Abstract
The tobacco-related species Nicotiana benthamiana has recently emerged as a promising host for the manufacturing of protein therapeutics. However, the production of recombinant proteins in N. benthamiana is frequently hampered by undesired proteolysis. Here, we show that the expression of the human anti-HIV antibodies 2F5, 2G12, and PG9 in N. benthamiana leaves leads to the accumulation of discrete heavy chain-derived degradation products of 30–40 kDa. Incubation of purified 2F5 with N. benthamiana intercellular fluid resulted in rapid conversion into the 40-kDa fragment, whereas 2G12 proved largely resistant to degradation. Such a differential susceptibility to proteolytic attack was also observed when these two antibodies were exposed to various types of proteinases in vitro. While serine and cysteine proteinases are both capable of generating the 40-kDa 2F5 fragment, the 30-kDa polypeptide is most readily obtained by treatment with the latter class of enzymes. The principal cleavage sites reside within the antigen-binding domain, the VH–CH1 linker segment and the hinge region of the antibodies. Collectively, these results indicate that down-regulation of endogenous serine and cysteine proteinase activities could be used to improve the performance of plant-based expression platforms destined for the production of biopharmaceuticals.
Keywords: Antibodies, Biopharmaceutical, Nicotiana, Proteinase, Protein stability
1 Introduction
Due to their outstanding specificities, monoclonal antibodies (mAbs) have become one of the most popular products of the biopharmaceutical industry. In recent years, plants have gained increasing attention as versatile expression platforms for recombinant therapeutic proteins such as mAbs [1]. Due to the large extent of similarity between the secretory pathways of plants and mammals, even highly complex molecules like mAbs can be efficiently produced in plant systems [2]. The possibility to produce grams of purified mAbs within days after generation of an appropriate DNA construct [3] provides unique advantages for plant-based expression platforms when production speed is of utmost importance, as necessary for the generation of individualized cancer vaccines [4].
Recent advances in the development of transient expression systems have placed Nicotiana benthamiana in a particularly favorable position since this tobacco-related plant species is well suited for the large-scale production of therapeutic proteins. Nevertheless, a major problem encountered with recombinant protein production in Nicotiana species remains to be solved: the proteolytic degradation of the target protein within the plants [5, 6]. Recent studies have shown that co-expression of proteinase inhibitors is a promising approach to alleviate unwanted proteolysis in plant cells and whole plants [7, 8]. Alternatively, down-regulation of endogenous proteinase activities by means of RNA interference has been attempted for improvement of the performance of plant-based expression platforms destined for the production of protein therapeutics [9]. For either strategy, substantial knowledge about the host enzymes involved in proteolytic breakdown of foreign proteins is required [10], but genetic and biochemical information on N. benthamiana proteinases is still scarce [11]. Alternatively, characterization of the cleavage sites within the protein of interest can provide hints about the proteinases involved in its degradation. Unfortunately, only one such cleavage site has been elucidated so far for mAbs produced in plants [12].
In this study, we have performed a detailed characterization of the degradation fragments observed upon expression of the three anti-HIV mAbs 2F5, 2G12, and PG9 [13–15] in N. benthamiana. Furthermore, the proteolytic susceptibility of 2F5 and 2G12 was tested in vitro with a series of representative proteinases. Collectively, these results suggest that mAb proteolysis in N. benthamiana is largely due to serine and cysteine proteinases.
2 Materials and methods
2.1 Construction of mAb expression vectors
The MagnICON expression vectors pICH26033 and pICH31160 (kindly provided by Viktor Klimyuk, Icon Genetics, Halle, Germany) were modified by insertion of the coding sequence for the signal peptide of barley α-amylase, yielding the plasmids pICHα26033 and pICHα31160. Codon-optimized PG9 heavy and light chain cDNAs (GeneArt, Regensburg, Germany; see Table S1 in Supporting information for protein sequences) were cloned with or without a C-terminal KDEL tag into the BsaI sites of pICHα31160 and pICHα26033, respectively. The resulting vectors pPG9HC, pPG9HC-KDEL, pPG9LC, and pPG9LC-KDEL were first transformed into Escherichia coli and then, after sequence confirmation, into the Agrobacterium tumefaciens strain GV3101::pMP90. The constructs used for the expression of 2F5, 2F5-KDEL, and 2G12 have been described in previous studies [16, 17]. All mAbs are human immunoglobulin G (IgG) antibodies of subclass IgG1 with either κ (2F5, 2G12) or λ (PG9) light chains.
2.2 mAb expression in N. benthamiana
N. benthamiana ΔXTFT plants lacking plant-specific α1,3-fucosylation and β1,2-xylosylation were grown at 24°C with a 16-h light:8-h dark photoperiod. Four- to five-week-old plants were used for agroinfiltration experiments as described previously [18]. Briefly, overnight cultures were pelleted and then resuspended in infiltration buffer (25 mM Mes buffer (pH 5.5), 25 mM MgSO4, 0.1 mM acetosyringone) at an OD600 of 0.2 (1.0 OD600 corresponds to 5 × 108 cells/mL). In the case of PG9 expression, equal amounts of the strains carrying the respective heavy and light chain constructs were used. Infiltrated N. benthamiana leaves were harvested after 3 days.
2.3 Preparation of leaf extracts and intercellular fluid
For total leaf extracts, 250 mg fresh material was snap-frozen in liquid nitrogen and then ground in a ball mill (Retsch, Haan, Germany). After addition of 500 μL of extraction buffer (100 mM sodium acetate (pH 5.5), 40 mM ascorbic acid), the samples were incubated for 10 min at 4°C prior to centrifugation (5 min, 14 000g, 4°C) to remove insoluble material. In some experiments, the extraction buffer was supplemented with a proteinase inhibitor cocktail (P9599; Sigma–Aldrich, St. Louis, MO). For the recovery of intercellular fluid, fresh leaves were submerged in extraction buffer prior to vacuum exposure in a desiccator. The leaves were then blotted dry before centrifugation for 15 min at 1000g and 4°C. The recovered solution was concentrated by ultrafiltration. The total protein content of leaf extracts and intercellular fluid was determined with the Bio-Rad Protein Assay kit (Bio-Rad, Hercules, CA), using bovine serum albumin (BSA) as a standard.
2.4 mAb purification
Small-scale antibody purification was performed essentially as described previously [17]. Briefly, frozen leaf material was first crushed in a ball mill as above and then extracted with 2 μL buffer (45 mM tris/HCl (pH 7.4), 1.5 M NaCl, 40 mM ascorbic acid, 1 mM EDTA) per mg of leaf material for 15 min at 0°C. After centrifugation (5 min, 14000g, 4°C), the supernatant was incubated with rProtein A Sepharose 4 Fast Flow (GE Healthcare, Little Chalfont, UK) for 90 min under constant agitation at 4°C. The beads were then collected by centrifugation and washed with phosphate-buffered saline (PBS) prior to elution of bound antibody with sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE) sample buffer.
2.5 mAb treatment with proteinases
Chinese hamster ovary (CHO)-derived mAbs 2F5 or 2G12 were provided by Polymun Scientific GmbH (Klosterneuburg, Austria) and tested for their susceptibility to the following proteinases: recombinant human cathepsin B [19]; recombinant human cathepsin L ([20]; from John S. Mort, Shriners Hospital for Children, Montreal, Canada); cathepsin D (from bovine spleen), chymotrypsin, subtilisin, trypsin (all from Sigma–Aldrich); recombinant human legumain ([21]; from Hans Brandstetter, Univer-sity of Salzburg, Austria). In general, 2F5 or 2G12 (200 μg/mL) were treated with the respective proteinase (10–250 μg/mL) in 100 mM sodium acetate (pH 5.5) at 37°C. mAb digests with cysteine proteinases were performed in the presence of 2 mM cysteine. In the case of cathepsin D, 10 μM E-64 and 1 mM phenylmethylsulfonyl fluoride (PMSF) (both from Sigma–Aldrich) were added to inhibit contaminating cysteine and serine proteinases. Assays with total soluble leaf extracts or intercellular fluid were done in 50 mM sodium acetate (pH 5.5) or 20 mM sodium citrate/40 mM sodium phosphate (pH 4.0–7.0) at 37°C in the absence or presence of selected proteinase inhibitors (all from Sigma–Aldrich). After incubation for up to 16 h, reactions were stopped by addition of SDS–PAGE sample buffer.
2.6 SDS–PAGE and immunoblotting
mAbs and their fragments were fractionated by 12.5% SDS–PAGE. The gels were then either stained with Coomassie Brilliant Blue R-250 or blotted on nitrocellulose membranes (GE Healthcare). After blocking for 1 h in PBS containing 3% BSA, the membranes were incubated with anti-human IgG (γ-chain-specific; Sigma–Aldrich) or anti-human IgG (CH + CL-specific; Promega, Madison, WI) conjugated to horseradish peroxidase (HRP) at a concentration of 0.2–0.5 μg/mL PBS containing 0.05% Tween 20 (PBST) and 0.5% BSA for 90 min prior to development using chemiluminescence reagents (Bio-Rad). Monoclonal anti-KDEL antibodies (Merck Millipore, Darmstadt, Germany) were used at a concentration of 0.2 μg/mL in PBST containing 3% BSA and detected with anti-mouse IgG-HRP (Jackson ImmunoResearch, West Grove, PA) as outlined above.
2.7 Cleavage site analysis
N-terminal sequence analysis of bands blotted on polyvinylidene difluoride membranes (Bio-Rad) as described previously [22] was performed by Edman degradation on an Applied Biosystems Procise 492 protein sequencer (Protein Micro-Analysis Facility, Medical University of Innsbruck, Austria). The N-termini of mAb degradation products were also characterized by liquid chromatography (LC)-electrospray ionization–MS/MS in a similar way as reported earlier [23]. Briefly, mAbs and their fragments were separated by SDS–PAGE. The heavy-chain band degradation products were excised, S-alkylated and subjected to tryptic or chymotryptic digestion. Peptides were eluted from the gel slices with 50% acetonitrile, concentrated in vacuo and then separated by nano-LC (150 mm × 0.32 mm BioBasic-18; Thermo Scientific, Waltham, MA) with a gradient of 1–80% acetonitrile. Data mining was conducted in positive ion mode on a maXis-4GQ-TOF mass spectrometer (Bruker, Billerica, MA). MS2 scans of dominant precursor peaks were acquired and manually analyzed with DataAnalysis software version 4.0 (Bruker).
3 Results
3.1 Expression of mAbs 2F5, 2G12, and PG9 in N. benthamiana
The human anti-HIV antibody 2G12 could be produced in N. benthamiana without observing substantial amounts of degradation products as contaminants (Fig. 1A). These results are in agreement with our previous studies on this mAb [18], although N-terminally truncated heavy-chain fragments have been detected in 2G12 preparations derived from other plant species [17, 24, 25]. However, the production of the anti-HIV mAbs 2F5 and PG9 in N. benthamiana was associated with more pronounced proteolysis. In either case, a 40-kDa fragment was found to co-purify with intact heavy and light chains, which was less abundant in 2G12 samples. In sporadic cases of reduced light-chain synthesis, 2F5 preparations contained a major 30-kDa polypeptide (Fig. 1A, lane 1). The 40- and 30-kDa degradation products were both recognized by antibodies to the heavy chain, indicating that they represent truncated forms of this mAb component (Fig. 1A). In contrast, degradation products derived from the light chain were not observed. Notably, supplementation of the extraction buffer with a proteinase inhibitor cocktail did not prevent proteolysis (Fig. 1B). However, 2F5 and PG9 degradation was reduced upon addition of the endoplasmic reticulum (ER)-retrieval sequence KDEL to the C-termini of their heavy and light chains (Fig. 1A). It has been reported previously that the stability of antibodies in planta can be increased by retention of the proteins in this compartment [16, 26].
Figure 1.
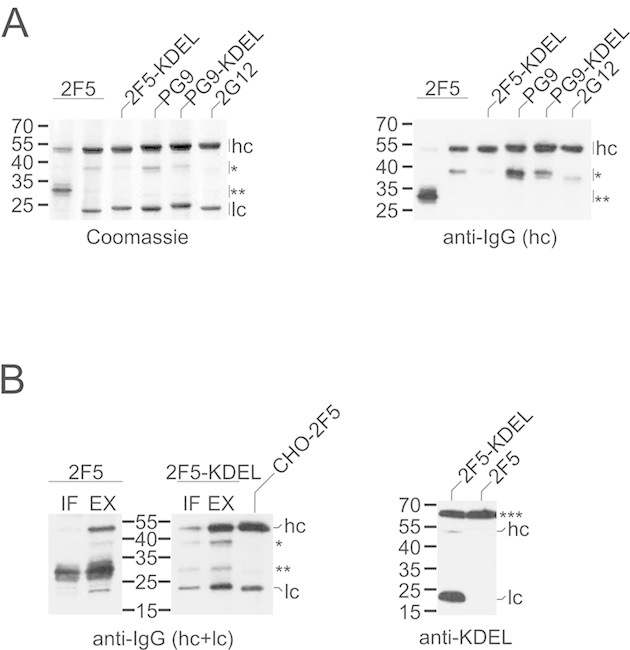
Expression of 2F5, 2G12, and PG9 in N. benthamiana. (A) mAbs were affinity-purified from total soluble leaf extracts (30 mg leaf wet weight) and then analyzed by SDS–PAGE/Coomassie blue staining (left panel) or immunoblotting with antibodies to the heavy chain of human IgG (right panel). (B) Left panel: mAbs affinity-purified from intercellular fluid (IF; 100 mg leaf wet weight) or total soluble leaf extracts prepared in the presence of proteinase inhibitors (EX; 50 mg leaf wet weight) were analyzed by immunoblotting with antibodies to the heavy and light chains of human IgG. CHO-derived 2F5 (100 ng) was loaded as control. Right panel: total soluble extracts (2 μg protein) of leaves infiltrated with different 2F5 constructs were analyzed by immunoblotting with anti-KDEL antibodies (right panel). hc, heavy chain; lc, light chain; *40-kDa fragment; **30-kDa degradation product. Endogenous N. benthamiana BiP (***) served as internal control in the case of KDEL detection. The migration positions of selected molecular mass standards are indicated, with their respective masses expressed in kDa. The results shown are representative of at least two independent experiments.
To characterize the degradation products in more detail, the N-terminal sequences of the 30- and 40-kDa fragments were determined by Edman degradation or mass spectrometry (MS). The 2F5 30-kDa fragment was found to be the result of a large N-terminal truncation since its N-terminus was identified as K247 (75%) or K248 (25%). This revealed that the main cleavage site (KVD246↓K247KVEP) was a few residues upstream of the hinge region connecting the CH1 and CH2 domains (see Table S1 of Supporting information for mAb heavy and light chain sequences). In the case of the 40-kDa fragment, cleavage occurred within the complementarity-determining region (CDR) H3 loop of the VH domain, yielding predominantly G108 as N-terminal amino acid (TLF107↓G108VPIA). However, it should be noted that molecules starting with the next residue (V109) were also found. The PG9 heavy chain was mainly cleaved within its CDR H3 loop (NYY111↓D112FYDG or YYD112↓F113YDGY), whereas the cleavage site identified within 2G12 was C-terminal to this structural element (GPG116↓T117VVTV).
To test whether the apoplast contains mAb degradation products, intercellular fluid was isolated from leaves infiltrated with 2F5 or 2F5-KDEL constructs. Comparison with total leaf extracts revealed that the fractions of the heavy-chain fragments and full-length mAb molecules accumulating in this compartment were similar (Fig. 1B). Interestingly, secretion of 2F5-KDEL was at best marginally lower than that of its counterpart lacking an ER retrieval sequence. Hence, we tested the heavy and light chains of 2F5-KDEL for the presence of the tag by means of immunoblotting with anti-KDEL antibodies. Although a specific signal could be obtained for both polypeptides, the heavy chain reacted much weaker than the light chain. This indicates a much higher proteolytic vulnerability of the targeting sequence when attached to the C-terminus of the heavy chain and suggests that this selective loss of the ER retrieval motif probably accounts for the inefficient intracellular retention of 2F5-KDEL (Fig. 1B).
3.2 Differential sensitivity of 2F5 and 2G12 to N. benthamiana proteinases
To test for the presence of mAb-degrading activities in N. benthamiana tissues, CHO-derived 2F5 and 2G12 were incubated in vitro with total soluble leaf extracts or intercellular fluid prepared from non-infiltrated plants. In the case of 2F5, a 40-kDa heavy-chain fragment was rapidly generated, in particular upon treatment with intercellular fluid (Fig. 2). In some instances, further processing into a 30-kDa fragment was also detected (data not shown). In contrast, 2G12 proved largely resistant to leaf proteinases under the same experimental conditions (Fig. 2). Similar results were obtained with intercellular fluid prepared from leaves infiltrated with A. tumefaciens carrying an expression vector for an unrelated recombinant protein (human transferrin) or the parental bacterial strain, indicating that the observed proteolysis is due to constitutively expressed plant proteinases (Supporting information, Fig. S1). The 2F5-hydrolyzing activity present in intercellular fluid was further characterized with respect to its pH profile and cleavage site. 2F5 heavy-chain processing into the 40-kDa form was strongest at pH 5.5–6.0, which conforms to the apoplastic pH in situ (Fig. 3A; Supporting information, Fig. S2A). To determine its N-terminal sequence, the 40-kDa degradation product was affinity-purified and then subjected to MS analysis. The most N-terminal peptide found was half-tryptic, commencing with residue S132. Hence, cleavage occurred within the linker segment connecting the VH and CH1 domains (TIS131↓S132TSTK) and not in the CDR H3 loop as observed in planta. We also tested the effect of various synthetic proteinase inhibitors on 2F5 processing by N. benthamiana proteinases in vitro. 2F5 proteolysis by total soluble leaf extracts and intercellular fluid could be inhibited by addition of PMSF, a serine proteinase inhibitor that also displays some reactivity with cysteine proteinases. In contrast, inhibitors of aspartic and metalloproteinases (pepstatin, phenanthroline) as well as trypsin- and papain-like enzymes (leupeptin, E-64) did not exert detectable effects on 2F5 fragmentation. Interestingly, addition of a commercially available proteinase inhibitor cocktail designed for prevention of unwanted proteolysis in plant extracts proved less effective than treatment with PMSF. In this context, it should be pointed out that this proteinase inhibitor cocktail does not contain PMSF (Fig. 3B; Supporting information, Fig. S2B).
Figure 2.
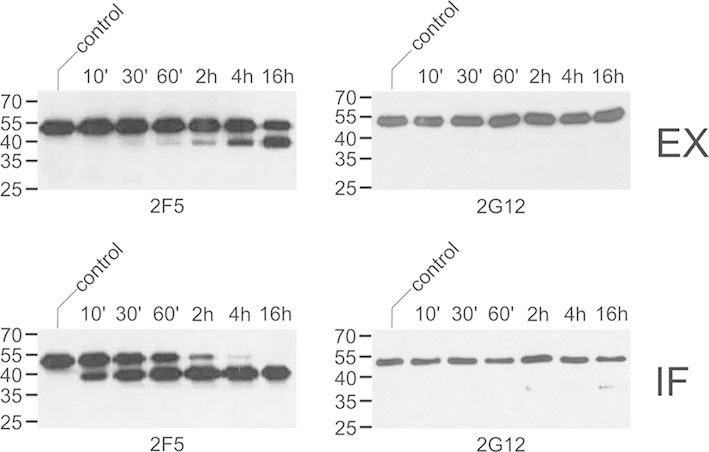
Processing of 2F5 and 2G12 by N. benthamiana proteinases in vitro. CHO-derived 2F5 or 2G12 (400 ng) was incubated with intercellular fluid (IF; 650 ng protein) or total soluble leaf extracts (EX; 600 ng protein) for the indicated times and then analyzed by immunoblotting with antibodies to the heavy chain of human IgG. Untreated antibody was loaded as control. The migration positions of selected molecular mass standards are indicated, with their respective masses expressed in kDa. The results shown are representative of at least three independent experiments.
Figure 3.
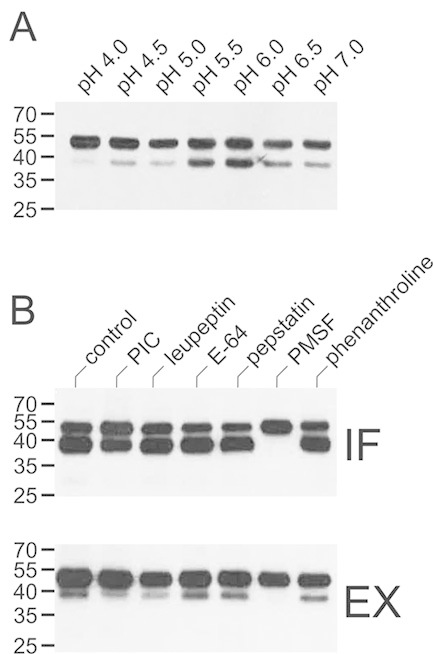
Effect of pH and proteinase inhibitors on in vitro processing of 2F5. (A) CHO-derived 2F5 (200 ng) was incubated with intercellular fluid (130 ng protein) for 16 h at the indicated pH and then analyzed by immunoblotting with antibodies to the heavy chain of human IgG. (B) CHO-derived 2F5 (200 ng) was incubated with intercellular fluid (IF; 650 ng protein) or total soluble leaf extracts (EX; 200 ng protein) for 16 h in the absence (control) or presence of the indicated proteinase inhibitors (10 μM: leupeptin, E-64, pepstatin; 2 mM: PMSF, phenanthroline) and then analyzed as above. PIC, proteinase inhibitor cocktail. The migration positions of selected molecular mass standards are indicated, with their respective masses expressed in kDa. The results shown are representative of at least two independent experiments.
3.3 Susceptibility of 2F5 and 2G12 to representative proteinases
To get a better picture of which type(s) of proteinases could be involved in mAb degradation in planta, CHO-derived 2F5 and 2G12 were treated with representatives of different classes of proteolytic enzymes under conditions mimicking the milieu in the apoplast (pH 5.5). The cysteine proteinase papain is frequently used to prepare antibody fragments [27]. However, commercial papain can be contaminated with other papain-like cysteine proteinases displaying different specificities [28]. We therefore decided to test purified recombinant versions of two human papain-like cysteine proteinases, cathepsin B, and cathepsin L, for their ability to digest 2F5. Cathepsin L quickly converted the 2F5 heavy-chain into a 40-kDa fragment, which was further processed into a 30-kDa polypeptide upon prolonged incubation. Cathepsin B was much slower in generating the 40-kDa degradation product while formation of the 30-kDa form was not detected at all (Fig. 4). Analysis of either 40-kDa fragment by Edman degradation led to the identification of V109 as N-terminal amino acid, indicating that both proteinases cleave at the same position within the CDR H3 loop of the antibody. Interestingly, this cleavage site (LFG108↓V109PIAR) is also found in planta (see Section 3.1). The N-terminus of the 30-kDa fragment produced by cathepsin L was determined by MS as T240, located within the hinge region connecting the CH1 and CH2 domains. This cleavage site (KTH239↓T240CPPC) is identical to that previously described for papain [29].
Figure 4.
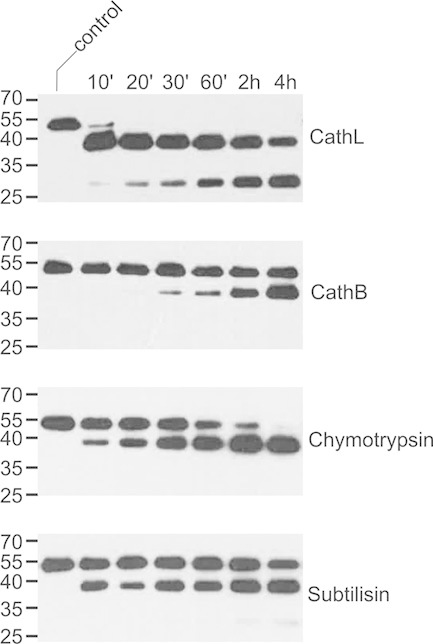
Processing of 2F5 by serine and cysteine proteinases. CHO-derived 2F5 (200 ng) was incubated with selected proteinases (10 ng) at pH 5.5 for the indicated times and then analyzed by immunoblotting with antibodies to the heavy chain of human IgG. Untreated antibody was used as control. CathB, cathepsin B; CathL, cathepsin L. The migration positions of selected molecular mass standards are indicated, with their respective masses expressed in kDa. The results shown are representative of at least two independent experiments.
Incubation of 2F5 with the serine proteinases chymotrypsin and subtilisin also led to rapid appearance of a 40-kDa fragment. Prolonged treatment with subtilisin yielded small amounts of a 30-kDa processing product, while this was not observed for chymotrypsin (Fig. 4). The N-terminus of the 40-kDa fragment produced by subtilisin could not be identified unambiguously due to heterogeneity. However, the N-terminal sequence of the 40-kDa polypeptide generated by digestion with chymotrypsin was found to correspond to G108VPIA, identical to the main cleavage site within the CDR H3 loop of 2F5 observed in planta.
In contrast to 2F5, 2G12 was hardly sensitive to treatment with cathepsin B, chymotrypsin, and subtilisin. The latter antibody also proved far more resistant to digestion with cathepsin L. However, the sequential appearance of the 40- and 30-kDa fragments still occurred, albeit much delayed as compared to 2F5 (Fig. 5).
Figure 5.
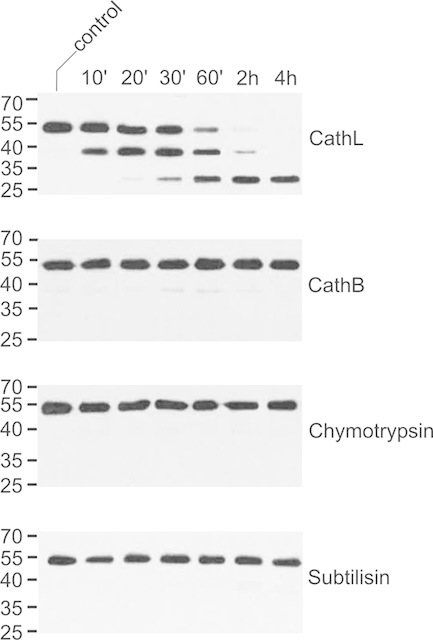
Processing of 2G12 by serine and cysteine proteinases. CHO-derived 2G12 (200 ng) was incubated with selected proteinases (10 ng) at pH 5.5 for the indicated times and then analyzed by immunoblotting with antibodies to the heavy chain of human IgG. Untreated antibody was used as control. CathB, cathepsin B; CathL, cathepsin L. The migration positions of selected molecular mass standards are indicated, with their respective masses expressed in kDa. The results shown are representative of at least two independent experiments.
A number of enzymes was found to display no or very little activity toward 2F5 when tested under apoplastic conditions. This includes the serine proteinase trypsin, the aspartic proteinase cathepsin D and the cysteine proteinase legumain (Supporting information, Fig. S3A). The same was observed when cathepsin D and legumain were used at pH 4.5, which is more favorable for their enzymatic activities (data not shown). However, prolonged incubation of 2F5 with high amounts of legumain resulted in the formation of a 30-kDa degradation product (Supporting information, Fig. S3B). The N-terminus of this fragment was identified by Edman degradation as K237. Interestingly, the same cleavage site (SCD236↓K237THTC) was observed in the case of a mAb fragment isolated from tobacco leaves [12].
4 Discussion
Fragments of mAbs heterologously expressed in tobacco (Nicotiana tabacum) or N. benthamiana have been observed in a number of previous studies [11, 25, 30–32]. However, the molecular characterization of the cleavage products has been superficial so far [12, 33]. Although it is possible that some degradation products might have escaped detection in our studies, the presented data indicate that proteolytic fragmentation of 2F5 and PG9 occurs mainly by cleavage in the CDR H3 loop of the antibodies. This coincides with the site of 2G12 proteolysis when produced in transgenic maize, although the exact processing sites could not be identified [24]. Different findings have been reported for two other mAbs. In the case of the tumor-targeting antibody H10, MS-based peptide fingerprinting indicated that proteolysis took place in the CH1–CH2 hinge region [33]. The same was observed for the IgM-neutralizing antibody LO-BM2 expressed in tobacco BY-2 cells [12]. Interestingly, such a 30-kDa degradation product was observed for 2F5, but not for PG9. This could be related to different efficiencies in heavy-light chain assembly, which could possibly expose the hinge region to proteolytic attack. Such a scenario would be in agreement with our observation that formation of the 30-kDa 2F5 fragment was most prominent when light-chain synthesis was reduced.
The presented in vitro experiments were all performed with CHO-derived mAbs as substrates, which differ from their plant-made counterparts with respect to their N-glycan structures. Since the single N-glycan attachment site is located in the Fc part and thus sterically remote from the cleavage regions, it is unlikely that differences in N-glycosylation would affect the proteinase sensitivity of the antibodies. This notion is supported by the observation that the proteolytic susceptibility of 2F5 produced in N. benthamiana and CHO cells is comparable. Furthermore, 2G12 isolated from N. benthamiana leaves was equally resistant to apoplastic proteinases as its CHO-derived counterpart (data not shown).
Although the antigen-binding sites of 2F5 and 2G12 have been grafted onto the same IgG1κ framework, the two antibodies differ markedly in their sensitivity to proteolysis in planta and in vitro. It is likely that this is at least partially due to the unusual architecture of 2G12, which features a swap between the variable domains of its two heavy-chain molecules, resulting in the formation of a third antigen-binding site at the newly created interface [34]. Thus, the domain-exchange possibly restricts access to the CDR H3 loop, the primary proteinase-sensitive region in the far more susceptible conventional mAbs 2F5 and PG9. Furthermore, 2G12 contains a particularly adaptable linker region between the VH and CH1 domains, which adopts a buried conformation as compared to a 2G12 variant with a canonical tertiary structure [35]. Hence, the relative inaccessibility of the CDR H3 loop and the VH–CH1 linker segment could account for the high resistance of 2G12 to proteolytic enzymes.
It should be also noted that 2F5 and PG9 are mAbs with very long CDR H3 loops. PG9 has a 28-residue CDR H3, which equals the longest in human antibody sequences known to date [36]. This structural element is only slightly shorter in the case of 2F5 (22 residues). In contrast, the CDR H3 loop of 2G12 is more compact with a length of 14 amino acids. Hence, the length of the CDR H3 loop correlates well with the sensitivity of these mAbs to proteolytic degradation in N. benthamiana. This notion is further supported by the positions of the primary cleavage sites with the CDR H3 loops of 2F5 and PG9, which are invariably located close to the loop tip and therefore sterically exposed.
It has been recognized already some time ago that proteolytic degradation in planta poses a serious constraint for the production of mAbs in tobacco and other Nicotiana species, with this undesired turnover limiting both yield and quality of the protein of interest [37]. Previous studies have consistently reported acidic pH optima for mAb-hydrolyzing activities in tissue extracts [10, 37]. This argues in favor of the involvement of proteolytic enzyme(s) accumulating in the intercellular fluid, in line with the apoplast being a proteinase-rich environment [5]. Support for this notion is also derived from the finding that fragments of the mAb Guy's 13 expressed in tobacco largely accumulate in the intercellular fluid [10]. However, the issue of the identity of the Nicotiana proteinases accounting for mAb degradation is still unresolved. Interestingly, co-expression of tomato cystatin 9, an inhibitor of papain- and legumain-like cysteine proteinases, was reported to stabilize the murine mAb C5-1 in N. benthamiana. The same was observed for tomato cathepsin D inhibitor, which also targets serine proteinases [11]. These results are in good agreement with our findings that serine and cysteine proteinases are largely responsible for mAb degradation in N. benthamiana, thus providing important clues for future studies aiming at the identification of the detrimental host proteinases.
Acknowledgments
We gratefully acknowledge Jakub Jez and Johannes Stadlmann for excellent technical support. We also thank Hans Brandstetter, Viktor Klimyuk, John S. Mort, and Polymun Scientific GmbH for reagents. The amaZon speed ion trap and the maXis-4GQ-TOF mass spectrometer were provided by EQ-BOKU-VIBT-GmbH. The work was supported by the Laura Bassi Centres of Expertise (Grant No. 822757) and the Austrian Science Fund (Grant No. W1224-B09).
The authors declare no financial or commercial conflict of interest.
Glossary
Abbreviations
- BSA
bovine serum albumin
- CDR
complementarity-determining region
- CHO
Chinese hamster ovary
- ER
endoplasmic reticulum
- IgG
immunoglobulin G
- LC
liquid chromatography
- mAb
monoclonal antibody
- MS
mass spectrometry
- PAGE
polyacrylamide gel electrophoresis
- PBS
phosphate-buffered saline
- PMSF
phenylmethylsulfonyl fluoride
- SDS
sodium dodecyl sulfate
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
suppinfo
References
- 1.Melnik S, Stöger E. Green factories for biopharmaceuticals. Curr. Med. Chem. 2013;20:1038–1046. [PubMed] [Google Scholar]
- 2.de Muynck B, Navarre C, Boutry M. Production of antibodies in plants: Status after twenty years. Plant Biotechnol. J. 2010;8:529–563. doi: 10.1111/j.1467-7652.2009.00494.x. [DOI] [PubMed] [Google Scholar]
- 3.Marillonnet S, Thoeringer C, Kandzia R, Klimyuk V, et al. Systemic Agrobacterium tumefaciens-mediated transfection of viral replicons for efficient transient expression in plants. Nat. Biotechnol. 2005;23:718–723. doi: 10.1038/nbt1094. [DOI] [PubMed] [Google Scholar]
- 4.Bendandi M, Marillonnet S, Kandzia R, Thieme F, et al. Rapid, high-yield production in plants of individualized idiotype vaccines for non-Hodgkin's lymphoma. Ann. Oncol. 2010;21:2420–2427. doi: 10.1093/annonc/mdq256. [DOI] [PubMed] [Google Scholar]
- 5.Doran PM. Foreign protein degradation and instability in plants and plant tissue cultures. Trends Biotechnol. 2006;24:426–432. doi: 10.1016/j.tibtech.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 6.Benchabane M, Goulet C, Rivard D, Faye L, et al. Preventing unintended proteolysis in plant protein biofactories. Plant Biotechnol. J. 2008;6:633–648. doi: 10.1111/j.1467-7652.2008.00344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Komarnytsky S, Borisjuk N, Yakoby N, Garvey A, et al. Cosecretion of protease inhibitor stabilizes antibodies produced by plant roots. Plant Physiol. 2006;141:1185–1193. doi: 10.1104/pp.105.074419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rivard D, Anguenot R, Brunelle F, Le VQ, et al. An in-built proteinase inhibitor system for the protection of recombinant proteins recovered from transgenic plants. Plant Biotechnol. J. 2006;4:359–368. doi: 10.1111/j.1467-7652.2006.00187.x. [DOI] [PubMed] [Google Scholar]
- 9.Kim N-S, Kim T-G, Kim O-H, Ko E-M, et al. Improvement of recombinant hGM-CSF production by suppression of cysteine proteinase gene expression using RNA interference in a transgenic rice culture. Plant Mol. Biol. 2008;68:263–275. doi: 10.1007/s11103-008-9367-8. [DOI] [PubMed] [Google Scholar]
- 10.Delannoy M, Alves G, Vertommen D, Ma J, et al. Identification of peptidases in Nicotiana tabacum leaf intercellular fluid. Proteomics. 2008;8:2285–2298. doi: 10.1002/pmic.200700507. [DOI] [PubMed] [Google Scholar]
- 11.Goulet C, Khalf M, Sainsbury F, D'Aoust M-A, et al. A protease activity-depleted environment for heterologous proteins migrating towards the leaf cell apoplast. Plant Biotechnol. J. 2012;10:83–94. doi: 10.1111/j.1467-7652.2011.00643.x. [DOI] [PubMed] [Google Scholar]
- 12.de Muynck B, Navarre C, Nizet Y, Stadlmann J, et al. Different subcellular localization and glycosylation for a functional antibody expressed in Nicotiana tabacum plants and suspension cells. Transgenic Res. 2009;18:467–482. doi: 10.1007/s11248-008-9240-1. [DOI] [PubMed] [Google Scholar]
- 13.Trkola A, Purtscher M, Muster T, Ballaun C, et al. Human monoclonal antibody 2G12 defines a distinctive neutralization epitope on the gp120 glycoprotein of human immunodeficiency virus type 1. J. Virol. 1996;70:1100–1108. doi: 10.1128/jvi.70.2.1100-1108.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kunert R, Rüker F, Katinger H. Molecular characterization of five neutralizing anti-HIV type 1 antibodies: Identification of nonconventional D segments in the human monoclonal antibodies 2G12 and 2F5. AIDS Res. Hum. Retroviruses. 1998;14:1115–1128. doi: 10.1089/aid.1998.14.1115. [DOI] [PubMed] [Google Scholar]
- 15.Walker LM, Phogat SK, Chan-Hui PY, Wagner D, et al. Broad and potent neutralizing antibodies from an African donor reveal a new HIV-1 vaccine target. Science. 2009;326:285–289. doi: 10.1126/science.1178746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sack M, Paetz A, Kunert R, Bomble M, et al. Functional analysis of the broadly neutralizing human anti-HIV-1 antibody 2F5 produced in transgenic BY-2 suspension cultures. FASEB J. 2007;21:1655–1664. doi: 10.1096/fj.06-5863com. [DOI] [PubMed] [Google Scholar]
- 17.Schähs M, Strasser R, Stadlmann J, Kunert R, et al. Production of a monoclonal antibody in plants with a humanized N-glycosylation pattern. Plant Biotechnol. J. 2007;5:657–663. doi: 10.1111/j.1467-7652.2007.00273.x. [DOI] [PubMed] [Google Scholar]
- 18.Strasser R, Stadlmann J, Schähs M, Stiegler G, et al. Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant Biotechnol. J. 2008;6:392–402. doi: 10.1111/j.1467-7652.2008.00330.x. [DOI] [PubMed] [Google Scholar]
- 19.Mach L, Mort JS, Glössl J. Maturation of human procathepsin B. Proenzyme activation and proteolytic processing of the precursor to the mature proteinase, in vitro, are primarily unimolecular processes. J. Biol. Chem. 1994;269:13030–13035. [PubMed] [Google Scholar]
- 20.Adams-Cioaba MA, Krupa JC, Xu C, Mort JS, et al. Structural basis for the recognition and cleavage of histone H3 by cathepsin L. Nat. Commun. 2011;2:197. doi: 10.1038/ncomms1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dall E, Brandstetter H. Mechanistic and structural studies on legumain explain its zymogenicity, distinct activation pathways, and regulation. Proc. Natl. Acad. Sci. USA. 2013;110:10940–10945. doi: 10.1073/pnas.1300686110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowan AD, Mason P, Mach L, Mort JS. Rat procathepsin B. Proteolytic processing to the mature form in vitro. J. Biol. Chem. 1992;267:15993–15999. [PubMed] [Google Scholar]
- 23.Pabst M, Chang M, Stadlmann J, Altmann F. Glycan profiles of the 27 N-glycosylation sites of the HIV envelope protein CN54gp140. Biol. Chem. 2012;393:719–730. doi: 10.1515/hsz-2012-0148. [DOI] [PubMed] [Google Scholar]
- 24.Ramessar K, Rademacher T, Sack M, Stadlmann J, et al. Cost-effective production of a vaginal protein microbicide to prevent HIV transmission. Proc. Natl. Acad. Sci. USA. 2008;105:3727–3732. doi: 10.1073/pnas.0708841104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hehle VK, Paul MJ, Drake PM, Ma JK-C, et al. Antibody degradation in tobacco plants: A predominantly apoplastic process. BMC Biotechnol. 2011;11:128. doi: 10.1186/1472-6750-11-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petruccelli S, Otegui MS, Lareu F, Tran Dinh O, et al. A KDEL-tagged monoclonal antibody is efficiently retained in the endoplasmic reticulum in leaves, but is both partially secreted and sorted to protein storage vacuoles in seeds. Plant Biotechnol. J. 2006;4:511–527. doi: 10.1111/j.1467-7652.2006.00200.x. [DOI] [PubMed] [Google Scholar]
- 27.Adamczyk M, Gebler JC, Wu J. Papain digestion of different mouse IgG subclasses as studied by electrospray mass spectrometry. J. Immunol. Methods. 2000;237:95–104. doi: 10.1016/s0022-1759(00)00135-6. [DOI] [PubMed] [Google Scholar]
- 28.Buttle DJ, Ritonja A, Dando PM, Abrahamson M, et al. Interactions of papaya proteinase IV with inhibitors. FEBS Lett. 1990;262:58–60. doi: 10.1016/0014-5793(90)80153-a. [DOI] [PubMed] [Google Scholar]
- 29.Yan B, Valliere-Douglass J, Brady L, Steen S, et al. Analysis of post-translational modifications in recombinant monoclonal antibody IgG1 by reversed-phase liquid chromatography/mass spectrometry. J. Chromatogr. A. 2007;1164:153–161. doi: 10.1016/j.chroma.2007.06.063. [DOI] [PubMed] [Google Scholar]
- 30.Grohs BM, Niu Y, Veldhuis LJ, Trabelsi S, et al. Plant-produced trastuzumab inhibits the growth of HER2 positive cancer cells. J. Agric. Food Chem. 2010;58:10056–10063. doi: 10.1021/jf102284f. [DOI] [PubMed] [Google Scholar]
- 31.Lombardi R, Villani ME, Di Carli M, Brunetti P, et al. Optimisation of the purification process of a tumour-targeting antibody produced in N. benthamiana using vacuum-agroinfiltration. Transgenic Res. 2010;19:1083–1097. doi: 10.1007/s11248-010-9382-9. [DOI] [PubMed] [Google Scholar]
- 32.Lombardi R, Donini M, Villani ME, Brunetti P, et al. Production of different glycosylation variants of the tumour-targeting mAb H10 in N. benthamiana: Influence on expression yield and antibody degradation. Transgenic Res. 2012;21:1005–1021. doi: 10.1007/s11248-012-9587-1. [DOI] [PubMed] [Google Scholar]
- 33.Villani ME, Morgun B, Brunetti P, Marusic C, et al. Plant pharming of a full-sized, tumour-targeting antibody using different expression strategies. Plant Biotechnol. J. 2009;7:59–72. doi: 10.1111/j.1467-7652.2008.00371.x. [DOI] [PubMed] [Google Scholar]
- 34.Calarese DA, Scanlan CN, Zwick MB, Deechongkit S, et al. Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science. 2003;300:2065–2071. doi: 10.1126/science.1083182. [DOI] [PubMed] [Google Scholar]
- 35.Doores KJ, Fulton Z, Huber M, Wilson IA, et al. Antibody 2G12 recognizes di-mannose equivalently in domain- and nondomain-exchanged forms but only binds the HIV-1 glycan shield if domain exchanged. J. Virol. 2010;84:10690–10699. doi: 10.1128/JVI.01110-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pejchal R, Walker LM, Stanfield RL, Phogat SK, et al. Structure and function of broadly reactive antibody PG16 reveal an H3 subdomain that mediates potent neutralization of HIV-1. Proc. Natl. Acad. Sci. USA. 2010;107:11483–11488. doi: 10.1073/pnas.1004600107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens LH, Stoopen GM, Elbers IJW, Molthoff JW, et al. Effect of climate conditions and plant developmental stage on the stability of antibodies expressed in transgenic tobacco. Plant Physiol. 2000;124:173–182. doi: 10.1104/pp.124.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
suppinfo


