Abstract
Purpose:
Simultaneous multi-slice (SMS) echo planar imaging (EPI) is incorporated into two-dimensional (2D) arterial spin labeling (ASL) imaging to produce more slices for measuring perfusion in a larger region of the brain than currently possible with multi-slice EPI.
Methods:
Pulsed ASL (PASL) preparations using FAIR and QUIPSS II techniques were combined with SMS-EPI. Testing was performed in four subjects at 3 Tesla. Multiband slice acceleration factors (MB) from MB-2 to MB-5 using 40 averages were evaluated. Comparisons were made quantitatively to PASL 2D EPI and qualitatively to PASL 3D GRASE.
Results:
In the 12 slice data set, spatial SNR for the perfusion weighted images averaged across subjects was 3.28 and 3.44 for the two sequential MB-1 acquisitions as control comparison, 3.25 for MB-2 and 2.98 for MB-3. The temporal SNR averaged 1.01 and 0.99 for MB-1, 0.89 for MB-2, and 0.78 for MB-3. For whole-brain spatial coverage, the 20 slice data sets could be acquired in narrower time windows, from 874 ms using EPI (MB-1) down to 196 ms using MB-5. SMS-EPI ASL differed from 3D GRASE ASL, which can use background suppression and has less susceptibility artifact as a CPMG SE sequence.
Conclusion:
SMS-EPI has a major advantage over EPI-based ASL imaging by increasing slice coverage without lengthening the acquisition time window.
Keywords: EPI, multiband, simultaneous, ASL, GRASE, PASL
INTRODUCTION
Arterial spin labeling (ASL) is a noninvasive MRI technique invaluable for measuring tissue perfusion (1). ASL is based upon subtraction of images to reveal the perfused blood signal separated from static brain tissue. ASL can be performed with several different blood labeling schemes and combined with two-dimensional (2D) or 3D imaging readout sequences. One challenge for successful ASL measurement is the relatively low blood volume fraction in tissue and this typically has necessitated signal averaging to raise signal-to-noise ratio (SNR) at the cost of longer scan times. There is also a limitation on the number of image slices for spatial coverage possible within the time between delivery of labeled water to capillaries with exchange into the brain parenchymal and T1 decay. This limitation can be mitigated with innovations in pulse sequence design to allow useful ASL imaging for medical and scientific applications. Improvements have been made to both the labeling methodology (2,3) and to the image readout component of ASL pulse sequence (4,5). With the advent of efficient approaches to blood labeling and image readout, the sensitivity of ASL perfusion techniques has increased while acquisition times have decreased, contributing to ASL’s recent success in reaching clinical practice.
Arterial Spin Labeling
In general, the ASL labeling scheme tags intra-arterial blood near the perfused tissue and the inflow inversion time (TI) is sufficient for labeled blood to arrive in the capillary network of the brain parenchyma. There is a limited time interval before the decay of labeled water by means of T1 relaxation places a time constraint on image readout. Parallel imaging can be used to reduce the echo train length to increase the obtainable number of slices (6) and concurrently shortening the effective echo time (TE) while also increasing g-factor and reducing SNR from fewer acquired signals in the echo train (7). Dissimilar to 2D readout, the use of 3D techniques, gradient-and-spin echo (GRASE), TSE, and stack-of-spiral trajectories, use a single TI for all slices recorded within one echo train. The 3D sequences inherently raise SNR by means of more signal averaging in the 3D FT compared with 2D FT image reconstructions. Compared to 2D, the 3D imaging sequences have two distinct advantages for ASL related to the single excitation. The 3D sequences excite all slices with each excitation pulse so there are no sequential time delays between the slice acquisitions. This permits double inversion pulse suppression of background brain signal to improve SNR (8) and reduce scan times. Secondly, with no slice dependent variation of inflow time all measurements are at an identical time in the capillary phase.
Simultaneous Multi-slice EPI
Simultaneous multi-slice (SMS) EPI (9–12) is a parallel imaging technique that simultaneously excites multiple slices with a multiband excitation pulse and reads out these images in an echo train. In this work we use the term MB to describe slice acceleration factors created by the multiband pulses, e.g., MB-4; for conventional EPI, we denote MB-1 in places thereafter. Unlike 3D sequences, the multiple slices are not achieved with a slice axis of phase encoding but instead are achieved with multiband excitation pulses and signal recorded with array coils. Spatial sensitivity profiles from coil arrays are used to unalias signal acquired from different image planes. A significant improvement in reducing g-factor penalty in SMS-EPI has been achieved by performing controlled aliasing with the blipped-CAIPI technique (13).
SMS-EPI has recently been developed and optimized for fMRI (11–13) and diffusion imaging (14). In fMRI, the time of whole-brain imaging is reduced by the number of simultaneous images in each EPI echo train, with up to MB-12 factor and repetition time (TR) reductions from 3 s to below 300 ms can be achieved. SMS-EPI has been used in diffusion imaging enabling faster scanning of HARDI data sets.
This report describes the incorporation of SMS-EPI in ASL with the aim of increasing spatial coverage without lengthening the time window of image readout within the capillary phase of blood circulation. It should be possible to use SMS-EPI to record several of the ASL images simultaneously. This motivated our belief that simultaneous 2D EPI ASL perfusion mapping could have an ability to scan the entire brain instead of a more limited number of slices performed with 2D EPI ASL to date.
METHODS
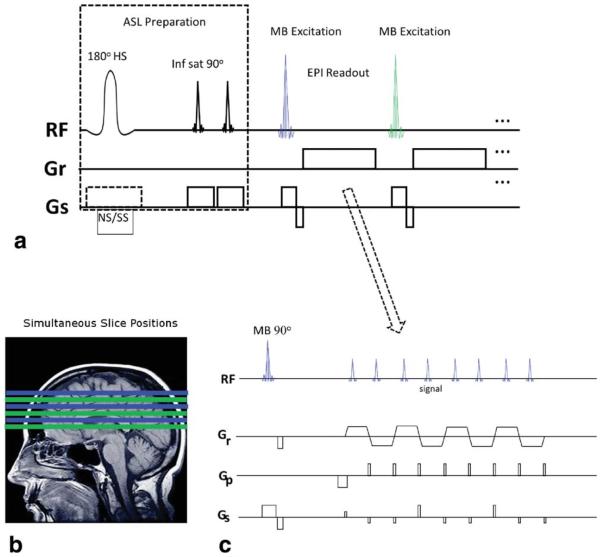
Experiments were performed on a 3 Tesla scanner (Siemens Trio) with a 32-channel head coil. Four subjects were scanned under institutional guidelines with informed consent. Figure 1 shows the pulse sequence diagram of pulsed ASL (PASL) with SMS-EPI. The FAIR labeling scheme (15) was combined with QUIPSSII (2) with conventional pulse timing and spatial positioning as described in (16). The image readout used 1/MB fewer echo trains than multi-slice EPI to produce the same number of images. The blipped-CAIPI gradient pulse scheme was used to achieve image shifts by applying Gs pulses at the time of the blipped phase encoding pulses as shown in Figure 1c where, 1/3 FOVSHIFT is used for illustrative purpose. The directionality of slice order in all the multi-slice acquisitions was from bottom to top of head in para-axial slice orientation. The simultaneously acquired slices are spaced at maximum distance from each other within the total covered volume to effect coil sensitivity variance and improve g-factor. Single band (SB) reference EPI images are acquired at the beginning for MB separation, not shown in Figure 1.
FIG. 1.
Diagram of SMS-EPI ASL pulse sequence. a: the initial ASL preparation of the sequence (left) uses nonselective and slab selective (NS/SS) inversion pulses in consecutive TRs, and QUIPSSII saturation in regions inferior to the image volume. The image readout (right) is similar to conventional multi-slice 2D EPI using multibanded (MB) excitation pulses to reduce the number of echo trains. More slices may be readout within a time window than possible with 2D EPI. b: Slice positioning with MB-3. Each MB pulse and echo train creates three slices (same color) which are widely separated to improve slice unaliasing. c: SMS-EPI echo train with blipped-CAIPI gradient pulses on Gs slice axis for controlled aliasing.
The acquisition parameters are as below unless indicated: TR = 3000 ms for all EPI/SMS-EPI, TE = 16–18 ms, 4 × 4 mm2 in plane resolution, slice thickness = 5 mm, 20% slice gap; field of view (FOV) = 256 × 256 mm2, matrix = 64 × 64, partial Fourier = 7/8, signal averages = 40, echo spacing = 0.41–0.46 ms with ramp sampling on, flip angle = 90°, sinc excitation pulse width = 2.56 ms for MB-2 and up to 6.71 ms for MB-5 to reduce peak RF power. Three navigator echoes at the beginning of the echo train (not shown) acquired without phase encoding are used for phase correction. In-plane parallel imaging was not used. MB factors ranged from 2 to 5.
The FAIR sequence had inflow time = 1800 ms for the earliest acquired slices. The slice selective (SS) pulse slab thickness = 20 mm + total image slice coverage (mm), the nonselective (NS) pulse slab thickness = 220 mm + total image slice coverage (mm). The TI1 and TI2 parameter of the QUIPSS II are 800 and 1800 ms, respectively.
The NS labeling scheme was used to generate reference data to calculate the kernel with slice-GRAPPA algorithm (13). For MB-2 and MB-3, the kernel size of 3 was used and for MB > 3 the kernel size of 5 was used. Controlled aliasing FOVSHIFT created by blipped-CAIPI differed with MB factor, and was FOV/2 with MB-2, FOV/3 with MB-3, FOV/4 for MB-4 and MB-5.
For a qualitative comparison to SMS-EPI ASL in whole-brain coverage, segmented 3D GRASE ASL (17) was acquired. To achieve equivalent slice positions and coverage as the SMS-EPI that used 5-mm slice and 20% (1 mm) gap, the 3D GRASE used 6-mm contiguous slices. Sequence parameters: TR = 3000 ms, TE = 19.9 ms, total scan time = 48 s, matrix 64 × 64 × 20, resolution 4 × 4 × 6 mm3; partial Fourier = 6/8 in slice axis; four segments (two phase axis × two slice axis), centric k-space ordering, EPI factor = 33, SE factor = 9, slice over-sampling = 20%, average = 2, bandwidth = 2790 Hz/pixel, echo spacing = 0.5 ms, ramp sampling on, slab selective thickness= 120 mm, post IR delay (TI) = 1800 ms, QUIPSS II duration 60 ms. Background suppression was used in 3D GRASE as earlier described (4,17).
Image Acquisition
Two pairs of single band (SB) reference scans with SS and NS labeling pulses were followed with the MB data acquisition using the same TR and only the last acquired NS reference scan was used for kernel calculation. Identical sequence gradient pulses were used in the SB and MB data acquisitions and four total repeats (two SS and two NS ASL preparations) are used to establish steady state in both SB and MB acquisitions, adding .5 to 1.0 min to the total acquisition time. Both the SB and the MB acquisitions alternated between the SS and NS excitations and the MB acquisition was repeated for 40 signal averages, using 80 excitations total, not including the SB reference acquisition. The NS and SS image pairs were subtracted and then averaged across the image series to get the mean ASL perfusion weighted images. CBF images were calculated as in (16,18).
To make conventional PASL 2D EPI, the same sequence code was used, only excluding the initial SB reference data by setting MB to be 1 and removal of blipped-CAIPI gradient pulses. The conventional EPI (MB-1) ASL acquisition had 1 ms shorter TE made possible by the absence of the blipped-CAIPI gradient pulses.
For image quality and CBF comparisons, 12 slice scans are acquired with MB-1 (conventional EPI), MB-2 and MB-3. To obtain a measure of scan–scan variability as a control when comparing variability across different MB factors, two sequential MB-1 scans were acquired. For increased spatial coverage comparison, 20 slices in each scan were acquired with MB-1, MB-2, MB-4, MB-5, and 21 slices in MB-3.
Statistical Analysis
The images were motion-corrected, and the skull and scalp were excluded from the analysis using the FSL toolbox (http://www.fmrib.ox.ac.uk/fsl). Perfusion weighted images were constructed from the difference between the nonselective and slice-selective images.
For constant TR comparisons, voxel-wise temporal SNR (tSNR) was calculated as the mean value divided by the standard deviation of the 40-image perfusion weighted time-series. The mean tSNR was calculated across the whole-brain and the signal intensity changes due to TI variation were corrected. For image quality assessment, SNR was calculated using the method of Glover and Lai (19). The even- and odd-numbered image time points from the perfusion weighted time-series were separately averaged, and the sum and difference of these two images were calculated. The SNR was calculated as the mean value across the brain in the sum image, divided by the standard deviation across the same region in the difference image.
Agreement between the different scans was measured using intra-class correlation coefficient (ICC) and the repeatability coefficient (RC). The ICC measures how strongly the values measured for a given voxel using two different sequences resembled each other, taking into account the variation between measurements in all measured voxels, with a value close to 1 indicating good agreement. RC represents the variation between voxel-wise measurements across 2 sequences.
RESULTS
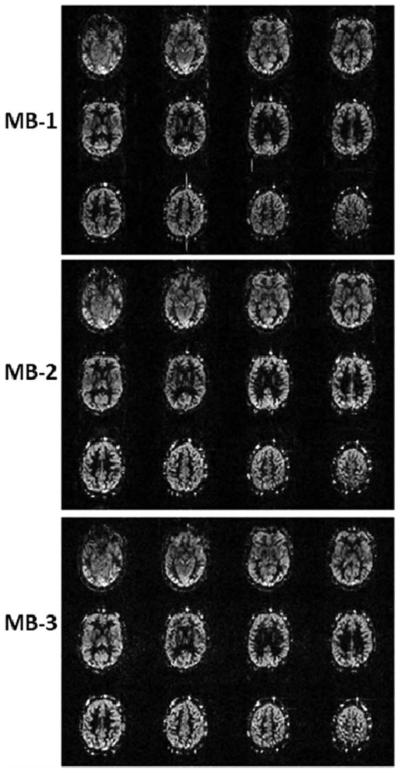
Figure 2 shows PASL perfusion images using a 12 slice acquisition. SMS-EPI has similar image quality as 2D EPI except for the increased noise level in the SMS-EPI ASL images. The susceptibility artifacts and distortions are the same in all three image sets. The spatial SNR calculated in perfusion weighted images in four subjects are 3.28 ± 0.42, 3.44 ± 0.78, 3.25 ± 0.27, and 2.98 ± 0.80 for MB-1, MB-1, MB-2, and MB-3, respectively. The tSNR calculated from perfusion weighted images in 4 subjects are 1.01 ± 0.10, 0.99 ± 0.15, 0.89 ± 0.13 and 0.78 ± 0.15 for MB-1, MB-1, MB-2, and MB-3, respectively. The time window of image acquisition was reduced from 523 ms to 261 ms and 178 ms in normal EPI (MB-1), MB-2 and MB-3, respectively.
FIG. 2.
Comparison of perfusion weighted images acquired with 12 slices covering a 72-mm slice volume using MB-1, MB-2, and MB-3 from one subject. The scaling factors are set to be the same for all three MB factors.
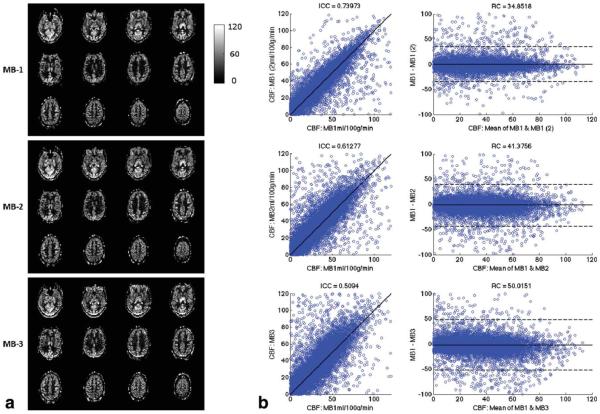
Quantitative CBF maps shown in Figure 3a were calculated with the data acquired from subject 2. Comparison between MB-1, MB-2, and MB-3 maps across 12 slices is shown in scatter and Bland-Altman plots, in Figure 3b. Bland-Altman plots show the difference between two measures versus the mean of those two measurements. The dotted lines on the plots indicate the repeatability coefficient (RC) for that comparison, which in this case is 1.96 times the standard deviation of the difference measures. Agreement between different sequences is calculated using the intraclass correlation coefficient (ICC). The ICC from one subject between MB-1 and MB-2 is .61 and between MB-1 and MB-3 is .5, in comparison to .74 between two MB-1 scans acquired with the same parameters sequentially. Mean ICC across subjects for the two MB-1 acquisitions across all 12 slices was .71 with .57 ICC for MB-1_MB-2 and .56 ICC for MB-1_MB-3. For the first four slices, which have the same TI for each slice across sequences, the values were .73 for MB-1_MB-1, .62 for MB-1_MB-2, and .61 for MB-1_MB-3. Scatter and Bland-Altman plots for the additional subjects for both 4 slice and 12 slice comparisons are included in the Supp. Material, which is available online.
FIG. 3.
Quantitative CBF maps acquired with MB-1, MB-2, and MB-3 SMS-EPI ASL with 12 slices covering a 72-mm slice volume from a second subject. a) CBF maps. b) Scatter plot and Bland-Altman plots showing the pixel-wise comparison of MB-1 versus MB-1, MB-1 versus MB-2, and MB-1 versus MB-3. Two separate MB-1 scans were used to establish a relative measure for comparisons.
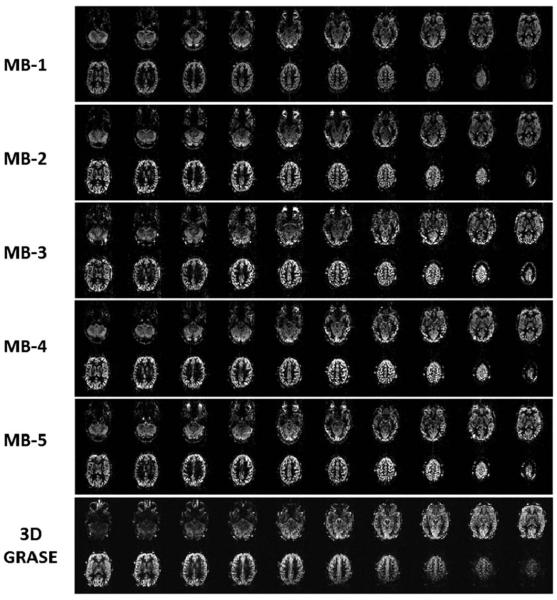
The RC in a single subject for MB-1 and MB-2 was 41.38, and the RC for MB-1 and MB-3 was 50.02, compared with an RC of 34.85 between the two MB-1 acquisitions. Mean RC across subjects for all 12 slices was 43.29 for MB-1_MB-1, 62.74 for MB-1_MB-2, and 58.80 for MB-1_MB-3. For the first four slices, mean RC was 38.30 for MB-1_MB-1, 50.94 for MB-1_MB-2, and 48.21 for MB-1_MB-3. The higher RC value for the MB1-MB2 comparison seems surprising given we would expect greater variability between MB1 and MB3. The standard deviation across subjects of the RC for the MB1-MB2 comparison was much higher than for the other subjects, so this different may just be driven by inter-subject variability. Alternatively, this difference may be due to varying sequence parameters, such as using FOV/2 in MB2, or other differences that we cannot currently explain. Twenty slices coverage is obtained with SMS-EPI as shown in Figure 4. The normal EPI and SMS-EPI used the same image data acquisition time = 4 min (with additional 0.5 to 1.0 min for the reference SB scans in SMS-EPI) for 20 slices (21 slices for MB-3). The time window of image acquisition was reduced from 874 ms using normal EPI (MB-1) to 436 ms, 312 ms, 237 ms, and 196 ms with MB-2, MB-3, MB-4, and MB-5, respectively. Figure 5 shows temporal SNR measurements in the perfusion weighted time series using the 20 slice acquisitions. TSNR increases by around 15% between MB-1 and MB-2, and then decreases at higher MB factors.
FIG. 4.
Comparison of perfusion weighted images covering 120-mm slice volume, whole-brain coverage, in the second subject. The 20 slice coverage with MB-1 (standard EPI), MB-2, MB-3, MB-4, and MB-5 slice acceleration factors. The 3D GRASE ASL images are shown offset by one image to the right of the corresponding positioned SMS-EPI ASL images.
FIG. 5.
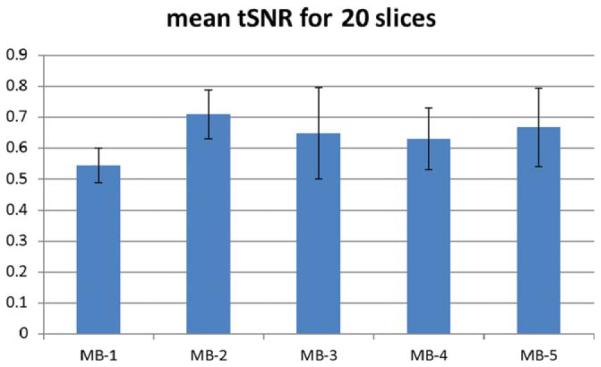
Temporal SNR for 20 slice perfusion weighted images in four subjects.
DISCUSSION
The physiological constraint on the variation of TI within the perfusion window ultimately limits the total number of echo trains in 2D EPI ASL techniques. In this respect, the ability of SMS-EPI to increase the slice coverage by as much as five-fold without substantially increasing the readout time is an important gain. In these experiments, the image SNR loss are in part due to g-factor related noise with higher g-factor penalty as MB increases. The g-factor (SNR ~1/g-factor) would become very high at MB-5 if controlled aliasing with blipped-CAIPI were not utilized.
The SNR losses in SMS-EPI are less than with in-plane parallel imaging accelerations for which SNR losses result from g-factor and from signal reduction (fewer echoes) not occurring in SMS. A somewhat shorter TE can be achieved with partial Fourier and in-plane parallel imaging; however, the blipped-CAIPI approach to controlled aliasing greatly reduces g-factor losses in SNR and, therefore, should give advantage to SMS-EPI compared with in-plane acceleration in EPI. A direct comparison and incorporation of in-plane acceleration is beyond the scope of this work. It can be anticipated that combining in-plane parallel imaging to reduce distortions in blipped-CAIPI SMS-EPI might be useful at lower accelerations without greatly increasing g-factor from the combined accelerations.
An alternative approach to SMS-EPI used to increase slice coverage with EPI ASL is to scan adjacent regions of the brain in two or more separate scans. Using this approach, if in the same scanning time and slice coverage, the SNR of EPI ASL is greater than SMS-EPI ASL, there would be no advantage to using SMS-EPI ASL. To test this, a 12 slice MB-2 acquisition with 40 averages was compared with two acquisitions of 6 slices MB-1 with half the number of averages, 20 averages. In the test on a single subject, the SNR across 12 slices where the two MB-1 image sets were combined was 1.65, and SNR was 2.90 for the MB-2 acquisition. The much higher SNR for the MB-2 seems surprising given that we would only expect a benefit of 1.41 from increasing the number of averages. It may be that using MB2 has additional denoising effects due to the slice-GRAPPA algorithm, although we cannot be sure of this due to the comparison being done in only one subject. Therefore, collecting multiple MB-1 acquisitions with fewer averages appears to be a less desirable approach.
One surprising finding is that MB-2 has higher tSNR than MB-1 in the 20 slice comparison study which could be due to head motion or other factors. Both g-factor and slice leakage artifact caused by residual aliasing of images increase with higher MB factor but this should only lower SNR. Considering hemodynamics, we compared tSNR using only the first four acquired lower brain slices which had identical inflow time (TI) for all MB factors and found the difference between MB-1 and MB-2 was decreased but it remained (0.61 ± 0.16 versus 1.71 ± 0.14). Another possible reason for the higher SNR in MB-2 SMS-EPI than in normal EPI ASL is that the slice-GRAPPA algorithm using matrix multiplication to separate the SMS images has a denoising effect and this will contribute to higher SNR. Comparing tSNR across slices (Supp. Material) showed that there was an advantage in terms of tSNR for the later acquired slices covering higher brain regions when using higher MB factors. This deficit in tSNR for MB-1 is most likely due to a decrease in signal in these slices, which can clearly be seen, Figure 5.
The greater slice coverage of SMS-EPI–based ASL may be useful for both medical and biological research purposes and for improving quantitation using narrower TI variability within the data. This ability to perform SMS-EPI ASL scanning of whole brain could have applicability in ASL-based fMRI (20) and for basic research on various metabolic and physiologic conditions. It will be possible to combine other spin labeling preparations with SMS-EPI including pseudo-continuous ASL (pCASL) and to further improve the image reconstruction processes to raise SNR. In retrospect, it should be possible to acquire less data for the reference scans to reduce the time by half and with using four TRs of MB dummy scans to establish steady state, it would take between 15 and 30 s.
The 2D SMS-EPI ASL achieved similar slice coverage as 3D GRASE ASL; however, there were apparent differences in image quality, Figure 4. The 3D GRASE used a CPMG sequence for spin echo refocusing to greatly reduce signal loss and distortions from susceptibility artifact, which are present in SMS-EPI in the temporal and frontal lobes and appear the same as in normal EPI.
One major advantage of 3D GRASE ASL over SMS-EPI ASL is its ability to incorporate background suppression which raises SNR to give faster scanning. This is possible because the inversion pulse timing is identical in all slices created by the single excitation of a 3D volume, whereas the multiband excitation pulses encode only a few slices and, therefore, must be repeated at different times relative to the inversion pulses which give incorrect timing for suppression. Based on these initial results using up to MB-5, it should be possible to use even higher MB factors (21) and can be combined with SIR in multiplexed EPI (11) to simultaneously record a larger number of images to possibly enable the use of background suppression at the price of higher g-factors.
CONCLUSIONS
SMS-EPI has a major advantage over EPI ASL with its ability to increase the number of slices for volume coverage. SMS-EPI ASL differs from 3D GRASE ASL which can use background suppression and has less susceptibility artifact being a CPMG-based imaging sequence. With fewer echo trains required for SMS-EPI than EPI, the narrower acquisition time window allows more images to be closer to an optimal TI for perfusion measurement.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Steen Moeller of University of Minnesota for helpful discussions on image reconstruction, Daniel JJ Wang of UCLA for providing PASL labeling scheme code, and Matthias Günther of Fraunhofer Institute, Bremen for PASL 3D GRASE code. The authors wish to thank both reviewers for their constructive comments.
Grant sponsor: NIH; Grant numbers: 1R44 NS073417, 1R44 NS084788.
Footnotes
Additional Supporting Information may be found in the online version of this article.
REFERENCES
- 1.Detre JA, Leigh JS, Williams DS, Koretsky AP. Perfusion imaging. Magn Reson Med. 1992;23:37–45. doi: 10.1002/mrm.1910230106. [DOI] [PubMed] [Google Scholar]
- 2.Wong EC, Buxton RB, Frank LR. Quantitative imaging of perfusion using a single subtraction (QUIPSS and QUIPSS II) Magn Reson Med. 1998;39:702–708. doi: 10.1002/mrm.1910390506. [DOI] [PubMed] [Google Scholar]
- 3.Dai W, Robson PM, Shankaranarayanan A, Alsop DC. Modified pulsed continuous arterial spin labeling for labeling of a single artery. Magn Reson Med. 2010;64:975–982. doi: 10.1002/mrm.22363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gunther M, Oshio K, Feinberg DA. Single-shot 3D imaging techniques improve arterial spin labeling perfusion measurements. Magn Reson Med. 2005;54:491–498. doi: 10.1002/mrm.20580. [DOI] [PubMed] [Google Scholar]
- 5.Fernandez-Seara MA, Wang Z, Wang J, Rao HY, Guenther M, Feinberg DA, Detre JA. Continuous arterial spin labeling perfusion measurements using single shot 3D GRASE at 3 T. Magn Reson Med. 2005;54:1241–1247. doi: 10.1002/mrm.20674. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z, Wang J, Connick TJ, Wetmore GS, Detre JA. Continuous ASL (CASL) perfusion MRI with an array coil and parallel imaging at 3T. Magn Reson Med. 2005;54:732–737. doi: 10.1002/mrm.20574. [DOI] [PubMed] [Google Scholar]
- 7.Pruessmann KP, Weiger M, Scheidegger MB, Boesiger P. SENSE: sensitivity encoding for fast MRI. Magn Reson Med. 1999;42:952–962. [PubMed] [Google Scholar]
- 8.Ye FQ, Frank JA, Weinberger DR, McLaughlin AC. Noise reduction in 3D perfusion imaging by attenuating the static signal in arterial spin tagging (ASSIST) Magn Reson Med. 2000;44:92–100. doi: 10.1002/1522-2594(200007)44:1<92::aid-mrm14>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 9.Larkman DJ, Hajnal JV, Herlihy AH, Coutts GA, Young IR, Ehnholm G. Use of multicoil arrays for separation of signal from multiple slices simultaneously excited. J Magn Reson Imaging. 2001;13:313–317. doi: 10.1002/1522-2586(200102)13:2<313::aid-jmri1045>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 10.Nunes RG, Hajnal JV, Golay X, Larkman DJ. Simultaneous slice excitationand reconstruction for single shot EPI. Proceedings of the 14th Annual Meeting of ISMRM; Seattle, Washington, USA. 2006. Abstract 293. [Google Scholar]
- 11.Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Gunther M, Glasser MF, Miller KL, Ugurbil K, Yacoub E. Multiplexed echo planar imaging for sub-second whole brain FMRI and fast diffusion imaging. PLoS One. 2010;5:e15710. doi: 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Ugurbil K. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn Reson Med. 2010;63:1144–1153. doi: 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Setsompop K, Gagoski BA, Polimeni JR, Witzel T, Wedeen VJ, Wald LL. Blipped-controlled aliasing in parallel imaging for simultaneous multislice echo planar imaging with reduced g-factor penalty. Magn Reson Med. 2012;67:1210–1224. doi: 10.1002/mrm.23097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Setsompop K, Cohen-Adad J, Gagoski BA, Raij T, Yendiki A, Keil B, Wedeen VJ, Wald LL. Improving diffusion MRI using simultaneous multi-slice echo planar imaging. Neuroimage. 2012;63:569–580. doi: 10.1016/j.neuroimage.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim SG. Quantification of relative cerebral blood flow change by flow-sensitive alternating inversion recovery (FAIR) technique: application to functional mapping. Magn Reson Med. 1995;34:293–301. doi: 10.1002/mrm.1910340303. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Licht DJ, Jahng GH, Liu CS, Rubin JT, Haselgrove J, Zimmerman RA, Detre JA. Pediatric perfusion imaging using pulsed arterial spin labeling. J Magn Reson Imaging. 2003;18:404–413. doi: 10.1002/jmri.10372. [DOI] [PubMed] [Google Scholar]
- 17.Feinberg D, Ramanna S, Guenther M. Evaluation of new ASL 3D GRASE sequences using parallel imaging, segmented and interleaved k-space at 3T with 12- and 32-channel Coils. Proceedings of the 17th Annual Meeting of ISMRM; Honolulu, Hawaii, USA. 2006. Abstract 0622. [Google Scholar]
- 18.Wang J, Alsop DC, Li L, Listerud J, Gonzalez-At JB, Schnall MD, Detre JA. Comparison of quantitative perfusion imaging using arterial spin labeling at 1.5 and 4.0 Tesla. Magn Reson Med. 2002;48:242–254. doi: 10.1002/mrm.10211. [DOI] [PubMed] [Google Scholar]
- 19.Glover GH, Lai S. Self-navigated spiral fMRI: interleaved versus single-shot. Magn Reson Med. 1998;39:361–368. doi: 10.1002/mrm.1910390305. [DOI] [PubMed] [Google Scholar]
- 20.Vidorreta M, Wang Z, Rodriguez I, Pastor MA, Detre JA, Fernandez-Seara MA. Comparison of 2D and 3D single-shot ASL perfusion fMRI sequences. Neuroimage. 2012;66C:662–671. doi: 10.1016/j.neuroimage.2012.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feinberg DA, Setsompop K. Ultra-fast MRI of the human brain with simultaneous multi-slice imaging. J Magn Reson. 2013;229:90–100. doi: 10.1016/j.jmr.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.