Abstract
In the resent study, a diazotrophic bacterial isolate JPA2 having the ability to tolerate salinity (6 % NaCl) and plant growth-promoting features was isolated from rhizospheric soil of weed Chenopodium murale growing in saline soil of Pindara (EC 11.47 dS m−1), district Jind (Haryana). The nitrogen fixing ability of the isolate was confirmed by nifH gene amplification and acetylene reduction assay (38.9 nmol ethylene h−1 mg−1 protein). The potential of strain JPA2 to promote growth of pearl millet was investigated by inoculation experiment which showed significant increase in plant height (51.1, 39.9 and 28.8 %) and dry weight (55.9, 36.4 and 35.5 %) over uninoculated control plants at EC 0, 6, 8 dS m−1, respectively. The strain JPA2 was Gram +ve and identified as Gordonia sp. on the basis of partial 16S rRNA gene sequencing and biochemical characterization. It is concluded that salt tolerant diazotrophic Gordonia sp. can be considered as a beneficial microbe for agriculture in saline soils.
Keywords: Salt tolerant, Plant growth-promoting rhizobacterium, nifH, Pearl millet, Gordonia sp.
Introduction
Saline soils are extensive in the plains of Haryana state (Choubey et al. 2009) particularly in arid and semi-arid regions with ~2,32,985 hectares of area is considered as salt affected. Soils containing excess amount of water soluble salts are called saline soils. The concentration of salt in the root-zone of soil goes so high that it badly affects growth of the most crop species. But some cereal species are well adapted to semi-arid and arid tropics, where salinity and drought are the major problems due to limited water supply. Among the cereals, pearl millet (Pennisetum glaucum) is the only major crop that has high level of tolerance to saline soils. It can be cultivated even in the most sandy infertile soils and saline environments where no other cereal crop can survive (Kumar et al. 2010). However, to survive in saline soils, the increased demand of nitrogen in pearl millet could be met by the exploitation of diazotrophic bacteria (Latake et al. 2009). Saline rhizosphere habitats are blessed with some free-living nitrogen fixing bacteria which siphon out appreciable amount of nitrogen from reservoirs and enrich the soil. So, it is important to exploit rhizobacteria which often play crucial role in increasing crop productivity of the plants they colonize due to their close proximity with the roots they inhabit. Plant growth-promoting rhizobacteria (PGPR) stimulate the plant growth by producing Indole-3-acetic acid (IAA) and synthesizing siderophores that chelate iron and make it available to plant root (Kumar and Gera 2013). The application of PGPR as crop inoculants for biofertilization would be an attractive alternative to decrease the use of chemical fertilizers which effect environmental pollution.
The genus Gordonia, a coryneform bacterial member, is widespread in nature and comprises over 33 species, with broader characteristics such as the ability to degrade hydrocarbons (Xue et al. 2003), toxic environmental pollutants, xenobiotics (Hernandez et al. 2001), and natural compounds that are not readily biodegradable. Gordonia sp. may play an important role during waste water treatment (Bendinger et al. 1992) and also used as PGPR for Zea mays (Hong et al. 2010). Even though the higher number of Gordonia species described, to the best of our knowledge, there are no in-depth studies on the occurrence of nitrogen fixing species within the genus in saline soils and their possible role in growth promotion of pearl millet. Therefore, in the present investigation, diazotrophic Gordonia sp. was isolated from rhizospheric soil of weed growing in saline soils and was further characterized for its PGP features. In addition, the inoculation effects of Gordonia sp. on the growth of pearl millet were evaluated in artificially created saline conditions under pot house experiment.
Materials and methods
A salt affected barren field of Pindara village (latitude: 29°30′ N, longitude: 76°34′ E), district Jind, Haryana was chosen for sampling which harbors only few weeds such as C. murale. The weed rhizospheric soil sample was collected carefully by uprooting the root system and soil was placed in the sterile plastic bags for transport to the laboratory. Physiochemical analysis of soil properties, including pH, EC, organic carbon (Kalembasa and Jenkinson 1973) and total N (Bruel et al. 1946) was estimated. Three morphotypes were obtained on Jensen’s medium (Jensen 1951) and maintained on respective medium slants at 4 °C. To investigate their nitrogen fixing ability, the genomic DNA of cultures was subjected to nifH gene amplification using primers: nifH for (5′-TAY GGN AAR GGN GGHATY GGY ATC-3′) and nifH rev (5′-ATR TTR TTN GCN GCR TAV ABB GCC ATC AT-3′) (Sarita et al. 2007). The PCR reaction mixture (25 μl) contained 0.5 μl of dNTP mixture (10 mM), 0.5 μl of Taq DNA polymerase (3 U μl−1), 10× PCR assay buffer with 25 mM MgCl2 (2.5 μl), 2 μl of each primer (10 pmol each), 2 μl template DNA (0.1–0.14 μg/μl) and 15.5 μl sterile water. PCR protocol conditions were: initial denaturation at 95 °C for 4 min, 35 cycles of denaturation at 94 °C for 30 s, annealing at 54 °C for 1 min and extension at 72 °C for 1 min followed by a final extension at 72 °C for 5 min. The PCR was then set on hold at 4 °C. Amplified PCR product was separated by electrophoresis on 2 % agarose gel stained with ethidium bromide and photographed under UV illumination using Gel Documentation system (DNR Bio-Imaging Systems). The isolate having nifH gene designated as JPA2 was further examined for acetylene reduction according to the method of Bridage et al. (1982).
For the identification of the strain JPA2, its 16S rRNA region was amplified and partially sequenced (Bioserve, Hyderabad, India) by primer walking using five different internal primers (16SEQ2R, 16SEQ3F, INS16SREV, 16SEQ4R and 16SEQ4F). The resulting 16S rRNA gene sequence was compared with sequences in GenBank database using the BLAST-N algorithm to identify sequences with a high degree of similarity. Comparable reference sequences were downloaded in FASTA format from NCBI database (http://www.ncbi.nlm.nih.gov). All the sequences were aligned with ClustalW program and phylogenetic tree was constructed using MEGA4 software through neighbor-joining method. Bootstrap analysis was used to evaluate the tree topology of the neighbor-joining data with 1,000 resamplings. The partial 16S rRNA gene sequence determined in this study was deposited in the GenBank database (http://www.ncbi.nlm.nih.gov/Genbank/index.html) under the accession number JQ437540. Bacterial culture JPA2 was further characterized morphologically and subjected to a battery of biochemical tests such as nitrate reduction, hydrolysis of Aesculin and starch (Cowan and Steel 1965). Sugar utilization pattern was assayed according to Collins and Lyne (1984).
The ability of the strain JPA2 to produce IAA was detected using the rapid assay (Bric et al. 1991), ammonia production examined using Nessler’s reagent (Cappuccino and Sherman 1992) while siderophore production tested by CAS assay (Schwyn and Neilands 1987). Jensen’s medium modified to the saline concentration of 2–10 % NaCl was used to examine the capability of strain JPA2 to tolerate different concentrations of salinity.
In vivo pot test for plant growth promotion was carried out at CCS Haryana Agricultural University, Hisar to evaluate the effect of salt tolerant PGPR isolate using pearl millet as test crop. Seeds of pearl millet (HHB 146: a salt tolerant variety) obtained from Bajra section, Agriculture college, CCS HAU, Hisar, were surface sterilized by exposing to 95 % ethanol and immersing in 0.2 % of HgCl2 solution for 3 min. The seeds were then subjected to five washings with sterile distilled water. Earthen pots with drainage hole were filled with 5 kg of soil and two salinity levels (EC 6, 8 dS m−1) were maintained through irrigation water by addition of different concentration of salts such as NaCl, CaCl2·2H2O, MgCl2 and MgSO4·7H2O while pots of EC 0 dS m−1 were irrigated with normal distilled water. Recommended dose of commercial fertilizer at the rate of 80 N and 40 P kg ha−1 was supplied to each pot through urea and single super phosphate, respectively. Nitrogen was applied in two splits, while full dose of phosphorus was applied as basal dose. Experiment was laid out in randomized block design with three replications per treatment. Four seeds were planted per pot at a uniform depth of 1.5 cm. For preparation of bacterial formulation, the strain JPA2 was cultured in LB medium and incubated on an orbital shaker at 150 rpm at 30 °C. 1 ml of this overnight grown bacterial suspension (106–107 cells ml−1, 0.5 O.D. at 540 nm) was applied to each pearl millet seed. However, in control, seeds were treated with sterile non-inoculated LB medium. The pots were kept in sunlight, irrigated and observed everyday for 45 days. On 46th day, plants were harvested carefully at vegetative stage. Shoot length was measured after harvesting and samples were dried in oven at 75 °C. After 4 days, shoot dry weight (g/pot) was calculated and statistical significance of the data was determined by ANOVA (Analysis of Variance) using MINITAB version 15.0 and Tukey’s tests (P = 0.05).
Results and discussion
In the present investigation, a salt tolerant PGPR strain was isolated from rhizosphere of weed Chenopodium murale grown in saline fields of Pindara village. The rhizospheric soil sample has the following physiochemical properties: pH 6.24; 11.47 dS m−1 EC; 0.13 % organic carbon and 0.12 % total N. Three different morphotypes were selected on the basis of their colony color, shape and size on Jensen’s nitrogen free media. Amplification of nifH gene segment yielded the product of expected size (420 bp) from DNA template of only one isolate JPA2. Further, considerable acetylene reduction of 38.9 nmol ethylene h−1 mg−1 protein confirmed the presence of nitrogenase activity.
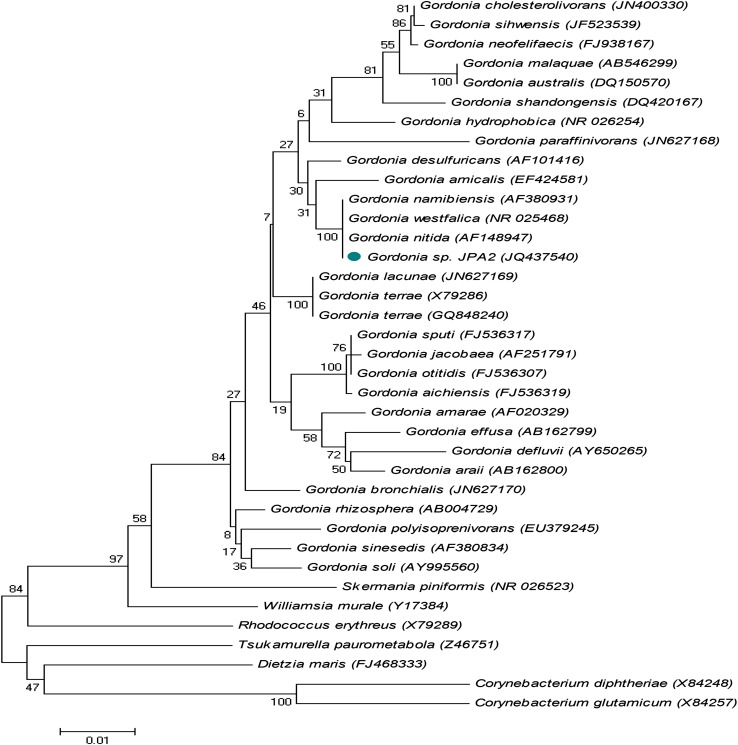
Identification of the strain JPA2 was done on the basis of partial 16S rRNA gene sequencing from Bioserve, Hyderabad (India). Figure 1 represents the phylogenetic position of the detected partial 16S rRNA sequence of JPA2 compared to sequences published in GenBank database. Phylogenetic analyses of the strains based on the NJ method with 1,000 bootstrap sampling resulted in two major clusters and strain JPA2 fell within the cluster comprising the Gordonia sp. The bootstrap resampling value for the major cluster comprising distantly related Gordonia sp. was 84 % and Rhodococcus erythreus (X79289) was used as an outgroup. The identification was further confirmed by morphological and biochemical characterization of the isolate. Colony color of JPA2 was yellowish and cells were Gram +ve cocci as observed in other Gordonia sp. Exhibition of biochemical traits such as nitrate reduction, aesculin hydrolysis and starch hydrolysis, assimilation of sugars like glycerol, d- cellobiose and N-Acetyl d-glucosamine confirmed it as Gordonia sp.
Fig. 1.
Phylogenetic tree showing the position of strain JPA2 Gordonia species and some other related taxa based on 16S rRNA gene sequences. Distances were calculated by neighbor-joining method. Bootstrap values (expressed as percentages of 1,000 replications) are shown at branch points. Genbank accession numbers are given in parentheses. The scale bar indicates 0.01 substitutions per nucleotide position
The isolate was able to produce significant amount of IAA (193.27 μg ml−1), ammonia (0.085 μg ml−1) and positive siderophore production was also detected. Strain JPA2 was found to be salt tolerant as it could tolerate up to 6 % NaCl.
To study the effect of bacterial inoculation, a pot house experiment was conducted and JPA2 was used as an inoculant for pearl millet. JPA2 had more stimulating effect on plant growth and showed significant increase in plant height (51.1, 39.9 and 28.8 %) and plant dry weight (55.9, 36.4 and 35.5 %) over uninoculated control plants at EC 0, 6, 8 dS m−1, respectively (Table 1). The higher plant height and shoot dry weight response to inoculant compared to control under different salinity levels clearly showed the beneficial role of this rhizobacterium. Such an improvement might be attributed to N2-fixing capacity as well as the ability of this PGPR strain to produce growth-promoting substances. The enhancing effect of seeds treated with bioinoculant on growth and yield of pearl millet has been reported by many researchers. (Latake et al. 2009). Tanawy (2009) isolated bacteria from highly saline soil (EC 23 dS m−1) that showed a high ability for promoting plants establishment under adverse saline conditions by producing plant growth promoter substances and fixing a high amount of nitrogen in plant ecosystem. Several studies have also shown that application of PGPR Azospirillum, Bacillus and Enterobacter strains as bioinoculants improves plant growth (Tahir et al. 2013). PGPR Gordonia sp. S2RP-17 showed plant growth promotion in Zea mays (Hong et al. 2010). Similar to these reports, our findings also demonstrated that strain JPA2 may be used as a bioinoculant for plant growth promotion.
Table 1.
Effect of JPA2 on plant height and dry weight of the pearl millet plants
| Treatment | EC-0 (dS m−1) | EC-6 (dS m−1) | EC-8 (dS m−1) | |||
|---|---|---|---|---|---|---|
| Plant height (cm) | Dry weight (g/plant) | Plant height (cm) | Dry weight (g/plant) | Plant height (cm) | Dry weight (g/plant) | |
| Control | 67.83 ± 2.061 | 3.70 ± 0.080 | 59.85 ± 1.507 | 2.85 ± 0.063 | 53.88 ± 1.882 | 2.14 ± 0.069 |
| 100 % RDF + JPA2 | 102.5 ± 2.425*** | 5.77 ± 0.051*** | 83.75 ± 3.152** | 3.89 ± 0.040*** | 69.42 ± 0.842** | 2.90 ± 0.046*** |
Data represent the mean ± SEM in each group. Group 1: plant height; Group 2: dry weight
** P < 0.01
*** P < 0.001
In conclusion, all data in the current work emphasize that the inspected strain JPA2 fixed the nitrogen biologically producing substances of plant growth promoters (IAA, ammonia and siderophores) that enhanced growth and productivity of pearl millet plants when inoculated with this strain, under normal and saline conditions. Also this work proved that the isolated strain JPA2 is saline tolerant (6 % NaCl tolerance) and had the ability to tolerate high soil salinity. Therefore, this strain proved to be potential candidate for the development of bioinoculant for crop plants growing in saline soils.
Acknowledgments
This research work is a part of the AMAAS project funded by National Bureau of Agriculturally Important Microorganisms (NBAIM) and Mau Nath Bhanjan. The authors are grateful to these funding agencies.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- Bendinger B, Kroppenstedt RM, Klatte S, Altendorf K. Chemotaxonomic differentiation of coryneform bacteria isolated from biofilters. Int J Syst Bacteriol. 1992;42:474–486. doi: 10.1099/00207713-42-3-474. [DOI] [PubMed] [Google Scholar]
- Bric JM, Bostock RM, Silvestone SE. Rapid in situ assay for indole acetic acid production by bacteria immobilized on a nitrocellulose membrane. Appl Environ Microbiol. 1991;57:535–538. doi: 10.1128/aem.57.2.535-538.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridage GL, Davidson MS, Sparling GP. Critical evaluation of the acetylene reduction test for estimating the nitrogenase activity of nitrogen fixing bacteria associated with the roots of wheat and barley. Soil Biol Biochem. 1982;14:27–35. doi: 10.1016/0038-0717(82)90073-6. [DOI] [Google Scholar]
- Bruel D, Holter H, Linderstrom-Lang K. C R Lab Carlsberg. Scr Chim. 1946;25:289. [Google Scholar]
- Cappuccino JG, Sherman N. Microbiology: a laboratory manual. New York: Addison-Wesley; 1992. [Google Scholar]
- Choubey VK, Singh O, Srivastava SL. Study of hydrological soil properties of salt affected areas around Gohana, Sonipat district, Haryana. Earth Sci India. 2009;2(3):211–223. [Google Scholar]
- Collins CH, Lyne PM. Microbiological methods. 5. London: Butterworth; 1984. [Google Scholar]
- Cowan ST, Steel KJ. Manual for the identification of medical bacteria. London: Cambridge University Press; 1965. [Google Scholar]
- Hernandez P, Fayolle GF, Vandecasteele JP. Biodegradation of ethyl t-butyl ether (ETBE), methyl t-butyl ether (MTBE) and t-amyl methyl ether (TAME) by Gordonia terrae. Appl Microbiol Biotechnol. 2001;55:117–121. doi: 10.1007/s002530000482. [DOI] [PubMed] [Google Scholar]
- Hong SH, Ryu HW, Kim J, Cho KS. Rhizoremediation of diesel-contaminated soil using the plant growth-promoting rhizobacterium Gordonia sp. S2RP-17. Biodegradation. 2010 doi: 10.1007/s10532-010-9432-2. [DOI] [PubMed] [Google Scholar]
- Jensen V. Notes on biology of Azotobacter. Proc Soc Appl Bacteriol. 1951;74:89–93. doi: 10.1111/j.1365-2672.1951.tb01997.x. [DOI] [Google Scholar]
- Kalembasa SJ, Jenkinson DS. A comparative study of titrimetric and gravimetric methods for the determination of organic carbon in soil. J Sci Food Agric. 1973;24:1089–1090. doi: 10.1002/jsfa.2740240910. [DOI] [Google Scholar]
- Kumar V, Gera R (2013) Isolation of a multi-trait plant growth promoting Brevundimonas sp. and its effect on the growth of Bt-cotton. 3Biotech. doi:10.1007/s13205-013-0126-4 [DOI] [PMC free article] [PubMed]
- Kumar A, Kumar R, Yadav VPS, Kumar R. Impact assessment of frontline demonstrations of Bajra in Haryana state. Indian Res J Ext Education. 2010;10(1):105–108. [Google Scholar]
- Latake SB, Shinde DB, Bhosale DM. Effect of inoculation of beneficial microorganisms on growth and yield of Pearl millet. Indian J Agric Res. 2009;43(1):61–64. [Google Scholar]
- Sarita S, Priefer U, Prell J, Sharma PK. Diversity of nifH gene amplified from rhizosphere soil DNA. Curr Sci. 2007;94:109–114. [Google Scholar]
- Schwyn B, Neilands JB. Universal chemical assay for the detection and determination of siderophores. Anal Biochem. 1987;160:47–56. doi: 10.1016/0003-2697(87)90612-9. [DOI] [PubMed] [Google Scholar]
- Tahir M, Mirza MS, Zaheer A, Dimitrov MR, Smidt H, Hameed S. Isolation and identification of phosphate solubilizer Azospirillum, Bacillus and Enterobacter strains by 16S rRNA sequence analysis and their effect on growth of wheat (Triticum aestivum L.) Aust J Crop Sci. 2013;7(9):1284–1292. [Google Scholar]
- Tanawy EA. Acquainting with salt tolerant endophytic bacteria isolated from rice plant grown in highly saline soil in Egypt. Int J Acad Res. 2009;1(2):72–79. [Google Scholar]
- Xue Y, Xuesong X, Zhou R, Liu R, Liang F, Ma Y. Gordonia paraffinivorans sp. nov., a hydrocarbon-degrading actinomycete isolated from an oil-producing well. Int J Syst Evol Micriobiol. 2003;53:1643–1646. doi: 10.1099/ijs.0.02605-0. [DOI] [PubMed] [Google Scholar]