Abstract
Chlorite is a serious environmental concern, as rising concentrations of this harmful anthropogenic compound have been detected in groundwater, drinking water, and soil. Chlorite dismutases (Clds) are therefore important molecules in bioremediation as Clds catalyze the degradation of chlorite to chloride and molecular oxygen. Clds are heme b-containing oxidoreductases present in numerous bacterial and archaeal phyla. This review presents the phylogeny of functional Clds and Cld-like proteins, and demonstrates the close relationship of this novel enzyme family to the recently discovered dye-decolorizing peroxidases. The available X-ray structures, biophysical and enzymatic properties, as well as a proposed reaction mechanism, are presented and critically discussed. Open questions about structure-function relationships are addressed, including the nature of the catalytically relevant redox and reaction intermediates and the mechanism of inactivation of Clds during turnover. Based on analysis of currently available data, chlorite dismutase from “Candidatus Nitrospira defluvii” is suggested as a model Cld for future application in biotechnology and bioremediation. Additionally, Clds can be used in various applications as local generators of molecular oxygen, a reactivity already exploited by microbes that must perform aerobic metabolic pathways in the absence of molecular oxygen. For biotechnologists in the field of chemical engineering and bioremediation, this review provides the biochemical and biophysical background of the Cld enzyme family as well as critically assesses Cld's technological potential.
Keywords: Bioremediation, Chlorite degradation, Chlorite dismutase, Enzymology, Metalloprotein
1 Introduction
In 1996 van Ginkel and co-workers [1] discovered, in perchlorate-reducing bacteria (PCRB), the heme b-containing oxidoreductase chlorite dismutase (Cld), which catalyzes the decomposition of chlorite (ClO2–) into chloride (Cl–) and molecular oxygen (O2). During turnover a covalent oxygen-oxygen bond is formed, an uncommon biochemical reaction that is also catalyzed by the water-splitting manganese complex of photosystem II of oxygenic organisms (cyanobacteria and plants) and an enzyme of an anaerobic methane-oxidizing bacterium [2].
Perchlorate-reducing bacteria are facultative anaerobes that can utilize perchlorate (ClO4–) and chlorate (ClO3–) as terminal electron acceptors [3] in the absence of oxygen. In this way they intracellularly produce the strong oxidant chlorite [E°' (ClO2–/ClO–) = 1.175 V] [4], which exhibits strong cell-damaging effects [5]. The high two-electron reduction potentials of perchlorate [E° (ClO4–/ClO3–) = 0.79 V] and chlorate [E°' (ClO3–/ClO2–) = 0.71 V] make them ideal electron acceptors for microbial respiratory electron chains [6]. The transiently formed chlorite is degraded to harmless chloride and O2 by chlorite dismutases. As will be discussed in section 4, the denomination “dismutase” is chemically incorrect and should be eliminated in future terminology.
The first biophysical and biochemical studies of Clds were performed on the perchlorate-reducing bacteria Azospira oryzae (GR-1) [1, 4], Ideonella dechloratans [7], Dechloromonas aromatica [8], and Pseudomonas chloritidismutans [9]. Additionally, homologous enzymes with chlorite dismutase activity have been found and characterized in the nitrite-oxidizing bacteria Candidatus Nitrospira defluvii [10] and Nitrobacter winogradskyi [11]. Further phylogenetic analysis showed that cld genes are present in numerous bacterial and archaeal phyla indicating that they represent ancient sequences [12]. The question regarding the natural substrate for Clds and Cld-like proteins, as well as their physiological role(s), remains unanswered, since (i) except in PCRBs chlorite is not a metabolic intermediate in prokaryotes, (ii) reservoirs of chlorite on Earth are very rare [13], and (iii) most chlorite present in our environment is of anthropogenic origin [6, 14].
Structurally, Clds and Cld-like proteins form a superfamily together with recently discovered dye-decolorizing peroxidases (DyPs) suggesting common phylogenetic roots (see section 2) [15]. DyPs are heme b-containing peroxidases with histidine as the proximal ligand but secondary and tertiary structures, as well as heme cavity architecture, show no homology to the two main heme peroxidase superfamilies, i.e. the peroxidase-catalase and the peroxidase-cyclooxygenase superfamilies [16, 17]. DyPs exhibit a very broad substrate range. Originally, they were found to degrade anthraquinone derivatives, such as Reactive Blue 5, a synthetic dye, as well as relatively bulky compounds used in the textile industry [18]. From these early observations the denomination dye-decolorizing peroxidase was derived. Later on, DyPs were found also to be able to metabolize artificial electron donors such as ABTS [19], non-phenolic lignin model compounds [20], azo-dyes [21], aromatic sulfides [22], and manganese [23]. It was also reported that DyPs efficiently degrade β-carotene [24]. DyPs are found in a variety of prokaryotes and – in contrast to Clds – also in eukaryotes (mainly fungi). DyPs cluster into four subfamilies [25].
In recent years, insightful findings have been published regarding both chlorite dismutases and dye-decolorizing peroxidases including: protocols of recombinant production; mutational analyses; three-dimensional structures; and biochemical and biophysical properties. This information will enable rational engineering strategies to be devised in the near future in order to apply these novel iron enzymes in chemical and biotechnological processes as well as in bioremediation. This review focuses on chlorite dismutases and discusses – where necessary – relationships with DyPs. We present an updated phylogenetic tree of Clds and DyPs. We analyze, compare, and critically discuss all available biophysical and biochemical data. Based on this information, the biotechnological potential of these oxidoreductases as well as engineering strategies will be discussed.
2 Phylogeny of chlorite dismutases and dye-decolorizing peroxidases
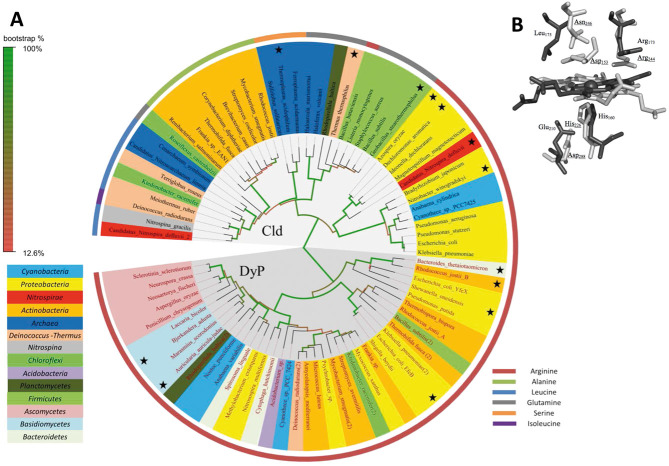
In 2010, Kostan and co-workers presented a Maximum Likelihood tree of Clds and Cld-like proteins that demonstrated a wide distribution of the cld gene across Bacteria and Archaea [12]. It showed that proteins from the same phylogenetic lineage (phylum) – based on 16S rRNA-phylogeny – group together to a high degree and that several lateral gene transfer events occurred during evolution, reflecting functional diversification. We have updated this analysis using a selection of 120 Clds and Cld-like proteins as well as 86 DyP sequences (July 2013) collected from public databases (Uniprot, NCBI). After separate multiple sequence alignments [26] and reconstruction of phylogenetic trees for Cld/Cld-like proteins and DyPs, a common phylogenetic Maximum Likelihood tree was constructed (Fig. 1A) [27].
Figure 1.
Phylogeny and active site architecture of clorite dismutase (Cld) and dye-decolorizing peroxidase (DyP). (A) Maximum likelihood tree based on amino acid sequences of Clds, Cld-like proteins and DyPs. Sequences marked with an asterisk represent proteins of known X-ray structure. Colors highlighting the species name indicate the affiliations of the respective organisms to bacterial and archaeal phylae based on 16S rRNA phylogeny. Color of the stripe, bordering the figure, define the residues at the position which is homologous to Arg173 of NdCld, in the respective proteins. This residue is fully conserved in functional (i.e. chlorite degrading) Clds. The circular tree was drawn using FigTree v1.4 (http://tree.bio.ed.ac.uk/software/figtree/). (B) Overlay of active site of Cld and DyP. Cld from “Candidatus Nitrospira defluvii” (NdCld, PDB: 3NN1) is depicted in dark gray, and DypB from Rhodococcus jostii (PDB: 3QNS) in light gray; Amino acid numbering follows NdCld and DypB from Rhodococcus jostii (underlined). Figure was generated using PyMOL (http://www.pymol.org/).
Figure 1A shows that all Clds with chlorite decomposition activity (i.e. functional Clds) deriving from different phyla (Proteobacteria, Cyanobacteria, Nitrospirae) cluster together. The fact that bacterial phyla with distinct metabolism group together, and Clds are not randomly distributed over Bacteria, might suggest that these metalloenzymes play similar physiological role(s) in these organisms. This hypothesis is underlined by mapping the homologous amino acid residues at position 173 (Cld from Candidatus Nitrospira defluvii [NdCld] numbering), which was shown to be important for efficient degradation of chlorite [12, 15]. However, it is important to note that the physiological substrate of these heme enzymes is unknown. For example, Cyanobacteria and Nitrospirae possess functional Clds but do not produce chlorite intracellularly. Possible clues for physiological role(s) of Clds might result from future comparative studies of Clds and DyPs. The phylogeny of the latter has been studied recently using different algorithms and four subfamilies have been defined [25, 28]. Moreover, in 2011 Goblirsch and co-workers showed, for the first time, the phylogenetic relationship between Clds and DyPs suggesting a common ancestor [15].
The updated phylogenetic tree presented in Fig. 1A represents all relevant branches of DyPs, chlorite dismutases, and chlorite-dismutase-like proteins, i.e. Clds missing the distal arginine (Arg173 in NdCld) known to be important for efficient chlorite degradation. The overall categorization into Cld and DyP sequences is obvious and each of these protein families can be rooted against the other, suggesting a common ancestor. All functional Clds have an arginine residue at the distal side of heme b and can be further divided into two lineages [11] differing in overall sequence length. From the group of “short Clds” (Lineage II) only Cld from Nitrobacter winogradskyi has been studied so far, whereas several representatives from the group of “long Clds” (Lineage I) were characterized in more detail (see Sections 3–6).
Little is known about the physiological role of the Cld-like proteins. Cld-like proteins from Bacillus subtilis, Mycobacterium tuberculosis [29], and Staphylococcus aureus [30] have been reported to play a (yet undefined) role in heme biosynthesis. The Cld-like protein from Haloferax volcanii was shown to have a role in antibiotic biosynthesis. Its gene is located together with a monooxygenase-like protein within a single open reading frame [31]. It was interesting to see that in the updated tree the sequence of Bacillus bataviensis, which represents a functional Cld, clusters together with all Firmicutes and sequences from other phyla having a glutamine residue on the distal side of the heme b. But this seems to be an exception. Typically, Cld-like proteins having the distal arginine exchanged for a distinct amino acid, mainly occur in one phylum. Figure 1A clearly demonstrates that Cld-like proteins in Firmicutes and Actinobacteria have a glutamine and an alanine, respectively, instead of the Arg173 in NdCld. By contrast, enzymes having leucine at this position are found in diverse phyla. This might indicate a more general functionality for this group of enzymes.
In contrast to Clds, the family of DyPs shows minimal grouping of organisms from the same phylogenetic lineage (with the exception of subfamily D), indicating numerous lateral gene transfer events. The sequences show a higher degree of similarity throughout the whole family and all sequences have the catalytic arginine residue (present in functional chlorite dismutases) structurally conserved (Fig. 1). The previously described four subfamilies (A-D) [15] can be roughly identified but the pattern of distribution seems to be more complicated and needs further evaluation in the future. Bacterial representatives of subfamily B seem to be closely related to the common ancestor of Clds and DyPs. The first node divides the representatives of subfamily B from other DyPs. The next node divides bacterial sequences of subfamily A from subfamilies C and D. Subfamily C is comprised of bacterial heme proteins, whereas subfamily D also contains sequences of fungal origin. In subfamily D, sequences of distinct phyla (e.g. Ascomycetes and Basidiomycetes) clearly cluster together.
3 Crystal structures of chlorite dismutases
All currently available (up to July 2013) crystal structures in the Protein Data Bank (PDB)-database (www.rsbc.org) of Clds and Cld-like proteins are listed in Supporting information, Table S1. Figure 1B shows a comparison of the active site of the Cld from Candidatus Nitrospira defluvii (PDB-code 3NN1) [12] and of DyP from Rhodococcus jostii (PDB-code 3QNS) [19]. The latter is a member of subfamily B and thus closest to the common origin of Clds and DyPs. Both structures show the conserved catalytically important distal arginine at similar positions [12, 19]. In both enzyme families, a histidine is the proximal heme ligand. Due to hydrogen bonding with an acidic amino acid (glutamate in Clds and aspartate in DyPs) the histidine has some imidazolate characteristics. In functional Clds, the distal arginine is the only charged amino acid [12], whereas in DyPs a fully conserved aspartate is found at the distal heme cavity. Furthermore, the orientation of the prosthetic group and its substituents is different (Fig. 1B) with the propionate-, vinyl- and methyl-substituents being tilted by 90° with respect to each other.
Figure 2 shows all of the structures of Clds and Cld-like proteins solved so far, with those having the same set of amino acids in the heme cavity being grouped and represented by a single structure (distal heme ligands deriving from crystallization liquids are omitted for clarity). To date three-dimensional structures of Clds and Cld-like proteins from seven different organisms have been published, revealing a high structural conservation of the subunit fold. Two distinct subunit topologies have been identified. The first topology (“long Clds”) includes Clds from Lineage I and Cld-like representatives from Azospira oryzae strain GR-1 (AoCld) [32], Dechloromonas aromatica (DaCld) [33], Candidatus Nitrospira defluvii (NdCld) [12], Thermus thermophilus (TtCld) [34], Thermoplasma acidophilum (TaCld) (PDB-code: 3DTZ), and Geobacillus stearothermophilus (GsCld) (PDB-code: 1T0T). These proteins have a subunit topology consisting of an N-terminal and a C-terminal ferredoxin-like fold. The N-terminal fold is always heme free, whereas in functional Clds the C-terminal fold has a heme b bound. The second subunit topology (“short Clds”) is only found in functional Clds of Lineage II. The only representative with known X-ray structure is the Cld from Nitrobacter winogradskyi (NwCld) [11]. It has a smaller subunit size, lacking almost the entire N-terminal domain of the first group described above. However, the heme-binding ferredoxin-like fold is highly similar to the C-terminal domain of “long Clds”.
Figure 2.
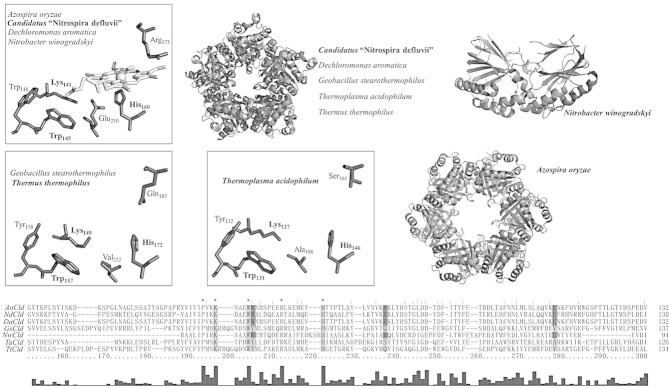
Crystal structures and sequence alignment of Clds and Cld-like proteins. Oligomeric structures and active site residues of Clds from Azospira oryzae (2VXH), Candidatus Nitrospira defluvii (3NN1), Dechloromonas aromatica (3Q08, 3Q09), and Nitrobacter winogradskyi (3QPI). Oligomeric structures and heme cavity residues in Cld-like proteins from Geobacillus stearothermophilus (1T0T), Thermus thermophilus (1VDH) and Thermoplasma acidophilum (3DTZ). Note that these X-ray structures do not contain the prosthetic group. Cld structures from species written in bold are depicted. Residues in the sequence alignment of the C-terminal domains, which are depicted in the structures, are highlighted in light gray; depicted residues unique for Clds with chlorite decomposition activity are highlighted in dark gray. Bar graph shows conserved amino acids. Figures were generated using PyMOL (http://www.pymol.org/).
All Clds and Cld-like proteins form oligomers. NwCld forms a dimer [11], AoCld [32] crystallizes as a hexamer, but appears to be a pentamer in solution. All other Clds show pentameric crystal structures stabilized by hydrogen bonds and salt bridges. Neither inter- nor intra-subunit disulphide bonds are found, due to the complete absence of cysteines in “long Clds” and the presence of a maximum of a single cysteine in “short Clds”. Oligomerization of some Clds depends on conditions like pH and ionic strength [35]. AoCld was reported to be a homotetramer [1] or homopentamer in solution [32] and to crystallize as a hexamer [32]. Cld from Pseudomonas chloritidismutans (PcCld) [9] and DaCld were described as homotetramers in solution [8] but DaCld crystallized as a pentamer [33]. Recently, Blanc and co-workers proposed the existence of dimeric DaCld depending on buffer conditions and protein concentration [35]. It is worth mentioning, that AoCld and DaCld have a sequence identity of 94.3 % (Supporting information, Table S2) with just a few amino acids differing in the heme free N-terminal domain and 100% identity in the heme-bound catalytic C-terminal domain (Fig. 2). Additionally, there are two structures of NdCld variants having point mutations of the catalytically important arginine 173. They exhibit subunit and oligomeric structures almost identical to the wild-type protein.
The distal ligand of ferric Clds is typically a water molecule. In the crystal structures it can be exchanged with molecules of the crystallization liquid. In some structures thiocyanate, nitrite and cyanide act as the distal ligand. Whereas cyanide is a typical low-spin ligand that inhibits heme proteins, SCN– and NO2– was found to act as a ligand and substrate of heme oxidoreductases. Whether Clds can use thiocyanate and/or nitrite as a substrate is unknown.
4 Spectral properties
Several Clds and Cld-like proteins have been analyzed by UV-vis electronic absorption, electron paramagnetic resonance (EPR), electronic circular dichroism (ECD), and resonance Raman (RR) spectroscopy. Despite the fact of almost identical heme cavity architecture (see Section 3), large differences in spectral signatures have been reported at comparable pH-values. This becomes obvious by inspection of Table 1 that summarizes the UV-vis absorbance maxima of Clds and Cld-like proteins in the respective ferric and ferrous state and in the low-spin cyanide complex.
Table 1.
Spectral parameters of recombinant wild-type chlorite dismutases and variants in the ferric and ferrous states as well as low-spin complex with cyanide
| ferric Soret (nm) | ferrous Soret (nm) | CN– Soret | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| pH | Soret (nm) | CT2 (nm) | Qv (nm) | Q0 (nm) | CT1 (nm) | Soret (nm) | Qv (nm) | Q0 (nm) | Soret (nm) | reference | |
| AoCld | 7.2 | 392 | – | – | – | – | 432 | 560 | – | – | [1, 4, 32] |
| 7.0 | 394 | ||||||||||
| DaCld | 6.8 | 392 | 506 | 536 | – | 644 | 433 | 556 | 587 | 419 | |
| 8.0 | 400 | – | 538 | – | 650 | – | – | – | – | [8, 35, 36, 38] | |
| 10 | 408 | – | 539 | 576 | 608 | – | – | – | – | ||
| IdCld | 7.0 | 392 | 509 | – | – | 648 | 434 | 555 | 586 | – | [7] |
| NdCld | 7.0 | 408 | – | 533 | 570 | 640 | 435 | 556 | 590 | 422 | [12, 37] |
| NwCld | 7.0 | 405 | 506 | 543 | – | 640 | 435 | 556 | 590 | 420 | [11, 37] |
| PcCld | 6.0 | 411 | – | – | – | – | 433 | – | – | 420 | [9] |
| SaHemQ (Cld) | 6.8 | 406 | 510 | – | – | 630 | 433 | 558 | – | 420 | [30] |
| TtCld | 7.0 | 403 | 495 | – | – | 634 | 429 | 558 | – | 420 | [34] |
| DaCld R183A | 6.8 | 391 | 509 | 530 | – | 644 | [38] | ||||
| 10 | 391 | 530 | – | 625 | |||||||
| DaCld R183K | 6.8 | 410 | – | 530 | 560 | – | [38] | ||||
| 10 | 410 | – | 530 | 560 | – | ||||||
| DaCld R183Q | 6.8 | 403 | 509 | 530 | – | 635 | [38] | ||||
| 10 | 403 | 509 | 530 | – | 640 | ||||||
| DaCld W227F | 6.0 | 402 | – | 538 | – | 650 | [35] | ||||
| 8.0 | 409 | – | 540 | 576 | 650 | ||||||
| DaCld W156F | 6.0 | 411 | – | 534 | – | – | [35] | ||||
| 8.0 | 412 | – | 535 | 575 | 650 | ||||||
| DaCld W155F | 6.0 | 413 | – | – | – | 660 | [35] | ||||
| 8.0 | 413 | – | 535 | 565 | 660 | ||||||
| NdCld R173A | 7.0 | 410 | – | 535 | 570 | 640 | [12] | ||||
| NdCld R173K | 7.0 | 412 | – | 535 | 565 | – | [12] | ||||
Closely related AoCld, DaCld and IdCld (Table 1) exhibit a relatively broad Soret maximum at around 393 nm and charge transfer bands at around 510 and 645 nm with a Qv band at approximately 535 nm at neutral pH. These bands indicate the presence of 5-coordinated high-spin (5cHS) heme iron. All three proteins show a clear pH dependence of the UV-visible spectral signatures. At alkaline pH, the resulting spectra indicate the presence of a hydroxyl 6-coordinated low-spin (6cLS) complex with Q-bands around 540 and 575 nm and a sharpened red-shifted Soret band. This alkaline transition is stated to be irreversible [36] and has pKa – values of 8.2 for AoCld [4], 8.5 for IdCld [7], and 8.7 for DaCld [36]. These findings are also reflected by the RR data of DaCld. In the acidic pH region DaCld loses its heme at around pH 4, exhibiting a spectrum of free heme with a Soret maximum at 375 nm [36]. Resonance Raman spectroscopy of wild-type DaCld also demonstrated that the hydrogen bond between the proximal histidine and the conserved glutamate is rather weak compared to other His-coordinated heme proteins. RR data on carbon monoxide binding suggest no significant electrostatic interactions of CO with residues in the distal heme pocket.
Interestingly, ferric PcCld, which has more than 90% sequence identity to AoCld and DaCld (Table 1), has a Soret maximum at 411 nm [9]. Furthermore, NdCld, NwCld, SaCld, and TtCld show Soret maxima between 403–408 nm and CT1 bands between 630 and 640 nm, indicating dominating high-spin iron heme species at pH 7. The Soret bands of these proteins are sharper than those of AoCld, IdCld and DaCld and comparable with other His-ligated heme b protein families. One reason for the significant differences in spectral properties between these two groups of Clds could be the relatively low conformational and thermal stability of AoCld, IdCld and DaCld, which will be discussed in Section 6.
Spectra of the ferrous state of Clds are characteristic of 5cHS Fe(II), with the exception of ferrous TtCld, which has a blue-shifted Soret maximum at 429 nm, indicative of a 6cLS ferrous heme iron. TtCld and SaCld are Cld-like proteins with no or very limited chlorite dismutase activity [30, 34].
In recombinant heme proteins, occupancy of the active site with the prosthetic group is reflected by the Reinheitszahl (Rz-value or purity number, ASoret/A280nm). Typical Rz-values for functional recombinant Clds are 2.1 (NdCld) [37], 1.1 (DaCld) [35], and 1.5 (AoCld) [4]. Low Rz-values are often found in Cld-like proteins and might reflect weaker or hindered binding of the prosthetic group compared to Clds. Most probably this was the reason why the crystal structures of GsCld, TaCld and TtCld were obtained without the prosthetic group (Supporting information, Table S1).
Mutations at the active site were shown to have an impact on spectral properties in DaCld [35, 38]. UV-visible electronic absorption spectroscopy as well as Soret excited RR spectroscopy show the importance of the distal arginine residue for keeping the enzyme in the catalytically active 5cHS Fe(III) state (Table 2). Especially the R183K variant of DaCld exhibits spectral features indicative of a 6cLS heme iron at pH 6.8 [38]. Exchange of proximal amino acids in DaCld severely diminished heme-binding, and also modified the oligomerization and the conformational stability of the protein [35].
Table 2.
Steady-state kinetic parameters of chlorite degradationa)
| pH | KM (μM) | kcat (s–1) | kcat/KM (M–1 s–1) | E°' (mV) | reference | |
|---|---|---|---|---|---|---|
| AoCld | 7.2b) | 170 | 1200 | 7.1 × 106 | –23 ± 9 | [1, 4, 32] |
| –158 ± 9 | ||||||
| 5.2c) | 620 | 20000 | 3.2 × 107 | |||
| DaCld | 6.8d) | 212 | 7500 | 3.5 × 107 | –23 | [8, 36, 38] |
| 7.6c) | 430 | 3000 | 6.9 × 106 | |||
| IdCld | 7.0e) | 260 | 1800 | 6.9 × 106 | –21 | [7] |
| NdCld | 7.0f) | 69 | 43.0 | 6.2 × 105 | –113 ± 5 | [12, 37] |
| NwCld | 7.0g) | 90 | 190 | 2.1 × 106 | –119 ± 5 | [11, 37] |
| PcCld | 6.0h) | 84 | 230 | 2.7 × 106 | – | [9] |
| SaHemQ (Cld) | 6.8i) | NA | NA | NA | – | [30] |
| TtCld | 7.0j) | 13 | 0.77 | 59 | – | [34] |
| DaCld R183A | 6.8k) | 14600 | 91 | 6.2 × 103 | –4 | [38] |
| DaCld R183K | 6.8k) | 42000 | 1000 | 2.5 × 104 | –18 | [38] |
| DaCld R183Q | 6.8k) | 50000 | 350 | 6.9 × 103 | –34 | [38] |
| DaCld W227F | 6.0j) | 210 | 1300 | 6.1 × 106 | – | [35] |
| 8.0j) | 620 | 440 | 7.1 × 105 | |||
| DaCld W156F | 6.0j) | 160 | 2100 | 1.3 × 107 | – | [35] |
| 8.0j) | 130 | 470 | 3.7 × 106 | |||
| DaCld W155F | 6.0j) | NA | NA | NA | – | [35] |
| 8.0j) | NA | NA | NA | |||
| NdCld R173A | 7.0l) | 90 | 2.8 | 3.1 × 104 | – | [12] |
| NdCld R173K | 7.0l) | 898 | 14.0 | 1.5 × 104 | – | [12] |
Please note (i) the varying pH-values; (ii) that chlorite dismutases are inactivated during chlorite degradation; and (iii) the different chlorite concentration regimes used in the various laboratories. Additionally, standard reduction potentials of the Fe(III)/Fe(II) couple for some Clds have been published. NA, no activity detected.
[ClO2–] up to 15 mM
[ClO2–] 0.08–21 mM
[ClO2–] 0.08–2 mM
[ClO2–] 0.2–1.5 mM
[ClO2–] 0.05–80 mM
[ClO2–] 0.05–100 mM
[ClO2–] 0.001–0.5 mM
[ClO2–] up to 20 mM
[ClO2–] not reported
[ClO2–] 0.08–2 mM
[ClO2–] 0.05–80 mM
Electronic circular dichroism spectroscopy in the far UV-region of ferric NdCld, apo-NdCld, DaCld [39], and proximal tryptophan variants of DaCld [35] show minima at 208 and 222 nm typical for a-helical proteins (NdCld and DaCld). In Lineage II Clds such as NwCld, the four N-terminal a-helices present in “long Clds” are lacking [40], which is reflected by the ECD spectrum in the far-UV region. Consequently, the dichroic minimum typical for β-sheets between 212 and 214 nm is observed [40]. For recombinant SaCld, minima in the ECD-spectrum at 227 nm for the apo-form and at 234 nm for the holo-form were reported [30]. These minima cannot be assigned to secondary structure elements and are probably resulting from misfolded protein [41].
ECD spectra in the near UV- and visible regions are only reported for NdCld and NwCld [40]. These spectra show positive ellipticity maxima for the heme region at 417 (NdCld) and 416 nm (NwCld), and clear differences in the fingerprint region suggesting slightly different heme environments in solution.
Electron paramagnetic resonance (EPR) spectroscopy was also used to determine the spin-state and electron distribution at the heme iron. IdCld shows a rhombically distorted high-spin spectrum at neutral pH and a low-spin dominated spectrum at alkaline pH [7]. AoCld and DaCld show a similar rhombic high-spin signal at pH 6 and pH 9 [4, 42]. The EPR spectrum of NdCld shows an axial and a rhombic high-spin form and also a low-spin form at pH 7, whereas the high-spin species of NwCld gives a completely axial signal next to a weak low-spin signal [37]. These data clearly show that the electron distribution of the ferric heme iron might differ in Clds, which cannot be seen in the respective crystal structures. Still, one has to be cautious about comparing the above mentioned results since EPR spectroscopy is a very sensitive method and results can be influenced by buffer- and cryo-conditions [43] as well as experimental setup. Buffer or cryo-components can interact with the active site. Furthermore, the ferric low-spin signal saturates at much lower microwave power (i.e. approximately 1 mW at 10 K) than the high-spin signal [37]. Spectra discussed above have been taken at significantly different micro-wave powers; 0.2 mW for NdCld and NwCld [37], 2 mW for IdCld [7], 50 mW for DaCld [42], and 80 mW for AoCld [4].
5 Enzymatic activity and redox chemistry
Originally, chlorite dismutases were discovered in PCRBs and were shown to degrade chlorite to chloride and O2. This reactivity attracted the attention of environmental biotechnologists, since chlorite is an anthropogenic environmental pollutant abundant in bleaching agents. Its occurrence in ground waters is a severe problem in the USA [6, 14]. Additionally, functional Clds fascinated enzymologists, since they represent the only soluble enzyme family known that is able to catalyze the formation of a covalent oxygen-oxygen bond, which has been proven by 18O-labeling studies [42]. This can also be of interest from a biotechnological point of view, since it enables the local and controlled generation of O2 in an oxygen-free environment. In studies on O2-utilizing enzymes such as monooxygenases, Dassama and co-workers [44] applied DaCld for in situ generation of molecular oxygen.
In order to use Clds for bioremediation and biotechnological application, it is important to understand structure-function relationships. Interesting and still open questions concern substrate specificity, electron structure of the catalytic redox intermediates, substrate and intermediate binding and oxidation/reduction, as well as the origin of the mechanism-based inactivation of Clds during chlorite degradation.
The ability to degrade chlorite to chloride and produce molecular oxygen has been shown for several functional Clds that all have the distal arginine residue fully conserved (Fig. 1B). All currently published steady-state kinetic parameters of functional Clds are listed in Table 2. Cld-like proteins (SaCld, TaCld, GsCld) do not show any or very weak (TtCld) chlorite dismutase activity. It is important to mention that catalytic efficiencies are problematic to compare, since the various enzymes were probed at different chlorite concentrations, pH-values, and temperatures. Especially at high chlorite concentrations, Clds are irreversibly inactivated which does not allow the determination of reliable kinetic parameters [8]. We strongly suggest working with micromolar up to 1 mM chlorite concentrations and only using the initial linear phase of polarographically measured O2 release within the first 30 s for rate calculation. The inhibition mechanism itself is not yet understood, but heme bleaching is observed when Clds are treated with high molar (∼1000 eq) excess of chlorite [8]. Formation of a tryptophanyl radical at the proximal heme site was hypothesized [33] but later it was shown that mutations of tryptophans close to the catalytic center do not prevent inhibition [35]. So far, published temperature optima for characterized Clds are between 20–30°C, and pH optima for chlorite degradation activity are between pH 5.0–6.0 [10, 11, 36]. The published KM values at pH 7.0 vary from 69 to 260 μM, kcat values from 43 to 7500 s–1 and kcat/KM values from 6.2 × 105 to 3.5 × 107 M–1 s–1 (Table 2). Generally, exchange of the distal arginine decreased the affinity for chlorite as well as the catalytic efficiency, but these effects were more pronounced in DaCld compared to NdCld. In DaCld, the effects of mutations of tryptophans at the proximal heme cavity on the chlorite degradation activity were relatively small (Table 2).
The redox potential of the Fe(III)/Fe(II)-couple of the heme protein determines the stable oxidation state of the native protein in solution. In addition, it reflects the redox properties of higher intermediate oxidation states of the respective protein family and therefore gives hints on substrate specificities [45]. Performing these redox measurements at variable temperatures allows determination of the enthalpic and entropic contribution to the reduction reaction [46]. Determined redox potentials of Clds are listed in Table 2.
AoCld, DaCld, and IdCld have a redox potential of the Fe(III)/Fe(II) couple of about –20 mV [4, 7, 38], which has been determined via indirect dye-mediated [47, 48] or dye- and enzyme-mediated [49] methods. Recombinant AoCld shows a redox potential of –158 mV. DaCld variants R173A, R173K, and R173Q have reduction potentials of –4, –18, and –34 mV, respectively, similar to the wild-type protein. The redox potentials and redox thermodynamics of pentameric NdCld (Lineage I) and dimeric NwCld (Lineage II) were determined spectroelectrochemically under identical conditions. In this method, the sample is directly reduced and oxidized by applied potentials and not by addition of dithionite or xanthine oxidase for reduction and ferricyanide for oxidation. Redox potentials for both proteins are around –120 mV for the ferric high-spin enzymes and about -400 mV for the respective cyanide complexes [37]. The ferric forms of the high-spin enzymes are enthalpically favored, while the entropic contribution partly compensates for enthalpic stabilization. This suggests that despite different subunit and oligomeric architecture of NdCld and NwCld, the redox properties and active site architectures are very similar in solution, as was suggested by the crystal structures [11, 12]. Differences in E°' values as described above might be derived from different applied electrochemical methods. It would be interesting to measure the E°' values of the Fe(III)/Fe(II) couple of DaCld and closely related AoCld under the same conditions used for NdCld and NwCld.
A few mechanistic investigations of AoCld and DaCld have been performed. AoCld was shown to react with H2O2 possibly forming Compound I- or Compound II-like intermediates [4]. Similarly, DaCld reacted with peracetic acid to the same redox states. Compound II was reduced back to the ferric state using ascorbate, whereas putative Compound I was reported to interact weakly with the two-electron reductant thioanisole in a monooxygenase-like reaction [42]. Freeze-quench EPR using chlorite as substrate showed the formation of radicals at remote sites associated with Compound I and Compound II formation [35, 42]. Chlorite at higher concentrations, but also hypochlorite, hydrogen peroxide, and peracetic acid promote heme bleaching [4, 42].
Based on all these findings, a reaction mechanism was proposed by Lee and co-workers [42] and is summarized in Fig. 3. The conserved and (as the X-ray structures suggest) mobile distal arginine supports binding of the anionic chlorite to the ferric resting state of Cld. Chlorite immediately oxidizes the enzyme to the Compound I state, which, most probably, is an oxoiron(IV) porphyryl radical intermediate. Thereby, hypochlorite is formed as an intermediate that is kept in the reaction sphere by the distal arginine. Finally, hypochlorite attacks the ferryl oxygen and chloride and O2 are released. It is important to note that so far the formation of Compound I and hypochlorite is not fully proven. Moreover, the role of a Compound II state in this reactivity as well as the mechanism of inactivation are unknown so far.
Figure 3.
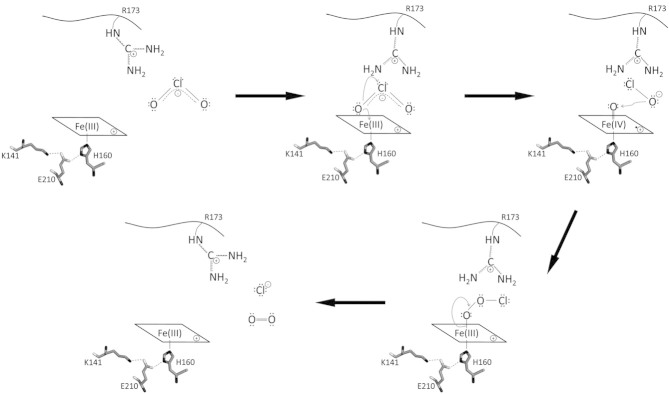
Proposed reaction mechanism of chlorite dismutase. The reaction starts with the attack of anionic chlorite at ferric heme b. After formation of the Fe(III)-chlorite complex, heterolytic cleavage of one covalent oxygen-chlorine bond in chlorite leads to the formation of hypochlorite and the redox intermediate compound I [oxoiron(IV) porphyrin cation radical]. Finally, upon nucleophilic attack of intermediate anionic hypochlorite at the ferryl oxygen, compound I is reduced to the resting state and dioxygen and chloride are released. In addition, the scheme shows the putative role of distal arginine (fully conserved in functional Clds) in the orientation and stabilization of the substrate as well as the postulated intermediate(s). On the proximal site the conserved triad H160-E210-K141 that forms a H-bonding network is depicted.
Figure 1 demonstrates the close relationship between Clds and DyPs. Nevertheless, so far there is no study on the use of chlorite as substrate for DyPs and only a few data are available about the peroxidase activity of Clds. In the presence of hydrogen peroxide, DaCld and SaCld show weak peroxidase reactivity towards artificial one-electron donors such as guaiacol and ABTS with catalytic efficiencies around 102 – 103 M–1 s–1 [30, 35, 38, 42]. A very poor catalase activity has been reported for Clds from Mycobacterium tuberculosis, Bacillus subtilis [29], and Staphylococcus aureus [30]. As outlined above, the endogenous substrate and/or physiological role of most Clds and Cld-like proteins is yet unknown.
From both, an enzymatic and mechanistic point of view, the structural and phylogenetic relationship between Clds and DyPs is interesting and needs further investigation. Recently mutational analysis has demonstrated that the distal arginine in DyP (subfamily B) from Rhodococcus jostii, which is structurally conserved in all functional Clds and DyPs, is essential for the peroxidase activity of this enzyme (just as the distal arginine in Clds is crucial for chlorite-dismutase activity) [50]. Since recombinant enzymes from both families can be produced in high yields, these biochemical relationships should be addressed now in more detail.
6 Conformational and thermal stability
Biotechnologically applied enzymes are often required to withstand elevated temperatures, acidic or alkaline environments, and the presence of denaturing agents, while at the same time maintaining their catalytic efficiency. In the case of chlorite dismutases, studies on conformational and thermal stability as well as enzymatic activity at higher temperatures have mainly been performed with NdCld and NwCld. Pentameric NdCld of Lineage I has been shown to be a highly thermostable enzyme with a melting temperature at around 90°C compared to less stable dimeric NwCld (Lineage II) which denatures at around 50°C [40]. Moreover, NdCld also shows high stability in acidic and alkaline conditions. Furthermore, NdCld is able to detoxify chlorite even at 80°C [40].
Compared to NwCld, the pentameric protein also has a higher chemical stability probed by guanidinium hydrochloride [40]. Both NdCld and NwCld have similar activities and redox properties as discussed above but differ in subunit and oligomeric structure. Heme b bound to the heme cavity has a stabilizing effect, since apo-NdCld has a significantly lower Tm-value than the holoprotein.
DaCld was incubated at different pH values prior to activity measurements at pH 6.8 and showed loss of activity below pH 4.5 and above pH 10 [36]. Also spectroscopic data suggest inactivation due to an irreversibly formed 6cLS heme-iron of DaCld [36] and closely related AoCld [4]. Mutational studies of conserved tryptophan residues of DaCld showed a significant impact on protein stability [35]. The authors hypothesized that in solution oligomerization state of DaCld is dependent on different buffer conditions and protein concentrations [35].
Although the crystal structures of DaCld and NdCld are almost superposable, there appears to be a difference in stability, indicated by the fact that DaCld changes its oligomeric structure depending on pH or ionic strength, a phenomenon not observed with NdCld [12, 35, 37, 40]. Examination of the crystal structures of DaCld (PDB-code: 3Q09) and NdCld (PDB-code: 3NN1) by PDBePISA (Protein Interfaces, Surfaces and Assemblies; http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver) of EMBL-EBI shows that 17 residues at the N-terminus of NdCld are involved in inter-subunit hydrogen bonds and salt bridges, whereas only 11 residues at the N-terminus of DaCld stabilize the oligomeric structure. The contribution of the C-terminal region to subunit interaction is similar in both proteins. This observation might indicate that the heme free N-terminal domain of functional Lineage I proteins (“long Clds”) is important for modulation of the overall stability of the enzyme. Clearly, more detailed studies need to be performed to fully understand the interplay between oligomerization and stability, which could then be the basis for rational stability engineering.
7 Conclusion
Since their discovery in 1996 [1], several Clds and Cld-like proteins from different organisms have been investigated. Functional Clds cluster into two lineages with Lineage I containing (mainly) homopentameric metalloproteins with subunits consisting of an N-terminal and a C-terminal heme b containing ferredoxin-like fold, and Lineage II containing homodimeric enzymes with subunits lacking almost the entire N-terminal domain of Lineage I proteins. The heme cavity architecture of both lineages is almost identical and this is reflected by similar redox properties and catalytic efficiencies. Oligomerization seems to have an impact on conformational and thermal stability. All functional Clds have only one prominent and fully conserved arginine residue at the distal heme side that supports chlorite binding and oxidation, and (possibly) keeps the postulated intermediate hypochlorite in the reaction sphere (Fig. 3). Since the final product of ClO2– (Cl: +III) degradation is Cl– (Cl: –I) with OCl– (Cl: +I) as intermediate, this reaction is clearly not a dismutation reaction as the misleading name suggests. Due to the occurrence of Clds in many different bacterial and archaeal phyla (and not only in PCRBs), it is reasonable to assume that chlorite is not the endogenous substrate. Future studies on the biochemistry and physiology of Cld-like proteins (that lack the distal arginine and have no or very poor chlorite degradation activity) as well as a better understanding of the phylogenetic and biochemical relationship between Clds and DyPs will contribute to identifying the endogenous substrate of these enzymes.
Our comparative analyses of biochemical and biophysical data on Clds and Cld-like proteins show that, at the moment, NdCld is the best candidate for use in biotechnological applications. NdCld has a high conformational and thermal stability over a broad pH range and shows chlorite degradation activity even at very high temperatures. The oligomerization state of NdCld is independent of pH and ionic strength, and a high resolution crystal structure is available. Moreover, NdCld is the only functional Cld where X-ray structures of mutants have been solved. Nevertheless, before NdCld can be efficiently used in bioremediation of chlorite, several questions must be solved. It is important to understand: (i) the proposed reaction sequence and the involved redox intermediates (Fig. 3); and (ii) the mechanism of inhibition during chlorite degradation. This knowledge will provide the basis for rational enzyme engineering and/or modification of reaction conditions. Simultaneously, the heterologous expression and purification in E. coli must be improved to obtain high yields. Principally, as demonstrated in the recent years, this will not be the bottleneck for application in biotechnology. Successful overexpression and purification of recombinant chlorite dismutases in E. coli has been reported for AoCld [32], IdCld [51, 52], DaCld [8, 36], NdCld [10, 12, 37], and NwCld [11].
The biotechnological potential of Cld is very high due to its unique chlorite detoxifying activity. Together with perchlorate and chlorate, chlorite is a serious environmental concern as rising concentrations of these harmful compounds have been detected in ground water, drinking water, and soils [6]. Environmental contamination with perchlorate and chlorate results from its extensive use as oxidizer in pyrotechnics and rocket fuel, and its presence in certain fertilizers. Chlorite is used as bleaching agent in the textile, pulp and paper industries, as a disinfectant and component of cleaning solutions, and in pesticides. Intake of chlorite by humans occurs mainly via drinking water, milk and consuming certain plants, and should be minimized. Due to its oxidative nature cholrite reacts easily with organic material and thus has toxic effects on living cells [5].
Besides chloride, the second reaction product of chlorite degradation is molecular oxygen. Nature seems to use Cld as O2 generator. For example, in halophilic archaea a fusion protein consisting of a Cld and a monooxygenase was found [31]. Moreover, it has been suggested that PCRBs, which are facultative anaerobes, and other microbes, use Clds for metabolisms in anoxia. Several examples of aerobic pathways, e.g. of aromatic or aliphatic hydrocarbon degradation, have been reported that function in microbes in the absence of external O2. It might be possible that these organisms use molecular oxygen formed by Cld. Exploitation of this Cld activity in various biochemical or biotechnological applications depends on further investigation of the reaction mechanism and the use of alternative oxidants instead of chlorite.

Stefan Hofbauer holds an M.Sc. degree in Biotechnology from BOKU – University of Natural Resources and Life Sciences, Vienna, where he is currently a PhD student at the Department of Chemistry within the international PhD-Program BioToP (Biomolecular Technology of Proteins; http://biotop.boku.ac.at). His research focuses on heme enzymes, and he is particularly interested in structure-function relationships and the reaction mechanism of chlorite dismutases.

Christian Obinger studied Chemistry and Biochemistry and performed his doctoral thesis at the Institute of Physical Chemistry at the University of Vienna. In 1992 he became Assistant Professor at the Department of Chemistry at BOKU–University of Natural Resources and Life Sciences. In 1999 he was promoted to Associate Professor (habilitation) for Biochemistry, and in 2011 to Full Professor for Protein Biochemistry. He is interested in structure-function relationships of heme-containing oxidoreductases including peroxidases, peroxidasins, catalases, chlorite dismutases and related systems. His reseach focuses on the impact of the protein matrix and post-translational modifications on catalytically relevant redox intermediates and electron-transfer reactions.
Acknowledgments
This project was supported by the Austrian Science Foundation, FWF [Doctoral program BioToP–Molecular Technology of Proteins (W1224) and the FWF-project P25270-B22].
All authors declare no commercial or financial conflict of interest.
Glossary
Abbreviations
- AoCld
Cld from Azospira oryzae
- Cld
chlorite dismutase
- DaCld
chlorite dismutase from Dechloromonas aromatica
- DyP
dye-decolorizing peroxidase
- E°'
standard reduction potential
- ECD
electronic circular dichroism
- EPR
electron paramagnetic resonance
- GsCld
Cld from Geobacillus stearothermophilus
- HS
high-spin
- IdCld
Cld from Ideonella dechloratans
- LS
low-spin
- NdCld
Cld from “Candidatus Nitrospira defluvii”
- NwCld
Cld from Nitrobacter winogradskyi
- PcCld
Cld from Pseudomonas chloritidismutans
- PDB
Protein Data Bank
- RR
resonance Raman
- TaCld
Cld from Thermus acidophilum
- TtCld
Cld from Thermus thermophilis
- 5c
five-coordinated
- 6c
six-coordinated
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
suppinfo
References
- 1.van Ginkel CG, Rikken GB, Kroon AGM, Kengen SWM. Purification and characterization of chlorite dismutase: a novel oxygen-generating enzyme. Arch. Microbiol. 1996:321–326. doi: 10.1007/s002030050390. [DOI] [PubMed] [Google Scholar]
- 2.Ettwig KF, Butler MK, Le Paslier D, Pelletier E, et al. Nitrite-driven anaerobic methane oxidation by oxygenic bacteria. Nature. 2010;464:543–548. doi: 10.1038/nature08883. [DOI] [PubMed] [Google Scholar]
- 3.Rikken GB, Kroon AGM, van Ginkel CG. Transformation of (per) chlorate into chloride by a newly isolated bacterium: reduction and dismutation. Appl. Microbiol. Biotechnol. 1996;45:420–426. [Google Scholar]
- 4.Hagedoorn PL, de Geus DC, and Hagen WR. Spectroscopic characterization and ligand-binding properties of chlorite dismutase from the chlorate respiring bacterial strain GR-1. Eur. J. Biochem. 2002;269:4905–4911. doi: 10.1046/j.1432-1033.2002.03208.x. [DOI] [PubMed] [Google Scholar]
- 5.Ueno H, Oishi K, Sayato Y, Nakamuro K. Oxidative cell damage in Kat-sod assay of oxyhalides as inorganic disinfection by-products and their occurrence by ozonation. Arch. Environ. Contam. Toxicol. 2000;38:1–6. doi: 10.1007/s002449910001. [DOI] [PubMed] [Google Scholar]
- 6.Coates JD, Achenbach LA. Microbial perchlorate reduction: rocket-fueled metabolism. Nat. Rev. Microbiol. 2004;2:569–580. doi: 10.1038/nrmicro926. [DOI] [PubMed] [Google Scholar]
- 7.Stenklo K, Thorell D, Bergius H, Aasa R, Nilsson T. Chlorite dismutase from Ideonella dechloratans. J. Biol. Inorg. Chem. 2001;6:601–607. doi: 10.1007/s007750100237. [DOI] [PubMed] [Google Scholar]
- 8.Streit BR, DuBois JL. Chemical and steady-state analysis of a heterologously expressed heme dependent chlorite dismutase. Biochemistry. 2008;47:5271–5280. doi: 10.1021/bi800163x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehboob F, Wolterink AF, Vermeulen AJ, Jiang B, et al. Purifcation and characterization of a chlorite dismutase from Pseudomonas chloritidismutans. FEMS Microbiol. Lett. 2009;293:115–121. doi: 10.1111/j.1574-6968.2009.01517.x. [DOI] [PubMed] [Google Scholar]
- 10.Maixner F, Wagner M, Lücker S, Pelletier E, et al. Environmenteal genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”. Environ. Microbiol. 2008;10:3043. doi: 10.1111/j.1462-2920.2008.01646.x. Environmenteal genomics reveals a functional chlorite dismutase in the nitrite-oxidizing bacterium 3056. [DOI] [PubMed] [Google Scholar]
- 11.Mlynek G, Sjöblom B, Kostan J, Füreder S, et al. Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi. J. Bacteriol. 2011;193:2408–2417. doi: 10.1128/JB.01262-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kostan J, Sjoeblom B, Maixner F, Mlynek G, et al. Structural and functional analysis of the chlorite dismutase from the nitrite-oxidizing bacterium “Candidatus Nitrospira defluvii”: identification of a catalytically important amino acid residue. J. Struct. Biol. 2010;172:331–342. doi: 10.1016/j.jsb.2010.06.014. [DOI] [PubMed] [Google Scholar]
- 13.Eriksen GE. The Chilean nitrate deposits. Am. Sci. 1983:366–374. [Google Scholar]
- 14.Bardiya N, Bae JH. Dissimilatory perchlorate reduction: a review. Microbiol. Res. 2011;166:237–254. doi: 10.1016/j.micres.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Goblirsch B, Kurker RC, Streit BR, Wilmot CM, DuBois JL. Chlorite dismutases, DyPs and EfeB: 3 microbial heme enzyme families comprise the CDE structural superfamily. J. Mol. Biol. 2011;408:379–398. doi: 10.1016/j.jmb.2011.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zámocký M, Furtmüller PG, Obinger C. Two distinct groups of fungal catalase/peroxidases. Biochem. Soc. Trans. 2009:772–777. doi: 10.1042/BST0370772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zámocký M, Furtmüller PG, Obinger C. Evolution of structure and function of Class I peroxidases. Arch. Biochem. Biophys. 2010;500:45–57. doi: 10.1016/j.abb.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 18.Kim SJ, Shoda M. Purification and characterization of a novel peroxidase from Geotrichum candidum Dec 1 involved in decolorization of dyes. Appl. Environ. Microbiol. 1999;65:1029–1035. doi: 10.1128/aem.65.3.1029-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts JN, Singh R, Grigg JC, Murphy ME, et al. Molecular characterization of dye-decolorizing peroxidases from Rhodococcus jostii RHA1. Biochemistry. 2011;50:5108–5119. doi: 10.1021/bi200427h. [DOI] [PubMed] [Google Scholar]
- 20.Liers C, Pecyna MJ, Kellner H, Worrich A, et al. Substrate oxidation by dye-decolorizing peroxidases (DyPs) from wood- and litter-degrading agaricomycetes compared to other fungal and plant heme-peroxidases. Appl. Microbiol Biotechnol. 2013;97:5839–5849. doi: 10.1007/s00253-012-4521-2. [DOI] [PubMed] [Google Scholar]
- 21.Sezer M, Santos A, Kielb P, Pinto T, et al. Distinct structural and redox properties of the heme active site in bacterial dye decolorizing peroxidase-type peroxidases from two subfamilies: resonance Raman and electrochemical study. Biochemistry. 2013;52:3074–3084. doi: 10.1021/bi301630a. [DOI] [PubMed] [Google Scholar]
- 22.van Bloois E, Torres Pazmiño DE, Winter RT, Fraaije MW. A robust and extracellular heme-containing peroxidase from Thermobifida fusca as prototype of a bacterial peroxidase superfamily. Appl. Microbiol. Biotechnol. 2010;86:1419–1430. doi: 10.1007/s00253-009-2369-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown ME, Barros T, Chang MC. Identification and characterization of a multifunctional dye peroxidase from a lignin-reactive bacterium. ACS Chem. Biol. 2012;7:2074–2078. doi: 10.1021/cb300383y. [DOI] [PubMed] [Google Scholar]
- 24.Scheibner M, Hülsdau B, Zelena K, Nimtz M, et al. Novel peroxidases of Marasmius scorodonius degrade beta-carotene. Appl. Microbiol. Biotechnol. 2008;77:1241–1250. doi: 10.1007/s00253-007-1261-9. [DOI] [PubMed] [Google Scholar]
- 25.Ogola HJO, Kamiike T, Hashimoto N, Ashida H, et al. Molecular Characterization of a Novel Peroxidase from the Cyanobacterium Anabaena sp. Strain PCC 7120. Appl. Environ. Microbiol. 2009;75:7509–7518. doi: 10.1128/AEM.01121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K, Peterson D, Peterson N, Stecher G, et al. MEGA5: Molecular Evolutionary Genetics Analysis using Maximum Likelihood, Evolutionary Distance, and Maximum Parsimony Methods. Mol. Biol. Evol. 2011:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ahmad M, Roberts JN, Hardiman EM, Singh R, et al. Identification of DypB from Rhodococcus jostii RHA1 as a lignin peroxidase. Biochemistry. 2011:5096–5107. doi: 10.1021/bi101892z. [DOI] [PubMed] [Google Scholar]
- 29.Dailey TA, Boynton TO, Albetel AN, Gerdes S, et al. Discovery and Characterization of HemQ: an essential heme biosynthetic pathway component. J. Biol. Chem. 2010;285:25978–25986. doi: 10.1074/jbc.M110.142604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mayfield JA, Hammer ND, Kurker RC, Chen TK, et al. Chlorite Dismutase (HemQ) from Staphylococcus aureus Has a Redox-Sensitive Heme and is Associated with the Small Colony Variant Phenotype. J. Biol. Chem. 2013;288:23488–23504. doi: 10.1074/jbc.M112.442335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bab-Dinitz E, Shmuely H, Maupin-Furlow J, Eichler J, Shaanan B. Haloferax volcanii PitA: an example of functional interaction between the Pfam chlorite dismutase and antibiotic biosynthesis monooxygenase families? Bioinformatics. 2006;22:671–675. doi: 10.1093/bioinformatics/btk043. [DOI] [PubMed] [Google Scholar]
- 32.de Geus DC, Thomassen EAJ, Hagedoorn P-L, Pannu NS, et al. Crystal structure of chlorite dismutase, a detoxifying enzyme producing molecular oxygen. J. Mol. Biol. 2009;387:192–206. doi: 10.1016/j.jmb.2009.01.036. [DOI] [PubMed] [Google Scholar]
- 33.Goblirsch BR, Streit BR, DuBois JL, Wilmot CM. Structural features promoting dioxygen production by Dechloromonas aromatica chlorite dismutase. J. Biol. Inorg. Chem. 2010;15:879–888. doi: 10.1007/s00775-010-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebihara A, Okamoto A, Kousumi Y, Yamamoto H, et al. Structure-based functional identification of a novel heme-binding protein from Thermus thermophilis HB8. J. Struct. Func. Gen. 2005;6:21–32. doi: 10.1007/s10969-005-1103-x. [DOI] [PubMed] [Google Scholar]
- 35.Blanc B, Rodgers KR, Lukat-Rodgers GS, DuBois JL. Understanding the roles of strictly conserved tryptophan residues in O2 producing chlorite dismutases. Dalton Trans. 2013;42:3156–3169. doi: 10.1039/c2dt32312e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streit BR, Blanc B, Lukat-Rodgers BS, Rodgers KR, DuBois JL. How active-site protonation state influences the reactivity and ligation of the heme in chlorite dismutase. J. Am. Chem. Soc. 2010;132:5711–5724. doi: 10.1021/ja9082182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hofbauer S, Bellei M, Sündermann A, Pirker KF, et al. Redox thermodynamics of high-spin and low-spin forms of chlorite dismutases with diverse subunit and oligomeric structures. Biochemistry. 2012;51:9501–9512. doi: 10.1021/bi3013033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blanc B, Mayfield JA, McDonald CA, Lukat-Rodgers GS, et al. Understanding how the distal environment directs reactivity in chlorite dismutase: spectroscopy and reactivity of Arg183 mutants. Biochemistry. 2012;51:1895–1910. doi: 10.1021/bi2017377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goblirsch BR, Streit BR, DuBois JL, Wilmot CM. Crystallization and preliminary X-ray diffraction of chlorite dismutase from Dechloromonas aromatica RCB. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2009;65:818–821. doi: 10.1107/S1744309109026785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hofbauer S, Gysel K, Mlynek G, Kostan J, et al. Impact of subunit and oligomeric structure on the thermal and conformational stability of chlorite dismutases. Biochim. Biophys. Acta. 2012;1824:1031–1038. doi: 10.1016/j.bbapap.2012.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly SM, Jess TJ, Price NC. How to study proteins by circular dichroism. Biochim. Biophys. Acta. 2005:119–139. doi: 10.1016/j.bbapap.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 42.Lee AQ, Streit BR, Zdilla MJ, Abu-Omar MM, DuBois JL. Mechanism of and exquisite selectivity for O-O bond formation by the heme dependent chlorite dismutase. Proc. Natl. Acad. Sci. USA. 2008;105:15654–15659. doi: 10.1073/pnas.0804279105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Svistunenko DA, Worrall JA, Chugh SB, Haigh SC, et al. Ferric haem forms of Mycobacterium tuberculosis catalase-peroxidase probed by EPR spectroscopy: Their stability and interplay with pH. Biochimie. 2012;94:1274–1280. doi: 10.1016/j.biochi.2012.02.021. [DOI] [PubMed] [Google Scholar]
- 44.Dassama LM, Yosca TH, Conner DA, Lee MH, et al. O(2)-evolving chlorite dismutase as a tool for studying O2-utilizing enzymes. Biochemistry. 2012;51:1607–1616. doi: 10.1021/bi201906x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Battistuzzi G, Bellei M, Bortolotti CA, Sola M. Redox properties of heme peroxidases. Arch. Biochem. Biophys. 2010;500:21–36. doi: 10.1016/j.abb.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 46.Farhangrazi ZS, Fossett ME, Powers LS, Ellis WR., Jr Variable-temperature spectroelectrochemical study of horseradish peroxidase. Biochemistry. 1995;34:2866–2871. doi: 10.1021/bi00009a017. [DOI] [PubMed] [Google Scholar]
- 47.Dutton PL. Redox potentiometry: determination of midpoint potentials of oxidation-reduction components of biological electron-transfer systems. Methods Enzymol. 1978:411–35. doi: 10.1016/s0076-6879(78)54026-3. [DOI] [PubMed] [Google Scholar]
- 48.Pierik AJ, Hagen WR, Redeker JS, Wolbert RB, et al. Redox properties of the iron-sulfur clusters in activated Fe-hydrogenase from Desulfovibrio vulgaris (Hildenborough) Eur. J. Biochem. 1992;209:63–72. doi: 10.1111/j.1432-1033.1992.tb17261.x. [DOI] [PubMed] [Google Scholar]
- 49.Massey V. Curti B, Ronchi S, Zanetti G, editors. A simple method for the determination of redox potentials. Flavins and Flavoproteins, Walter de Gruyter, Berlin. 1991, 66–6.
- 50.Singh R, Grigg C, Armstrong Z, Murphy ME, Eltis LD. Distal heme pocket residues of B-type dye-decolorizing peroxidase: arginine but not aspartate is essential for peroxidase activity. J. Biol. Chem. 2012;287:10623–10630. doi: 10.1074/jbc.M111.332171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thorell HD, Karlsson J, Portelius E, Nilsson T. Cloning, characterisation, and expression of a novel gene encoding chlorite dismutase from Ideonella dechloratans. Biochim. Biophys. Acta. 2002;1577:445–451. doi: 10.1016/s0167-4781(02)00446-3. [DOI] [PubMed] [Google Scholar]
- 52.Danielsson Thorell H, Beyer NH, Heegaard NH, Ohman M, Nilsson T. Comparison of native and recombinant chlorite dismutase from Ideonella dechloratans. Eur. J. Biochem. 2004;271:3539–3546. doi: 10.1111/j.0014-2956.2004.04290.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
suppinfo