c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schlax PJ, Capp MW, Record MT. Inhibition of transcription initiation by lac repressor. Journal of Molecular Biology. 1995;245:331–350. doi: 10.1006/jmbi.1994.0028. [DOI] [PubMed] [Google Scholar]
- 32.Browning DF, Busby SJW. The regulation of bacterial transcription initiation. Nature Reviews Microbiology. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 33.Lamb BM, Mercer AC, Barbas CF., 3rd Directed evolution of the TALE N-terminal domain for recognition of all 5′ bases. Nucleic Acids Res. 2013 doi: 10.1093/nar/gkt754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meckler JF, Bhakta MS, Kim MS, Ovadia R, Habrian CH, Zykovich A, Yu A, Lockwood SH, Morbitzer R, Elsäesser J, et al. Quantitative analysis of TALE-DNA interactions suggests polarity effects. Nucleic Acids Research. 2013;41:4118–4128. doi: 10.1093/nar/gkt085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christian ML, Demorest ZL, Starker CG, Osborn MJ, Nyquist MD, Zhang Y, Carlson DF, Bradley P, Bogdanove AJ, Voytas DF. Targeting G with TAL effectors: a comparison of activities of TALENs constructed with NN and NK repeat variable di-residues. PLoS ONE. 2012;7:e45383. doi: 10.1371/journal.pone.0045383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cong L, Zhou RH, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nature Communications. 2012;3 doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Streubel J, Blücher C, Landgraf A, Boch J. TAL effector RVD specificities and efficiencies. Nat Biotechnol. 2012;30:593–595. doi: 10.1038/nbt.2304. [DOI] [PubMed] [Google Scholar]
- 38.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liang J, Chao R, Abil Z, Bao Z, Zhao H. FairyTALE: a high-throughput TAL effector synthesis platform. ACS Synthetic Biology. 2013 doi: 10.1021/sb400109p. [DOI] [PubMed] [Google Scholar]
- 40.Schmid-Burgk JL, Schmidt T, Kaiser V, Höning K, Hornung V. A ligation-independent cloning technique for high-throughput assembly of transcription activator-like effector genes. Nat Biotechnol. 2012 doi: 10.1038/nbt.2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Briggs AW, Rios X, Chari R, Yang L, Zhang F, Mali P, Church GM. Iterative capped assembly: rapid and scalable synthesis of repeat-module DNA such as TAL effectors from individual monomers. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Garg A, Lohmueller JJ, Silver PA, Armel TZ. Engineering synthetic TAL effectors with orthogonal target sites. Nucleic Acids Research. 2012 doi: 10.1093/nar/gks1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Horvath P, Barrangou R. CRISPR/Cas, the Immune System of Bacteria and Archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 44.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482:331–338. doi: 10.1038/nature10886. [DOI] [PubMed] [Google Scholar]
- 45.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, Thakore PI, Glass KA, Ousterout DG, Leong KW, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nat Meth. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nat Meth. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48**.Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. This is the first paper to demonstrate the use of an engineered sgRNA type II S. pyogenes CRISPR/Cas system to repress transcription initiation and elongation in E. coli. RNA-seq data and first order multiplexing studies are presented to demonstrate specificity and utility of the CRISPR/Cas sytem. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49**.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Research. 2013 doi: 10.1093/nar/gkt1520. This is the first paper to demonstrate the use of the native tracrRNA/crRNA type II S. pyogenes CRISPR/Cas system for gene repression in E. coli. The system was also used to activate target gene expression by fusing RpoZ to dCas9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50*.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. A seminal study elucidating the role of tracrRNAs in mediating Cas9 nuclease activity. This paper demonstrates that a fusion of a crRNA and a tracrRNA into a synthetic sgRNA still enables Cas9 activity. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51*.Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–2196. doi: 10.1038/nprot.2013.132. A methods paper describing how to implement the sgRNA-based CRISPR/dCas9 system for repression of gene expression in prokaryotes, as described in Qi et al. 2013, and eukaryotes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mojica FJ, Diez-Villasenor C, Garcia-Martinez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology. 2009;155:733–740. doi: 10.1099/mic.0.023960-0. [DOI] [PubMed] [Google Scholar]
- 53.Semenova E, Jore MM, Datsenko KA, Semenova A, Westra ER, Wanner B, van der Oost J, Brouns SJ, Severinov K. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci U S A. 2011;108:10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carroll D. Staying on target with CRISPR-Cas. Nat Biotech. 2013;31:807–809. doi: 10.1038/nbt.2684. [DOI] [PubMed] [Google Scholar]
- 55.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends in Genetics. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 56.De Lay N, Schu DJ, Gottesman S. Bacterial Small RNA-based Negative Regulation: Hfq and Its Accomplices. Journal of Biological Chemistry. 2013;288:7996–8003. doi: 10.1074/jbc.R112.441386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 58.Ji Y, Woodnutt G, Rosenberg M, Burnham MKR. Identification of essential genes in Staphylococcus aureus using inducible antisense RNA. Methods in Enzymology. 2002;358:123–128. doi: 10.1016/s0076-6879(02)58084-8. [DOI] [PubMed] [Google Scholar]
- 59.Sharma V, Sakai Y, Smythe KA, Yokobayashi Y. Knockdown of recA gene expression by artificial small RNAs in Escherichia coli. Biochemical and Biophysical Research Communications. 2013;430:256–259. doi: 10.1016/j.bbrc.2012.10.141. [DOI] [PubMed] [Google Scholar]
- 60.Kang Z, Wang X, Li Y, Wang Q, Qi Q. Small RNA RyhB as a potential tool used for metabolic engineering in Escherichia coli. Biotechnol Lett. 2012;34:527–531. doi: 10.1007/s10529-011-0794-2. [DOI] [PubMed] [Google Scholar]
- 61.Tummala SB, Welker NE, Papoutsakis ET. Design of antisense RNA constructs for downregulation of the acetone formation pathway of Clostridium acetobutylicum. Journal of Bacteriology. 2003;185:1923–1934. doi: 10.1128/JB.185.6.1923-1934.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62**.Na D, Yoo SM, Chung H, Park H, Park JH, Lee SY. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat Biotechnol. 2013;31:170–174. doi: 10.1038/nbt.2461. The E. coli MicC sRNA is shown to be amenable to re-programming and use as a general scaffold for post-transcriptional gene repression in E. coli. Engineered sRNAs are used to successfully increase tyrosine and cadaverine production. [DOI] [PubMed] [Google Scholar]
- 63.Sharma V, Yamamura A, Yokobayashi Y. Engineering artificial small RNAs for conditional gene silencing in Escherichia coli. ACS Synth Biol. 2012;1:6–13. doi: 10.1021/sb200001q. [DOI] [PubMed] [Google Scholar]
- 64.Man S, Cheng R, Miao C, Gong Q, Gu Y, Lu X, Han F, Yu W. Artificial trans-encoded small non-coding RNAs specifically silence the selected gene expression in bacteria. Nucleic Acids Res. 2011;39:e50. doi: 10.1093/nar/gkr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65*.Yoo SM, Na D, Lee SY. Design and use of synthetic regulatory small RNAs to control gene expression in Escherichia coli. UNKNOWN. 2013;8:1694–1707. doi: 10.1038/nprot.2013.105. A methods paper detailing how to successfully implement the sRNA-based post-transcriptional repression strategy described in Na et al. 2013. [DOI] [PubMed] [Google Scholar]
- 66.Updegrove TB, Correia JJ, Chen Y, Terry C, Wartell RM. The stoichiometry of the Escherichia coli Hfq protein bound to RNA. RNA. 2011;17:489–500. doi: 10.1261/rna.2452111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perez-Pinera P, Ousterout DG, Brunger JM, Farin AM, Glass KA, Guilak F, Crawford GE, Hartemink AJ, Gersbach CA. Synergistic and tunable human gene activation by combinations of synthetic transcription factors. Nat Meth. 2013;10:239–242. doi: 10.1038/nmeth.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qi L, Haurwitz RE, Shao WJ, Doudna JA, Arkin AP. RNA processing enables predictable programming of gene expression. Nature Biotechnology. 2012;30:1002–+. doi: 10.1038/nbt.2355. [DOI] [PubMed] [Google Scholar]
- 69.Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Levine E, Zhang Z, Kuhlman T, Hwa T. Quantitative characteristics of gene regulation by small RNA. Plos Biology. 2007;5:1998–2010. doi: 10.1371/journal.pbio.0050229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mercer AC, Gaj T, Sirk SJ, Lamb BM, Barbas CF., Iii Regulation of endogenous human gene expression by ligand-inducible TALE transcription factors. ACS Synth Biol. 2013 doi: 10.1021/sb400114p. [DOI] [PMC free article] [PubMed] [Google Scholar]


