Abstract
The recalcitrance of lignocellulose makes enzymatic hydrolysis of plant biomass for the production of second generation biofuels a major challenge. This work investigates an efficient and economic approach for the enzymatic hydrolysis of sugar beet pulp (SBP), which is a difficult to degrade, hemicellulose-rich by-product of the table sugar industry. Three fungal strains were grown on different substrates and the production of various extracellular hydrolytic and oxidative enzymes involved in pectin, hemicellulose, and cellulose breakdown were monitored. In a second step, the ability of the culture supernatants to hydrolyze thermally pretreated SBP was tested in batch experiments. The supernatant of Sclerotium rolfsii, a soil-borne facultative plant pathogen, was found to have the highest hydrolytic activity on SBP and was selected for further hydrolyzation experiments. A low enzyme load of 0.2 mg g–1 protein from the culture supernatant was sufficient to hydrolyze a large fraction of the pectin and hemicelluloses present in SBP. The addition of Trichoderma reesei cellulase (1–17.5 mg g–1 SBP) resulted in almost complete hydrolyzation of cellulose. It was found that the combination of pectinolytic, hemicellulolytic, and cellulolytic activities works synergistically on the complex SBP composite, and a combination of these hydrolytic enzymes is required to achieve a high degree of enzymatic SBP hydrolysis with a low enzyme load.
Keywords: Cellulases, Hemicellulases, Neurospora crassa, Sclerotium rolfsii, Trametes multicolor
1 Introduction
Enzymatic hydrolysis of lignocellulosic biomass is a widely employed strategy to obtain soluble sugars for the production of second generation biofuels [1–3]. Potential raw materials include fast growing energy crops such as switchgrass or Miscanthus, but the agricultural remnants sugar cane bagasse or sugar beet pulp (SBP) have also received attention [4, 5]. SBP is a by-product of the table sugar industry, is of low economic value, and is available in large quantities. In 2011, 271.6 million tons of sugar beets were harvested worldwide (Europe: 195.6 million tons) (http://faostat.fao.org/), which after saccharose extraction resulted in the production of approximately 68 million tons of wet SBP or 17 million tons of dried biomass [6]. The main carbohydrate constituents of SBP are glucose (19–23%), arabinose (16–24%) and galacturonic acid (19–23%), while the lignin content is comparatively low (ca. 2%). The chemical composition of SBP has been analyzed by several groups and differs annually and regionally (Supporting information, Table S1). SBP hydrolysis releases glucose, which can be fermentatively converted to ethanol, and hemicellulose-, or pectin-derived sugars, such as xylose and arabinose, for which efficient fermentation protocols and strains are currently being developed. The efficiency of enzymatic SBP hydrolysis is low and restricted by the complex composite structure, which hinders access of enzymes to their specific substrates. Current strategies designed to achieve fast and complete hydrolysis involve thermal or acid/base pretreatments in combination with high cellulase activities [6–8]. In previous studies hydrolyzation of SBP into soluble sugars was also accomplished by treatment with pectinase and cellulases [9, 10]. Despite intensive pretreatment and high enzyme dosages hydrolyzation efficiencies usually reach only 40–80% and the highest reported fermentative yield is 0.4 g ethanol per g SBP [11–14]. The main goals for SBP hydrolysis are: (i) to reduce the amount of process energy by optimizing enzymatic treatment; (ii) improved fermentative conversion of the main carbohydrate constituents into ethanol; and (iii) development of a low energy ethanol extraction method.
The hydrolytic power of wood-degrading fungi arises from a complex interplay of extracellular hydrolytic and redox enzymes activities classified in the carbohydrate-active enzyme (CAZy) database [15, 16]. Aside from the well-studied hydrolases, new research suggests that enzymatic attack on crystalline cellulose is initiated by the redox enzymes cellobiose dehydrogenase (CDH) and lytic polysaccharide monooxygenase (LPMO) forming a bi-enzymatic system [17–19], which introduces random cleavages into cellulose through monooxygenation [20]. The action of several polysaccharide hydrolases was found to be increased up to 2-fold after monooxygenase action [21]. As an alternative approach to increase the efficiency of hydrolysis of pectin and hemicellulose in SBP we propose to exploit the hydrolytic secretomes of fungi, which can adapt their metabolism to SBP degradation and secrete the enzymes needed for efficient breakdown of these polysaccharides.
In this study we investigate the secretomes from three fungal strains grown on SBP as the main carbon source. The aim was to identify a fungus equipped with an array of extracellular hydrolases and oxidases allowing the degradation of the major constituents of SBP: cellulose, hemicelluloses, and pectin. Trametes multicolor, a circumglobal hardwood-colonizing fungus, was chosen as a representative of lignocellulose-degrading white-rot fungi, which are a versatile source of the redox enzymes thought to increase the breakdown of recalcitrant polymers in SBP [22]. Sclerotium rolfsii is a soil borne facultative plant pathogen that causes considerable crop losses worldwide. Its broad host range includes more than 500 plant species including sugar beet. This fungus was chosen for its broad spectrum of secreted hemicellulases and pectinases [23]. Neurospora crassa, a saprotrophic organism commonly found on dead or burnt plant matter in tropical and subtropical climates [24] throughout the world was chosen for its broad substrate spectrum.
It is the objective of this work to provide an economic and efficient strategy for enzymatic SBP hydrolysis, aiming to minimize the pretreatment effort, enzyme load, and process time required for efficient SBP hydrolysis. In cultivated fungal strains the hydrolytic activities of various extracellular enzymes, the secretome, was differently induced by varied media components and the potential of the resulting culture supernatants was investigated with regards to their ability to hydrolyze SBP.
2 Materials and methods
2.1 Chemicals and raw material
Chemicals were purchased from Sigma (Steinheim, Germany), VWR (Darmstadt, Germany) or Roth (Karlsruhe, Germany). Dried SBP with a moisture content of 10.4% was obtained from the Sladorana d.o.o. sugar factory (ZŽupanja, Croatia) and was crushed with a blender to a particle size of 0.2–0.4 mm. Cellulase (liquid crude extract) from Trichoderma reesei ATCC 26921 was obtained from Sigma.
2.2 Fungal strains and culture conditions
S. rolfsii CBS 151.31 and N. crassa CBS 232.56 were obtained from the Centraalbureau voor Schimmelcultures (Utrecht, The Netherlands) and T. multicolor MB 49 was obtained from the culture collection of the Austrian Center of Biological Resources and Applied Mycology (Vienna, Austria). Strains were maintained and periodically subcultured on potato dextrose agar at 25°C. Submerged cultures were grown in unbaffled Erlenmeyer flasks filled with 100 mL of medium inoculated with 3 cm2 of sliced agar plugs taken from 4-day old agar plates. Cultures were grown for 8 to 12 days at 25°C (T. multicolor, N. crassa ) and 30°C (S. rolfsii ) in a shaking incubator operated at 120 rpm. The basic medium contained 1.5 g L–1 MgSO4 · 7H2O and 0.3 mL L–1 trace element solution [23]. Three different carbon sources (10 g L–1 α-cellulose, 10 g L–1 glucose, or 10 g L–1 SBP) in combination with two different nitrogen sources (5 g L–1 peptone from meat or a mix of 1 g L–1 NH4NO3, 1.2 g L–1 KH2PO4 and 0.6 g L–1 KCl) were investigated. Prior to sterilization (121°C, 20 min) cultivation media were adjusted to pH 5.5 with phosphoric acid. Culture supernatants from S. rolfsii and T. multicolor were harvested after 11 days and N. crassa after 6 days. Coarse particles and adhering mycelium were removed with a sieve (mesh size 2 mm), residual solids were then separated by fast vacuum filtration using a Whatman disc filter with a pore size of 8–12 μm. Clear culture supernatants were used for hydrolysis experiments without further treatment.
2.3 Enzymatic assays
Hydrolytic activities were determined by quantifying the reducing sugars liberated from 1% (w/v) solutions of hemicellulosic substrates using the 3, 5-dinitrosalicylic acid (DNSA) assay procedure [25]. Substrates used were Birchwood xylan (xylanases), mannan from Saccharomyces cerevisiae (mannanases), red-beet arabinan (arabinases), carboxymethyl cellulose (cellulases), and pectin from citrus peel (pectinase). Culture samples were centrifuged (10,000 × g, 5 min) and the resulting supernatant was mixed with the substrates in 50 mM citrate buffer, pH 5.0 (1 mL final volume) and incubated at 40°C for 60 minutes under constant agitation (800 rpm) in an Eppendorf Thermoshaker (Eppendorf, Germany). Diluted sample aliquots (600 μL final volume) resulting in absorption values within the range of the calibration standards were mixed with 600 μL of DNSA reagent containing 10.0 g L–1 DNSA, 0.5 g L–1 sodium sulfite, 10 g L–1 sodium hydroxide, and 2 mL L–1 phenol. The mixtures were incubated for 15 minutes at 95°C before adding 200 μL of a 40 g L–1 potassium sodium tartrate solution (1.4 mL final volume) and immediately measuring the absorbance at 575 nm. Product concentrations were calculated from calibration curves generated with the corresponding reducing sugars. One unit of enzymatic activity is defined as the amount of enzyme that releases 1 μmol of reducing sugar per minute under the specified assay conditions.
Laccase (EC 1.10.3.2) activity was assayed by following the oxygen-dependent oxidation of 2, 2'-azinobis-(3-ethylbenzthiazoline-6-sulfonic acid) (ABTS) at 30°C and at a wavelength of 420 nm (ε420 = 36 mM–1 cm–1). The assays (1 mL final volume) contained 1 mM ABTS in 86 mM acetate buffer, pH 4.0. One unit of laccase activity catalyzes the formation of 1 μmol ABTS cation radical per minute under the specified assay conditions.
Manganese peroxidase (MnP; EC 1.11.1.13) activity was assayed by the hydrogen peroxide-dependent oxidation of 2,6-dimethoxyphenol (syringol) at 30°C and at a wavelength of 469 nm (ε469 = 27.5 mM–1 cm–1) [26]. Assays (1 mL final volume) contained 1 mM syringol, 0.5 mM manganese sulfate and 0.1 mM hydrogen peroxide in 77 mM sodium tartrate buffer, pH 4.5. One unit of MnP activity catalyzes the oxidation of 1 μmol syringol per minute under the specified assay conditions.
CDH (EC 1.1.99.18) activity was measured by the lactose-dependent reduction of 2,6-dichloroindophenol (DCIP) at 30°C and at a wavelength of 520 nm (ε520 = 6.80 mM–1 cm–1) [27]. Assays (1 mL final volume) contained 300 μM DCIP and 30 mM lactose in 78 mM sodium acetate buffer, pH 4.0. In addition, sodium fluoride (10 mM final concentration) was added to suppress laccase activity, which would regenerate reduced DCIP and mask CDH activity. One unit of CDH activity catalyzes the oxidation of 1 μmol of lactose per minute under the specified assay conditions.
Protein concentrations were determined by the dye-binding method of Bradford using a prefabricated assay from Bio-Rad Laboratories (Hercules, CA) and bovine serum albumin as calibration standard.
2.4 Measurement of H2O2 formation
Culture samples were analyzed for total oxygen-reducing activity by measuring the time-dependent formation of hydrogen peroxide with an Amplex Red/horseradish peroxidase assay [28, 29]. For each hydrogen peroxide molecule generated one equivalent of Amplex Red is cleaved to the highly fluorescent resorufin (λEx = 569 nm; λEm =585 nm) in a peroxidase dependent reaction. All assays were performed in 96-well plates (200 μL total volume) using a plate reader (EnSpire Multimode, Perkin Elmer, Waltham, MA). Assays were started by mixing 20 μL of sample solution with 180 μL of a reaction mix containing Amplex Red (50 μM) and peroxidase (7.14 U mL–1) in 75 mM phosphate buffer, pH 6.0. Sugar substrates (glucose, cellobiose, xylose, and galactose) were added to a final concentration of 500 μM. To elucidate the contribution of media components on the overall peroxide production, sample aliquots were centrifuged in mini-spin columns (cut-off 10 kDa) and flow throughs measured as blanks. To detect lytic polysaccharide monooxygenase activity, CDH IIA from N. crassa was employed at a concentration of 0.01 or 0.03 mg mL–1 to serve as an electron donor. One unit of oxygen-reducing enzymatic activity corresponds to the formation of 1 μmol of H2O2 per min.
2.5 SDS-PAGE and zymogram
Fungal secretomes were visualized by SDS-PAGE and staining for cellulase activity. A 12% SDS-PAGE separation gel was co-polymerized with 0.1% carboxymethyl cellulose (CMC). Samples were mixed with Laemmli-buffer without heating and loaded onto the gel. One half of the gel was stained with Coomassie Brilliant Blue; the other was used for the zymogram. For activity staining the gel was washed for 60 minutes with a 1% Triton X-100 solution to remove SDS. After rinsing with MilliQ-water the gel was incubated for 60 minutes in 50 mM citrate buffer, pH 5.5, to degrade CMC. Staining was performed for 30 min in a 0.3% Congo red solution. After washing with 1 M NaCl clear, diffuse zones within a red background indicated cellulase activity.
2.6 HPLC analysis
Carbohydrates released were quantified by HPLC using a DIONEX DX 500 system (DIONEX, Sunnyvale, CA) and a CarboPac PA1 column coupled to an electrochemical ED40 detector (DIONEX). Samples (20 μL) were applied with a GP50 gradient pump and separated at a flow rate of 1 mL min–1 at a constant temperature of 30°C and verified by using standard substances and the standard addition method. Eluents were 18 mM NaOH (A), 200 mM NaOH (B) and 100 mM NaOH containing 150 mM sodium acetate (C). Initially, the column was equilibrated with 100% A for 15 min before sample application; then a gradient from 0 to 30% B was applied for 16 min. Next, a gradient from 30 to 82% B and from 0 to 12% C was applied for 2 min followed by a gradient from 82 to 0% B and from 12 to 100% C within 3 min, which was maintained for 4 min before changing to 100% B within 0.5 min. Finally, a gradient from 100 to 0% B was run for 3 min before the column was re-equilibrated with 100% A.
2.7 SBP hydrolysis
Hydrolysis of SBP was investigated in Erlenmeyer flasks containing 66 g L–1 beet pulp in 50 mM citrate buffer, pH 5.0 (200 mL total volume). Flasks were sterilized by hot steam (121°C, 20 min), which had a swelling effect on SBP particles, but did not cause the release of notable amounts of reducing sugars (∼0.002 g g–1 SBP). This is in good agreement with a previous study using the same method [14]. One mL of the fungal crude extracts was added to the flasks, regardless of their enzyme or protein concentration. Experiments were run for 48 hours at a temperature of 30°C and under constant agitation (120 rpm). Subsequent optimization experiments were performed as above, but SBP concentrations and enzyme loads were varied. Liberated sugars were quantified at the beginning (no hydrolyzation) and at the end of the experiments (120 hours) by HPLC analysis. Yields were calculated as described previously [30] (Supporting information, Table S1).
3 Results and discussion
3.1 Hydrolytic activities secreted by fungi
The fungal strains were cultivated in submerged culture using different media components to investigate the production of fungal pectinolytic, hemicellulolytic, and cellulolytic enzymes on different substrates. Glucose-based media were used as a control for secretion under carbon-catabolite repressing conditions. A high degree of physiological heterogeneity was observed (Supporting information, Fig. S1). Mycelia grew well in all media with the exception of N. crassa, which showed little growth on cellulose/NH4NO3 and glucose/NH4NO3 lacking biotin. Mycelium was observed only after three weeks of incubation and the level of extracellular protein was below the detection limit of the Bradford assay. In the other media N. crassa grew faster than T. multicolor or S. rolfsii as was obvious from the accumulation of mycelium, the rapid increase in extracellular protein and enzymatic activities. In media containing SBP or cellulose small pellets formed around the particles, whereas in glucose containing media large pellets (5-20 mm) were found. The production of enzymatic activities was followed by periodic sampling throughout the cultivation; examples of the three fungi grown on SBP/peptone-supplemented media are shown in Supporting information, Fig. S2. Maximum enzymatic activities found in cultures of N. crassa, S. rolfsii and T. multicolor in different media are summarized in Table 1.
Table 1.
Extracellular enzymatic activities of fungal cultures grown on different carbon and nitrogen sources
| Cultured organism | C-Source | N-Source | Hydrolytic activity (U L–1)a) | Extracellular protein (mg L–1) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Pectinase | Xylanase | Arabinase | CMCase | Laccase | MnP | CDH | ||||
| S. rolfsii | SBP | Peptone | 713 ± 25 (11) | 447 ± 17 (4) | 1333 ± 15 (7) | 364 ± 102 (11) | 200 ± 23 (7) | 0 | 49 ± 6 (9) | 277 ± 6 (7) |
| NH4NO3 | 563 ± 119 (11) | 613 ± 4 (4) | 784 ± 116 (7) | 170 ± 16 (11) | 74 ± 22 (9) | 0 | 39 ± 13 (11) | 72 ± 15 (9) | ||
| Cellulose | Peptone | 372 ± 3 (11) | 842 ± 23 (4) | 32 ± 9 (11) | 167 ± 56 (9) | 364 ± 7 (11) | 0 | 209 ± 50 (11) | 536 ± 24 (11) | |
| NH4NO3 | 35 ± 11 (11) | 76 ± 15 (11) | 0 | 0 | 83 ± 20 (11) | 0 | 0 | <LODb) | ||
| Glucose | Peptone | 64 ± 9 (11) | 19 ± 2 (11) | 0 | 0 | 287 ± 33 (9) | 8 ± 1 (7) | 0 | 150 ± 19 (11) | |
| NH4NO3 | 0 | 229 ± 4 (9) | 0 | 0 | 17 ± 6 (11) | 22 ± 6 (11) | 0 | <LODb) | ||
| T. multicolor | SBP | Peptone | 311 ± 55 (11) | 499 ± 20 (7) | 977 ± 18 (7) | 275 ± 24 (11) | 223 ± 44 (11) | 188 ± 34 (11) | 0 | 200 ± 20 (7) |
| NH4NO3 | 435 ± 133 (11) | 372 ± 41 (9) | 826 ± 166 (7) | 203 ± 50 (11) | 39 ± 14 (11) | 46 ± 13 (11) | 140 ± 5 (11) | 125 ± 37 (11) | ||
| Cellulose | Peptone | 67 ± 3 (9) | 326 ± 59 (7) | 20 ± 9 (9) | 358 ± 105 (11) | 43 ± 8 (11) | 110 ± 7 (11) | 0 | 194 ± 10 (7) | |
| NH4NO3 | 72 ± 3 (9) | 0 | 0 | 0 | 2 ± 1 (9) | 2 ± 1 (9) | 0 | <LODb) | ||
| Glucose | Peptone | 19 ± 9 (11) | 0 | 0 | 0 | 234 ± 9 (9) | 243 ± 10 (9) | 0 | 31 ± 1 (9) | |
| NH4NO3 | 0 | 0 | 0 | 0 | 7 ± 2 (9) | 0 | 0 | <LODb) | ||
| N. crassa | SBP | Peptone | 119 ± 2 (4) | 400 ± 40 (4) | 11 ± 5 (6) | 117 ± 43 (4) | 0 | 0 | 22 ± 1 (11) | 295 ± 35 (8) |
| NH4NO3 | 84 ± 2 (4) | 32 ± 3 (4) | 0 | 21 ± 1 (4) | 0 | 0 | 0 | 16 ± 1 (8) | ||
| Cellulose | Peptone | 70 ± 5 (6) | 396 ± 1 (4) | 10 ± 5 (6) | 76 ± 11 (6) | 0 | 0 | 21 ± 2 (11) | 319 ± 20 (8) | |
| Glucose | Peptone | 71 ± 1 (6) | 0 | 0 | 0 | 0 | 0 | 35 ± 17 (6) | 72 ± 3 (8) | |
day of maximum activity is shown in brackets
<LOD = Measurement was below level of detection
Pectinolytic activity, represented by pectinase and arabinase activities, was highest in S. rolfsii cultures, followed by T. multicolor. The highest pectinase activities were found in media based on SBP with a 1.5 to 5-fold higher activity than cellulose-based media. In S. rolfsii and N. crassa cultures peptone increased pectinase production, whereas for T. multicolor there was no difference in peptone containing cultures compared to those containing NH4NO3. In SBP media enzymatic activity did not decrease when peptone was replaced by NH4NO3, but did in cellulose-based media.
Hemicellulolytic activity showed a similar, culture medium-dependent pattern. SBP and cellulose induced the formation of xylanase activity, whereas glucose repressed its formation. S. rolfsii and T. multicolor both produced high xylanase activities, whereas little xylanase activity was present in N. crassa cultures. In combination with SPB, NH4NO3 supplemented cultures produced similar or even higher activities than those supplemented with peptone, whereas the combination cellulose/NH4NO3 showed no or very low enzymatic activities.
Cellulolytic activity was found in cultures of all three fungi. The dependence of CMCase production on the medium composition followed that of pectinolytic and hemicellulolytic enzymes. The activities observed were roughly equal for S. rolfsii and T. multicolor, but 5 to 10-fold lower for N. crassa cultures. The production of extracellular enzyme activities was consistent with the extracellular protein concentration measured in culture supernatants. The amount of enzyme bound to solid substrates was not determined. Culture supernatants with high enzymatic activities were evaluated for their potential to hydrolyze SBP in small batch conversions (Table 2). From the consistently high concentrations of released galacturonic acid, arabinose, and rhamnose, it can be deduced that all culture supernatants possess sufficient pectinase activity, even the N. crassa cultures which had a substantially lower activity. Pectinase has been used in previous studies to synergistically increase the hydrolysis of cellulose [10]. Hemicelluloses are most efficiently hydrolyzed by the S. rolfsii secretome, followed by T. multicolor. Only a low concentration of hemicellulose constituents was found in samples treated with N. crassa culture supernatants. The cellulolytic activity, judged by the final glucose concentration, was highest after treatment with S. rolfsii culture supernatant. Here, cellulose degradation was about 1.7-fold higher for cellulose/peptone supernatants over SBP/peptone supernatants as judged by the amount of glucose released. This higher cellulolytic activity of the cellulose/peptone culture supernatant over SBP/peptone was also observed for N. crassa cultures, but not for T. multicolor. Culture supernatants of the two latter fungi were much less efficient in cellulose degradation and released only 12 to 42% of the glucose when compared to SBP treated with the S. rolfsii secretome. The amount of extracellular protein found in SBP/peptone media was comparable for all three fungi. In the hydrolysis experiments fructose was also detected and the concentration measured increased over time. Tightly bound residual saccharose, which is extracted from SBP by hydrolysis and cleaved into glucose and fructose by invertase presumably contributes to the release of fructose.
Table 2.
Carbohydrate release from SBP by different fungal culture supernatants after 48 hours
| Cultured organism | Culture supernatant specification | Hydrolyzed sugar (mg g–1 SBP)a) | Total yield (g g–1 SBP) | |||||
|---|---|---|---|---|---|---|---|---|
| Hexonic acid | Rhamnose | Arabinose | Galactose | Fructose | Glucose | |||
| S. rolfsii | SBP/peptone | 59.0 (29) | 1.4 | 84.9 (44) | 40.9 (82) | 26.4 | 77.8 (36) | 0.29 |
| Cellulose/peptone | 72.4 (35) | 1.9 | 91.8 (47) | 36.7 (73) | 36.9 | 133.9 (62) | 0.37 | |
| T. multicolor | SBP/peptone | 67.3 (33) | 1.1 | 56.1 (29) | 22.3 (45) | 19.1 | 32.3 (15) | 0.20 |
| Cellulose/peptone | 67.5 (33) | 0.9 | 6.5 (3) | 19.1 (38) | 15.2 | 16.4 (8) | 0.13 | |
| N. crassa | SBP/peptone | 64.3 (31) | 0.4 | 16.0 (8) | n.d.b) | 7.6 | 22.7 (11) | 0.11 |
| Cellulose/peptone | 67.3 (33) | 1.1 | 56.1 (29) | n.d.b) | 19.1 | 32.3 (15) | 0.18 | |
Yields of individual sugars (in %) are shown in brackets.
n.d. = not detected
A visualization of the secretomes from S. rolfsii, T. multicolor, and N. crassa cultures grown on SBP/peptone is shown in Fig. 1. SDS-PAGE shows differences in the extracellular proteins present in culture supernatants obtained from cultures grown on cellulose or SBP. The patterns of the protein bands observed for all three fungi differ strongly between both carbon sources. The highest number of protein bands was counted in the S. rolfsii samples. Zymogram analysis on carboxymethyl cellulose (CMC) reveals a multitude of different cellulolytic enzymes secreted by the fungi, however, the pattern differs little between cellulose or SPB culture supernatants.
Figure 1.
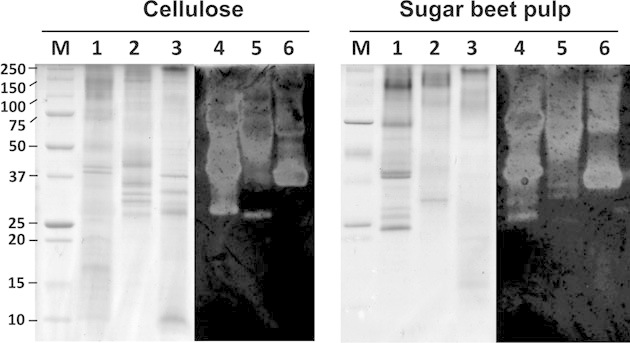
SDS-PAGE and zymogram of fungal culture supernatants grown on cellulose and SBP media. SDS-PAGE lanes show Mass Standard (M), and culture supernatants of S. rolfsii (1), T. multicolor (2) and N. crassa (3); CMC-zymogram lanes show culture supernatants of S. rolfsii (4), T. multicolor (5), and N. crassa (6).
The profiles of secreted hydrolytic activities demonstrate the adaptation of the fungi to growth on the respective substrates. In the case of S rolfsii, pectinase and arabinase activities were strongly induced by growth on SBP, which is rich in pectin. Enhanced cellulase activities were found in cultures supplied with cellulose as the main carbon source. However, it has to be noted that many hydrolases actively bind to their substrates via carbohydrate-binding modules and therefore can only be partially detected in culture supernatants.
3.2 Oxidoreductase activities secreted by fungi
Culture supernatants of all three fungi were also analyzed for laccase-, MnP and CDH activity and hydrogen peroxide production as an indicator for oxidase and lytic polysaccharide monooxygenase activity (Table 1). Oxidoreductase activities are reported to increase both lignin and cellulose degradation. Laccase activity was found in S. rolfsii and T. multicolor cultures and was 3 to 10-fold higher in peptone-supplemented media. Interestingly, laccase activity was also observed in media containing glucose, which is known to repress laccase induction. It is possible that laccase was formed by mycelium in the inner part of the large pellets where the glucose concentration is very low, or that laccase was formed after complete metabolization of glucose in the medium [31]. High MnP activity was only found in T. multicolor cultures; N. crassa did not produce either enzyme activity. CDH activity was produced by all fungi. The large differences in CDH activity observed for the different media may have resulted from different enzyme production under the different conditions. Alternatively as CDH binds to cellulose, differences in the amount of residual cellulose present at the time of sampling may account for differences in the enzyme activities observed.
Total hydrogen peroxide-producing activities of crude fungal extracts were measured alone or after supplementation with different sugars (glucose, xylose, galactose, cellobiose, final concentrations 500 μM) to estimate the contribution of specific sugar oxidizing enzymes such as CDH, lytic polysaccharide mono-oxygenase (LPMO), glucose oxidase, or galactose oxidase on peroxide formation (Fig. 2A). Negative controls were conducted by centrifuging sample aliquots in mini-spin columns with a cut-off of 10 kDa to obtain protein-free permeates (controls are shown in Supporting information, Fig. S3). T. multicolor showed the highest H2O2 formation rates among the fungi. Addition of glucose increased H2O2 formation four-fold in supernatants derived from cellulose-grown cultures, but only ca. 1.2-fold in supernatants from SBP, which had already a higher initial H2O2 formation. The effect of sugars other than glucose and cellobiose was insignificant. The culture supernatants from S. rolfsii showed a high H2O2-producing activity and the addition of sugars did not increase the peroxide-producing activity significantly. Peroxide production was, however, more than three-fold higher in cultures grown on SBP when compared to cultures grown on cellulose. In contrast to the two other fungi, supernatants derived from N. crassa showed only weak oxygen-reducing activity, which was 14 to 240 times lower compared to S. rolfsii or T. multicolor supernatants. There was, however, a detectable reaction upon addition of cellobiose, most likely due to the presence of CDH. The H2O2-producing activity observed in fungal crude supernatants illustrates the presence of different oxygen-utilizing redox enzymes. Hydrogen peroxide formation by the basidiomycetes S. rolfsii and T. multicolor was notably higher when compared to the ascomycete N. crassa. The oxygen-reducing activity was generally higher in cultures grown on SBP, and might be induced by the presence of lignin. However, it cannot be stated with certainty whether the role of the detected oxidoreductases is to degrade the lignin in SBP or enhance the breakdown of other polymers.
Figure 2.
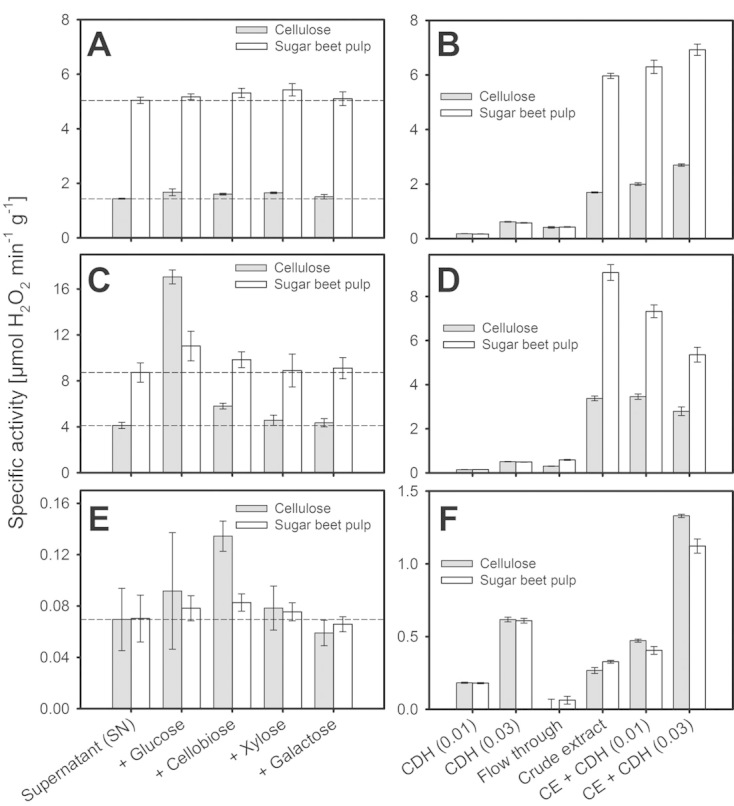
Hydrogen peroxide-forming enzymatic activities in crude extracts from (A) S. rolfsii alone, or (B) upon addition of CDH and 500 μM cellobiose, (C, D) T. multicolor and (E, F) N. crassa. Protein-free supernatants (flow through) obtained by centrifugal ultracentrifugation (cut-off 10 kDa) were used as controls. Sugar substrates were added to a final concentration of 500 μM. Dashed lines indicate the activity of the crude extract without addition of sugars. Error bars represent the standard deviation from 4 independent experiments. Control experiments are shown in Supporting information, Fig. S3.
To obtain an indication for LPMO activity, the electron donor CDH (recombinant CDH IIA from N. crassa; produced and purified as described in [18]) was added to culture supernatants. In the absence of a cellulosic substrate, LPMOs were found to generate H2O2 in a futile reaction [29]. H2O2 formation was observed after addition of CDH to cultures of N. crassa, which is consistent with the literature in which N. crassa has been reported to secrete CDH and LPMOs when grown on cellulosic substrates [32, 33]. The amount of H2O2 observed correlated with the amount of CDH added. S. rolfsii cultures showed a similar effect upon the addition of CDH, but when compared to the already high level of H2O2 formation, the effect of CDH was not as prominent as in the N. crassa samples. For both cultures this indicates the presence of LPMO in the culture supernatants. The opposite effect was observed for T. multicolor culture supernatants. Samples from cellulose or SBP grown cultures showed a high H2O2 formation rate and unexpectedly this was reduced by addition of CDH. Currently we have no explanation of this effect, but we assume that the addition of N. crassa CDH inhibits the efficient T. multicolor CDH/LPMO system. It is worth noting that the CDH IIA used in these experiments was from N. crassa and may not work equally well with LPMOs from other organisms.
3.3 Hydrolysis of SBP
As S. rolfsii was the best producer of pectinase, hemicellulase, and cellulase activities, and showed the highest level of SBP hydrolysis, this fungus was chosen for small batch hydrolysis experiments. In a first series of experiments the effect on the cellulase-secretome produced in the presence of SBP or cellulose was investigated. Culture supernatants from SBP/peptone and cellulose/peptone media and blends thereof were used in different protein to SBP weight ratios to investigate the release of monosaccharides and aldonic acids. The total yield of major hydrolysis products is reported in Tables 1, 2, and 3. Additional minor products such as mannose, di- and trisaccharide products, aldonic acids, methanol, and acetic acid are not listed, and were not included in the calculation of the yield. The cumulated peak area of these minor/unidentified products made up to about 10% of the total peak area. Based on a fixed SBP concentration of 66 g L–1 (59 g L–1 dry weight) enzyme loads were gradually increased from 0.05 to 0.2 mg protein per g of SBP (Table 3).
Table 3.
Effect of S. rolfsii culture supernatant enzyme load and cellulose addition on SBP hydrolysis after 120 hours
| Culture supernatant specification | Concentration (mg protein g–1 SBP) | Hydrolyzed sugar (mg g–1 SBP)a) | Total yield (g g–1 SBP) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Hexuronic acids | Rhamnose | Arabinose | Galactose | Fructose | Xylose | Glucose | |||
| S. rolfsii SBP/peptone | 0 | 0 (0) | 0 | 1.4 ± 0.1 (0) | 0.1 ± 0.1 (0) | 6.9 ± 0.7 | 0 | 8.4 ± 0.4 (2) | 0.02 |
| SBP/peptone | 0.05 | 64.9 ± 0.7 (32) | 3.2 ± 0.2 | 104.6 ± 2.3 (54) | 12.5 ± 0.6 (25) | 14.7 ± 1.1 | 0.7 ± 0.5 (4) | 23.1 ± 0.4 (11) | 0.22 |
| SBP/peptone | 0.1 | 76.0 ± 4.9 (37) | 4.2 ± 0.2 | 123.1 ± 2.3 (63) | 15.4 ± 0.1 (31) | 13.6 ± 1.1 | 0.8 ± 0.5 (5) | 22.9 ± 0.3 (11) | 0.26 |
| SBP/peptone | 0.2 | 88.9 ± 7.9 (43) | 5.6 ± 0.5 | 126.6 ± 7.2 (65) | 20.2 ± 1.0 (40) | 13.5 ± 1.5 | 0.4 ± 0.1 (3) | 24.7 ± 1.4 (12) | 0.28 |
| S. rolfsii cellulose/peptone | 0 | 0 | 0 | 1.2 ± 0.8 (0) | 0.1 ± 0.1 (0) | 5.5 ± 3.4 | 0 | 7.5 ± 5.2 (2) | 0.01 |
| Cellulose/peptone | 0.05 | 22.9 ± 1.4 (11) | 0.9 ± 0.1 | 29.1 ± 2.3 (15) | 6.6 ± 0.5 (13) | 14.3 ± 1.7 | 0.1 ± 0.1 (1) | 25.9 ± 2.0 (12) | 0.10 |
| Cellulose/peptone | 0.1 | 39.8 ± 1.6 (19) | 1.5 ± 0.1 | 51.7 ± 2 (26) | 8.9 ± 0.5 (18) | 14.6 ± 1.2 | 0.1 ± 0.1 (1) | 34.1 ± 1.1 (16) | 0.15 |
| Cellulose/peptone | 0.2 | 56.2 ± 4.0 (27) | 2.0 ± 0.1 | 77.4 ± 2.8 (40) | 10.8 ± 0.4 (22) | 13.5 ± 1.0 | 0.4 ± 0.1 (2) | 43.8 ± 1.0 (20) | 0.20 |
| SBP/peptone +cellulose/peptone | 0.1 / 0.1 | 79.3 ± 3.9 (39) | 4.34 ± 0.2 | 119.7 ± 3.3 (61) | 17.0 ± 0.1 (34) | 13.5 ± 0.2 | 0.9 ± 0.3 (6) | 36.9 ± 0.5 (17) | 0.27 |
| SBP/peptone + cellulose/peptone + cellulase | 0.1 / 0.1 / 1.0 | 79.6 ± 5.1 (39) | 4.4 ± 0.3 | 118.8 ± 5.1 (61) | 17.1 ± 1.8 (34) | 12.7 ± 0.8 | 3.5 ± 0.1 (22) | 101.3 ± 4.4 (47) | 0.34 |
| SBP/peptone + cellulose/peptone + cellulase | 0.1 / 0.1 / 10 | 77.3 ± 2.8 (38) | 4.5 ± 0.2 | 117.5 ± 3.9 (60) | 18.2 ± 1.6 (36) | 12.4 ± 0.6 | 4.1 ± 0.1 (26) | 142.9 ± 4.0 (66) | 0.38 |
| SBP/peptone + cellulase | 0.2 / 1.0 | 83.8 ± 1.4 (41) | 5.3 ± 0.2 | 119.2 ± 1.9 (61) | 20.6 ± 0.3 (41) | 12.5 ± 0.4 | 3.5 ± 0.2 (22) | 97.2 ± 1.3 (45) | 0.34 |
| SBP/peptone + cellulase | 0.2 / 10 | 83.8 ± 3.3 (41) | 5.4 ± 0.1 | 118.1 ± 3.12 (61) | 21.1 ± 2.0 (42) | 12.5 ± 0.6 | 4.7 ± 0.1 (30) | 143.2 ± 0.8 (67) | 0.39 |
| Cellulase | 1 | 2.6 ± 1.4 (1) | 0 | 6.8 ± 3.4 (3) | 0.8 ± 0.4 (2) | 7.3 ± 2.8 | 2.5 ± 0.3 (15) | 86.1 ± 9.0 (40) | 0.11 |
| Cellulase | 10 | 1.1 ± 1.0 (1) | 0.4 ± 0.1 | 17.1 ± 0.1 (9) | 1.0 ± 0.2 (2) | 4.6 ± 0.2 | 4.6 ± 1.8 (29) | 131.9 ± 0.9 (61) | 0.16 |
Yields of individual sugars (in %) are shown in brackets
The degradation of pectin and hemicellulose can be followed by the formation of hexuronic acids, arabinose, galactose, rhamnose, and xylose. Degradation of pectin and hemicellulose was found to be more efficiently catalyzed by the SBP/peptone culture supernatant than by the cellulose/peptone supernatant. An exception was the release of fructose, which was similar for both supernatants and all concentrations, which points towards the release of residual saccharose. The formation of glucose by the cellulose/peptone supernatant was higher at all supernatant concentrations tested. A 1:1 mixture of SBP/peptone and cellulose/peptone supernatant exhibited the same characteristics as the identical concentrations of SBP/peptone for pectin degradation, but released a lower amount of glucose when compared to the cellulose/peptone supernatant alone.
To overcome low cellulose hydrolysis, commercial cellulase from T. reesei was added to the final concentrations of 1 and 10 mg g–1 SBP. Cellulase addition increased hydrolysis of cellulose and xylan, but had no apparent effect on pectin hydrolysis. Interestingly, addition of cellulase alone resulted in the release of notable amounts of xylose, which was higher than the levels of xylose released by crude extracts alone. This points toward a synergistic effect of cellulose and hemicellulose degradation. Highest yields (0.39 g g–1 SBP) were obtained using 0.2 mg g–1 SBP/peptone supernatant in combination with 10 mg g–1 cellulase. Time-resolved hydrolyses show that under these conditions maximum conversion was achieved after 24 hours, whereas SBP treated solely with S. rolfsii supernatant shows a slower increase of sugar constituents over time (Supporting information, Fig. S5). Lowering the SBP concentration to 20 g L–1 (17.9 g L–1 dry weight) while increasing the enzyme load to 0.4 mg g–1 protein and 17.5 mg cellulase g–1 SBP resulted in almost complete cellulose hydrolysis and increased the yield to 0.65 g g–1 SBP (Supporting information, Table S2, and Fig. S4). These data suggest that pectin and hemicelluloses are the primary targets of the S. rolfsii secretome, while cellulose cannot be degraded with equal efficiency by the cellulases produced by this fungal strain. Higher glucose yields were obtained from cellulose/peptone media, which have a higher cellulolytic activity. This indicates that the cellulolytic activity present in the SBP/peptone supernatant is not sufficient and supplementation with cellulases is necessary. Indeed, the addition of cellulase from T. reesei increased the hydrolysis of SBP considerably.
The enzyme loads used in this study are low compared to those reported in the literature, but resulted in a highly efficient hydrolyzation of SBP polymers. In accordance with the findings of [10], and Leijdekkers et al. [34] we can state that a concerted action of many enzymes is needed to hydrolyze the components of cellulose, hemicellulose, and pectin. Several commercial enzyme mixtures have been investigated, showing similar hemicellulose hydrolyzation yields of 65 to 90% [10, 35] but required a much higher enzyme load (usually 1%).
Typically, a high cellulase load is needed to obtain sufficient enzymatic hydrolysis of cellulose. Certainly the amount reported by Sutton and Peterson et al. [13] is very high (60 mg g–1 Celluclast 1.5 L (Novozymes) plus 30 mg g–1 pectinase) and was used to investigate the maximum ethanol yield, but other groups also report high enzyme dosages, e.g. 234.8 μL g–1 [8] or 10.66 mL kg–1 [34]. Also, Foster et al. [6] stated that the “level of commercially available fungal enzymes required to increase ethanol yields [from SBP] to distillable levels may be too high” due to the high cost of enzymes. Ammonium pre-treatment was suggested to increase yields, but actually the yield of hydrolysed carbohydrates was higher in untreated samples. With acid pre-treatment Zheng et al. achieved a sugar yield of 62% and subsequently an ethanol yield of 0.4 g EtOH/g SBP [8]. Earlier, Kühnel et al. [7] obtained with a combination of thermal and acid pretreatment a solubilization of 62 – 90% of glucose. Our results suggest that enzymatic hydrolysis by optimized enzyme preparations can replace pre-treatment procedures and is an applicable strategy to reduce the cost of ethanol production from SBP.
4 Concluding remarks
We identified the plant pathogenic fungus S. rolfsii as a versatile producer of a mix of pectinolytic, hemicellulolytic, cellulolytic, and polymer-degrading redox enzymes when grown on a cheap SBP medium, which supports the observation that the composition of the exoenzyme system of a fungus reflects the metabolic adaption to the respective substrate [36]. The data obtained in this study show that S. rolfsii, a plant pathogen with a host range including green plants, grasses and vegetables, possesses a potent hemicellulolytic arsenal of enzymes which, when grown on cellulose, adapts to produce more cellulolytic enzymes. The white-rot fungus T. multicolor produces less hemicellulases on SBP than S. rolfsii, but more of the oxidoreductases also required to degrade this substrate. The saprotroph N. crassa produces hemicellulases and cellulases on SBP, but less oxidoreductases. The latter two secretomes were less efficient in SBP hydrolysis. Analysis of the conversion products of SBP by S. rolfsii culture supernatants showed that this fungus is a very efficient degrader of hemicellulose, but degradation of cellulose appears to be a limitation. Supplementation with a cheap cellulose mixture from T. reesei dramatically increased cellulose degradation. Even under non-optimized conditions and without pre-treatment of SBP a large fraction of pectin, hemicelluloses and cellulose present in SBP were hydrolyzed by very low enzyme loads.
Acknowledgments
The authors thank Cindy Lorenz for excellent technical support. This work has been supported by the doctoral program “BioTop – Biomolecular Technology of Proteins” (FWF W1224) of the Austrian Science Fund FWF, the European Commission (project BIOENERGY FP7-PEOPLE-2013-ITN-607793), the Austrian Agency for International Mobility, OEAD (bilateral scientific project Austria-Croatia, grant Nr. 13-2010), and the Ministry of Science, Education and Sport Republic of Croatia (Grant No. 058-0581990-1997 and 058-0581990-2004).
The authors declare no financial or commercial conflict of interest.
Glossary
Abbreviations
- ABTS
2,2'-azinobis-(3-ethylbenzthiazoline-6-sulfonic acid)
- CDH
cellobiose dehydrogenase
- DCIP
dichloroindophenol
- DNSA
3,5-dinitrosalicylic acid
- LPMO
lytic polysaccharide monooxygenase
- MnP
manganese peroxidase
- SBP
sugar beet pulp
Supporting Information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re-organized for online delivery, but are not copy-edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
suppinfo
References
- 1.Jordan DB, Bowman MJ, Braker JD, Dien BS, et al. Plant cell walls to ethanol. Biochem. J. 2012;442:241–252. doi: 10.1042/BJ20111922. [DOI] [PubMed] [Google Scholar]
- 2.Gírio FM, Fonseca C, Carvalheiro F, Duarte LC, et al. Hemicelluloses for fuel ethanol: A review. Bioresource Tech. 2010;101:4775–4800. doi: 10.1016/j.biortech.2010.01.088. [DOI] [PubMed] [Google Scholar]
- 3.Kumar R, Singh S, Singh OV. Bioconversion of lignocellulosic biomass: biochemical and molecular perspectives. J. Ind. Microbiol. Biotechnol. 2008;35:377–391. doi: 10.1007/s10295-008-0327-8. [DOI] [PubMed] [Google Scholar]
- 4.Cardona CA, Sánchez ÓJ. Fuel ethanol production: Process design trends and integration opportunities. Bioresource Tech. 2007;98:2415–2457. doi: 10.1016/j.biortech.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Edwards MC, Doran-Peterson J. Pectin-rich biomass as feedstock for fuel ethanol production. Appl. Microbiol. Biotechnol. 2012;95:565–575. doi: 10.1007/s00253-012-4173-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foster BL, Dale BE, Doran-Peterson JB. Enzymatic hydrolysis of ammonia-treated sugar beet pulp. Appl. Biochem. Biotechnol. 2001;91:269–282. doi: 10.1385/abab:91-93:1-9:269. [DOI] [PubMed] [Google Scholar]
- 7.Kühnel S, Schols HA, Gruppen H. Aiming for the complete utilization of sugar-beet pulp: Examination of the effects of mild acid and hydrothermal pretreatment followed by enzymatic digestion. Biotechnol. Biofuels. 2011;4:14. doi: 10.1186/1754-6834-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Lee C, Yu C, Cheng Y-S, et al. Dilute acid pretreatment and fermentation of sugar beet pulp to ethanol. Appl. Energy. 2013;105:1–7. [Google Scholar]
- 9.Micard V, Renard CMGC, Thibault J-F. Influence of pretreatments on enzymatic degradation of a cellulose-rich residue from sugar-beet pulp. Lebensmittel-Wissenschaft und -Technologie. 1997;30:284–291. [Google Scholar]
- 10.Spagnuolo M, Crecchio C, Pizzigallo MDR, Ruggiero P. Synergistic effects of cellulolytic and pectinolytic enzymes in degrading sugar beet pulp. Bioresource Tech. 1997;60:215–222. [Google Scholar]
- 11.Doran J, Foster B. Ethanol production from sugar beet pulp using engineered bacteria. Intl. Sugar J. 2000;102:336–340. [Google Scholar]
- 12.Rorick R, Nahar N, Pryor SW. Ethanol production from sugar beet pulp using Escherichia coliKO11 and Saccharomyces cerevisiae. Biol. Eng. 2011;3:199–209. [Google Scholar]
- 13.Sutton MD, Peterson JBD. Fermentation of sugarbeet pulp for ethanol production using bioengineered Klebsiella oxytoca strain P2. J. Sugar Beet Res. 2001;38:19–34. [Google Scholar]
- 14.Rezic T, Oros D, Markovic I, Kracher D, et al. Integrated hydrolysation and fermentation of sugar beet pulp to bioethanol. J. Microbiol. Biotechnol. 2013;23:1244–125. doi: 10.4014/jmb.1210.10013. [DOI] [PubMed] [Google Scholar]
- 15.Henrissat B. A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 1991;280:309–316. doi: 10.1042/bj2800309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levasseur A, Drula E, Lombard V, Coutinho PM, Henrissat B. Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes. Biotechnol. Biofuels. 2013;6:41. doi: 10.1186/1754-6834-6-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips CM, Beeson WT, Cate JH, Marletta MA. Cellobiose dehydrogenase and a copper-dependent polysaccharide monooxygenase potentiate cellulose degradation by Neurospora crassa. ACS Chem. Biol. 2011;6:1399–1406. doi: 10.1021/cb200351y. [DOI] [PubMed] [Google Scholar]
- 18.Sygmund C, Kracher D, Scheiblbrandner S, Zahma K, et al. Characterization of the two Neurospora crassa cellobiose dehydrogenases and their connection to oxidative cellulose degradation. Appl. Environ. Microbiol. 2012;78:6161–6171. doi: 10.1128/AEM.01503-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hemsworth GR, Davies GJ, Walton PH. Recent insights into copper-containing lytic polysaccharide mono-oxygenases. Curr. Opin. Struct. Biol. 2013;23:660–668. doi: 10.1016/j.sbi.2013.05.006. [DOI] [PubMed] [Google Scholar]
- 20.Beeson WT, Phillips CM, Cate JHD, Marletta MA. Oxidative cleavage of cellulose by fungal copper-dependent polysaccharide monooxygenases. J. Am. Chem. Soc. 2011;134:890–892. doi: 10.1021/ja210657t. [DOI] [PubMed] [Google Scholar]
- 21.Langston JA, Shaghasi T, Abbate E, Xu F, et al. oxidoreductive cellulose depolymerization by the enzymes cellobiose dehydrogenase and glycoside hydrolase 61. Appl. Environ. Microbiol. 2011;77:7007–7015. doi: 10.1128/AEM.05815-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nyanhongo GS, Gübitz G, Sukyai P, Leitner C, et al. Oxidoreductases from Trametes spp. in biotechnoogy: A wealth of catalytic activity. Food Technol. Biotechnol. 2007;45:250–268. [Google Scholar]
- 23.Sachslehner A, Haltrich D, Nidetzky B, Kulbe KD. Production of hemicellulose- and cellulose-degrading enzymes by various strains of Sclerotium rolfsii. Appl. Biochem. Biotechnol. 1997;63:189–201. doi: 10.1007/BF02920424. [DOI] [PubMed] [Google Scholar]
- 24.Znameroski EA, Glass NL. Using a model filamentous fungus to unravel mechanisms of lignocellulose deconstruction. Biotechnol. Biofuels. 2013;6:6. doi: 10.1186/1754-6834-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miller GL. Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. [Google Scholar]
- 26.Wariishi H, Valli K, Gold MH. Manganese(II) oxidation by manganese peroxidase from the basidiomycete Phanerochaete chrysosporium. Kinetic mechanism and role of chelators. J. Biol. Chem. 1992;267:23688–23695. [PubMed] [Google Scholar]
- 27.Baminger U, Subramaniam SS, Renganathan V, Haltrich D. Purification and characterization of cellobiose dehydrogenase from the plant pathogen SclerotiumAtheliarolfsii. Appl. Env. Microbiol. 2001;67:1766–1774. doi: 10.1128/AEM.67.4.1766-1774.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhou M, Diwu Z, Panchuk-Voloshina N, Haugland RP. A stable nonfluorescent derivative of resorufin for the fluorometric determination of trace hydrogen peroxide: applications in detecting the activity of phagocyte NADPH oxidase and other oxidases. Anal. Biochem. 1997;253:162–168. doi: 10.1006/abio.1997.2391. [DOI] [PubMed] [Google Scholar]
- 29.Kittl R, Kracher D, Burgstaller D, Haltrich D, Ludwig R. Production of four Neurospora crassa lytic polysaccharide monooxygenases in Pichia pastoris monitored by a fluorimetric assay. Biotechnol. Biofuels. 2012;5:79. doi: 10.1186/1754-6834-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Micard V, Renard CMGC, Thibault J-F. Enzymatic saccharification of sugar-beet pulp. Enzyme Microb. Tech. 1996;19:162–170. [Google Scholar]
- 31.Galhaup C, Wagner H, Hinterstoisser B, Haltrich D. Increased production of laccase by the wood-degrading basidiomycete Trametes pubescens. Enzyme Microb. Techn. 2002;30:529–536. [Google Scholar]
- 32.Phillips CM, Iavarone AT, Marletta MA. Quantitative proteomic approach for cellulose degradation by Neurospora crassa. J. Proteome Res. 2011;10:4177–4185. doi: 10.1021/pr200329b. [DOI] [PubMed] [Google Scholar]
- 33.Tian C, Beeson WT, Iavarone AT, Sun J, et al. Systems analysis of plant cell wall degradation by the model filamentous fungus Neurospora crassa. Proc. Natl Acad. Sci. USA. 2009;106:22157. doi: 10.1073/pnas.0906810106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leijdekkers AGM, Bink JPM, Geutjes S, Schols HA, Gruppen H. Enzymatic saccharification of sugar beet pulp for the production of galacturonic acid and arabinose; a study on the impact of the formation of recalcitrant oligosaccharides. Bioresour. Technol. 2013;128:518–525. doi: 10.1016/j.biortech.2012.10.126. [DOI] [PubMed] [Google Scholar]
- 35.Micard V, Renard CMGC, Thibault J-F. Enzymatic saccharification of sugar-beet pulp. Enzyme Microb. Tech. 1996;19:162–170. [Google Scholar]
- 36.Elisashvili V, Kachlishvili E, Penninckx M. Effect of growth substrate, method of fermentation, and nitrogen source on lignocellulose-degrading enzymes production by white-rot basidiomycetes. J. Ind. Microbiol. Biotechnol. 2008;35:1531–1538. doi: 10.1007/s10295-008-0454-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
suppinfo


