Abstract
Cells transmit information through molecular signals that often show complex dynamical patterns. The dynamic behavior of the tumor suppressor p53 varies depending on the stimulus; in response to double-strand DNA breaks it shows a series of repeated pulses. Using a computational model, we identified a sequence of precisely timed drug additions that alter p53 pulses to instead produce a sustained p53 response. This leads to the expression of a different set of downstream genes and also alters cell fate: cells that experience p53 pulses recover from DNA damage, whereas cells exposed to sustained p53 signaling frequently undergo senescence. Our results show that protein dynamics can be an important part of a signal, directly influencing cellular fate decisions.
Cells use molecular signaling networks to sense, interpret, and respond to stimuli. Recent advances in time-lapse microscopy have revealed that many signaling molecules show complex dynamical behaviors (1–13). In some instances, dynamical properties such as oscillation frequency or signal duration have been shown to alter gene expression (1, 3, 6, 8, 11, 13–15) or to control cellular differentiation (7, 12, 16). These examples point to a rich mode of regulation that is largely unexplored for most biological pathways. Here, we develop a mathematically-designed perturbation of p53 dynamics in response to DNA damage and show experimentally that p53 dynamics determine cellular responses.
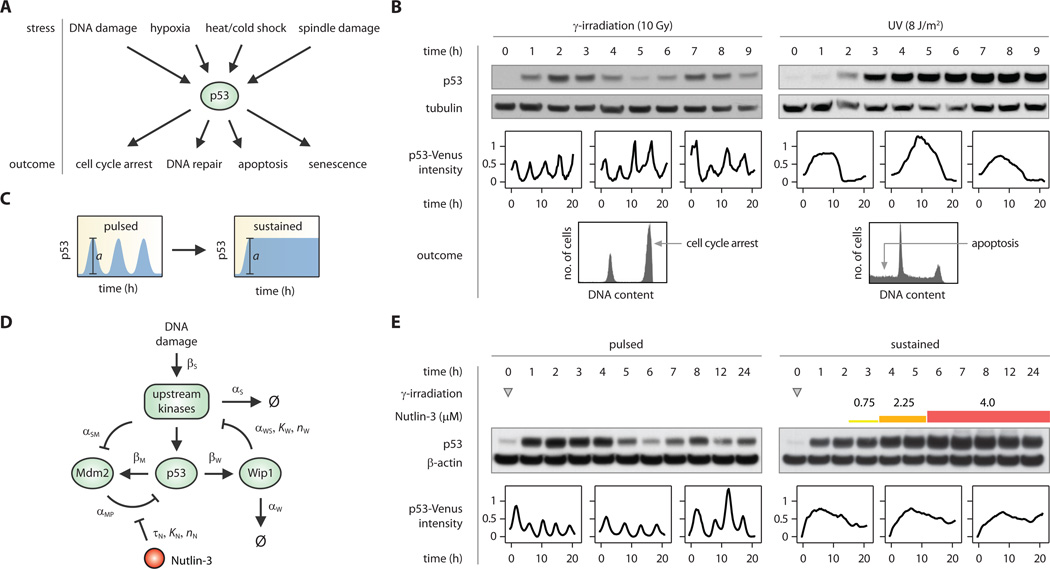
p53 is a tumor suppressor activated in response to cellular stress (17, 18). Induction of p53 triggers multiple cellular programs ranging from transient responses, such as DNA repair and cell cycle arrest, to terminal fates such as cell death (apoptosis) and permanent cell cycle arrest (senescence) (Fig. 1A). Recently it was shown that different stresses evoke different dynamic patterns of p53 protein levels (Fig. 1B, ref. (19)). In response to DNA breaks caused by γ-irradiation, the levels of p53 exhibit a series of pulses with fixed amplitude and frequency (4, 20). Higher radiation doses increase the number of pulses without affecting their amplitude or duration. These p53 pulses were observed in a live mouse model (21) and in various transformed and non-transformed human cell lines (22–24). In contrast, UV radiation triggers a single p53 pulse with a dose-dependent amplitude and duration (19). Although much insight has been gained into the molecular mechanisms that control these differential p53 dynamics in response to γ and UV radiation (19, 24, 25), the effect of p53 dynamics on downstream responses remains unknown. γ and UV radiation activate distinct targets of p53 (26) and lead to different cellular outcomes (Fig. 1B), suggesting that downstream elements in the p53 network may respond to the dynamic profiles of p53. However, γ and UV radiation also lead to many p53-independent events in cells, which could contribute to the differential outcomes. A definitive conclusion about the role of p53 dynamics on cellular outcomes may come from experimentally perturbing p53 dynamics in response to the same stress and observing the effect on downstream responses.
Figure 1. Generation and perturbation of p53 dynamics.
A, p53 mediates the response to multiple cellular stresses and evokes diverse cellular outcomes. B, γ-irradiation leads to p53 pulses and cell cycle arrest; UV radiation induces a single pulse and leads to apoptosis. C, p53’s natural pulses were perturbed to produce a sustained response with equal amplitude, a. D, A diagram capturing the main species and parameters in the mathematical model of p53 dynamics following DNA damage (25). This model was used to predict the optimal sequence of Nutlin-3 additions needed to generate a sustained p53 response following γ-irradiation (see Supporting Online Material). E, p53 dynamics under naturally pulsed (left) or pharmacologically sustained (right) conditions. The sequence of Nutlin-3 treatments is denoted by differently colored bars. Pulses in immunoblots appear as damped oscillations because of the asynchronous responses of single cells. Representative single-cell traces show average nuclear p53-Venus intensities that were normalized to the median value and zeroed to the minimum value. Sequential Nutlin-3 treatment did not alter the amplitude of p53 (Fig. S3).
We developed a method for altering p53 dynamics following γ-irradiation. Our goal was to switch p53 natural pulses into a sustained p53 signal held at the peak pulse amplitude (Fig. 1C). We used the small molecule Nutlin-3, which binds to the p53 inhibitor, Mdm2, thereby inhibiting degradation of the p53 protein (27). Nutlin-3 is selective for p53 as p53−/− cells show no change in genome-wide expression profiling upon Nutlin-3 treatment (28). Achieving a sustained signaling response with a single Nutlin-3 treatment proved to be difficult: MDM2 is activated by p53 and therefore addition of Nutlin-3 not only stabilizes p53 but also causes an increase in Mdm2 levels that eventually overcomes Nutlin-3 inhibition, resulting in down-regulation of p53 (Fig. S1). Treatment with a higher dose of Nutlin-3 led to prolonged induction of p53 but also to an overshoot in p53 levels (Fig. S1). To overcome this obstacle, we trained our model of p53 dynamics (25) to predict the optimal sequence of Nutlin-3 additions necessary to sustain p53 at a constant level (Fig. 1D, Fig. S2, Table S1, Table S2). The model predicted that three sequential treatments of Nutlin-3 at 2.5 h (0.75 µM), 3.5 h (2.25 µM), and 5.5 h (4 µM) following γ-irradiation would produce a sustained p53 response with an amplitude equal to p53 natural pulses. This prediction was validated experimentally in both cell populations and single cells (Fig. 1E, Fig. S3). These two dynamical ‘inputs’—naturally pulsed and pharmacologically sustained p53 signaling (hereafter, ‘pulsed’ and ‘sustained’)—were then used to study the downstream effects of p53 dynamics on target gene expression and cellular outcome.
To understand how p53 dynamics control gene expression, we selected a panel of wellstudied p53 target genes representing different functional pathways and cellular outcomes (29). A subset of genes showed a clear oscillatory response that mirrored p53 protein dynamics (Fig. 2A,B, Table S3). This group included genes involved in cell cycle arrest and DNA repair (CDKN1A, GADD45A, XPC), as well as genes known to regulate p53 levels (MDM2, PPM1D). In contrast, transcripts encoding apoptotic proteins (APAF1, BAX, TP53AIP) or involved in p53- dependent senescence (30, 31) (PML and YPEL3) were not induced by p53 pulses (Fig. 2C,D).
Figure 2. Pulsed and sustained p53 signaling activate different sets of target genes.
Expression of p53 target genes was measured under pulsed or sustained conditions after γ-irradiation. Genes are grouped according to function: A, cell cycle arrest and DNA repair; B, control of p53 levels; C, apoptosis; D, senescence. For reference, p53 protein levels are shown in the background as light blue (pulsed) or red (sustained) bars. p53 levels are normalized to the peak (t = 2 h) p53 concentration. Note that a base-2 logarithmic scale is used for CDKN1A, GADD45A, and MDM2. Data are mean +/− SD. Significance of correlation between target genes and p53 protein levels under pulsed conditions is reported in Table S3.
We next measured expression of these transcripts using our dynamic drug treatment (Fig.1E) to sustain p53 signaling. Oscillating genes (e.g., MDM2, CDKN1A) showed sustained increases in expression. Genes involved in apoptosis or senescence—which were not induced by p53 pulses—showed either no induction (APAF1, TP53AIP1) or a delayed increase in expression (BAX, PML, YPEL3) under sustained p53 signaling. These trends were p53-dependent (Fig. S4). Taken together, these results indicate that p53 pulses selectively activate genes involved in transient responses to DNA damage, while sustained p53 signaling allows induction of genes associated with terminal fates.
We next asked whether changes in p53 dynamics lead to different cell fates—specifically, whether sustained p53 will trigger irreversible fates such as apoptosis or senescence while pulsed p53 will allow recovery and growth. DNA content analysis by flow cytometry revealed only a small amount of cell death under both pulsed and sustained p53 conditions (Fig. S5). We therefore pursued the alternate possibility that sustained p53 signaling promotes cellular senescence, a state of permanent cell cycle arrest (32, 33). Cells were subjected to pulsed or sustained p53 signaling at several γ-irradiation doses and then assayed for senescence-associated β-galactosidase (β-gal) activity and their ability to proliferate in fresh growth media (Fig. S6). At lower doses of γ-irradiation (2.5 and 5 Gy), sustained p53 signaling led to large increases in β-gal-positive cells (Fig. 3A,B). At these levels of DNA damage, the majority of cells exposed to pulsed p53 were able to undergo multiple rounds of growth and division after recovery (Fig. 3C–E and Fig. S7) while sustained p53 signaling reduced this fraction significantly, leading to a characteristically flattened morphology and an apparent inability to divide (Fig. 3C). These differences were most pronounced following 5 Gy γ-irradiation—a dose at which nearly all cells showed a permanent arrest after sustained p53 signaling (Fig. 3D,E). Sustained p53 signaling at 2.5 Gy led to a greater fraction of senescent cells than did pulsed p53 at 5 Gy (Fig. 3B,E), suggesting that it is not the extent of DNA damage that induces senescence, but rather the dynamics of p53 signaling.
Figure 3. Pulsed and sustained p53 signaling lead to different cell fates.
A, Cells were subjected to pulsed (upper) or sustained (lower) conditions for 3 days and then stained for β-gal activity 1 day after recovery. Blue color and flattened morphology are indicative of senescence. B, Percentage of β-gal-positive cells under pulsed (P) or sustained (S) p53 signaling at various γ-irradiation doses. n ≥ 100 cells per condition per experiment. *P < 0.05; **P < 0.01. C, Typical images of single cells that were recovered after 3 days of pulsed (upper) or sustained (lower) p53 signaling at 5 Gy γ-irradiation. White boxes are drawn to show the fate of an individual cell. D, Number of cell divisions for single cells after recovery from pulsed or sustained p53 signaling at 5 Gy. E, (left) Percentage of cells that did not divide under resting conditions or sequential Nutlin-3 treatment alone (no γ-irradiation). (right) Percentage of non-dividing cells under pulsed (P) or sustained (S) p53 signaling. F, Fold change in expression of CDKN1A, PML and YPEL3 after 24, 48, and 72 h under pulsed (blue) or sustained (red) p53 signaling (10 Gy γ-irradiation). Expression levels were normalized to ACTB. Data are mean +/− SD.
Interestingly, we did not observe a large difference in β-gal activity or in proliferative ability at the highest dose of γ-irradiation (10 Gy). This suggested that prolonged p53 pulsing (4, 20, 24) caused by extensive DNA damage might eventually lead to expression of senescence genes. Indeed, we found that after 3 days under pulsed conditions the p53-dependent senescence genes were induced to similar levels reached under sustained conditions after 1 day (Fig. 3F). CDKN1A, which is involved in both cell cycle arrest and p53-dependent senescence (32, 33), showed the most dramatic increase in expression (>100 fold) under sustained p53 signaling.
Thus, sustained p53 signaling appears to accelerate the expression of senescence genes, while pulsed p53 delays gene expression and thereby protects cells from prematurely committing to an irreversible fate. However, by the time a cell commits to senescence the total amount of p53 accumulated over time (‘cumulative p53’) is much higher under sustained conditions than in pulsing cells. To determine whether this change in cell fate is due to p53 dynamics or merely to an increase in cumulative p53, we compared expression of senescence genes between pulsed and sustained p53 at equivalent levels of cumulative p53 (Fig. 4A). We found that even for similar cumulative p53, sustained p53 signaling led to higher expression of its target genes than pulsed p53, suggesting that it is the dynamics of p53 rather than its accumulated levels that control gene expression.
Figure 4. p53 dynamics, and not its cumulative level, control cell fate.
A, (inset) Time points for which pulsed and sustained p53 signaling show equivalent cumulative p53 levels (∫ p53(t) dt) lie along the gray line. Gene expression under pulsed (blue dots) or sustained (red dots) p53 signaling are plotted as a function of cumulative p53 using the data presented in Fig. 1E. The time integral of p53 protein levels was computed using trapezoidal integration of p53 levels over time. Gene expression was normalized to ACTB. The last time point in sustained conditions (24 h) was omitted since there is no comparable data point under pulsed conditions. B, p53 dynamics were recorded by live-cell microscopy under pulsed and sustained conditions. C–D, Representative single-cell traces of p53 levels under pulsed (blue) or sustained (red) conditions. Time-lapse imaging was terminated by fixing cells 21 hr after irradiation (pulsed conditions, t1) or 12 h after irradiation (sustained conditions, t2), and probing for expression of CDKN1A or PML by FISH. E, CDKN1A and F, PML expression versus cumulative p53 in individual cells. a.u., arbitrary units; r.f.u., relative fluorescence units (see Supporting Online Material). G, Model for p53 dynamics controlling cell fate. Transient damage encountered under low radiation dose or physiological conditions is repaired quickly and generates a small number of p53 pulses, allowing the cell to continue dividing. Persistent damage—whether from a large number of initial DNA lesions or a small number of irreparable breaks—generates repeated p53 pulses that ultimately trigger cellular senescence. t1 and t2 represent time points in which the cumulative level of p53 is equal between pulsed and sustained conditions. However the probability of entering senescence differs significantly between these two types of dynamics. Pulsed p53 allows more time for recovery from DNA damage while sustained p53 accelerates this process.
We further tested this in single cells. We first showed that individual senescent cells have significantly higher levels of CDKN1A and PML transcripts than proliferating cells (Fig. S8), confirming that these transcripts are reliable markers for the induction of senescence. We then quantified p53 dynamics under pulsed and sustained conditions and used FISH to compare the level of CDKN1A and PML (Fig. 4B,C). To achieve comparable cumulative p53 levels between pulsed and sustained p53, we terminated pulsed p53 21 h after irradiation (t1) and sustained p53 12 h after irradiation (t2) (Fig. 4D). We found that expression of both CDKN1A and PML was significantly higher under sustained p53 than under pulsed p53 even at similar cumulative p53 levels (Fig. 4E, F). These results suggest that the decision of whether and when to enter senescence is communicated to cells through the temporal pattern of p53 (Fig. 4G).
It is well established that different post-translational modifications of p53 or different cofactors that bind p53 affect the choice of downstream gene programs (34). Here we have shown another mechanism for specificity in this network—the dynamics of p53. Our work suggests that p53 dynamics are important but do not act alone: while p53 signaling produced by γ-irradiation and Nutlin-3 leads to senescence, a comparable dynamical profile in response to UV radiation leads to apoptosis (Fig. 1B). Future studies are required to explore the combined effect of p53 dynamics and modifications on cellular outcomes.
What molecular mechanism may decode p53 dynamics to determine cell fate? One hypothesis is that p53 pulses periodically exceed a threshold concentration for transcriptional activation of senescence genes. In this scenario, sustained p53 or a greater number of p53 pulses increases the probability that p53 will activate downstream targets involved in the induction of senescence. A similar mechanism was previously reported (14) in which the frequency of calcium oscillations controls specificity for activation of proinflammatory transcription factors. Another plausible mechanism involves a feed-forward loop motif that discriminates transient and persistent p53 signaling. This type of mechanism was identified in the extracellular signal-related kinase (ERK) signaling pathway, in which the early gene product c-Fos functions as a sensor for sustained ERK levels (16). By analogy, an early gene induced by p53 and essential for activating senescence with p53, may decay with a time scale close to the time scale of p53 pulses and therefore accumulates slowly during p53 pulses, but more rapidly during a sustained p53 response. It is also possible that expression of senescence genes is initially repressed for example by epigenetic silencing or antisense RNA, and that these factors are deactivated by persistent p53 signaling.
Other signaling pathways have been shown to encode information through the dynamics of their signaling molecules (1–3, 5–13), suggesting that varying protein dynamics may offer a functional advantage in certain contexts. For example, information encoded in the dynamics, rather than the absolute concentration of a signaling molecule, may be less sensitive to spontaneous fluctuations in the cellular environment. In addition, certain dynamical patterns may allow neighboring cells to synchronize their responses to produce emergent multicellular behaviors. A better understanding of how signaling dynamics are regulated and how they affect cellular responses will provide new insights for manipulating them in a controlled way. In addition, targeted perturbation of protein dynamics, such as the one illustrated in this study, may enable new pharmacological strategies for altering cell fate in a range of diseases.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Agami for the MCF7/p53shRNA cell line; B. Fang for the MCF7/Casp3 cell line; A. Raj and Z. Waks for help with mRNA FISH. R. Kishony, R. Ward, E. O’Shea, T. Mitchison, A. Klein, J. Orth and U. Alon for comments and discussions; and the Nikon Imaging Center at Harvard Medical School for help with light microscopy. This research was supported by the National Institutes of Health grant GM083303 and fellowship F32GM095168 (J.E.P), a National Science Foundation graduate fellowship (K.W.K), the American Cancer Society, California Division, Pamela and Edward Taft Postdoctoral Fellowship (E.B.), and fellowships from the German Research Foundation and the Charles A. King Trust (A.L.). J.E.P and G.L. conceived the study, designed the experiments, and wrote the paper. J.E.P modeled and designed perturbation experiments; performed gene expression and cell fate assays, live-cell microscopy, single-cell mRNA detection, and image analysis. K.W.K. performed live-cell microscopy, single-cell mRNA detection, and image analysis. C.M. characterized p53 and p53-Venus dynamics under radiation and various drug treatments. E. B. and A. L. performed UV treatment experiments and provided preliminary results and foundational concepts.
Footnotes
SUPPORTING ONLINE MATERIAL
www.sciencemag.org/cgi/content/full/science.1218351/DC1
Materials and Methods
Figs. S1 to S8
Tables S1 to S5
References (35–36)
REFERENCES
- 1.Cai L, Dalal CK, Elowitz MB. Frequency-modulated nuclear localization bursts coordinate gene regulation. Nature. 2008;455:485. doi: 10.1038/nature07292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hao N, et al. Regulation of cell signaling dynamics by the protein kinase-scaffold Ste5. Mol Cell. 2008;30:649. doi: 10.1016/j.molcel.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IkappaB-NF-kappaB signaling module: temporal control and selective gene activation. Science. 2002;298:1241. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 4.Lahav G, et al. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat Genet. 2004;36:147. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- 5.Mettetal JT, Muzzey D, Gomez-Uribe C, van Oudenaarden A. The frequency dependence of osmo-adaptation in Saccharomyces cerevisiae. Science. 2008;319:482. doi: 10.1126/science.1151582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson DE, et al. Oscillations in NF-kappaB signaling control the dynamics of gene expression. Science. 2004;306:704. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 7.Suel GM, Garcia-Ojalvo J, Liberman LM, Elowitz MB. An excitable gene regulatory circuit induces transient cellular differentiation. Nature. 2006;440:545. doi: 10.1038/nature04588. [DOI] [PubMed] [Google Scholar]
- 8.Tay S, et al. Single-cell NF-kappaB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ventura JJ, et al. Chemical genetic analysis of the time course of signal transduction by JNK. Mol Cell. 2006;21:701. doi: 10.1016/j.molcel.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 10.Lee TK, et al. A noisy paracrine signal determines the cellular NF-kappaB response to lipopolysaccharide. Sci Signal. 2009;2:ra65. doi: 10.1126/scisignal.2000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner SL, Barken D, Hoffmann A. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science. 2005;309:1857. doi: 10.1126/science.1113319. [DOI] [PubMed] [Google Scholar]
- 12.Santos SD, Verveer PJ, Bastiaens PI. Growth factor-induced MAPK network topology shapes Erk response determining PC-12 cell fate. Nat Cell Biol. 2007;9:324. doi: 10.1038/ncb1543. [DOI] [PubMed] [Google Scholar]
- 13.Ashall L, et al. Pulsatile stimulation determines timing and specificity of NF-kappaB-dependent transcription. Science. 2009;324:242. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dolmetsch RE, Xu K, Lewis RS. Calcium oscillations increase the efficiency and specificity of gene expression. Nature. 1998;392:933. doi: 10.1038/31960. [DOI] [PubMed] [Google Scholar]
- 15.Sung MH, et al. Sustained oscillations of NF-kappaB produce distinct genome scanning and gene expression profiles. PLoS One. 2009;4:e7163. doi: 10.1371/journal.pone.0007163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy LO, Smith S, Chen RH, Fingar DC, Blenis J. Molecular interpretation of ERK signal duration by immediate early gene products. Nat Cell Biol. 2002;4:556. doi: 10.1038/ncb822. [DOI] [PubMed] [Google Scholar]
- 17.Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- 18.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 19.Batchelor E, Loewer A, Mock C, Lahav G. Stimulus-dependent dynamics of p53 in single cells. Mol Syst Biol. 2011;7:488. doi: 10.1038/msb.2011.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geva-Zatorsky N, et al. Oscillations and variability in the p53 system. Mol Syst Biol. 2006;2:2006 0033. doi: 10.1038/msb4100068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamstra DA, et al. Real-time evaluation of p53 oscillatory behavior in vivo using bioluminescent imaging. Cancer Res. 2006;66:7482. doi: 10.1158/0008-5472.CAN-06-1405. [DOI] [PubMed] [Google Scholar]
- 22.Hu W, et al. A single nucleotide polymorphism in the MDM2 gene disrupts the oscillation of p53 and MDM2 levels in cells. Cancer Res. 2007;67:2757. doi: 10.1158/0008-5472.CAN-06-2656. [DOI] [PubMed] [Google Scholar]
- 23.Lev Bar-Or R, et al. Generation of oscillations by the p53-Mdm2 feedback loop: a theoretical and experimental study. Proc Natl Acad Sci U S A. 2000;97:11250. doi: 10.1073/pnas.210171597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Batchelor E, Mock CS, Bhan I, Loewer A, Lahav G. Recurrent initiation: a mechanism for triggering p53 pulses in response to DNA damage. Mol Cell. 2008;30:277. doi: 10.1016/j.molcel.2008.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao R, et al. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981. [PMC free article] [PubMed] [Google Scholar]
- 27.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 28.Tovar C, et al. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riley T, Sontag E, Chen P, Levine A. Transcriptional control of human p53-regulated genes. Nat Rev Mol Cell Biol. 2008;9:402. doi: 10.1038/nrm2395. [DOI] [PubMed] [Google Scholar]
- 30.de Stanchina E, et al. PML is a direct p53 target that modulates p53 effector functions. Mol Cell. 2004;13:523. doi: 10.1016/s1097-2765(04)00062-0. [DOI] [PubMed] [Google Scholar]
- 31.Kelley KD, et al. YPEL3, a p53-regulated gene that induces cellular senescence. Cancer Res. 2010;70:3566. doi: 10.1158/0008-5472.CAN-09-3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Campisi J, d'Adda di Fagagna F. Cellular senescence: when bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 33.d'Adda di Fagagna F. Living on a break: cellular senescence as a DNA-damage response. Nat Rev Cancer. 2008;8:512. doi: 10.1038/nrc2440. [DOI] [PubMed] [Google Scholar]
- 34.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 35.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.