Abstract
A polymerase chain reaction (PCR) assay was developed for discrimination of Bacillus subtilis from other members of B. subtilis group as well as rapid identification from environmental samples. Primers ENIF and EN1R from endoglucanase gene were used to amplify a1311 bp DNA fragment. The specificity of the primers was tested with seven reference strains and 28 locally isolated strains of endoglucanase positive Bacillus species. The PCR product was only produced from B. subtilis. The results demonstrated high specificity of two oligonucleotides for B. subtilis. This species-specific PCR method provides a quick, simple, powerful and reliable alternative to conventional methods in the detection and identification of B. subtilis. To our knowledge this is the first report of a B. subtilis specific primer set.
Keywords: Endoglucanase gene, PCR, Specific primers, B. subtilis
Introduction
Genus Bacillus is a Gram-positive, spore-forming, fermentative, aerobic and rodshaped bacteria. Several species of this group are non-pathogenic, simple to cultivate and secrete enzymes such as proteases, amylases and cellulases that are useful for a number of industrial applications (Arbige et al. 1993). The Bacillus subtilis group contains the closely related taxa Bacillus subtilis subsp. subtilis (Smith et al. 1964; Nakamura et al. 1999), Bacillus licheniformis (Skerman et al. 1980), Bacillus amyloliquefaciens (Priest et al. 1987), Bacillus atrophaeus (Nakamura 1989), Bacillus mojavensis (Roberts et al. 1994), Bacillus vallismortis (Roberts et al. 1996), Bacillus subtilis subsp. spizizenii (Nakamura et al. 1999). Classical identification methods based on biochemical tests or fatty acid methyl ester profiling were laborious and hence not applicable for the purpose of a rapid screening. These taxa can be differentiated from one another by fatty acid composition analysis, restriction digest analysis and DNA–DNA hybridization analysis, but are quite difficult to differentiate by phenotypic characteristics (Roberts et al. 1994; Nakamura et al. 1999). The use of the 16S rRNA sequence as a target for genetic detection was therefore considered. Numerous Bacillus species described so far have been found to display rather conserved 16S rRNA sequences compared to other genera. Thus the use of this taxonomic marker is sometimes inadequate for species definition according to generally accepted criteria (Stackebrandt and Goebel 1994). Such unusual similarities exist for members of the ‘Bacillus 16S rRNA groupI’, including B. subtilis, which displays 99.3 % similarity at the 16S rRNA level to B. atrophaeus and 98.3 % to B. licheniformis and B. amyloliquefaciens (Ash et al. 1991). In the present study, it has been shown that specific primers for detection of endoglucanase gene could be used for identification of B. subtilis.
Materials and methods
Bacterial strains and culture conditions
A total 35 Bacillus strains were used in this study (Table 1). Of 12 B. subtilis, ten strains were isolates from pond sediments. For Bacillus cereus, one ATCC strain and nine pond sediment isolates were tested. Five pond isolates that had been classified as Bacillus pumilus and two as B. amyloliquefaciens were also included in the test. For Bacillus megaterium,B. licheniformis and Bacillus thuringiensis, ATCC strains were analysed. All the pond sediment isolates were identified by 16S rDNA sequencing and available in our laboratory.
Table 1.
Bacterial strains used
| Species | Total no. of strains | Source | Accession number of sediment isolates |
|---|---|---|---|
| Bacillus subtilis | 14 | ATCC 11,774, ATCC 6,051 and 12 pond sediment | GQ214130, GQ21413 HQ388810–HQ388813 JX438679–JX438684 |
| Bacillus cereus | 10 | ATCC 13,061 and 9 pond sediment | GQ214131 HQ388814–HQ388817 JX438685–JX438788 |
| Bacillus pumilus | 6 | ATCC 14,884 and 5 pond sediment | HQ388808 JX438694–JX438697 |
| Bacillus megaterium | 1 | ATCC 9,885 | |
| Bacillus thuringiensis | 1 | ATCC 10,792 | |
| Bacillus licheniformis | 1 | ATCC 13,061 | |
| Bacillus amyloliquefaciens | 2 | Pond sediment | JX438692–JX438693 |
Pond sediment isolates were confirmed by 16s rDNA sequencing
Soil samples
24 Soil samples collected from agriculture field and fish culture ponds were also included.
Primers
The endoglucanase gene sequences (EC, 3.2.1.4) of B. subtilis were retrieved from GenBank nucleotide database and were aligned using Clustal W (1.82) Multiple Alignment Program. Two sets of primers EN1F (103–124 bp) 5′-CCAGTAGCCAAGAATGGCCAGC-3′, EN1R (1,413–1,393 bp) 5′-GGAATAATCGCCGCTTTGTGC-3′) were designed by analyzing the conserved regions of the aligned sequences.
DNA isolation
The total genomic DNA was extracted from bacterial suspension (after 12 h incubation in LB) using DNA extraction kit (Merck Bioscience, India) following the manufacture’s instruction. Soil genomic DNA was extracted by using ultra clean soil DNA isolation kits (MoBio, USA).
Polymerase chain reaction
The PCR reaction mixtures (50 μl) contained, dNTPs each 100 μmol; 1X PCR buffer (10 mM Tris Cl, 50 mM KCl, 1.5 mM MgCl2 and 0.01 % gelatin); each primer 10 pmol; Taq DNA polymerase (NEB) 0.75U and bacterial DNA 100 ng. The touch down PCR in a volume of 50 μl was carried out with initial denaturation of 94 °C for 5 min followed by ten cycles of touch down program (94 °C for 30 s, 70 °C for 20 s and 74 °C for 45 s, followed by a 1 °C decrease of the annealing temperature every cycle). After completion of the touch down program, 25 cycles were subsequently performed (94 °C for 30 s, 60 °C for 20 s and 74 °C for 45 s) and ending with a 10 min extension at 74 °C. PCR reactions were run on a 1.5 % agarose gel in 1X TBE.
Cloning and sequencing
Band was excised from the gel and PCR product was purified by using the QIAquick gel purification kit according to the manufacturer’s instructions (QIAGEN, Germany). The purified PCR product was cloned in pGEM®-T Easy vector following manufacturer’s protocol (promega) and transformed into DH5α cells. Sequencing of the positive clones were done by Sanger method using 96 capillary high through put sequencer; ABI 3,730 XL (Xcelris, India) with T7 and SP6 universal primer.
Results and discussion
BlastN seach of endoglucanase gene of B. subtilis (accession numbers HM470252.1, AF355629.1 and CP002906.1) revealed 93 % similarity with B. amyloliquefaciens, B. megaterium, B. pumilus and B. licheniformis 90 % with B. subtilis subsp. spizizenii and 98–99 % with Geobacillus stearothermophilus and Paenibacillus campinasensis. Based on multiple alignments of endo-β-1,4-glucanase genes, a specific consensus motif was identified in the endoglucanase gene of B. subtilis, G. stearothermophilus and P. campinasensis. Two PCR primers, EN1F and EN1R, were chosen that were predicted to specifically amplify a 1,311 bpDNA fragment of the B. Subtilis, G. stearothermophilus and P. campinasensis. The Genbank database (NCBI) search for complimentary sequences revealed 100 % homology between the primers and the gene encodes endo-β-1,4-glucanase of B. subtilis as well as G. stearothermophilus and P. campinasensis. No homologous sequences were found for other members of genus Bacillus indicating an excellent specificity of the primers for B. subtilis.
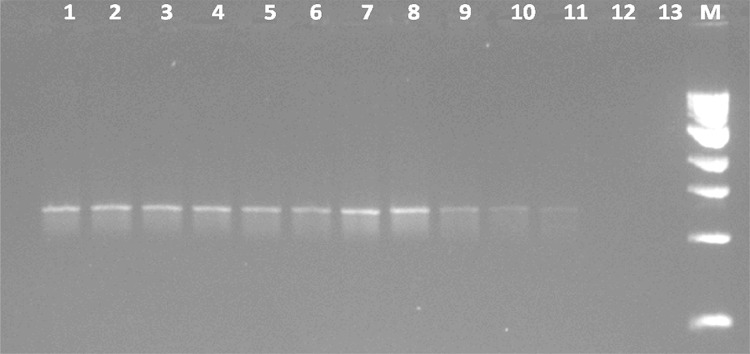
As expected, the test turned out to be positive only for B. subtilis among the different species of Genus Bacillus PCR amplification with genomic DNA isolated from in vitro cultured B. subtilis resulted in a reproducible amplification of 1,311 bp product with primer combinations EN1F/EN1R. To determine the sensitivity of PCR, endpoint titration with serial dilutions of genomic DNA isolated from the standard strain of B. subtilis was carried out and positive results obtained as little as 500 ficogram of DNA (Fig. 1).
Fig. 1.
Limit of detection of endoglucanase gene in different concentration of DNA. Lane 1 100 ng, lane 2 50 ng, lane 3 10 ng, lane 4 5 ng, lane 5 1 ng, lane 6 100 pg, lane 7 50 pg, lane 8 10 pg, lane 9 5 pg, lane 10 1 pg, lane 11 500 fg, lane 12 100 fg, lane 13 negative control, lane M size marker (1 kb ladder, NEB)
To assess the range of specificity of the PCR test, a number of endoglucanase positive Bacillus species were assayed. Given the considerable number of species established to date under B. subtilis group, our choice to assess the range of specificity was restricted to B. subtilis subsp. subtilis that was representative of the B. subtilis group.
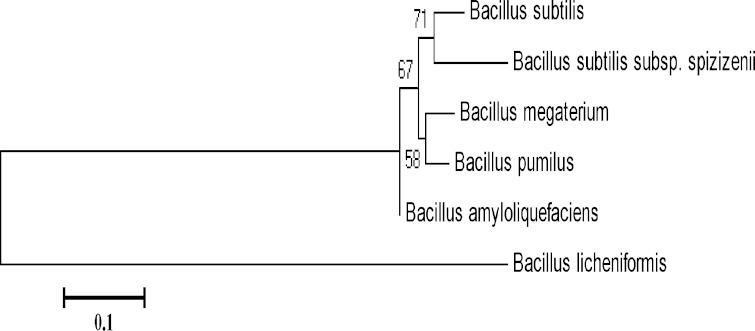
To test the specificity of the amplified products, control experiments were performed under the same conditions with DNA from different members of B. subtilis group as well as B. cereus group. The test found to be positive only for B. Subtilis (Fig. 2). It is noteworthy that the species detected as positive with this test are very close from a taxonomic point of view when phylogenetic tree was constructed on the basis of endoglucanase gene sequences of different species of Genus Bacillus (Fig. 3). Attempt to detect B. subtilis directly from soil samples collected from agriculture field and fish culture ponds were successful, out of 24 soil samples collected from fish pond and agricultural field ten samples were positive for amplification. Cloning and sequencing confirmed the amplicon to be endoglucanase gene of B. subtilis. However, after enrichment of negative soil samples on TSB 20 % increased positivity rate was obtained by PCR, demonstrating that the initial concentration of B. subtilis was at a proportion below the detection limit. The primers not only differentiated B. subtilis from other species but also differentiated at subspecies level as expected product size could not be predicted from B. subtilis subsp. spizizenii by primer blast (NCBI).
Fig. 2.
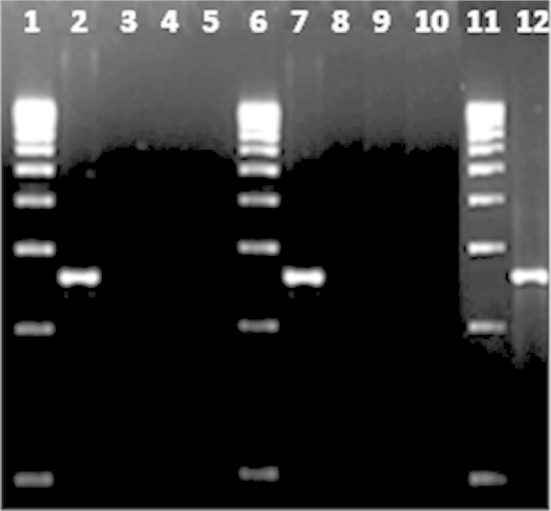
PCR amplification for endoglucanase gene in different Bacillus spp. Lane 1 size marker (500 bp ladder); lanes 2–5B. subtilis ATCC-6,051, B. cereus-ATCC 13,061, B. pumilus ATCC-14,884, B. megaterium ATCC-9,885; lane 6 size marker (500 bp ladder); lanes 7–10B. subtilis ATCC-11,774, B. thuringiensis ATCC-10,792, B. licheniformis ATCC-13,061, B. amyloliquefaciens CF8; lane 11 size marker (500 bp ladder); lane 12B. subtilis C11B1
Fig. 3.
Phylogenetic tree of Bacillus spp. based on endoglucanase gene sequence data
In this report, a PCR method has been established for identification of B. subtilis. Endo-β-1,4-glucanase gene has been chosen to design primers that could be useful for identification and direct detection of B. subtilis from environmental samples. Detection of B. subtilis has been shown to be specific, although the primers showed specificity for G. stearothermophilus and P. campinasensis in primer blast, and predicted amplicon to be 1,311 bp. However, B. subtilis could be differentiated from G. stearothermophilus and P. campinasensis by sequencing of the pcr product. 16S rRNA gene sequence analysis is the most commonly used method for identifying bacteria or for constructing bacterial phylogenetic relationships (Woese 1987; Vandamme et al. 1996; Joung and Cote 2002); however, its usefulness is limited because of the high percentage of sequence similarity between closely related species (Ash et al. 1991; Marti’nez-Murcia et al. 1992; Christensen et al. 1998). The use of protein-encoding genes as phylogenetic markers is now a common approach (Yamamoto and Harayama 1998; Ko et al. 2004; Chelo et al. 2007). Detailed investigations have demonstrated that sequences from protein-encoding genes can accurately predict genome relatedness and may replace DNA–DNA hybridization for species identification and delineation in the future (Stackebrandt et al. 2002; Zeigler 2003). Wang et al. (2007) clearly showed that in the B. subtilis group, within which species differentiation is very difficult, core genes such as gyrB allow differentiation on genetic basis. Compared to other genera, Bacillus species are having conserved 16s rRNA sequences and are difficult to identify at species level using this marker (Stackebrandt and Goebel 1994).
Conclusion
In the present study, the demonstrated specificity of the oligonucleotides used as PCR primers and results of experiments with soil samples provide the basis to develop a diagnostic assay for identification and detection B. subtilis form environment samples. As the primers used in this study have been found to be specific to endoglucanase gene of G. stearothermophilus and P. campinasensis, we suggest to use these primers as supplementary PCR assay to 16s rRNA sequencing for identification of B. subtilis.
Acknowledgments
The financial help received from AMAAS nodal centre ICAR, New Delhi, India for carrying out the work is dully acknowledged.
Conflict of interest
None.
References
- Arbige MV, Bulthuis BA, Schultz J, Crabb D. Fermentation of Bacillus. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillussubtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, DC: American Society for Microbiology; 1993. pp. 871–895. [Google Scholar]
- Ash C, Farrow JA, Dorsch M, Stackebrandt E, Collins MD. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- Chelo IM, Ze′-Ze′ L, Tenreiro R. Congruence of evolutionary relationships inside the Leuconostoc–Oenococcus–Weissella clade assessed by phylogenetic analysis of the 16S rRNA gene, dnaA, gyrB, rpoC and dnaK. Int J Syst Evol Microbiol. 2007;57:276–286. doi: 10.1099/ijs.0.64468-0. [DOI] [PubMed] [Google Scholar]
- Christensen H, Nordentoft S, Olsen JE. Phylogenetic relationships of Salmonella based on rRNA sequences. Int J Syst Bacteriol. 1998;48:605–610. doi: 10.1099/00207713-48-2-605. [DOI] [PubMed] [Google Scholar]
- Joung KB, Cote JC. Evaluation of ribosomal RNA gene restriction patterns for the classification of Bacillus species and related genera. J Appl Microbiol. 2002;92:97–108. doi: 10.1046/j.1365-2672.2002.01507.x. [DOI] [PubMed] [Google Scholar]
- Ko KS, Kim JW, Kim JM, Kim W, Chung SI, Kim IJ, Kook YH. Population structure of the Bacillus cereus group as determined by sequence analysis of six housekeeping genes and the plcR gene. Infect Immun. 2004;72:5253–5261. doi: 10.1128/IAI.72.9.5253-5261.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-T Wang, Lee F-L, Tai C-J, Kasai H. Comparison of gyrB gene sequences, 16S rRNA gene sequences and DNA–DNA hybridization in the Bacillus subtilis group. Int J Syst Evol Microbiol. 2007;57:1846–1850. doi: 10.1099/ijs.0.64685-0. [DOI] [PubMed] [Google Scholar]
- Martı′nez-Murcia AJ, Benlloch S, Collins MD. Phylogenetic interrelationships of members of the genera Aeromonas and Plesiomonas as determined by 16S ribosomal DNA sequencing: lack of congruence with results of DNA–DNA hybridization. Int J Syst Bacteriol. 1992;42:412–421. doi: 10.1099/00207713-42-3-412. [DOI] [PubMed] [Google Scholar]
- Nakamura LK. Taxonomic relationship of black-pigmented Bacillus subtilis strains and a proposal for Bacillus atrophaeus sp. nov. Int J Syst Bacteriol. 1989;39:295–300. doi: 10.1099/00207713-39-3-295. [DOI] [Google Scholar]
- Nakamura LK, Roberts MS, Cohan FM. Relationship of Bacillus subtilis clades associated with strains 168 and W23: aproposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int J Syst Bacteriol. 1999;49:1211–1215. doi: 10.1099/00207713-49-3-1211. [DOI] [PubMed] [Google Scholar]
- Priest FG, Goodfellow M, Shute LA, Berkeley RCW. Bacillus amyloliquefaciens sp. nov., nom. rev. Int J Syst Bacteriol. 1987;37:69–71. doi: 10.1099/00207713-37-1-69. [DOI] [Google Scholar]
- Roberts MS, Nakamura LK, Cohan FM. Bacillus mojavensis sp. nov., distinguishable from Bacillus subtilis by sexual isolation, divergence in DNA sequence, and differences in fatty acid composition. Int J Syst Bacteriol. 1994;44:256–264. doi: 10.1099/00207713-44-2-256. [DOI] [PubMed] [Google Scholar]
- Roberts MS, Nakamura LK, Cohan FM. Bacillus vallismortis sp. nov., a close relative of Bacillus subtilis, isolated from soil in Death Valley, California. Int J Syst Bacteriol. 1996;46:470–475. doi: 10.1099/00207713-46-2-470. [DOI] [PubMed] [Google Scholar]
- Skerman VBD, McGowan V, Sneath PHA. Approved lists of bacterial names. Int J Syst Bacteriol. 1980;30:225–420. doi: 10.1099/00207713-30-1-225. [DOI] [PubMed] [Google Scholar]
- Smith NR, Gibson T, Gordon RE, Sneath PH. Type cultures and proposed neotype cultures of some species in the genus Bacillus. J Gen Microbiol. 1964;34:269–272. doi: 10.1099/00221287-34-2-269. [DOI] [PubMed] [Google Scholar]
- Stackebrandt E, Goebel BM. Taxonomic note: a place for DNA–DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Bacteriol. 1994;44:846–849. doi: 10.1099/00207713-44-4-846. [DOI] [Google Scholar]
- Stackebrandt E, Frederiksen W, Garrity GM, Grimont PAD, Kampfer P, Maiden MCJ, Nesme X, Rossello-Mora R, Swings J, et al. Report of the ad hoc committee for the reevaluation of the species definition in bacteriology. Int J Syst Evol Microbiol. 2002;52:1043–1047. doi: 10.1099/00207713-52-3-1043. [DOI] [PubMed] [Google Scholar]
- Vandamme P, Pot B, Gillis M, De Vos P, Kersters K, Swings J. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol Rev. 1996;60:407–438. doi: 10.1128/mr.60.2.407-438.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Harayama S. Phylogenetic relationships of Pseudomonas putida strains deduced from the nucleotide sequences of gyrB, rpoD and 16S rRNA genes. Int J Syst Bacteriol. 1998;48:813–819. doi: 10.1099/00207713-48-3-813. [DOI] [PubMed] [Google Scholar]
- Zeigler DR. Gene sequences useful for predicting relatedness of whole genomes in bacteria. Int J Syst Evol Microbiol. 2003;53:1893–1900. doi: 10.1099/ijs.0.02713-0. [DOI] [PubMed] [Google Scholar]